Structural and functional analyses of artificial micro RNA (amiRNA) mutants reveal that the CSC plays a role not only in generating wild-type motility, but also in assembly of at least a subset of radial spokes. This study also produced the unexpected finding that, contrary to current belief, the radial spokes may not be homogeneous.
Abstract
The ubiquitous calcium binding protein, calmodulin (CaM), plays a major role in regulating the motility of all eukaryotic cilia and flagella. We previously identified a CaM and Spoke associated Complex (CSC) and provided evidence that this complex mediates regulatory signals between the radial spokes and dynein arms. We have now used an artificial microRNA (amiRNA) approach to reduce expression of two CSC subunits in Chlamydomonas. For all amiRNA mutants, the entire CSC is lacking or severely reduced in flagella. Structural studies of mutant axonemes revealed that assembly of radial spoke 2 is defective. Furthermore, analysis of both flagellar beating and microtubule sliding in vitro demonstrates that the CSC plays a critical role in modulating dynein activity. Our results not only indicate that the CSC is required for spoke assembly and wild-type motility, but also provide evidence for heterogeneity among the radial spokes.
INTRODUCTION
The motility of all eukaryotic cilia and flagella is modulated by the second messengers calcium and cAMP (for review, see Salathe, 2007). Substantial data from our laboratory and others have indicated that calmodulin (CaM) associated with the axoneme is a key calcium sensor; however, the mechanism by which CaM regulates motility in response to changes in intraciliary calcium concentration is unknown (Otter, 1989; Yang et al., 2001, 2004; Smith, 2002a, 2002b; Wargo and Smith, 2003; Patel-King et al., 2004; Wargo et al., 2004). To define a specific role for CaM in motility, we developed anti-CaM antibodies and identified three CaM-associated complexes by coimmunoprecipitation in the model organism Chlamydomonas; two complexes reside on the central apparatus (the PF6 and PCDP1 complexes) (Wargo et al., 2005; DiPetrillo and Smith, 2010), and one is associated with the radial spokes and named the CaM and Spoke associated Complex or CSC (Dymek and Smith, 2007). Importantly, the components of each complex appear to be conserved in mammals.
A large body of data from many laboratories has indicated that the central apparatus and radial spokes are essential for modulating the size and shape of ciliary bends (reviewed in Smith and Yang, 2004). Therefore, association of the CaM complexes with these axonemal structures suggests a role for them in regulating ciliary beating. Supporting this idea, functional data from our laboratory and others have demonstrated that the PF6 and PCDP1 complexes are required for motility (Dutcher et al., 1984; Rupp et al., 2001; Wargo and Smith, 2003; Wargo et al., 2005; Lee et al., 2008; DiPetrillo and Smith, 2010). The function of the CSC, however, remains unknown.
The CSC is composed of CaM and three polypeptides we originally named CaM-IP2, CaM-IP3, and CaM-IP4 (Dymek and Smith, 2007). Using several biochemical approaches, we determined that CaM-IP2 directly interacts with CaM as well as radial spoke protein 3, RSP3. Interestingly, RSP3 is an A-kinase anchoring protein (AKAP) (Gaillard et al., 2001), and CaM-IP2 shares homology with an AKAP binding protein originally described in studies of mouse sperm (Yukitake et al., 2002; Dymek and Smith, 2007). This finding suggests additional roles for the CSC in second messenger–mediated regulation of motility.
One question arising from our studies is the following: What is the localization of the CSC within the axoneme? To date, no mutants have been identified with defects in CSC components. Interestingly, Chlamydomonas pf14 mutants with defects in RSP3 fail to assemble the radial spokes and have paralyzed flagella (Luck et al., 1977; Huang et al., 1981; Piperno et al., 1981). This mutant phenotype tentatively localized RSP3 to the base of the spoke, presumably at a point of attachment to the axonemal microtubules (Diener et al., 1993). Because CaM-IP2 of the CSC interacts with RSP3, one prediction is that the CSC also localizes near the base of the spokes. This prediction is supported by the observation that the CSC assembles in the axoneme in the pf14 mutant in the absence of spoke assembly (Dymek and Smith, 2007).
A second question is the following: What is the relative stoichiometry of the CSC compared with the radial spokes? In Chlamydomonas the spokes repeat in pairs every 96 nm along the length of the axoneme. Biochemical characterization of spoke components has indicated that RSP3 is a dimer (Wirschell et al., 2008); therefore two copies of RSP3 may be associated with each of the two spokes (Wirschell et al., 2008; Kelekar et al., 2009). Although we have shown that CaM-IP2 interacts with RSP3, we do not know if the CSC is associated with each copy of RSP3 or with each radial spoke in a given spoke pair within the 96 nm axonemal repeat.
Finally, a key question is this: What is the function of the CSC? Given the putative localization of the CSC to the base of the spokes, close to the inner dynein arms and perhaps the dynein regulatory complex (see Nicastro et al., 2006; Bui et al., 2009; Heuser et al., 2009), we hypothesized that the CSC plays a role in regulating dynein activity and potentially spoke assembly. In our previous studies using an in vitro assay, we demonstrated that antibodies generated against CaM-IP2 modulate dynein-driven microtubule sliding in isolated axonemes (Dymek and Smith, 2007). These results provided the first indications that the CSC provides regulatory cues to the dynein arms, but they did not address a specific role for the CSC in generating the high beat frequency or complex waveforms typical of beating cilia and flagella.
Our understanding of CSC function would greatly benefit from mutants defective in CSC components, but targeted gene disruption has not been achieved for Chlamydomonas. Recently, we and other laboratories have successfully used an artificial microRNA (amiRNA) approach to reduce gene expression in Chlamydomonas (Zhao et al., 2009; Molnar et al., 2009; DiPetrillo and Smith, 2010). The resulting amiRNA mutants are stable and do not display the off-target effects observed with other RNA interference techniques. Here we report the acquisition of mutants with reduced expression of either CaM-IP2 or CaM-IP3. Our structural, biochemical, and functional analyses of these mutants reveal a unique role for the CSC in spoke assembly and in generating wild-type motility.
RESULTS
Reduced expression of either CaM-IP2 or CaM-IP3 results in reduced assembly of the entire CSC
Our initial attempts to reduce gene expression of CSC components by previously published methods for RNAi often generated mutants with clumpy phenotypes (failing to hatch from the mother cell wall following cell division), which appeared to be off-target effects of the RNAi procedures. In addition, knockdown of expression persisted for only several weeks before wild-type levels of protein expression resumed. We then adopted a new method to knock down expression in Chlamydomonas by using amiRNAs, reported to be highly efficient and stable (Zhao et al., 2009; Molnar et al., 2009). Because we have specific antibodies to assess levels of both CaM-IP2 and CaM-IP3, we designed oligonucleotides specific for the genes encoding each of these proteins (Supplemental Table 1) to generate amiRNA constructs. Construct preparation, transformation, and selection of transformants are described in Materials and Methods. Expression of CaM-IP4 was not targeted because we were unable to generate antibodies against CaM-IP4, and thus could not assess CaM-IP4 protein levels by Western blot.
