Caenorhabditis elegans pxl-1 is the orthologue of vertebrate paxillin and is expressed in body wall and pharyngeal muscle. In body wall muscle PXL-1 localizes to dense bodies, M-lines, and adhesion plaques, and in pharyngeal muscle PXL-1 localizes to podosome-like actin attachment complexes. PXL-1 is required in the pharynx for muscle contraction and viability.
Abstract
We have identified the gene C28H8.6 (pxl-1) as the Caenorhabditis elegans orthologue of vertebrate paxillin. PXL-1 contains the four C-terminal LIM domains conserved in paxillin across all species and three of the five LD motifs found in the N-terminal half of most paxillins. In body wall muscle, PXL-1 antibodies and a full-length green fluorescent protein translational fusion localize to adhesion sites in the sarcomere, the functional repeat unit in muscle responsible for contraction. PXL-1 also localizes to ring-shaped structures near the sarcolemma in pharyngeal muscle corresponding to podosome-like sites of actin attachment. Our analysis of a loss-of-function allele of pxl-1, ok1483, shows that loss of paxillin leads to early larval arrested animals with paralyzed pharyngeal muscles and eventual lethality, presumably due to an inability to feed. We rescued the mutant phenotype by expressing paxillin solely in the pharynx and found that these animals survived and are essentially wild type in movement and body wall muscle structure. This indicates a differential requirement for paxillin in these two types of muscle. In pharyngeal muscle it is essential for contraction, whereas in body wall muscle it is dispensable for filament assembly, sarcomere stability, and ultimately movement.
INTRODUCTION
Adhesion complexes are highly dynamic structures involved in the extension of a cell membrane for the purposes of locomotion. Movement of such cells involves polymerization of actin filaments and subsequent attachment to the extracellular matrix (Burridge et al., 1988; Ridley et al., 2003). Through studies on cultured murine fibroblasts, paxillin has been shown to be one of the key proteins required for proper maintenance and/or turnover of focal adhesion complexes (Hagel et al., 2002). Much of the work on paxillin has focused on its role in adhesion structures in cultured cells, but paxillin is also highly expressed in skeletal muscle, cardiac muscle, and smooth muscle (Turner et al., 1991). Because relatively little is known about the function of paxillin in muscle, the study of this protein in model organisms may help to elucidate its in vivo function (reviewed in Brown and Turner, 2004; Deakin and Turner, 2008). Caenorhabditis elegans is an appropriate organism with which to study the role of paxillin in muscle because nematode body wall muscles contain actin attachment structures analogous to Z-discs in vertebrate muscle called dense bodies, which also bear striking resemblance to adhesion complexes in terms of protein composition and function (Labouesse and Georges-Labouesse, 2003).
Paxillin is a well-conserved protein found in many organisms, including humans (Turner et al., 1990), zebrafish (Crawford et al., 2003), and fruitfly (Wheeler and Hynes, 2001). Human paxillin is a 591–amino acid protein with five LD motifs in its N-terminal region and four lin-11, isl-1, mec-3 (LIM) domains in its C-terminal half (Turner et al., 1990; Turner and Miller, 1994). LD motifs are short stretches of amino acids with a consensus sequence of LDXLLXXL (Brown et al., 1996), although variations from this sequence are possible across species and even within the third LD motif (reviewed in Tumbarello et al., 2002). The LD motifs are responsible for mediating many of the direct interactions that paxillin has with other proteins in focal adhesion complexes (Brown et al., 1996; Brown and Turner, 2004). LIM domains are tandem zinc fingers that are responsible for mediating interactions with other proteins (Schmeichel and Beckerle, 1994), and the four LIM domains in paxillin have been shown to be sufficient for its localization to adhesion complexes (Brown et al., 1996).
Paxillin is found in another type of adhesion complex called the podosome (Bowden et al., 1999). Similar to focal adhesion structures, podosomes are dynamic structures and include many of the same proteins but have a different structural arrangement of these proteins. Whereas focal adhesions comprise a complex of proteins extending inward from the cell membrane and anchoring actin filaments, podosomes have a core of F-actin (Marchisio et al., 1984), which has focal adhesion components such as paxillin, vinculin, and talin organized around the actin core in a ring-shaped formation (Bowden et al., 1999). Recently, podosome-like structures in cultured myotubes have been shown to play a role at neuromuscular junctions by guiding and remodeling the postsynaptic membrane as it matures (Proszynski et al., 2009). Podosomes are found in a variety of cell types, including smooth muscle cells (Hai et al., 2002; Kaverina et al., 2003) and migrating cells (Linder, 2007), and, like adhesion complexes, are able to play a role in cell movement (Linder, 2007).
Previously, little work has been done on the structural organization of actin attachment sites in C. elegans pharyngeal muscle. Much of the work directed on muscle in the worm has focused on the body wall muscle. Whereas body wall muscle is comprised of cells with multiple sarcomeres in register (reviewed in Moerman and Fire, 1997; Moerman and Williams, 2006), pharyngeal muscles contain a series of single sarcomeres that traverse the diameter of the cell (Albertson and Thomson, 1976). Pharyngeal muscle in the worm has been proposed as a possible model for cardiac muscle, albeit with limitations (Mango, 2007), whereas body wall muscle is generally used as a model for mammalian skeletal muscle, again with limitations (Moerman and Williams, 2006). As previously mentioned, dense bodies in body wall muscle are actin attachment sites functionally similar to Z-discs in mammalian muscle (Moerman and Williams, 2006), and transmission electron microscope (TEM) images of pharyngeal muscle cells also show similar electron-dense structures at the ends of actin filaments (Albertson and Thomson, 1976). Whereas green fluorescent protein (GFP)–tagged body wall muscle proteins that localize to dense bodies show a punctate pattern indicating localization throughout the sarcolemmal plane of the dense body, to date the specific organization of muscle proteins within the pharyngeal muscle dense bodies has not been established.
We have found that a single paxillin homologue is present in the genome of C. elegans and is expressed in body wall and pharyngeal muscle cells at sites of actin attachment. Animals homozygous for a pxl-1 deletion have paralyzed pharyngeal muscle and arrest and die as first-stage larvae. A GFP translational fusion for pxl-1 expressed exclusively in pharyngeal muscle and marginal cells restores pharyngeal muscle function in pxl-1 mutants, indicating that although pxl-1 is expressed in body wall muscle and pharyngeal muscle, its essential role is within the pharynx.
RESULTS
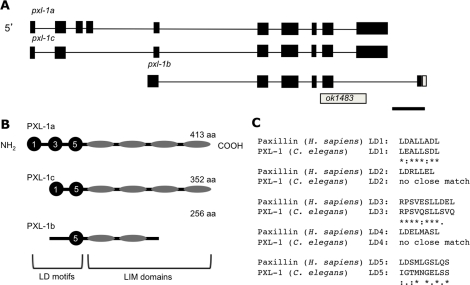
pxl-1 encodes three splice variants of paxillin
The gene C28H8.6 (pxl-1), found near the center of chromosome III, encodes three splice variants of a paxillin-like gene (Figure 1A). The largest of these splice variants encodes a 413–amino acid protein with sequence similarity to paxillin in other species, including humans. Human paxillin is a 557–amino acid protein containing an LD motif rich region in its N-terminal half and four LIM domains in its C-terminal half. (Brown et al., 1996). Splice variant a of C. elegans paxillin codes for a protein product with LD motifs in its amino half, albeit truncated as compared with its human counterpart, and four LIM domains in its C-terminal half (Figure 1B). An alignment of the two proteins using ClustalW2 (Chenna et al., 2003) reveals significant similarity between C. elegans and human paxillin throughout the protein, with some gaps in the C. elegans portion of the alignment in the LD motif region due to its shorter sequence length (Figure 2). In terms of the LD motifs, PXL-1 appears to share LD motifs 1, 3, and 5 but not 2 and 4 (Figure 1C). This overall similarity, and the fact that there are no other predicted nematode proteins with this combination of domains, supports the possibility that PXL-1 may be the nematode orthologue of human paxillin.
FIGURE 1:
C. elegans paxillin (pxl-1) gene and protein models. The pxl-1 gene in C. elegans has three alternatively spliced isoforms (A), which are all affected by the deletion allele ok1483. Exons are in black, with the UTR in white. Conservation of three of the five LD motifs found in the N-terminal half of paxillin in H. sapiens (PXN) is found in the a isoform of PXL-1, two of the 5 LD motifs in isoform c, and only one LD motif in isoform b. (B). Sequence homology between the LD motifs can be seen in C. Bar, 500 base pairs.
