Caenorhabditis elegans SNAP-29 is required for the proper morphology and functions of the Golgi and endosomes and general exocytosis.
Abstract
It is generally accepted that soluble N-ethylmaleimide–sensitive factor attachment protein receptors mediate the docking and fusion of transport intermediates with target membranes. Our research identifies Caenorhabditis elegans homologue of synaptosomal-associated protein 29 (SNAP-29) as an essential regulator of membrane trafficking in polarized intestinal cells of living animals. We show that a depletion of SNAP-29 blocks yolk secretion and targeting of apical and basolateral plasma membrane proteins in the intestinal cells and results in a strong accumulation of small cargo-containing vesicles. The loss of SNAP-29 also blocks the transport of yolk receptor RME-2 to the plasma membrane in nonpolarized oocytes, indicating that its function is required in various cell types. SNAP-29 is essential for embryogenesis, animal growth, and viability. Functional fluorescent protein–tagged SNAP-29 mainly localizes to the plasma membrane and the late Golgi, although it also partially colocalizes with endosomal proteins. The loss of SNAP-29 leads to the vesiculation/fragmentation of the Golgi and endosomes, suggesting that SNAP-29 is involved in multiple transport pathways between the exocytic and endocytic organelles. These observations also suggest that organelles comprising the endomembrane system are highly dynamic structures based on the balance between membrane budding and fusion and that SNAP-29–mediated fusion is required to maintain proper organellar morphology and functions.
INTRODUCTION
Intracellular membrane trafficking is regulated by vesicular transport from donor compartments to specific acceptor compartments. Vesicle fusion is mediated by soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs) (Jahn and Scheller, 2006). The SNAREs can be classified functionally into v-SNAREs, located on the vesicles/transport intermediates, and t-SNAREs, located on the target membrane. By their structures, SNAREs are classified as Q-SNAREs (syntaxins, synaptosomal-associated proteins [SNAPs]) and R-SNAREs (vesicle-associated membrane proteins [VAMPs]). It is thought that three different SNAREs (two Q-SNAREs and one R-SNARE) assemble into a twisted, parallel, four-helical bundle composed of two cognate parts (v-SNARE and t-SNARE), which catalyze the apposition and fusion between the vesicle and target membrane (Ungar and Hughson, 2003). The family of SNAP-25 proteins is made up of Q-SNARE proteins that have been shown to modulate membrane fusion in various systems (Ungar and Hughson, 2003). The mammalian protein SNAP-29 (SNAP, 29 kDa) is one of the SNAP-25 family proteins and is ubiquitously expressed in many tissues. In cultured mammalian cells, SNAP-29 localizes on many different internal membranes, including the Golgi apparatus, endosomes, and lysosomes, and it binds to a broad range of syntaxins (Steegmaier et al., 1998). SNAP-29 was reported to modulate synaptic transmission by interacting with syntaxin 1A and competing with α-SNAP for binding to the SNARE complex (Su et al., 2001). This interaction results in the modulation of synaptic transmission by inhibiting the dissociation of the SNARE complex. In addition to syntaxins, SNAP-29 interacts with clathrin, α-adaptin of adaptor complex 2, and EHD1 involved in the endocytic pathway (Rotem-Yehudar et al., 2001) and small GTPase Rab3A in myelinating glia (Schardt et al., 2009). The loss of SNAP-29 in humans causes cerebral disgenesis, neuropathy, ichthyosis, and keratoderma (CEDNIK) syndrome (Sprecher et al., 2005). In the skin of patients, SNAP-29 deficiency was found to prevent the maturation and secretion of lamellar granules, which are Golgi-derived vesicular structures responsible for the transport of lipids and proteases to the upper layers of the epidermis. Conversely, it has been reported that transport of the vesicular stomatitis virus G protein to the plasma membrane is almost normal in SNAP-29–deficient fibroblast cell lines from CEDNIK patients, even if the Golgi morphology is strongly affected (Rapaport et al., 2010).