For each of the amiRNA constructs, we picked between 800 and 1300 transformants. Of these, 80–90 transformants each for either CaM-IP2 or CaM-IP3 had motility defects and were screened by Western blot for reduced protein levels. From each transformation we also isolated flagella from 96 random transformants with no obvious motility defects and screened these by Western blot. In total, we identified two CaM-IP2 amiRNA strains and eight CaM-IP3 amiRNA strains with significantly reduced levels of CaM-IP2 and CaM-IP3, respectively (Figure 1 and Supplemental Figure S1). Importantly, all of these strains were identified in screens of mutants with obvious motility defects. Assessment of motility in 96-well dishes revealed that all of these strains swim slowly, in a manner similar to that of inner dynein arm–defective mutants. Unlike wild-type cells, which travel long distances in a straight trajectory, these mutants often pause or swim in wide circles. Both amiRNA mutants for CaM-IP2 (4D6 and 4A2) and two mutants for CaM-IP3 (6E6 and 1A3) were chosen for further analysis (Figure 1). The remaining CaM-IP3 amiRNA strains are shown in Supplemental Figure S1. All strains have retained the same degree of knockdown over many passages as well as through genetic crosses (later in this article). Therefore, knockdown of protein expression appears to be stable in these mutants.
FIGURE 1:

The flagella of CSC amiRNA mutants have reduced amounts of both CaM-IP2 and CaM-IP3. Western blots of flagella (8 μg) isolated from wild-type and CSC amiRNA mutants were probed with antibodies that recognize the CSC (CaM-IP2 and CaM–IP3), radial spokes (RSP1), inner dynein arm I1 (IC138), and central apparatus (PF20). CaM-IP2 amiRNA mutants 4A2 and 4D6 and the CaM-IP3 amiRNA mutants 6E6 and 1A3 have reduced amounts of both CaM-IP2 and CaM-IP3. 6E6 is the most efficient knockdown; no CaM-IP3 or CaM-IP2 is detectable by Western analysis. RSP1, IC138, and PF20 are present at near wild-type levels in each amiRNA mutant.
Western blots of flagella for each amiRNA mutant show concomitant reduction in assembly of other CSC components compared with wild type (Figure 1 and Supplemental Figure S1). Flagella of CaM-IP2 amiRNA mutant 4A2 have less CaM-IP2 than do flagella of 4D6, and also have comparatively less CaM-IP3 (Figure 1). Similarly, CaM-IP3 amiRNA mutant 6E6 shows virtually no assembly of CaM-IP3 and CaM-IP2 into flagella, whereas 1A3 has low levels of CaM-IP3 and correspondingly low levels of CaM-IP2 assembled (Figure 1).
In Chlamydomonas, integration of the transforming plasmid into the host's genome may disrupt genes required for motility (Tam and Lefebvre, 1993). In addition, transformants from any single transformation experiment may not represent independent isolates. To address this possibility, we probed blots of genomic DNA isolated from mutants using the plasmid vector sequence as a hybridization probe. Restriction fragment length polymorphisms (RFLPs) for all amiRNA mutants revealed that each strain represents an independent isolate and suggested that the plasmid integrated into a different region of the genome in each mutant strain (Figure 2A and unpublished data).
FIGURE 2:

CSC amiRNA mutants are independent isolates with reduced expression of both CaM-IP2 and CaM-IP3. (A) Southern blot of 5 μg of genomic DNA, digested with KpnI, probed with the APHVIII gene. Each amiRNA mutant has a different RFLP, indicating that the strains are independent isolates and that the amiRNA plasmid most likely inserted at different sites. (B) Relative transcript levels of wild-type and CSC amiRNA mutants. Transcript abundance was quantified by densitometry of Northern blot phosphorimages using Image J. S14 levels were used as a loading control, and wild-type levels were set to 100%. For CaM-IP2 amiRNA mutants, CaM-IP2 transcript levels (red) were ∼30–40% of wild-type levels. For CaM-IP3 amiRNA mutants, CaM-IP3 transcript levels (blue) were ∼30–50% of wild-type levels. Note that in CaM-IP2 amiRNA mutants, the CaM-IP3 transcript levels are also reduced, as well as the CaM-IP2 transcript levels in CaM-IP3 amiRNA mutants. RSP3 transcript levels (black) for all strains were near 100%.
To determine whether transcript levels were reduced, we probed Northern blots of mRNA isolated from each transformant by using cDNA corresponding to a fragment of the CaM-IP2 and CaM-IP3 genes as hybridization probes (Figure 2B). For strains 4A2 and 4D6, transcript levels of CaM-IP2 were reduced to ∼30 and 40%, respectively, of wild-type levels. This relative reduction in transcript levels correlates well with the relative reduction in protein levels. These two CaM-IP2 mutants also had a slight reduction of CaM-IP3 transcript, ∼70% of wild type.
For CaM-IP3 amiRNA strains 6E6 and 1A3, CaM-IP3 transcript levels were reduced to 50 and 30% of wild-type levels, respectively. In this case, 6E6 had more abundant transcript levels than 1A3, but significantly less protein assembled into flagella. We have found that with the amiRNA technique, relative transcript levels do not always strictly correspond to relative protein levels. These two CaM-IP3 amiRNA strains also showed marked reduction in CaM-IP2 transcript, 30–70% of wild-type levels. These combined analyses suggest that transcription of the genes encoding CaM-IP2 and CaM-IP3 may be coregulated.
To assess the assembly and integrity of the CSC and to determine if CaM-IP4 had assembled in the amiRNA strains, we performed immunoprecipitation experiments using anti-CaM and anti-CaM-IP2 antibodies and extracts isolated from wild-type and mutant axonemes. For extracts isolated from wild-type and pf14 (radial spokeless) axonemes, the anti-CaM antibodies precipitate all three components of the CSC (labeled, Figure 3A) and the PF6 central apparatus complex (circles, Figure 3A); in wild-type axonemes, spoke components are also precipitated as previously reported (squares, Figure 3A; Wargo et al., 2005; Dymek and Smith, 2007). In extracts isolated from 4A2 and 6E6 (the strains with the most significant reduction in assembly of CaM-IP2 and CaM-IP3, respectively) little if any CSC is observed in anti-CaM precipitates (Figure 3A). In strains 4D6 and 1A3, CSC components are more easily observed in the anti-CaM precipitate, including CaM-IP4.
FIGURE 3:

For all CSC amiRNA mutants, the remaining CSC subunits associate with the radial spokes. (A) Silver-stained gels of immunoprecipitates using anti-CaM antibodies and axonemal extracts. As previously reported (Dymek and Smith, 2007), anti-CaM antibodies precipitate central pair (circles), radial spoke (squares), and CSC proteins (labeled) from wild-type extracts. Only central pair and CSC proteins precipitate from radial spokeless extracts (pf14). In CSC amiRNA mutants only small quantities of CSC and radial spoke proteins are precipitated from axonemal extracts compared with wild type; in contrast, the amount of central pair proteins precipitated is the same as in wild type. For strain 6E6, which is the most efficient knockdown, only central pair proteins precipitate. Heavy chain (HC) from the precipitating antibody is present at 50 kDa. (B) Silver-stained gels of immunoprecipitates using anti-CaM-IP2 antibodies and axonemal extracts. As previously reported (Dymek and Smith, 2007), anti-CaM-IP2 antibodies precipitate CSC members and radial spoke proteins from wild-type extracts. Only the CSC is precipitated in pf14 extracts. Little if any protein precipitates from strain 6E6 axonemal extracts. For strains 4A2, 4D6, and 1A3, small amounts of CSC and radial spokes precipitate, indicating that the remaining CSC in the amiRNA mutants is associated with the radial spokes.
Because CaM is also a component of the radial spokes (Patel-King et al., 2004; Yang et al., 2004), we expected radial spoke components to precipitate using anti-CaM antibodies regardless of whether the CSC is present. Interestingly, fewer radial spoke proteins are observed in the anti-CaM precipitates performed using the amiRNA mutants (Figure 3A). To determine whether the small amount of CSC that assembles in the amiRNA mutants remains associated with the spokes, we used the anti-CaM-IP2 antibodies in immunoprecipitation experiments (Figure 3B). For strain 6E6, virtually no CSC is assembled into the axoneme; therefore, no proteins are observed in the precipitate. For the other amiRNA strains, however, the CaM-IP2 antibodies precipitated the CSC as well as radial spoke components (Figure 3B). Therefore, the CSC that assembles into the axoneme in amiRNA strains remains associated with the radial spokes.