FIGURE 2:
Comparison of PXL-1 protein sequence with paxillin (H. sapiens). PXL-1a aligns strongly with human, fly, and frog paxillin, especially in the LIM domain C-terminal half of the protein (A) using ClustalW2 software. Within the N-terminal half of PXL-1, homology is seen between species, with PXL-1 containing three of the five LD motifs found in the each of the compared proteins (B). Amino acid alignments were marked using ClustalW2 conventions (Larkin et al., 2007): asterisks (*), identical amino acids; colons (:), conserved substitutions; periods (.), semiconserved substitutions. Bars below the LIM domains mark their boundaries.
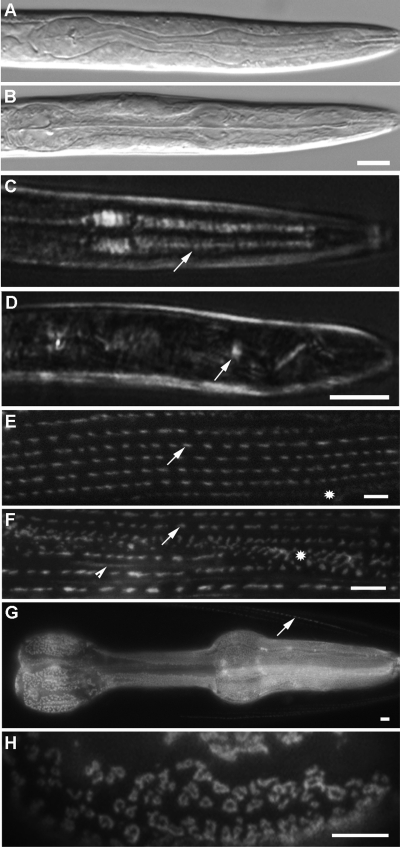
pxl-1 mutant animals die as young larvae
The C. elegans Gene Knockout Consortium provided the strain VC1012, which carries a 943-bp deletion within the coding sequence of C28H8.6 (Figure 1A). When homozygous for the mutant allele ok1483, animals arrest at the first larval stage of development (L1), which is ∼0.2 mm in length, and die. Whereas a wild-type worm grows to ∼1 mm in length at the adult stage after 3 d and begins to lay eggs, pxl-1 mutants remain at ∼0.2 mm in length. On close examination of the pharynx in mutant animals, a lack of pumping is clearly noticeable. The pharynx structure appears to be normal, and we detect no visible attachment defects (Figure 3B) like those seen in the α-integrin mutant ina-1 (Baum and Garriga, 1997), but the pharyngeal muscles do not contract. The internal pharyngeal muscle structure can be observed by using polarized light microscopy (Avery, 1993), and on closer examination, the birefringence of myofilaments in animals lacking PXL-1 is reduced throughout most of the pharyngeal muscle (Figure 3D). The reduced birefringence of pharyngeal myofilaments that we observe is very similar to that observed in an actin mutant (Avery, 1993; Figure 5C–5D). This lack of organized myofilament structure and thus pharyngeal pumping in pxl-1(ok1483) homozygotes means the animals cannot feed, so we presume that the animals die of starvation as L1 larvae. The interfering RNA (RNAi) phenotype of worms fed dsRNA for pxl-1 is similar in appearance to ok1483 lethal homozygotes. Whereas a balanced lethal animal contributes maternal transcripts due to its heterozygous genotype, RNAi knocks down the gene product for pxl-1 in the treated adult animals, and thus a maternal contribution of pxl-1 cannot occur in its progeny. The lack of a stronger phenotype using RNAi indicates that there is no maternal contribution of pxl-1 that is critical to the developing embryo.
FIGURE 3:
Phenotype of homozygous pxl-1 (ok1483) mutant animals and subcellular localization of PXL-1::GFP translational fusion products. The pharynx of pxl-1 mutants does not pump, but the physical structure of the pharynx in wild-type worms (A) appears similar to the structure in pxl-1 mutant animals (B). Within the pharyngeal muscle, polarized light microscopy was used to look at the myofilament structure. In worms with pxl-1 expression restored in the pharynx, myofilaments extend across the pharyngeal muscle cells, indicated by arrows highlighting the birefringence of the myofilaments (C). Animals not expressing pxl-1::GFP arrest at the L1 stage of development and have a lack of myofilament birefringence in pharyngeal muscle (D). pxl-1a(cDNA)::GFP is expressed strongly in body wall muscle (E) and pharyngeal muscle under control of its endogenous promoter. PXL-1a(cDNA)::GFP is localized to dense bodies (arrows) and adhesion plaques (star) and weakly to M-lines (not shown). A GFP fusion product containing only the four LIM domains of PXL-1 fused to GFP also localizes to dense bodies (arrows) and adhesion plaques (star) and weakly to M-lines (arrowhead) (F). pxl-1a/c::GFP under control of a minimal promoter lacking body wall muscle promoter elements is expressed strongly in the pharyngeal muscle and weakly in body wall muscle (arrow), solely at the anterior tip of the animal (G). Within pharyngeal muscle, PXL-1a/c::GFP and PXL-1a(CDNA)::GFP (not shown but identical to PXL-1a/c::GFP) is localized to ring-shaped structures near the muscle cell membrane (H). Bars: A–D, 10 μm; E–F, 2 μm.
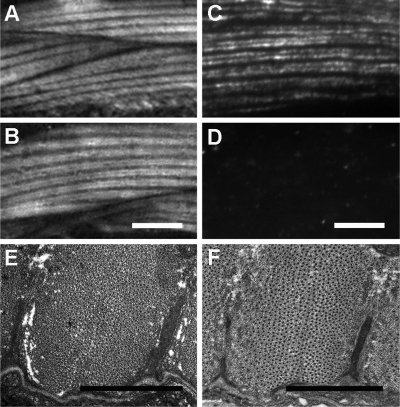
FIGURE 5:
Body wall muscle in animals with pxl-1 expression restricted to the pharynx. Animals that were transformed using a pxl-1a/c::GFP construct with only ∼150 base pairs of endogenous promoter have pxl-1 expression almost exclusively in the pharynx. Compared with the body wall muscle structure in wild-type worms observed using polarized light microscopy (A), the muscle structure looks identical in pharyngeal-specific pxl-1–rescued animals (B). In animals lacking paxillin in the body wall muscle, actin (antibody C4) is clearly seen (C), whereas GFP antibody does not detect any GFP-tagged paxillin, with only background fluorescence from the secondary antibody observed (D). TEM was also used to look at the body wall muscle structure in wild-type animals (E) and animals lacking pxl-1 expression in body wall muscle (F), also showing properly formed dense bodies (arrows) and no structural differences. Bars: A–D, 5 μm; E, F, 1 μm.
Isoform diversity leads to differential subcellular location and tissue expression of paxillin
We first rescued the pxl-1 mutant phenotype with a GFP translational fusion containing ∼2.4 kb of upstream promoter sequence, the full pxl-1 gene, and GFP sequence tagged to the terminal exon of pxl-1 in the expression vector pPD95.75 (provided by Andrew Fire, Stanford University, Stanford, CA). Because the 3′ exon for the b isoform is downstream of the terminal exon for pxl-1a and pxl-1c, non–GFP-tagged PXL-1a and PXL-1c are produced from the extrachromosomal array, along with GFP-tagged PXL-1b. This construct fully rescued the first larval stage arrest phenotype. In addition, we noted that PXL-1b::GFP is diffusely expressed in the pharyngeal epithelial cells and weakly so in pharyngeal muscles one and three.
Isoform a is most similar to human paxillin. We used a full-length cDNA for pxl-1a to create a GFP-tagged injectable marker, under the control of a ∼2.5-kb endogenous promoter. We found the a isoform of paxillin in all 95 body wall muscle cells in an adult animal, as well as in the pharyngeal muscle cells. Strong localization of PXL-1a(cDNA)::GFP is seen in adult body wall muscle at dense bodies, as well as adhesion plaques between adjacent muscle cells (Figure 3E). Weak GFP localization also can be observed at the M-line (not shown). In the pharynx, expression of PXL-1a(cDNA)::GFP can be seen in all muscle cells, localized to both the luminal and cuticular membranes. PXL-1a(cDNA)::GFP is localized in small punctae, as well in ring-like structures near muscle cell membranes (identical to Figure 3F). We were unable to design a c isoform–specific GFP reporter because it shares its 3′-terminal exon with the a isoform, so we do not know whether the distribution of this isoform overlaps or mimics the a and/or b isoforms.