Reduced expression of either CaM-IP2 or CaM-IP3 results in defects in spoke assembly
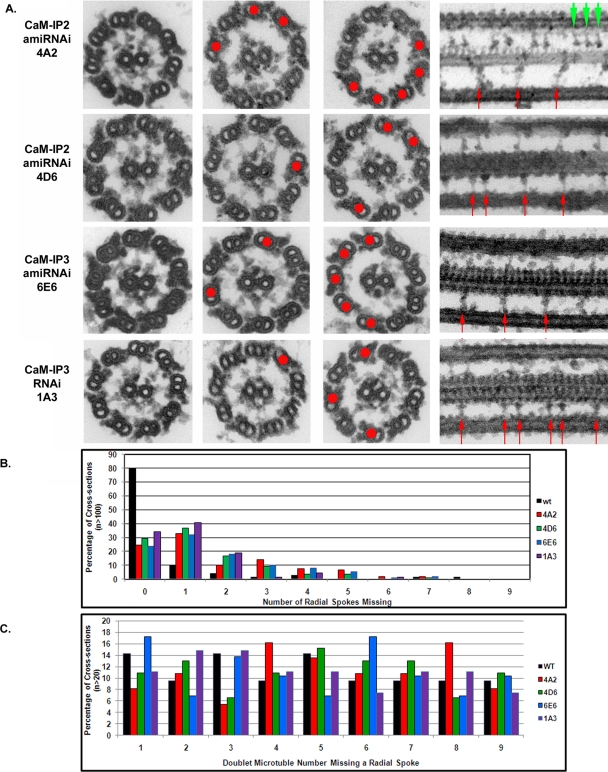
To determine if any structural defects result from knockdown of CaM-IP2 or CaM-IP3 expression, mutant axonemes were prepared for conventional transmission electron microscopy (EM). In cross-sections of each mutant, we observed that several doublet microtubules lacked spokes (red dots, Figure 4A). The number of doublets that lacked spokes is quantified in the histogram shown in Figure 4B; typically from one to four doublets lacked spokes. We observed no significant differences in the total number of cross-sections with missing spokes among the CSC amiRNA mutants. In addition, we did not detect any preference for spoke loss on specific doublet microtubules based on the doublet numbering system (Hoops and Witman, 1983; Figure 4C).
FIGURE 4:
CSC amiRNA mutants have reduced assembly and/or stability of the radial spokes. (A) Electron micrographs of cross- and longitudinal sections of chemically fixed axonemes isolated from CSC amiRNA mutants. Cross-sections show various numbers of doublet microtubules, which lack the radial spokes (red dots). In longitudinal sections, we observe both doublets with repeating pairs of radial spokes, as in wild type, and stretches of regularly repeating, single radial spokes (red arrows). Occasionally, a triplet of radial spokes was also observed (green arrowheads in 4A2 panel). (B) Histogram of the number of radial spokes missing in cross-sections of axonemes (n > 100 for each strain); ∼80% of cross-sections of wild-type axonemes (black bar) have spokes present on all nine doublets. In contrast, only ∼30% of cross-section for CSC amiRNA mutant sections have spokes present on all doublet microtubules. The majority of cross-sections for CSC amiRNA mutants have between one and five doublets that lack spokes. (C) Histogram indicating the specific doublet that lacked radial spokes in cross-sections (n > 20 for each strain). Doublet identity was determined by specific axonemal markers according to that established in Hoops and Witman (1983). In all strains, the distribution of doublets lacking radial spokes appeared random.
Longitudinal images of mutant axonemes revealed an astonishing pattern of spoke loss (arrows, Figure 4A). Normally in Chlamydomonas the spokes repeat every 96 nm in pairs along the length of the doublet microtubules, where the proximal spoke of each pair is spoke 1 and the more distal is spoke 2. In each of our four amiRNA strains, a subset of doublet microtubules showed axonemal repeats with only a single spoke instead of a pair (Figure 4A); often repeats with only a single spoke clustered in stretches along the doublet microtubules (Figure 4A). Occasionally, three spokes assembled (green arrowheads, Figure 4A), reminiscent of other ciliated organisms such as Tetrahymena or sea urchin sperm flagella in which the spokes assemble in triplet groups along the length of the axonemal doublet microtubules. This observation further implicates the CSC in directing proper spoke attachment to the microtubules.
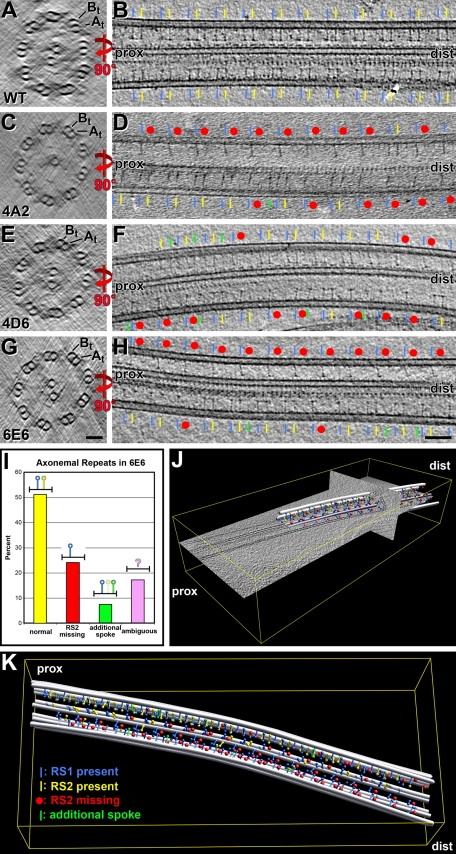
For a more detailed structural analysis, we performed cryo-electron tomography of frozen hydrated axonemes. Analysis of tomograms revealed two major groups of axonemal repeats (Figure 5 and Supplemental Video 1): In one group both radial spokes of the pair are present; in the other group only a single spoke per repeat is present. Often axonemal repeats that had only one spoke appeared in stretches along the doublet microtubules (Figure 5, D and H), similar to the EM observations of chemically fixed axonemes (Figure 4). Tomographic data, however, provide three-dimensional information, allowing for the unambiguous assignment of the proximal and distal ends of the doublet microtubules (Figure 5, J and K); therefore the missing spoke was identified as spoke 2 for all four amiRNA mutants (yellow label where spoke 2 is present, red dot where it is missing, Figure 5). This remarkable observation indicates not only that the CSC is potentially associated with only a subset of spokes, spoke 2, but also that the two spokes of the pair may not be identical, particularly with respect to their microtubule attachment. No spoke 1 defects (blue label, Figure 5) were observed, but occasionally more than two spokes were seen per axonemal repeat (green label, Figure 5, D, F, and H–K; Supplemental Video 1). In an effort to assign a specific axonemal structure to the CSC and to determine whether there are additional structural defects associated with reduced expression of CSC components, we are currently performing a complete structural analysis of axonemes isolated from each amiRNA strain by using cryo-electron tomography and subtomographic averaging.
FIGURE 5:
Radial spoke 2 is the missing spoke in CSC amiRNA mutant axonemes. (A–H) Slices through cryo-electron tomograms of axonemes from wild-type (WT; A and B) and amiRNA mutants 4A2 (C and D), 4D6 (E and F), and 6E6 (G and H), showing the three-dimensional (3D) structure in cross-sectional (A, C, E, and G) and longitudinal views (B, D, F, and H). By identifying A- and B-tubules (At, Bt) in cross-sectional slices, the proximal (prox) and distal (dist) end of the axoneme can be determined and thus radial spokes 1 and 2 (RS1/blue; RS2/yellow) assigned unambiguously. All three CSC amiRNA mutants show many axonemal repeat units with missing spoke 2 (red dots). Occasionally, repeat units have additional spokes (green) at locations different from the normal RS1 or RS2 position. No defects of RS1 were observed. Scale bars: 50 nm (G), 100 nm (H). (I) Histogram showing the analysis of 675 repeat units from six tomograms of 6E6 axonemes; approximately half of the repeat units were normal (yellow bar), one quarter were missing RS2 partly or completely (red bar), and the remaining repeats had either an additional spoke (green bar) or were too ambiguous to score (purple bar). (J and K) Graphical model and 3D visualization of a cryo-tomogram of a 6E6 axoneme, showing the 3D architecture of the axoneme and location of the radial spokes. The modeled microtubules (gray), spokes 1 (blue) and 2 (yellow), and occasional additional spokes (green) are shown both as partial overlaid on tomographic slices (J) or alone (K); transparent colors indicate ambiguous spokes (see also Supplemental Video 1).
CSC is required for wild-type motility
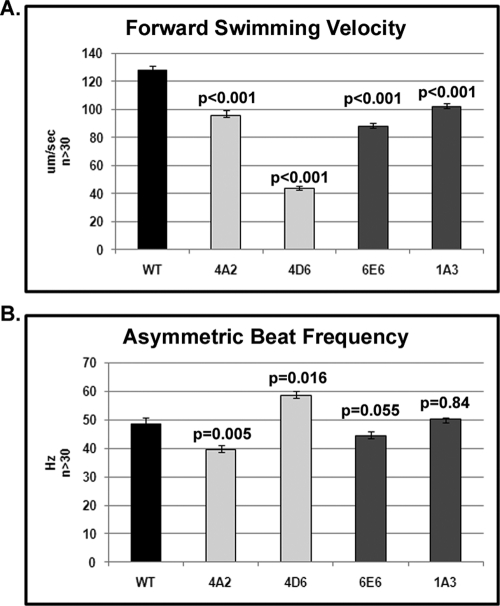
To quantify motility defects in amiRNA mutants, we used high-speed video capture to analyze flagellar beat frequency and waveform. For all mutants, swimming velocity was significantly reduced (p < 0.001 for all strains by Student's t test) compared to wild type with strain 4D6 showing the greatest reduction (Figure 6 and Supplemental Table 2). The average beat frequency for each mutant (between 40 and 58 Hz) was not drastically different, however (0.005 < p < 0.900 by Student's t test) from wild type (50 Hz). Analysis of flagellar waveforms revealed that, for all mutants, the two flagella for any single cell often lost coordination and began beating out of sync (Figure 7). Loss of coordination was due to frequent stalling at the end of the recovery stroke and a failure to initiate the next effective stroke. After the effective stroke is initiated, the mutant cells do not complete the effective stroke before switching to the recovery stroke. This behavior is particularly evident in 4D6. Thus, the reduced swimming speed is the product of uncoordinated flagella and shallow effective and recovery strokes.
FIGURE 6:
CSC amiRNA mutant cells have significantly slower swimming velocities than do wild-type cells; yet flagellar beat frequencies are nearly wild type. Swimming velocity (A) and beat frequency (B) were measured from swimming cells using high-speed video microscopy.
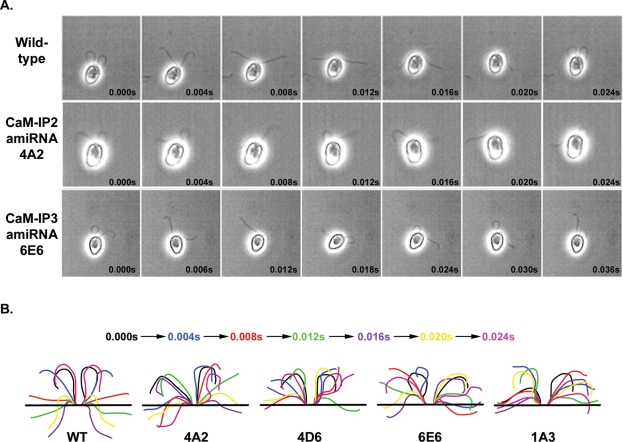
FIGURE 7:
All CSC amiRNA mutants exhibit defects in flagellar beating. (A) Montages of images from digitally recorded videos of wild-type, 4A2, and 6E6 cells. Wild-type cells swim forward with an asymmetric waveform, and the flagella are in sync through the entire beat. CSC amiRNA mutant cells swim forward with a few beats in sync, and then their flagella lose synchrony. (B) Traced diagrams of flagellar beats from wild-type, 4A2, 4D6, 6E6, and 1A3 cells. Colors correspond to traces taken from the same frame (see colored time sequence). Wild-type flagella complete a full asymmetric beat with flagella in sync. The two flagella for the CSC amiRNA strains are out of sync and do not exhibit complete recovery and effective strokes.
In Chlamydomonas, the changes in motility that accompany both the phototaxis and photoshock responses are calcium mediated. Therefore, we tested whether the amiRNA mutants could phototax or swim with a symmetric waveform (see Materials and Methods). All amiRNA mutants phototax and switch to a symmetric waveform in response to bright light (photoshock response).
Using an in vitro microtubule sliding assay, we measured sliding velocities for axonemes isolated from both 6E6 and 4D6; velocities for both strains were ∼17.3 μm/s and not significantly different from that previously reported for wild type (Smith, 2002a, 2002b). In these studies we also found that axonemes isolated from the central apparatus defective mutant pf18 have significantly slower sliding velocities compared with wild type (Smith, 2002a, 2002b). We then demonstrated that addition of either high calcium buffer or anti-CaM-IP2 antibodies to pf18 axonemes could restore sliding velocity to wild-type levels (Smith, 2002a; Dymek and Smith, 2007). We hypothesized that the CSC is involved in the high calcium-induced increase in dynein activity in pf18 axonemes. To test this hypothesis, we generated double mutants lacking the central apparatus and the CSC by crossing pf18 with amiRNA strains 4A2 and 6E6. These double mutants have slow sliding velocities in both low- and high-calcium buffers that are not significantly different (p > 0.25 by Student's t test) from those of the pf18 axonemes (Figure 8A and unpublished data; see Dymek and Smith, 2007, for pf18 velocities).
FIGURE 8:
Reconstitution of the CSC onto CSC amiRNA mutant axonemes rescues high calcium–induced dynein activity. (A) Microtubule sliding velocity of 6E6pf18 and 4A2pf18 mutant axonemes. All sliding experiments represent the result of three to five independent experiments with a total n for each mutant and a condition of between 60 and 160. Sliding was measured under low (pCa8) or high calcium (pCa4, “+Ca”) conditions. For both double mutants, no increase in dynein activity is observed in pCa4 buffer. When the CSC is reconstituted (+CSC), a significant increase (p < 0.001, by Student's t test) in dynein activity is observed in pCa4 buffer. (B) Western blot of CSC reconstituted onto mutant axonemes. KI (after NaCl) extracts (E) isolated from pf14 axonemes were added to 6E6pf18 axonemes (A) at various ratios of equivalent protein (see Materials and Methods). Axonemes were then pelleted, and the supernatant and pellet were analyzed by Western blot using anti–CaM-IP2 antibodies. The first four lanes to the left are controls using either extract or axonemes alone mixed with buffer (B). When extracts and axonemes are mixed at a ratio of 1:1, all of the CaM-IP2 binds to 6E6pf18 axonemes. As the ratio of extract to axoneme is increased, CaM-IP2 is observed in the supernatant, indicating that binding of CaM-IP2 saturates.