To reinforce our conclusions on paxillin subcellular localization within muscle, a polyclonal antibody was raised against the full-length PXL-1a protein. This antiserum is able to bind to PXL-1 in both nematode lysate and yeast lysate expressing hemagglutinin (HA)-tagged PXL-1 via Western blot (Figure 4D). The PXL-1 antiserum was also used to verify the localization pattern seen with our GFP constructs. Within adult body wall muscle the PXL-1a antibody localizes to dense bodies and adhesion plaques and very weakly to M-lines (Figure 4C), colocalizing with PAT-3/β-integrin. Within the pharynx, there is staining within the muscle cells, but the staining is somewhat fuzzy and punctate (unpublished data).
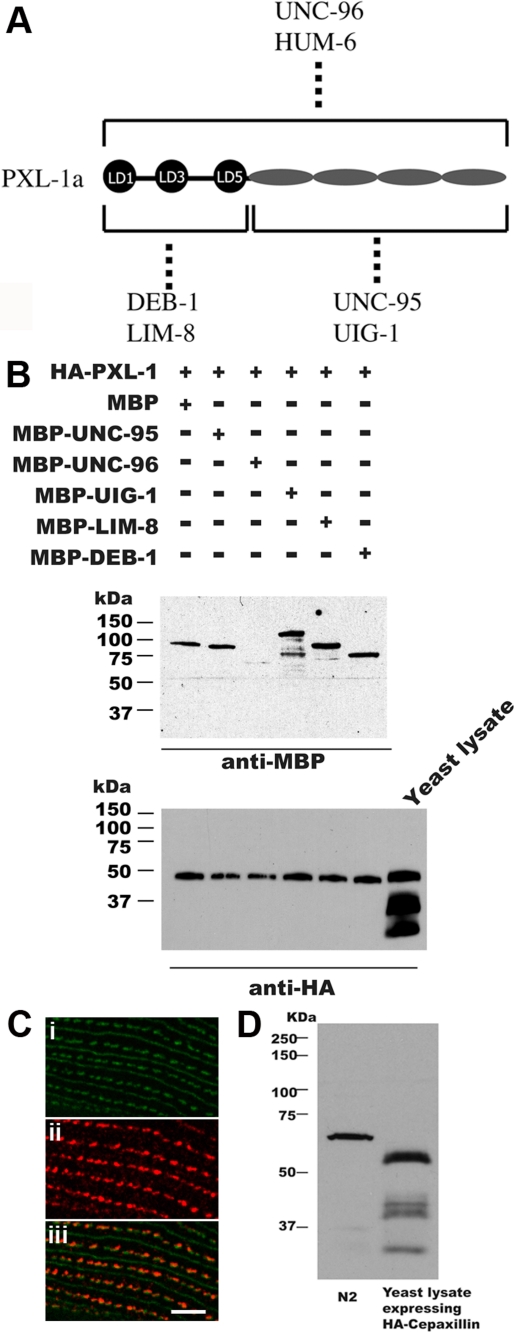
FIGURE 4:
Confirmation of protein interactions and immunostaining with a PXL-1 antibody (A). Screening by the yeast two-hybrid method of a collection of 30 known components of dense bodies and M-lines revealed that the six indicated proteins interact with PXL-1. By domain mapping, the LD motif region is sufficient for interaction with DEB-1 (vinculin) and LIM-8, and the LIM domain region is sufficient for interaction with UNC-95 and UIG-1 (a Cdc42 GEF). UNC-96 and HUM-6 are also able to bind to PXL-1. (B) These interactions, except that with HUM-6, were verified using purified proteins. Yeast-expressed HA-tagged PXL-1 was precipitated with anti-HA beads, washed, incubated with bacterially expressed MBP of the indicated MBP fusion proteins, and washed, and the eluted proteins were separated on a gel, blotted, and reacted with either anti-MBP or anti-HA. All tested proteins except UNC-96 showed conclusive results, with a faint band observed in 5B when reacted with anti-MBP. A paxillin (PXL-1) polyclonal rabbit antibody was produced to detect PXL-1 in C. elegans as well as in yeast expressing HA-PXL-1. PAT-3/β-integrin (GFP; Ci) is a major component of dense bodies, M-lines, and attachment plaques, and the PXL-1 antibody (Cii) clearly colocalizes with PAT-3/β-integrin in dense bodies and adhesion plaques (Ciii). The PXL-1 antibody also stains M-lines, but very weakly (not shown). Paxillin from the nematode lysate displays a slower mobility than that of nematode paxillin expressed in yeast (see Discussion) (D). Bar, 2 μm.
Pharyngeal muscle pxl-1 rescue
It was not clear why pxl-1 null animals die, although we suspected that it was a result of starvation, not a problem with the body wall muscle. To test this possibility, we designed a pxl-1 construct with expression only in the pharynx. Using the C. elegans muscle cis-regulatory module locator (Zhao et al., 2007) showed that the pxl-1 promoter has multiple body wall muscle–specific expression motifs between 250 base pairs upstream of the transcriptional start site and 3 kb upstream. We cloned the genomic region of pxl-1 covering the a and c isoforms along with the immediate 5′ ∼150 base pairs of the endogenous promoter in frame with sequence encoding GFP. The translational fusion construct produces GFP-tagged PXL-1a and PXL-1c, but because the terminal exon for pxl-1b is downstream of the terminal exon for pxl-1a/c, PXL-1b is not produced, at least not as a full-length protein (see Figure 1, A and B, for clarification). The L1 arrest phenotype was fully rescued using this construct. The tissue expression using the minimal promoter is almost solely within the pharyngeal muscle cells. Only a few body wall muscle cells around the anterior tip of the worm display a faint GFP signal, but otherwise expression from this PXL1a/c::GFP construct is pharyngeal specific (Figure 3G). Progeny carrying this extrachromosomal array display wild-type movement, and the body wall muscle structure appears wild type using both polarized light microscopy (Figure 5, A and B) and transmission electron microscopy (Figure 5, E and F). Within the pharynx, we again observe that paxillin is associated with ring-like structures similar to those previously described (Figure 3H). Rescue of mutant lethality by expressing paxillin only within the pharynx supports our earlier speculation that animals lacking paxillin starve to death. It also leads us to the unexpected conclusion that paxillin is not necessary for the function of body wall muscle in the nematode.
PXL-1 LIM domains are sufficient for proper localization in body wall muscle
Previous studies (Brown et al., 1996) showed that LIM domains (specifically LIM domain 3) are necessary and sufficient for the localization of paxillin to focal adhesion complexes. To test whether the LIM domains of pxl-1 are sufficient for proper localization of paxillin in the worm, we constructed a GFP expression construct coding for only the four LIM domains of PXL-1 fused to GFP and under the control of the body wall muscle–specific promoter from the gene T05G5.1 (unpublished data). The extrachromosomal array does not rescue the mutant phenotype, but the GFP-tagged protein containing only LIM domains is localized to dense bodies, adhesion plaques, and M-lines in body wall muscle (Figure 3F). The identical subcellular localization pattern seen with the LIM-only portion of PXL-1 and the full-length PXL-1 indicates that the LIM domains are sufficient for localization of the protein to its proper subcellular locations.