Because axonemes of amiRNA strains lack subsets of radial spokes and potentially other axonemal components that we have not detected, it is possible that slow sliding velocities of the double mutants in high calcium buffer are not specifically due to the loss of the CSC. To address this possibility, we isolated the CSC from radial spokeless mutants (pf14 axonemes, see Materials and Methods) and reconstituted the CSC onto axonemes isolated from the pf18:amiRNA double mutants. To assess the efficiency of reconstitution, extracts and axonemes were mixed at several ratios based on equivalent amounts of axonemes and extract (see Materials and Methods). The axonemes were pelleted, and the amount of CaM-IP2 in the resulting supernatant and pellet was visualized by Western blot; the results for reconstitution of 6E6pf18 double-mutant axonemes are shown in Figure 8B. Up to a ratio of 1:1, extract and axonemes, CaM-IP2 is only found in the pellet indicating that all of the added CaM-IP2 binds to the axoneme. At higher ratios, the amount of CaM-IP2 in the pellet remains constant, whereas increasing amounts of CaM-IP2 are observed in the supernatant; therefore, binding of the CSC saturates. These results demonstrate that the CSC is reconstituted onto the axoneme and that binding is most likely specific.
We then reconstituted the axonemes of both 6E6pf18 and 4A2pf18 double mutants, and repeated the microtubule sliding assays under low and high calcium conditions (Figure 8A). Microtubule sliding velocities of CSC-reconstituted double-mutant axonemes remained slow in low calcium as before. In high calcium buffer, however, the sliding velocities of reconstituted axonemes for both double mutants were restored to wild-type levels (Figure 8A). These data provide strong evidence that the CSC plays a role in regulating dynein-driven microtubule sliding in response to calcium.
DISCUSSION
Using a combination of biochemical and genetic approaches and in vitro functional assays, our previous studies demonstrated that calcium control of ciliary motility involves regulation of dynein-driven microtubule sliding and that CaM anchored to the axoneme is a key calcium sensor (Smith, 2002a, 2002b; Wargo et al., 2004; Dymek and Smith, 2007; DiPetrillo and Smith, 2010). The identification of a CSC that interacts with RSP3 raised the intriguing possibility that the CSC mediates regulatory signals between the radial spokes and dynein arms (Dymek and Smith, 2007). To define the in vivo function of the CSC, we have now obtained mutants with reduced expression of two different CSC components. Structural and functional analyses of these mutants reveal that the CSC plays a role not only in generating wild-type motility, but also in assembling at least a subset of radial spokes. This study also produced the unexpected finding that, contrary to current belief, the radial spokes may not be homogeneous.
Expression of CSC components is reduced using amiRNAs
Although transformation of Chlamydomonas may result in the generation of insertional mutants, our data indicate that the phenotypes of the amiRNA strains resulted from reduced expression of the targeted genes. Southern blots reveal that all transformants represent independent isolates. Given the RFLPs we observed between strains, it is highly unlikely that the transforming plasmid integrated into the same gene in multiple transformation experiments. It is also unlikely that the transforming plasmid integrated into multiple different genes producing the same novel phenotype (the regular absence of spoke 2 has not been previously reported). This same phenotype was obtained in multiple transformation experiments targeting two different genes. Finally, Northern blots indicate that the transcript levels of the targeted gene are reduced for both the CaM-IP2 and CaM-IP3 amiRNA mutants. These combined data strongly support the conclusion that the observed structural and motility defects are due to reduced expression of the targeted genes
CSC plays a role in spoke assembly and/or stability
In motile cilia and eukaryotic flagella, the radial spokes repeat in pairs or triplet groups every 96 nm along the length of axonemal doublet microtubules. Despite significant insights into spoke composition and structure, and their importance in motility, we still do not know how the unique spacing of the spokes is established, which proteins specifically attach the spokes to the doublet microtubules, and the precise mechanism for spoke modulation of motility.
On the basis of our finding that CaM-IP2 interacts with RSP3, as well as previous studies of RSP3 (Diener et al., 1993), we predicted that the CSC is localized near the base of the spokes, in a position to affect spoke assembly and possibly dynein arm activity. The pf14 mutation in the RSP3 gene leads to a failure in radial spoke assembly into axonemes. In addition, studies by Diener et al. (1993) using RSP3 expressed in vitro demonstrated that RSP3 binds to pf14 axonemes but does not bind to microtubules assembled in vitro from purified tubulin. The authors hypothesized that RSP3 may require an adaptor protein for microtubule binding that remains bound to pf14 axonemes. Our data suggest that the CSC may serve as an adaptor for at least a subset of radial spokes. In flagella isolated from our CSC amiRNA mutants, the most striking structural defect we observed was a defect in the assembly of radial spoke 2.
In one of our mutants, 6E6, virtually no CSC is detectable in Western blots of 6E6 axonemes, and yet, this mutant does not show a complete lack of spoke 2 assembly. There are several possible explanations for this result. One possibility is that the CSC is not an absolute requirement for spoke 2 to assemble but functions in stabilizing the attachment of spoke 2 to the A-tubule. A second possibility is that an undetectable amount of CSC remains in strain 6E6 that is sufficient to stabilize assembly of a subset of spoke 2. A third possibility is that the CSC is only required for assembly/stability of a subset of spoke 2. A final possibility is that the CSC interacts with yet unknown axonemal components required for spoke assembly/stability; in this case even in the complete absence of the CSC, this additional component may partially stabilize a subset of spokes. Our results also imply that a second, unidentified adaptor may be required for the assembly of spoke 1.
In Western blots of isolated flagella, we did not observe an obvious reduction in spoke protein RSP1. In longitudinal images, we observe short stretches along the microtubules where RS2 is missing and, in cross-section, not every doublet appears to have missing spokes. Therefore the reduction of RS2 may not be sufficient to discern a difference in protein levels by Western blot. As discussed later in this article, it is also possible that the composition of RS1 and RS2 are not identical, and, for either spoke, we still do not know the precise stoichiometry of radial spoke components. Finally, it is possible that the proteins comprising RS2 are mislocalized in our mutants.
This result raises the obvious question of whether the two radial spokes of the spoke pair are structurally and biochemically distinct, particularly with respect to their microtubule attachment. In three of our amiRNA mutants (4A2, 4D6, and 1A3), a small amount of CSC still assembles into flagella. If the two spokes are identical in their requirement for the CSC to assemble, we would have predicted a more random pattern of spoke loss in the absence of the CSC. The fact that we observed no spoke 1 defects, but a clear preference for reduced assembly of spoke 2, indicates that spokes 1 and 2 may require different proteins as adaptors for attachment to the doublet microtubules.
Although there are no direct data demonstrating that spoke 1 and 2 are biochemically distinct, it is interesting to note that in WT flagella a subset of spoke proteins is reduced in abundance (Yang et al., 2006). It is possible that this difference in abundance simply reflects differences in the stoichiometry of these polypeptides in any single radial spoke. It is also possible that some polypeptides are present only in a subset of radial spokes. Along these lines, we observed that in anti-CaM immunoprecipitation experiments, significantly fewer spoke proteins are precipitated in mutants compared with wild type. This result was somewhat surprising because CaM is associated with two spoke proteins, RSP2 and RSP23 (Yang et al., 2001; Patel-King et al., 2004). The reduction in the number of assembled radial spokes in amiRNA mutants is not sufficient to explain this loss of spoke immunoprecipitation. In the absence of the CSC, less CaM may associate with the radial spokes. Alternatively, CaM may predominantly associate with a subset of spokes that are missing or reduced in CSC mutants.
Localization of the CSC
Our results indicate that the CSC is likely to localize near the base of spoke 2. Cryo-electron tomography has revealed a number of never-before-seen structures at the base of the radial spokes as well as the precise localization of the nexin dynein regulatory complex (N-DRC) near spoke 2 (Nicastro et al., 2006; Heuser et al., 2009). Note that small differences between the base of RS1/2 have been reported for wild-type axonemes (see connection #8 between RS2 and nexin dynein regulatory complex, Supplemental Figure S2J in Heuser et al., 2009). To determine which, if any, of these structures correspond to the CSC, we are in the process of analyzing cryo-tomograms from the CSC mutants and pf14 to compare them with each other and with wild-type axonemes. Although cryo-tomography combined with subtomogram averaging of regularly repeating units has provided images of axonemes in unprecedented detail, a major requirement is that units to be averaged are homogeneous. Because the amiRNA CSC mutants exhibit marked heterogeneity with respect to spoke assembly, analysis of tomograms from these mutants has been particularly challenging. If we are successful in this analysis, the results will be presented in a subsequent publication.
A role for the CSC in modulating motility
In Chlamydomonas, both the phototaxis and photoshock response are mediated by an influx in intraflagellar calcium and result in changes in beat frequency and/or waveform. Although motility is defective in our CSC amiRNA mutants, they are still able to phototax, albeit slowly, and to switch waveforms. Therefore, either the CSC is not directly involved in these responses or the little CSC that assembles into flagella in these mutants is sufficient for these changes in motility.
Given the likely localization of the CSC at the base of spoke 2, this complex would certainly be in a position to mediate regulatory signals between the spokes and inner dynein arms. Our analysis of motility in the CSC amiRNA mutants supports this prediction. All of our mutants have reduced swimming speed that results largely from altered waveforms and lack of coordination between the two flagella. Based on studies of Chlamydomonas mutants, modulation of flagellar waveforms appears to be a function of the inner dynein arms (Brokaw and Kamiya, 1987; Luck and Piperno, 1989; Kamiya et al., 1991; Yagi et al., 2005), consistent with the probable localization of the CSC.
Additional data supporting a role for CSC regulation of motility include our analysis of dynein-driven microtubule sliding. We previously showed that microtubule sliding velocities are reduced in central pairless mutant axonemes (Smith, 2002b) and could be restored by addition of calcium or antibodies generated against CaM-IP2 (Smith, 2002a; Dymek and Smith, 2007). We also demonstrated that slow sliding velocities of radial spokeless axonemes, which still retain the CSC, could be restored by the addition of CaM-IP2 antibodies (Dymek and Smith, 2007). These results suggested that the regulation of dynein-driven microtubule sliding by the CSC is downstream of the central apparatus and radial spokes.
By generating double mutants with reduced expression of CSC components and that lack the central apparatus, we demonstrate here that the CSC is required for the increase in sliding velocity in high calcium. Although it is possible that the failure of our double mutants to respond to high calcium is due to missing radial spokes, we think it unlikely. We extracted the CSC from radial spokeless axonemes and reconstituted the CSC onto axonemes isolated from our double mutants; therefore no spokes were reconstituted in axonemes that showed wild-type sliding velocity under high calcium conditions. In addition, to isolate the CSC, axonemes were first extracted with NaCl, which removes most of the dynein arms as well as additional axonemal components, and then with KI for CSC extraction. Although we cannot completely rule out the possibility that additional axonemal components in the KI extract played a role in restoring sliding velocities, the combined data from these experiments and our previous publication support the conclusion that the CSC is required for high calcium-induced increase in sliding velocities in central apparatus–defective mutants.
In future experiments we would like to determine which dynein isoforms are the targets of this regulation. We previously reported that an anti-CaM-IP2 increase in sliding velocity in central pairless axonemes requires the presence of the I1 inner dynein arm (Dymek and Smith, 2007). The likely localization for the CSC is at or near the base of spoke 2, which is far distal to I1 within the 96 nm spoke repeat. Therefore, the relationship between the CSC and inner dynein arm I1 activity is most likely not direct. To further explore the relationship between the CSC and modulation of dynein-driven microtubule sliding, future experiments will include the generation of double mutants with reduced expression of the CSC as well as particular dynein arm subforms. We expect that extensive analysis of CSC amiRNA mutants by cryo-electron tomography should provide important clues that will inform our future functional studies.
MATERIALS AND METHODS
Chlamydomonas reinhardtii strains
A54-e18 (nit1–1, ac17, sr1, mt+) was obtained from Pete Lefebvre (University of Minnesota, St. Paul) and used as our wild-type strain. pf14 and pf18 were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). For amiRNA double mutants, cells were mated with pf18 (mt–). Flagella were isolated from tetrads, and Western blots were performed to confirm that mutants lacked the central apparatus and had reduced expression of CSC components; CaM-IP2 and CaM-IP3 antibodies were used as CSC markers (Dymek and Smith, 2007), PF20 antibodies were used as a central pair marker (Smith and Lefebvre, 1997), and IC138 antibodies (provided by Win Sale, Emory University, Atlanta, GA) were used as a loading control (for Western methods, see below). All cells were grown in constant light in Tris acetate phosphate (TAP) medium (Gorman and Levine, 1965).
RNAi constructs and Chlamydomonas transformations
amiRNAi constructs were made according to Zhao et al. (2009), Molnar et al., (2009), and DiPetrillo and Smith (2010). Briefly, the Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Ossowski et al., 2008) designated possible amiRNA sequences for CaM-IP2 and CaM-IP3 using their respective cDNA sequences, and designed oligonucleotides with flanking SpeI sites. For CaM-IP2, the amiRNA sequences target the 5´ region of the cDNA: nt25–44 TTTGGGGTCATAGAGAGCATC (#1) and nt113–132 TATTGTTGTACACCGGAGCAC (#2). CaM-IP3 amiRNA sequences targeted the 3´ end of the cDNA sequence: nt2948–2967; TGTAAACAAAGTAGTTCCCGC (#1), nt2125–2144; TGATCCGCGATCCAGGCCAAT (#2), and nt2827–2846; TGAGACAAGACTGGAAGCGAG (#3). See Supplemental Table 1 for complete oligonucleotide sequences.
Oligonucleotides were annealed, phosphorylated, and ligated into a SpeI-digested pChlamiRNA3int vector, which includes the APHVIII gene (Molnar et al., 2009). Colony PCR was used to identify bacteria transformed with plasmids with the correct insert and orientation. IP2 and IP3 amiRNA vectors were transformed into Chlamydomonas using the glass bead method (Kindle, 1990). Cells with integrated plasmid were selected for on 10 μg/ml paromomycin-TAP plates. Picked colonies were assessed for motility defects. Flagella were isolated from cells with motility defects as well as 96 random transformants for each experiment. Flagella were screened by Western blot analysis for the presence of CaM-IP2 or CaM-IP3 protein.
Isolation of flagella and axonemal extracts
The dibucaine method was used to sever flagella from cell bodies (Witman, 1986). Flagella were separated from the cell bodies using differential centrifugation and resuspended in NaLow (10 mM HEPES, pH 7.5, 5 mM MgSO4, 1 mM dithiothreitol [DTT], 0.5 mM EDTA, and 30 mM NaCl). For axonemes, NP-40 (Calbiochem, San Diego, CA) was added to a 0.5% wt/vol final concentration to remove the flagellar membrane. The resulting axonemes were either resuspended in NaLow or, for the examination, by cryo-electron tomography in 10 mM HEPES (pH 7.4, 25 mM NaCl, 4 mM MgSO4, 1 mM, EGTA, 0.1 mM EDTA) to a concentration of ∼1 mg/ml.