PXL-1 interacts with DEB-1/vinculin, UIG-1, LIM-8, UNC-96, and UNC-95
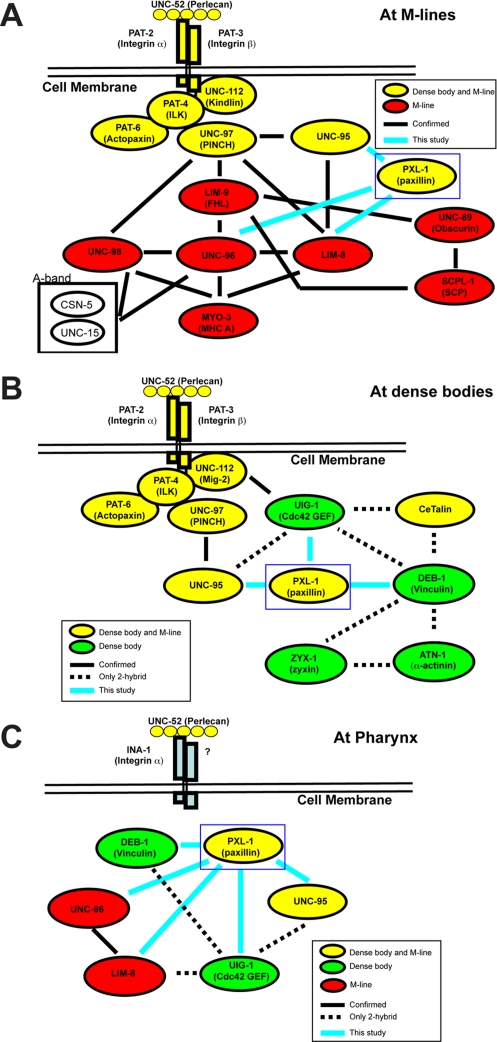
LIM-domain proteins, including paxillin, have been implicated as adapter molecules responsible for the recruitment of proteins and acting as a scaffold for protein complexes to assemble (Schmeichel and Beckerle, 1994; reviewed in Brown and Turner, 2004). We used the yeast two-hybrid method to screen a “bookshelf” of primarily known dense body and M-line proteins using PXL-1a as bait. Six interactors were detected: DEB-1/vinculin, UIG-1, LIM-8, UNC-96, UNC-95, and HUM-6 (Figure 4A). Except for HUM-6 (to be described elsewhere), each positive hit was then retested by fixing HA-tagged PXL-1a to anti-HA beads and incubating with a purified maltose-binding protein (MBP)–tagged sample of each putative interactor. Western blots confirm that each of the putative yeast two-hybrid interactors do bind to PXL-1 in vitro (Figure 4B). Vinculin is a major interactor of paxillin across species (reviewed in Brown and Turner, 2004), and this relationship is conserved in C. elegans. Along with DEB-1/vinculin, we also observe in vitro binding between the LD motifs of PXL-1 and the muscle protein LIM-8 and in vitro binding between the LIM domains of PXL-1 and the proteins UIG-1 and UNC-95 (Figure 4A). Two additional proteins, UNC-96 and HUM-6, can bind PXL-1 along the length of the protein. Each of UNC-96 (Mercer et al., 2006), UIG-1 (Hikita et al., 2005), and LIM-8 (Qadota et al., 2007) is expressed in body wall and pharyngeal muscle in vivo. In addition, DEB-1/vinculin is detected in pharyngeal and body wall muscle using the monoclonal antibody (mAb) MH24, and both DEB-1 and UNC-95 have serial analysis of gene expression (SAGE) data indicating expression in those two tissues (Meissner et al., 2009). Each of the interactions between paxillin and its binding partners can be further expanded by mapping out in vitro interactions between these partners and other proteins found in attachment complexes to show what is likely occurring in vivo (Figure 6). The protein–protein interactions at body wall muscle adhesion junctions have been mapped (reviewed in Moerman and Williams, 2006; Qadota and Benian, 2010). In pharyngeal muscle, however, the network of interactions is less well studied, as a number of body wall muscle proteins are not found in this tissue, including PAT-3/β-integrin. This means that there is not a one-to-one correspondence between attachment sites between these two types of muscle.
FIGURE 6:
Protein interactions with PXL-1. PXL-1 is found in dense bodies (A) and M-lines (B), as well as in attachment sites in the pharynx (C). When mapped out, the interactions found in each of these structures can be compared with previously documented interactions. A solid line indicates a confirmed in vitro interaction, and a dotted line indicates that an interaction has only been observed using a yeast two-hybrid method. Proteins highlighted in yellow are found in both dense bodies and M-lines. Proteins found only in dense bodies (green) and those found only in M-lines (red) have also been included. All of the interactions confirmed in this study are labeled with blue lines and confirmed as shown in Figure 4.
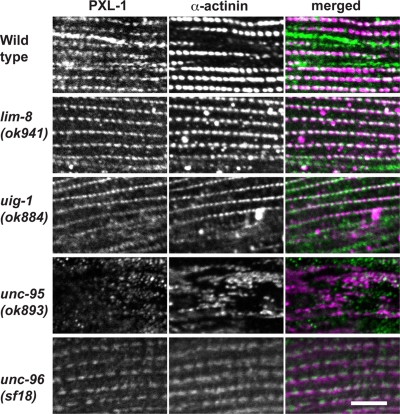
Comparing protein–protein interactions with genetic interactors of pxl-1 demonstrates unc-95 is necessary for pxl-1 localization in body wall muscle
To further characterize the interactions we discovered between PXL-1 and four of its binding partners (LIM-8, UIG-1, UNC-95, and UNC-96), we examined PXL-1 localization in body wall and pharyngeal muscle in the mutant background of each interactor. We concentrated our examination on the dense body localization of PXL-1 rather than the M-line localization because of the weak signal for PXL-1::GFP at M-lines. Although PXL-1 is properly localized at dense bodies in lim-8, uig-1, and unc-96 mutant animals, PXL-1 is diffuse in unc-95 mutants and does not integrate into the dense body structure (Figure 7). The same experiment was carried out testing pharyngeal muscle in the four mutant backgrounds. In lim-8, uig-1, unc-95, and unc-96 mutant worms, PXL-1 was properly localized to the pharyngeal muscle membrane (unpublished data). Although loss of unc-95 alters the localization of PXL-1 in body wall muscle, it does not affect its localization in the pharynx.
FIGURE 7:
Localization of PXL-1 in the mutant background of binding partners. PXL-1 immunostaining was used to observe the subcellular localization of PXL-1 in four mutant backgrounds. A monoclonal antibody for α-actinin was used to label dense bodies. Although PXL-1 is properly localized to dense bodies in the lim-8, uig-1, and unc-96 backgrounds, it is diffuse in the cytoplasm in the unc-95 background. Bar, 10 μm.
LIM domain redundancy in body wall muscle?
Due to the number of different LIM domain–containing proteins in the worm, it is possible that animals lacking pxl-1 expression have phenotypically wild-type body wall muscle due to another LIM domain protein compensating for its loss. To test this, we used the strain DM7335, which has pxl-1 expression in the pharynx but not body wall muscle, and used RNAi feeding to knock down the expression of eight genes coding for LIM domain proteins with known expression in body wall muscle (Meissner et al., 2009), including zyx-1/zyxin and alp-1/enigma (Table 1). Other than our pxl-1 RNAi control, each tested gene does not have an RNAi phenotype in wild-type animals, so we screened RNAi-treated DM7335 animals for uncoordinated movement, as well as observed the myofilament organization in muscle cells using polarized light microscopy. Only lim-8 had any significant effect, with RNAi-treated animals displaying uncoordinated movement and mildly disorganized myofilaments in body wall muscle cells.
TABLE 1:
RNAi phenotypes of genes coding for LIM domain proteins in DM7335 animals lacking pxl-1 body wall muscle expression.
| Gene | RNAi phenotype | Body wall muscle structure |
|---|---|---|
| pxl-1 | L1 arrest | Wild type |
| zyx-1 | Wild type | Wild type |
| alp-1 | Wild type | Wild type |
| mlp-1 | Wild type | Wild type |
| lim-9 | Wild type | Wild type |
| F42H10.3 | Wild type | Wild type |
| lim-8 | Uncoordinated | Myofilaments mildly disorganized |
Localization of PXL-1–interacting proteins in body wall muscle lacking PXL-1
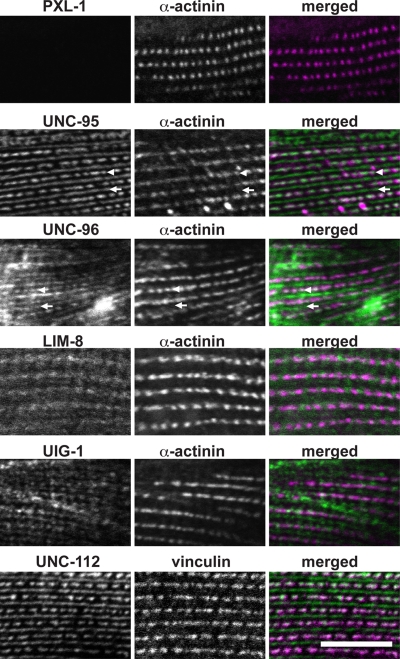
PXL-1 is localized properly in the mutant background of each of its interactors except for unc-95 (Figure 7), so we examined the localization of interacting proteins in body wall muscle lacking pxl-1 expression, using antibodies to stain the strain DM7335 to see whether PXL-1–interacting proteins would be mislocalized (Figure 8). ATN-1/α-actinin is normally localized in a pxl-1 background and was used to costain animals to mark dense body adhesion complexes (Figure 8). Each of the proteins tested that interact with PXL-1 are normally localized in body wall muscle lacking PXL-1. UNC-95 localization was normal, indicating that in body wall muscle PXL-1 requires UNC-95 for proper localization (Figure 7), but UNC-95 does not require PXL-1. UNC-112/kindlin and DEB-1/vinculin are essential muscle proteins upstream of nonessential muscle proteins (as reviewed in Moerman and Williams, 2006) and are normally localized in a pxl-1 background (Figure 8), as expected. As seen in Figure 7, each of the proteins with which PXL-1 interacts also interacts with multiple other proteins, so PXL-1 may not be solely responsible for their proper localization.
FIGURE 8:
Localization of PXL-1–interacting proteins in body wall muscle lacking pxl-1. Immunostaining for proteins that interact with PXL-1 was carried out using worms that are viable due to pxl-1 expression in the pharynx but lack body wall muscle expression of pxl-1. In body wall muscle lacking PXL-1, each of UNC-95, UNC-96, LIM-8, UIG-1, and UNC-112 is localized normally. UNC-96 is not normally observed in dense bodies with antibody staining in wild-type animals (Mercer et al., 2006), but in nematodes lacking PXL-1, dense body (arrowheads) and M-line (arrows) localization of UNC-96 is clearly seen in animals stained with anti–UNC-96. Bar, 10 μm.
Normally UNC-96 is observed at dense bodies and M-lines using an unc-96::GFP translational fusion but only at M-lines using antibody staining (Mercer et al., 2006). In animals lacking pxl-1 body wall muscle expression, however, we see M-line and dense body localization using the same UNC-96 antibody. It is possible that PXL-1 may normally sterically block the UNC-96 antibody from binding UNC-96 in dense bodies due to a physical interaction with UNC-96, but in dense bodies lacking PXL-1, the antibody is able to bind UNC-96 with better access to the molecule.
Paxillin is localized around a core of actin filaments in pharyngeal muscle
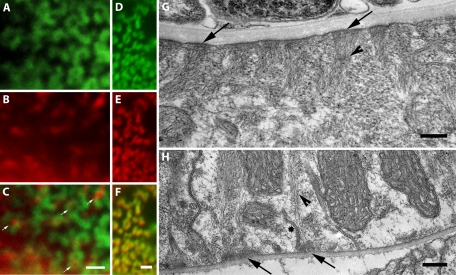
The ring-like localization pattern of paxillin in the pharynx is intriguing because the attachment structures in pharyngeal muscle have previously been assumed to be structurally similar to dense bodies in body wall muscle, albeit with a different complement of proteins. We looked at the distribution of both actin and vinculin in pharyngeal muscle to determine their location relative to paxillin. DEB-1/vinculin colocalizes with PXL-1 in the pharynx in a mix of ring-like and smaller punctate structures (Figure 9F). Because PXL-1 and DEB-1/vinculin are proteins that are in structures that anchor actin, we also looked at the distribution of actin in the pharynx. Antibody staining for actin in animals expressing PXL-1a/c::GFP shows that actin is distributed within the middle of the ring-like structures in pharyngeal muscle, as well as adjacent to the smaller punctate structures containing PXL-1 and DEB-1/vinculin (Figure 9C). Comparison of the actin-anchoring structures in the pharyngeal muscle indicates a structural difference between attachments at the basal membrane and those at the apical (luminal) membrane (Figure 9, G-H). The adhesion complexes at basal membrane have a dome-like shape with actin filaments extending from middle of the structure. Attachment complexes at the luminal membrane are comparatively flatter, with little extension into the cell. Each of these attachment structures differs from those in body wall muscle, which extend from the sarcolemma deep into the cell. The visible structural difference between each attachment and the intriguing pattern of localization for paxillin, vinculin, and actin raise questions about the organization of actin attachment structures found in pharyngeal muscle.
FIGURE 9:
Actin attachment complexes in pharyngeal muscle. Paxillin is localized to both full and partial ring-like structures in pharyngeal muscle. Full and partial ring-like PXL-1 localization using an anti-GFP antibody (A), along with punctate actin localization (B) using an antiactin antibody. (C) Actin at the middle of the PXL-1 ring-like pattern, with arrows indicating podosome-like attachment structures. The PXL-1 localization in pharyngeal muscle using an anti-GFP antibody (D) is identical to that of DEB-1/vinculin antibody staining (E), as shown in the overlay (F) of their localization patterns. The luminal face of the pharyngeal muscle (G) contains attachment structures (arrows) that differ in size and shape from those found in the cuticular face (H) as shown by TEM. Filaments can be seen projecting into the muscle cell from each type of attachment, as indicated by arrowheads. Membrane invaginations are also visible at the cuticular face of the muscle cell as marked by a star. Bar, 1 μm.
DISCUSSION
The pxl-1 gene in C. elegans encodes the orthologue of human paxillin
The initial observation that C. elegans paxillin has only three strong LD motifs left open the possibility that it is not a true paxillin homologue but rather another member of the paxillin superfamily, more similar perhaps to leupaxin or Hic-5. Leupaxin and Hic-5 are two proteins similar to paxillin, each with four LD motifs, compared with five in paxillin (reviewed in Tumbarello et al., 2002). Members of the paxillin superfamily have four LIM domains in the same synteny as paxillin (reviewed in Tumbarello et al., 2002), making them possible homologues of pxl-1. However, with the possible exception of zebrafish pxn (Crawford et al., 2003), only mammalian proteomes have paxillin and paxillin-related proteins. The third LD motif provides another convincing piece of data in support of PXL-1 being a true paxillin homologue rather than a homologue of the paxillin superfamily. LD motif 3 is not a conventional LD motif in paxillin, as it does not fit the consensus sequence. This sets it apart from all other members of the paxillin superfamily, which have an LD3 motif that strongly matches the consensus sequence (Tumbarello et al., 2002). The sequence surrounding LD3 in Homo sapiens paxillin aligns strongly with the LD3 sequence in C. elegans PXL-1 (Figure 1C), whereas PXL-1 is not similar to the LD3 of other members of the paxillin superfamily. Our protein interaction studies also support the idea that pxl-1 encodes a paxillin homologue. We found that PXL-1 binds DEB-1 (vinculin), a primary interacting protein of paxillin in adhesion complexes. DEB-1 is expressed in body wall muscle and pharyngeal muscle and is localized to actin attachment complexes (Barstead and Waterston, 1989).
The gene unc-95 has also been proposed as a homologue of vertebrate paxillin (Broday et al., 2004). UNC-95 is 350 amino acids long and contains one LIM domain near its carboxy terminus (Broday et al., 2004). Structurally, PXL-1 has much stronger domain conservation, with all four LIM domains and three of five LD motifs found in vertebrate paxillin. Of note, UNC-95 has an important role in the assembly of body wall muscle (Broday et al., 2004), whereas PXL-1 is necessary for the structural integrity of pharyngeal muscle. Both genes are expressed in each muscle tissue, but each appears to be necessary in only one type of muscle. Whether UNC-95 is a weak paxillin homologue with a role in body wall muscle or more similar to another LIM domain–containing protein requires further study.
Finally, it is known that vertebrate paxillin is heavily phosphorylated at tyrosines by Src family kinases (Turner, 1991); paxillin was originally identified as a phosphoprotein, 65–70 kDa. In the Western blot in Figure 4D, we noticed that nematode PXL-1 has a slower gel mobility than yeast-expressed nematode PXL-1. This would be compatible with PXL-1 being phosphorylated, since C. elegans has, but yeast lack, Src family tyrosine kinases. These combined observations lead us to conclude that PXL-1 is a paxillin homologue and is the nematode orthologue of paxillin.
Paxillin is required in the pharynx for muscle function
We have found that paxillin is expressed in all body wall and pharyngeal muscle cells in C. elegans. Antibody staining and GFP fusion experiments have shown that paxillin is localized to actin attachment structures in each of these muscle types, as well as weakly to the M-line (myosin attachment structure) in body wall muscle. In animals lacking paxillin the pharynx appears to be properly formed, but internally the myofilaments of the pharyngeal sarcomeres are disorganized. This lack of organized sarcomeres is likely what is leading to paralysis and lack of contraction of the pharyngeal muscle and in turn starvation due to an inability to feed (Figure 3, A–D). Expressing pxl-1 in pharyngeal muscle of pxl-1 animals is sufficient to rescue the L1 arrest phenotype, and pharyngeal muscle functions normally. Although the rescued animals display weak expression in the body wall muscle around the anterior tip of the worm, expression is almost fully restricted to the pharyngeal muscle. In body wall muscle cells posterior to the pharynx lacking paxillin, the cells are wild type in structure and movement is not impaired. It is unlikely that weak expression in a small subset of anterior body wall muscle cells is sufficient to rescue a body wall muscle–specific phenotype. Thus we conclude that PXL-1 has an integral role in the contraction of pharyngeal muscle, possibly in transmitting the forces generated by myosin/actin interaction, but no obvious functional role within body wall muscle.