For axonemal extracts, axonemes were extracted on ice at 6 mg/ml in NaHigh (10 mM HEPES, pH 7.5, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 0.6 M NaCl) once for 20 min. After pelleting, axonemes were resuspended in NaHigh and immediately pelleted. NaCl extracted axonemes were extracted again at 6 mg/ml for 30 min with KI (10 mM HEPES, pH 7.5, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 0.5 M KI). Axonemes were pelleted and the supernatant (KI extract) was dialyzed against NaLow buffer. After dialysis, KI extracts were clarified by centrifugation at 12,000 × g for 5 min.
Immunoprecipitation
Immunoprecipitation was performed according to Dymek and Smith (2007) with the following modifications: 250 μg of KI extract and 70 μg of affinity-purified antibody (anti-CaM or anti-CaM-IP2 antibody) were incubated for 2 h with 50 μl of protein A beads (Invitrogen, Carlsbad, CA) in TBST150 buffer (150 mM NaCl, 50 mM Tris-HCl, and 0.5 mM EDTA, pH 7.5). After four 5-min washes with TBST150, beads were resuspended in 90 μl of TBST150 and 50 μl of 5X SDS–PAGE sample buffer.
Rebinding the CSC to amiRNA mutant axonemes
KI extracts (from NaCl-extracted axonemes extracted at 6 mg/ml) from pf14 axonemes were isolated and dialyzed into NaLow buffer. Axonemes from 4A2pf18 and 6E6pf18 were isolated and resuspended at 1 mg/ml in NaLow. Equivalent volumes were used for rebinding; for example, axoneme equivalents of extracts were 6 times more concentrated than axonemes (i.e., for a 1:2 ratio of extract:axoneme, 3 μl of extract was mixed with 36 μl of axonemes; 1:1 ratio = 6 μl of extract:36 μl of axonemes, 2:1 ratio = 12 μl of extract:36 μl of axonemes, 2.5:1 ratio = 15 μl of extract:36 μl of axonemes). Each tube was brought to a total volume of 70 μl with NaLow buffer. Tubes were rotated at room temperature for 10 min then centrifuged at 12,000 × g for 10 min. Supernatants were moved to a new tube, pellets were resuspended in 70 μl of NaLow, and both were prepared for SDS–PAGE. For Western blots, 25 μl of sample was loaded on SDS–PAGE gels.
Protein gel electrophoresis, silver-stained gels, and Western blotting
For flagellar and immunoprecipitation Western blots, 8 μg of flagellar protein or 30 μl of immunoprecipitation sample was run on 7% polyacrylamide SDS–PAGE gels and transferred to polyvinylidene difluoride (Immobilon-P; Millipore, Billerica, MA). Membranes were blocked in 5% milk/TTBS (0.1% Tween/Tris-buffered saline, pH 7.5). Primary antibodies were diluted into TTBS. The following dilutions were used: affinity-purified anti-CaM-IP2, 1:500 (vol:vol); affinity-purified anti-CaM-IP3, 1:1000 (vol:vol); anti-RSP1 (provided by Joel Rosenbaum, Yale University, New Haven, CT), 1:10,000 (vol:vol); anti-IC138 (provided by Win Sale, Emory University), 1:20,000 (vol:vol); and anti-PF20, 1:5000 (vol:vol). Anti-rabbit–horseradish peroxidase (GE Healthcare, Piscataway, NJ) was used as the secondary antibody at 1:30,000 (vol:vol) in TTBS, and the ECL Plus Western Blotting Kit (GE Healthcare) was used for detection according to the manufacturer's instructions.
For silver staining, proteins were separated by SDS–PAGE on 7% polyacrylamide gels and fixed for 1 h in 0.1% acetic acid/50% methanol. Gels were rinsed briefly in dH2O and incubated with 0.001% DTT for 20 min then with 0.1% silver nitrate for 30 min. After two brief rinses with dH2O, the gels were placed in developing solution (50 μl of 37% formaldehyde per 100 ml of solution, 3% sodium carbonate), and the reaction was quenched with 2 M citric acid.
Southern blots
For each strain, 5 μg of genomic DNA was digested with KpnI and size fractionated on a 0.7% agarose gel. The DNA was transferred to MagnaGraph nylon membrane (GE Healthcare) and UV cross-linked using the Stratagene Stratlinker (La Jolla, CA). The membrane was probed with the APHVIII gene, which was labeled with 50 μCi of α-32P-dCTP using the Random Primed DNA Labeling Kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. The membrane was exposed to a phosphor screen and scanned with a Typhoon 9200 Imager (GE Healthcare).
Northern blots
Total RNA was isolated from cells 45 min after deflagellation. Poly-A+ RNA was isolated from 1 mg of total RNA using the Oligotex mRNA Midi Kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. Poly-A+ RNA (10 μg) was fractionated on a 1% agarose gel containing formaldehyde and transferred to Hybond-N+ nylon membrane (GE Healthcare). The membrane was probed with cDNA from CaM-IP2 (nt174–619), CaM-IP3 (nt1–630), or RSP3 (full length), which were labeled with 50 μCi of α-32P-dCTP using the Random Primed DNA Labeling Kit (Roche). The ribosomal S14 gene was also labeled and used to probe the membrane as a loading control. The membrane was then exposed to a phosphor screen and scanned with a Typhoon 9200 Imager (GE Healthcare). Densitometry on the scanned image was performed using ImageJ (W.S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij/, 1997–2009). To quantify transcript levels in mutants relative to wild-type cells, we first normalized the amount of transcript relative to the S14 signal for each strain; then wild-type levels were assigned a value of 100% for comparison with each amiRNA mutant.
Swimming velocity, phototaxis, and photoshock
A pco.1200HS camera and Camware software (The Cooke Corporation, Londonderry, NH) were used for high-speed video analysis. A red filter was used for asymmetric swimming analyses and was removed for symmetric swimming analyses. All data are presented as mean ± SEM. The Student's t test was used to determine the significance of differences between means. Images were analyzed and montages were created in ImageJ (http://rsb.info.nih.gov/ij/, 1997–2009). For phototaxis, cells grown in TAP were placed in a Petri dish. Approximately 75% of the plates were covered with dark plastic, leaving ∼25% exposed to light. After a few hours, plates were removed and compared with wild type. Cells that phototaxed accumulated at the end of the plate exposed to light.
EM
For classical EM, axonemal pellets were fixed in 1% glutaraldehyde and 1% tannic acid in 0.1 M sodium cacodylate (NaCac). Postfixation required 1% osmium tetroxide in 0.1 M NaCac. Samples were stained with 1% uranyl acetate, dehydrated, and embedded. Thin sections were stained with uranyl acetate and Reynold's lead citrate. A JEOL JEM-1010 transmission electron microscope (JEOL, Tokyo, Japan) was used for examination.
For cryo-electron tomography, details about sample preparation, imaging, and tomogram reconstruction were published previously (Nicastro et al., 2006; Heuser et al., 2009; Nicastro 2009). Briefly, isolated axonemes (see above) were vitrified on EM grids within 24 h of preparation. Copper EM grids with a carbon support film (Quantifoil, R2/2, 200 mesh; Quantifoil Micro Tools, Jena, Germany) were glow discharged for 30 s at –40 mA, coated with 10 nm of colloidal gold (Sigma-Aldrich, St. Louis, MO), and loaded into a homemade plunge freezer. Three microliters axoneme sample was applied to an EM grid and mixed with 1 μl of a 10 times concentrated 10 nm colloidal gold solution. The mixture was blotted from the front side with filter paper (Whatman #1, Piscataway, NJ) for ∼2 s, and immediately plunge-frozen in liquid ethane. Grids were then transferred to liquid nitrogen for storage.