Loss of β-integrin, ILK, UNC-112 or PINCH (Gettner et al., 1995; Hobert et al., 1999; Rogalski et al., 2000; Mackinnon et al., 2002; Norman et al., 2007) each leads to embryonic lethality in C. elegans. The diagnostic phenotype of mutations in these genes is a Pat animal (paralyzed and arrested at twofold) (Williams and Waterston, 1994). These proteins are all involved in integrin placement and organization of the adhesion complex to accept thick and thin filaments in the initial phase of myofilament formation during development. By contrast, PXL-1 is not required until later in development, as pxl-1 animals die after embryogenesis and after hatching during the first larval stage. Although the function of paxillin remains to be determined in body wall muscle, it is clearly important for pharyngeal muscle function. This difference in paxillin function in two different muscle tissues is intriguing and may be the result of how myofilaments are organized in body wall muscle versus the pharynx (see later discussion).
Paxillin's role in body wall muscle?
Although pxl-1 is expressed in both pharyngeal and body wall muscle, we have found it to be essential only in the pharynx. In the strain DM7335, which lacks pxl-1 expression in body wall muscle, the myofilament structure is wild type (Figure 5). It is surprising that pxl-1 appears to be dispensable in body wall muscle; however, other body wall muscle genes encoding LIM domain proteins lack a mutant or RNAi phenotype, including zyx-1/zyxin (Smith et al., 2002) and alp-1 (Han and Beckerle, 2009), which encodes homologues for both α-actinin–associated LIM protein and the LIM protein enigma (McKeown et al., 2006). The lack of a mutant or RNAi phenotype for these genes raises the question of whether there is redundancy, or genetic buffering, where more than one of these LIM domain proteins would need to be lost to elicit a phenotype. In our RNAi screen of genes encoding LIM domain proteins carried out using strain DM7335 that lacks pxl-1 expression in body wall muscle (Table 1), lim-8 knockdown resulted in uncoordinated movement and mild disorganization of the myofilaments. In a wild-type background, RNAi for lim-8 does not have such an effect (Qadota et al., 2007). Although pxl-1 does not have a clear role in body wall muscle and its loss does not cause any visual changes to the tissue, our observations with lim-8 indicate that muscle lacking pxl-1 is not as robust as wild-type muscle when exposed to additional gene loss.
Muscle attachment structures in C. elegans pharyngeal muscle are structurally similar to podosomes
The actin-anchoring dense bodies in the worm have attracted interest from researchers studying focal adhesion complexes due to the similarity in protein composition between these two dynamic structures. Less well studied are the attachment structures of the nematode pharynx, which we propose resemble podosomes. In podosomes, actin filaments form the core of the attachment complex and are surrounded by a ring of proteins found in focal adhesion proteins such as vinculin, paxillin, and integrin heterodimers (reviewed in Wernimont et al., 2008). Whereas the dense body structures in C. elegans body wall muscle appear punctate when observed using a fluorescent marker for paxillin, the attachment structures in the pharynx appear ring-like, which is what we would expect to see with a podosome-like structure. In EM images, the body wall muscle dense body (Figure 5, D–F) and pharyngeal attachment complex (Figure 9, G–H) appear as dense structures at their respective membranes. Membrane invaginations are also present at the cuticular face of the pharyngeal muscle as well; however they do not appear to be associated with the sarcomere itself (Figure 9H). The assumption has been that the attachment structure in pharyngeal muscle is similar to the dense body in body wall muscle, with actin filaments attaching outward from a core of attachment proteins, rather than actin forming a core on which attachment proteins form a ring-like structure to anchor it. Here we provide the first data that indicate otherwise.
PXL-1a/c::GFP localization in the pharynx is in ring-like structures within the muscle cell membranes, as well as in comparatively smaller punctate structures. Actin staining reveals that actin is localized within the middle of the paxillin-containing rings, as well as directly adjacent to paxillin punctae (Figure 9C). This raises an interesting question as to how ring-like adhesion structures are formed in pharyngeal muscle. It is possible that the initial accumulation of paxillin in the membrane leads to actin recruitment and attachment and subsequent encircling of the actin core by paxillin and other attachment proteins. In Figure 9C, both partial and full paxillin-containing rings can be seen, each encircling concentrated actin. Determining the exact mechanism of building a podosome-like structure will require future experimentation.
Defining the pharyngeal muscle attachment structure as podosome-like is supported by the expression and localization of a number of other podosome-associated proteins. DEB-1 (vinculin) colocalizes with PXL-1 in the pharyngeal muscle membrane in a ring-like pattern (Figure 9F). INA-1 (α-integrin), and ATN-1 (α-actinin) are expressed in pharyngeal muscle (Hresko et al., 1994; Baum and Garriga, 1997), but their exact localization pattern has not been established. Surprisingly, PAT-3 (β-integrin) is not expressed in developing and mature pharyngeal muscle. It is possible that there is another β-integrin in the C. elegans genome; however, a suitable candidate has not yet been identified. Whereas a null mutant for DEB-1 arrests embryonically and has paralyzed body wall muscle, the pharyngeal muscle is not completely paralyzed in all animals (Barstead and Waterston, 1991). In contrast, the pharyngeal muscle in pxl-1 null mutants is paralyzed, whereas the body wall muscle appears unaffected. The requirement of paxillin in the pharyngeal muscle for proper sarcomere organization and muscle contraction indicates that paxillin is a key component of the pharyngeal sarcomere. Further investigation into the requirement and localization of proteins found within pharyngeal muscle will lead, it is hoped, to a more complete understanding of the structure and function of the sarcomere in the pharynx.
MATERIALS AND METHODS
Strains
N2 (Bristol) is the principal wild-type strain used in C. elegans research and was grown using standard conditions (Brenner, 1974). The VC1012 (tag-327(ok1483)/mT1[dpy-10(e128)] III) strain was provided by the International C. elegans Gene Knockout Consortium (Vancouver, Canada). The ok1483 mutation is a 943–base pair deletion in the pxl-1 gene, and animals homozygous for ok1483 arrest development as L1 larvae. Balancing of the homozygous lethal genotype was accomplished by crossing in the chromosome III balancer mT1, a dpy-10 marked translocation. Strain GE24 (pha-1(e2123ts) III) was used for one microinjection because of its temperature-sensitive lethal phenotype (Granato et al., 1994). Animals homozygous for pha-1(e2123ts) arrest development during embryogenesis at 25C but are viable at 15C. Adult animals grown at 15C can be injected with DNA encoding a wild-type pha-1 gene and then transferred to 25C. Only progeny harboring the injected DNA as an extrachromosomal array survive. Strain NJ0784 (N2; rhIs1[pat-3::GFP; pRf4(rol-6)]) produces GFP-tagged PAT-3 and was used to confirm colocalization of PXL-1 with PAT-3 in body wall muscle.
Production of ok1483-rescuing GFP translational fusions
Construction of a GFP translational fusion able to rescue animals homozygous for the ok1483 allele was accomplished by using PCR to amplify the full sequence of C28H8.6b along with ∼2.4 kb of endogenous promoter region to clone into the pPD95.75 GFP expression vector provided by Andrew Fire (Stanford University). Similarly, the coding sequence of C28H8.6a along with ∼150 base pairs of its endogenous promoter was amplified and cloned into pPD95.75. Primers were designed to eliminate the stop codon in C28H8.6b and C28H8.6a while also allowing for it to be inserted into pPD95.75 such that it would be in-frame with GFP sequence downstream of the multiple cloning site. The 3′ untranslated region for unc-54 is downstream of the coding sequence for GFP in the pPD95.75 vector.
Construction of a Gateway GFP translational fusion
A GFP translational fusion for the LIM domain–coding portion of pxl-1 was produced by taking advantage of the ORFeome (Reboul et al., 2003) publicly available from Open Biosystems (Thermo Biosystems, Huntsville, AL). The C. elegans ORFeome consists of 12,625 open reading frames (ORFs) cloned into a Gateway (Walhout et al., 2000) donor vector (Invitrogen, Carlsbad, CA). On the basis of methodology described in Meissner et al. (2011), a GFP-tagged “‘LIM domain–only” paxillin construct was created, driven by the body wall muscle–specific promoter T05G5.1.
Identification of pxl-1 splice isoforms
Forward primers specific to the exons in the 5′ portion of the previously named C28H8.13 gene were used to attempt to amplify RT-PCR products using each of the two 3′ exons of C28H8.6. Three splice isoforms were confirmed by sequencing the isolated cDNAs and named C28H8.6a, C28H8.6b, and C28H8.6c, with GenBank accession numbers EU239658, EU239659, and EU239660, respectively. Our analysis demonstrates that C28H8.13 and C28H8.6 are one gene, now termed C28H8.6 or pxl-1. Our analysis of the alternative splicing of pxl-1 is also supported by data from whole-genome transcriptional tiling array experiments (He et al., 2007).
Isolation of a full-length pxl-1a cDNA
Primers flanking the 5′ and 3′ exons of pxl-1a were used to amplify the pxl-1a cDNA using RT-PCR. The forward primer contained a 5′ HindIII restriction site, and the reverse primer contained a 3′ EcoRV site to allow for cloning into the pBlueScript SK-plasmid vector. The cloned cDNA was sequenced to confirm identical sequence to the annotated National Center for Biotechnology Information sequence.
Microinjection procedure
Transformation of worms with GFP translational fusion constructs was carried out using microinjection. Two coinjection markers were used in this process: pRF4 [rol-6(su1006dm)], which contains a copy of rol-6 and results in a Rol phenotype of successful transformants, and pBx [pha-1::pha-1(+)], which is able to rescue the embryonic lethal pha-1 phenotype of the strain GE24. Injection mixes were prepared containing either 45 ng/μl pRF4, 45 ng/μl pBx, and 10 ng/μl of GFP-tagged construct for injection into GE24 animals or 90 ng/μl pRF4 rol-6(su1006dm) and 10 ng/μl GFP-tagged construct for injection into animals heterozygous for the ok1483 null allele of pxl-1 and phenotypically balanced with the recessive mutation dpy-17. All injections were carried out using a microinjection setup featuring a Zeiss inverted compound microscope (IM35) by conventional methods (Mello et al., 1991). The four transgenic strains obtained were DM7055 (pha-1(e2123) III; raEx55[pha-1(+); rol-6(su1006); pT05G5.1::C28H8.6a::GFP]), DM7082 (tag-327(ok1483) III; raEx82[rol-6(su1006); C28H8.6b::GFP]), DM7335 (tag-327(ok1483) III; raEx335[rol-6(su1006); C28H8.6a/c::GFP]), and DM7438 (tag-327(ok1483) III; raEx438[rol-6(su1006); pC28H8.6::C28H8.6a(cDNA)::GFP])
RNAi feeding
RNAi feeding clones from the frozen Geneservice feeding library (Source BioScience LifeSciences, Nottingham, United Kingdom) were picked and grown overnight in Luria (L) broth containing 50 μg/ml ampicillin. Specialized nematode growth medium plates were prepared containing 1 mM isopropyl-β-d-thiogalactopyranoside to induce production of dsRNA in bacteria and 50 μg/ml carbenicillin to select for bacteria containing an RNAi construct. These plates were then streaked with 50 μl of overnight culture and incubated at room temperature overnight to allow for dsRNA production. Approximately 20 L1 worms were spotted onto each RNAi plate by transferring a small aliquot of the previously prepared M9 solution containing the hatched animals. These plates were then incubated at 20ºC until the worms reached the young adult stage (∼60–68 h). Four worms from each plate were then transferred to fresh RNAi plates corresponding to the same feeding construct (two plates, each with two worms) and left for ∼18 h to lay embryos, at which time they were removed. The phenotype of affected worms was assayed over the next 2 d.
Antibody production
An MBP fusion of the PXL-1a N-terminus (residues 1–112) was used as immunogen for preparation of anti–PXL-1 antibodies. The cDNA fragment was cloned into pMAL-KK-1. BL21 (DE3) CodonPlus bacteria (Stratagene, Santa Clara, CA) was used for production of MBP-PXL-1 (residues 1–112), which was supplied to Spring Valley Laboratories (Woodbine, MD) for production of rabbit antibodies. After affinity purification using a column containing MBP-PXL-1 (residues 1–112), antibodies were evaluated by reaction to a Western blot containing a lysate of soluble C. elegans proteins and a lysate from yeast expressing HA-tagged PXL-1 full-length cDNA. A yeast expression plasmid for HA-tagged PXL-1 full-length (residues 1–413) was constructed by two-step cloning: first, the cDNA of PXL-1 full-length was cloned into pKA-HA(Nhex2); second, the NheI fragment containing HA-tagged PXL-1 full-length (residues 1–413) was cloned into pGAP-C-Nhe (Lin et al., 2003; Norman et al., 2007). The PJ69-4A yeast strain harboring pGAP-C-HA-PXL-1 plasmid was cultured in 2 ml of media until saturation, and the cells were collected. Yeast lysate was prepared by suspending yeast cells in Laemmli buffer, heating at 95°C for 5 min. The Western blot was performed as described previously (Mercer et al., 2006).
Yeast two-hybrid method
Yeast two-hybrid assays were performed as described previously (Mackinnon et al. 2002). The PCR-amplified cDNA fragments of PXL-1a full-length (residues 1–413), PXL-1a N-terminus (residues 1–112), and PXL-1a LIM domains (residues 113–413) tagged with BamHI and SalI were cloned into pGBDU-C1 (bait) and pGAD-C1 (prey) yeast two-hybrid vectors (James et al., 1996). Yeast host strain PJ69-4A harboring a combination of PXL-1 bait and “bookshelf” collection (Qadota et al., 2007) preys or PXL-1 prey and “bookshelf” collection bait plasmids was examined for HIS3 and ADE2 reporters on selective plates.
HA-pulldown assay
Bacterial expression plasmids for MBP fusions of UNC-95 full-length, UNC-96 C-terminal half (residues 201–411), and LIM-8 (residues 635–1004) were described previously (Mercer et al., 2006; Qadota et al., 2007). Bacterial expression plasmids for the MBP fusion of DEB-1 (residues 1–291) and UIG-1 (residues 1–638) were made by cloning each cDNA fragment a into pMAL vector. MBP and other MBP fusions were expressed in Escherichia coli and purified as described previously (Mercer et al., 2006). On the basis of a yeast lysate containing HA-tagged PXL-1 full-length (residues 1–413) and purified MBP or MBP fusion proteins, a HA-pulldown assay was carried out as described previously (Qadota et al., 2008).
Immunostaining
For the images shown in Figures 7 and 8, worms were fixed by the Nonet method (Nonet et al., 1993). Antibody staining with anti–PXL-1 (1:100 dilution) and anti–α-actinin (MH35, 1:200 dilution) was performed as described previously (Qadota et al., 2007). Secondary antibodies were anti–rabbit Alexa 488 (Invitrogen) for anti–PXL-1 and anti–mouse Alexa 594 (Invitrogen) for anti–α-actinin, each used at 1:200 dilution. Stained samples were mounted on a glass slide with a coverslip containing mounting solution (20 mM Tris, pH 8.0, 0.2 M 1,4-diazabicyclo[2.2.2]octane [DABCO], and 90% glycerol). Images were captured with a Zeiss confocal system (LSM510) equipped with an Axiovert 100M microscope using an Apochromat 63×/1.4 oil objective in 2.5× zoom mode. The color balances of the images were adjusted by using Adobe Photoshop (San Jose, CA). All additional antibody staining was carried out using a method modified from Albertson (1984). Worms were washed off plates and suspended in 4% sucrose, 1 mM EDTA, pH 7.4, solution. Some animals were dissected to expose the pharynx for images in Figure 9. The worm suspensions were transferred to glass slides coated with 2 mg/ml poly-l-lysine, covered by a rectangular glass coverslip, and frozen on an aluminum slab at −80°C overnight. The coverslips were removed from the slides using a razor blade, and slides were transferred to −20°C methanol for 4 min, −20°C acetone for 4 min, and subsequent 1-min intervals of 75% acetone at 20°C, 50% acetone, 25% acetone, and Tween/Tris-buffered saline (TBS) for 2 min. Primary antibodies solutions were added to the slides for 5 h, and included anti–PXL-1 (1:100 dilution), antiactin (AbCam8224) at 1:500 dilution, antivinculin (MH24; Francis and Waterston, 1985) at 1:250 dilution, and anti-GFP (AbCam13970) at 1:1000 dilution. After 1 h in TBS/Tween, slides were coated with secondary antibodies for 3 h, Alexa 488 (Invitrogen) for anti–PXL-1, Alexa 594 for antiactin and antivinculin, and Dylight 488 (703485155; Jackson ImmunoResearch, West Grove, PA) for anti-GFP. After 1 h in TBS/Tween, mounting solution was added (20 mM Tris, pH 8.0, 0.2 M DABCO, and 90% glycerol), and slides were sealed with nail polish.
Microscopic analysis
All phenotypic analysis of worms homozygous for the mutant allele ok1483 or affected by RNAi targeting C28H8.6 was carried out using a dissecting microscope (Wild Heerbrugg model) and a fluorescence dissecting microscope (Stemi SV11; Zeiss, Thornwood, NY). Whole-worm images and immunofluorescence images were taken using a Zeiss Axiophot compound microscope with a QImaging QICAM digital camera running Qcapture, version 1.68.4, and a Zeiss Axioplan 2 imaging compound microscope with 2.5× Optivar and a QImaging Retiga EXi digital camera. Images in Figure 4C were obtained using a Zeiss Axiovert inverted compound microscope with a Zeiss Pascal confocal setup (LSM5) using Pascal imaging software, version 3.2sp2. Fluorescence images in Figure 9 were processed with Image J software (National Institutes of Health, Bethesda, MD), using the Parallel Iterative Deconvolution 2D plug-in (version 1.11).