For imaging, the grid was transferred to an FEI Tecnai F30 transmission electron microscope in a cryo-holder (Gatan, Pleasanton, CA). Using the microscope control software SerialEM (Mastronarde, 2005), frozen-hydrated axonemes were imaged in low dose mode, at 300 keV extraction voltage and in zero-loss mode of the postcolumn energy filter (Gatan) with a 20 eV slit width. We used a 2k × 2k charge-coupled device camera (Gatan) to record images at 13,500-fold magnification resulting in a pixel size of 1 nm. Defocus values from −6 to −8 μm were used to enhance image contrast. Between 50 and 100 images were recorded per tilt series by stepwise rotation of the specimen from –65° to +65° with tilting increments of 1.5° to 2.5°. To minimize radiation damage to the sample, the accumulative electron dose was limited to <100 e/Å2. Cryo-electron tomograms were reconstructed using the IMOD software package (Kremer et al., 1996) and fiducial alignment. Data analysis was restricted to undistorted tomograms of intact axonemes with no or only minor compression (Figure 5A).
For the structural analysis, especially of the radial spokes, four to six cryo-tomograms (the equivalent of 600–1000 axonemal repeats) of each strain (CaM-IP2 amiRNA mutants 4A2 and 4D6, and the CaM-IP3 amiRNA mutant 6E6) were analyzed and compared with tomograms of wild-type axonemes (wild-type data were provided from a previous study; Heuser et al., 2009).
Microtubule sliding assay
Flagella were severed from cell bodies by using the dibucaine method (Witman, 1986) and isolated by differential centrifugation in buffer A (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, and 50 mM potassium acetate). Axonemes were isolated by adding NP-40 (Calbiochem) to flagella for a final concentration of 0.5% (wt/vol) to remove flagellar membranes. Measurement of sliding velocity between doublet microtubules was based on the methods of Okagaki and Kamiya (Okagaki and Kamiya, 1986). Microtubule sliding was initiated with buffer A containing 1 mM ATP and 2 μg/ml type VIII protease (Sigma-Aldrich). Sliding was observed and recorded using an AxioSkop 2 microscope (Carl Zeiss, Thornwood, NY) equipped for dark-field optics including a Plan-Apochromat 40× oil immersion objective with iris and ultra dark-field oil immersion condenser. Images were captured using a silicon-intensified target camera (VE-1000 SIT; Dage-MTI, Michigan City, IN) and converted to digital images using LabVIEW 7.1 software (National Instruments, Austin, TX). For some experiments, equivalent volumes of KI extracts (see methods above) were added to axonemes before ATP addition. Reconstitution proceeded for 15 min at room temperature before initiation of microtubule sliding. All data are presented as mean ± SEM. All sliding experiments represent the result of three to five experiments with a total n for each mutant and condition of between 60 and 160. The Student's t test was used to determine the significance of differences between means.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM083122 to D. Nicastro and GM66919 to E.F. Smith) and the National Science Foundation (DMR-MRSEC-0820492) and by a Pew Scholars Award (to D. Nicastro).
Abbreviations used:
- amiRNA
artificial microRNA
- CaM
calmodulin
- CSC
CaM and Spoke associated Complex
- DTT
dithiothreitol
- EM
electron microscopy
- RFLP
restriction fragment length polymorphism
- RSP
radial spoke protein
- TAP
Tris acetate phosphate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0271) on May 25, 2011.
REFERENCES
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo CG, Smith EF. Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J Cell Biol. 2010;189:601–612. doi: 10.1083/jcb.200912009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Huang B, Luck DJ. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J Cell Biol. 1984;98:229–236. doi: 10.1083/jcb.98.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J Cell Biol. 2001;153:443–448. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol. 1983;97:902–908. doi: 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J Cell Biol. 1981;88:80–88. doi: 10.1083/jcb.88.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelekar P, Wei M, Yang P. Isolation and analysis of radial spoke proteins. Methods Cell Biol. 2009;92:181–196. doi: 10.1016/S0091-679X(08)92012-4. [DOI] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lee L, Campagna DR, Pinkus JL, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, Pinkus GS, Fleming MD. Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol Cell Biol. 2008;28:949–957. doi: 10.1128/MCB.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck D, Piperno G, Ramanis Z, Huang B. Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc Natl Acad Sci USA. 1977;74:3456–3460. doi: 10.1073/pnas.74.8.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck DJ, Piperno G. Dynein arm mutants of Chlamydomonas. New York:: Alan R. Liss; 1989. pp. 49–60. [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Nicastro D. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol. 2009;91:1–39. doi: 10.1016/S0091-679X(08)91001-3. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol. 1986;103:1895–1902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Otter T. Cell Movement. Vol. 1. New York: Alan R. Liss; 1989. Calmodulin and the control of flagellar movement; pp. 281–298. [Google Scholar]
- Patel-King RS, Gorbatyuk O, Takebe S, King SM. Flagellar radial spokes contain a Ca2+-stimulated nucleoside diphosphate kinase. Mol Biol Cell. 2004;15:3891–3902. doi: 10.1091/mbc.E04-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Huang B, Ramanis Z, Luck DJ. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, O'Ooole E, Porter ME. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol Biol Cell. 2001;12:739–751. doi: 10.1091/mbc.12.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol. 2007;69:401–422. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by calcium and a role for an axonemal calmodulin and calmodulin-dependent kinase. Mol Biol Cell. 2002a;13:3303–3313. doi: 10.1091/mbc.E02-04-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil Cytoskeleton. 2002b;52:33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- Smith EF, Lefebvre PA. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol Biol Cell. 1997;8:455–467. doi: 10.1091/mbc.8.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Dymek EE, Smith EF. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J Cell Sci. 2005;118:4655–4665. doi: 10.1242/jcs.02585. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, McPeek MA, Smith EF. Analysis of microtubule sliding patterns in Chlamydomonas flagellar axonemes reveals dynein activity on specific doublet microtubules. J Cell Sci. 2004;117:2533–2544. doi: 10.1242/jcs.01082. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, Smith EF. Asymmetry of the central apparatus defines the location of active microtubule sliding in Chlamydomonas flagella. Proc Natl Acad Sci USA. 2003;100:137–142. doi: 10.1073/pnas.0135800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Zhao F, Yang C, Yang P, Diener D, Gaillard A, Rosenbaum JL, Sale WS. Building a radial spoke: flagellar radial spoke protein 3 (RSP3) is a dimer. Cell Motil Cytoskeleton. 2008;65:238–248. doi: 10.1002/cm.20257. [DOI] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Yagi T, Minoura I, Fujiwara A, Saito R, Yasunaga T, Hirono M, Kamiya R. An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J Biol Chem. 2005;280:41412–41420. doi: 10.1074/jbc.M509072200. [DOI] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, Sale WS. Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol. 2001;153:1315–1326. doi: 10.1083/jcb.153.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, et al. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Sale WS. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3:72–81. doi: 10.1128/EC.3.1.72-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukitake H, Furusawa M, Taira T, Iguchi-Ariga SM, Ariga H. AAT-1, a novel testis-specific AMY-1-binding protein, forms a quaternary complex with AMY-1, A-kinase anchor protein 84, and a regulatory subunit of cAMP-dependent protein kinase and is phosphorylated by its kinase. J Biol Chem. 2002;277:45480–45492. doi: 10.1074/jbc.M206201200. [DOI] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009 doi: 10.1111/j.1365-313X.2008.03758.x. 58, 157–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.