TEM microscopy
Worms were removed from growth media and immersed in fixative (1.5% paraformaldehyde, 1.5% glutaraldehyde, 0.1 M sodium cacodylate, pH 7.3, room temperature). Under a microscope, the anterior halves of the animals were removed and collected in a small vial. After fixing for ∼3 h, the fixative was replaced with buffer (0.1 M sodium cacodylate, pH 7.3) and the samples left at room temperature until the following day. Samples were washed twice with buffer, postfixed (1% OsO4 in 0.1 M sodium cacodylate) for 1 h on ice, washed three times with dH2O, en bloc stained for 1 h with 1% uranyl acetate (aqueous), and then washed again three times with dH2O. Samples were dehydrated through a graded series of ethanol and then infiltrated through propylene oxide into EMbed 812 (Electron Microscopy Sciences, Hatfield, PA). The worm halves were oriented in the embedding molds so that the midsection of the worm and the pharynx could be transversely cut when sectioned, and then the resin hardened at 60°C overnight. Thin sections were stained with uranyl acetate and lead citrate and photographed on a Philips 300 operated at 60 kV.
Acknowledgments
Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources of the National Institutes of Health. We thank the C. elegans Gene Knockout Consortium for providing the pxl-1 knockout strain VC1012. We also like thank Edward Hedgecock for use of the strain NJ0784 and David Hall and Zeynep Altun for analysis of pxl-1 expression in the pharynx. This research was supported by grants from the Canadian Institute for Health Research and the National Science and Engineering Research Council of Canada to D.G.M. and Grant AR052133 from the National Institutes of Health to G.M.B. D.G.M. also received support from the Canadian Institute for Advanced Research.
Abbreviations used:
- HA
hemagglutinin
- LIM domain, lin-11, isl-1
mec-3 domain
- L1
first larval stage
- MBP
maltose-binding protein
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0941) on June 1, 2011.
REFERENCES
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. Vinculin is essential for muscle function in the nematode. J Cell Biol. 1991;114:715–724. doi: 10.1083/jcb.114.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135:1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BD, Henry CA, Clason TA, Becker AL, Hille MB. Activity and distribution of paxillin, focal adhesion kinase, and cadherin indicate cooperative roles during zebrafish morphogenesis. Mol Biol Cell. 2003;14:3065–3081. doi: 10.1091/mbc.E02-08-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121:2435–2444. doi: 10.1242/jcs.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner SN, Kenyon C, Reichardt LF. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280:64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- Han HF, Beckerle MC. The ALP-Enigma protein ALP-1 functions in actin filament organization to promote muscle structural integrity in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2361–2370. doi: 10.1091/mbc.E08-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, et al. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17:1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T, Qadota H, Tsuboi D, Taya S, Moerman DG, Kaibuchi K. Identification of a novel Cdc42 GEF that is localized to the PAT-3-mediated adhesive structure. Biochem Biophys Res Commun. 2005;335:139–145. doi: 10.1016/j.bbrc.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko MC, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;4:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Stradal TE, Gimona M. Podosome formation in cultured A7r5 vascular smooth muscle cells requires Arp2/3-dependent de-novo actin polymerization at discrete microdomains. J Cell Sci. 2003;116:4915–4924. doi: 10.1242/jcs.00818. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Georges-Labouesse E. Cell adhesion: parallels between vertebrate and invertebrate focal adhesions. Curr Biol. 2003;13:R528–R530. doi: 10.1016/s0960-9822(03)00448-2. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin XY, Qadota H, Moerman DG, Williams BD. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr Biol. 2003;13:922–932. doi: 10.1016/s0960-9822(03)00372-5. [DOI] [PubMed] [Google Scholar]
- Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Mango SE. The C. elegans pharynx: a model for organogenesis. WormBook. 2007:1–26. doi: 10.1895/wormbook.1.129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisio PC, Cirillo D, Naldini L, Primavera MV, Teti A, Zambonin-Zallone A. Cell-substratum interaction of cultured avian osteoclasts is mediated by specific adhesion structures. J Cell Biol. 1984;99:1696–1705. doi: 10.1083/jcb.99.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown CR, Han HF, Beckerle MC. Molecular characterization of the Caenorhabditis elegans ALP/Enigma gene alp-1. Dev Dyn. 2006;235:230–238. doi: 10.1002/dvdy.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B, Rogalski T, Viveiros R, Warner A, Plastino L, Lorch A, Granger L, Segalat L, Moerman DG. Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS One. 2011;5:e19937. doi: 10.1371/journal.pone.0019937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KB, Miller RK, Tinley TL, Sheth S, Qadota H, Benian GM. Caenorhabditis elegans UNC-96 is a new component of M-lines that interacts with UNC-98 and paramyosin and is required in adult muscle for assembly and/or maintenance of thick filaments. Mol Biol Cell. 2006;17:3832–3847. doi: 10.1091/mbc.E06-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Fire A. In: Muscle: structure, function and development. elegans C. II, Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. pp. 417–470. [PubMed] [Google Scholar]
- Moerman DG, Williams BD. Sarcomere assembly in C. elegans muscle. WormBook. 2006:1–16. doi: 10.1895/wormbook.1.81.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Norman KR, Cordes S, Qadota H, Rahmani P, Moerman DG. UNC-97/PINCH is involved in the assembly of integrin cell adhesion complexes in Caenorhabditis elegans body wall muscle. Dev Biol. 2007;309:45–55. doi: 10.1016/j.ydbio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Proszynski TJ, Gingras J, Valdez G, Krzewski K, Sanes JR. Podosomes are present in a postsynaptic apparatus and participate in its maturation. Proc Natl Acad Sci USA. 2009;106:18373–18378. doi: 10.1073/pnas.0910391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Benian GM. Molecular structure of sarcomere-to-membrane attachment at M-lines in C. elegans muscle. J Biomed Biotechnol. 2010;2010:864749. doi: 10.1155/2010/864749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, McGaha LA, Mercer KB, Stark TJ, Ferrara TM, Benian GM. A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (Obscurin) in Caenorhabditis elegans. Mol Biol Cell. 2008;19:2424–2432. doi: 10.1091/mbc.E08-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/pinch to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–4326. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul J, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol. 2000;150:253–264. doi: 10.1083/jcb.150.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Smith P, Leung-Chiu WM, Montgomery R, Orsborn A, Kuznicki K, Gressman-Coberly E, Mutapcic L, Bennett K. The GLH proteins, Caenorhabditis elegans P granule components, associate with CSN-5 and KGB-1, proteins necessary for fertility, and with ZYX-1, a predicted cytoskeletal protein. Dev Biol. 2002;251:333–347. doi: 10.1006/dbio.2002.0832. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Brown MC, Turner CE. The paxillin LD motifs. FEBS Lett. 2002;513:114–118. doi: 10.1016/s0014-5793(01)03244-6. [DOI] [PubMed] [Google Scholar]
- Turner CE. Paxillin is a major phosphotyrosine-containing protein during embryonic development. J Cell Biol. 1991;115:201–207. doi: 10.1083/jcb.115.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE, Glenney JR, Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CE, Kramarcy N, Sealock R, Burridge K. Localization of paxillin, a focal adhesion protein, to smooth muscle dense plaques, and the myotendinous and neuromuscular junctions of skeletal muscle. Exp Cell Res. 1991;192:651–655. doi: 10.1016/0014-4827(91)90090-h. [DOI] [PubMed] [Google Scholar]
- Turner CE, Miller JT. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J Cell Sci. 1994;107(Pt 6):1583–1591. doi: 10.1242/jcs.107.6.1583. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wernimont SA, Cortesio CL, Simonson WT, Huttenlocher A. Adhesions ring: a structural comparison between podosomes and the immune synapse. Eur J Cell Biol. 2008;87:507–515. doi: 10.1016/j.ejcb.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GN, Hynes RO. The cloning, genomic organization and expression of the focal contact protein paxillin in Drosophila. Gene. 2001;262:291–299. doi: 10.1016/s0378-1119(00)00512-6. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Schriefer LA, Stormo GD. Identification of muscle-specific regulatory modules in Caenorhabditis elegans. Genome Res. 2007;17:348–357. doi: 10.1101/gr.5989907. [DOI] [PMC free article] [PubMed] [Google Scholar]