We identified Caenorhabditis elegans SNAP-29 as an essential regulator of membrane trafficking in the intestine and oocytes. We showed that a knockdown of SNAP-29 inhibits the transport of the apical- and basolateral-directed cargos and accumulates fine cargo-containing vesicles in the cytoplasm. Inhibition of SNAP-29 functions also causes vesiculation of the Golgi and endosomes, suggesting that SNAP-29 is involved in multiple steps of exocytic and endocytic pathways. These results also suggest that the organelles comprising the endomembrane system are highly dynamic structures, and constitutive membrane fusion activity is required to maintain certain organellar structures.
RESULTS
SNAP-29 is required for anterograde transport of plasma membrane–targeted and secretory proteins
To monitor membrane trafficking in the polarized intestinal cells of living animals, we chose the syntaxins UNC-64 and SYN-1, which are associated with the plasma membrane (PM) in C. elegans, as cargo molecules and expressed as proteins tagged with green fluorescent protein (GFP) in the intestine. Among eight syntaxin genes in the C. elegans genome, unc-64 encodes a mammalian syntaxin 1A homologue and is involved with presynaptic function in the nerve systems (Saifee et al., 1998). The unc-64 gene is also expressed in many secretory tissues, including the intestine (Wu et al., 2010). Syntaxin gene syn-1, the product of which is most similar to syntaxin 1, is expressed in the intestine and is involved in defecation behavior (Yamashita et al., 2009). N-terminal GFP-tagged SYN-1 is reported to be functional and to localize to the PM (Yamashita et al., 2009). We found that, in wild-type intestinal cells, GFP-UNC-64 and GFP-SYN-1 predominantly localized to the apical and basolateral PM, respectively (Figure 1, A and C).
FIGURE 1:
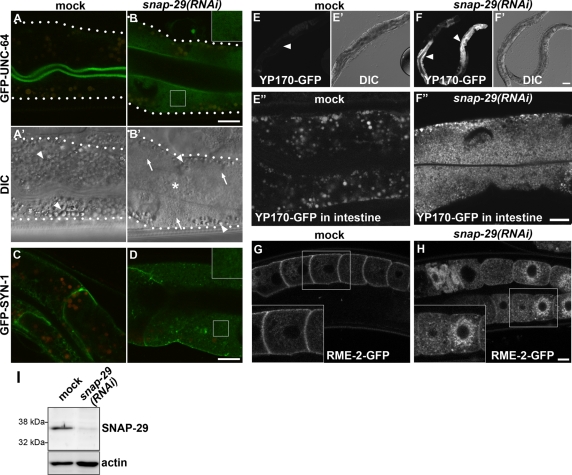
Knockdown of snap-29 causes accumulation of the transmembrane and secreted cargo proteins in cytoplasmic small vesicles. (A–D) In the wild-type intestine, GFP-UNC-64 and GFP-SYN-1 localize to the apical and basolateral PM, respectively. In the snap-29(RNAi) animals, GFP-UNC-64 and GFP-SYN-1 targeted to the PM are reduced and the GFP signals are observed in fine-vesicular cytoplasmic structures. Autofluorescence of lysosome-like organelles can be seen in both the green and red channels, whereas GFP appears only in the green channel. Merged images of green and red channels are shown in A–D, in which autofluorescence is seen as yellow. An enlarged (2×) image of the boxed area is shown in the inset (B and D). (A′, B′) Nomarski images corresponding to A and B, respectively. The wild-type intestine is filled with granules (A′, arrowheads). In the intestine of snap-29(RNAi) animals (B′), there are fewer granules (arrowheads) and the cytoplasm appears grainy (arrows). The lumen of the intestine accumulates undigested E. coli (asterisk). (E–F″) Accumulation of GFP-tagged yolk protein YP170-GFP in the intestine of snap-29(RNAi) animals. In the wild-type animal, YP170-GFP is secreted from the intestine and taken up by growing oocytes, resulting a weak GFP signal in the intestine (E, arrowhead). snap-29(RNAi) causes a high accumulation of YP170-GFP in the intestine (F, arrowheads). High-magnification images of the intestine are also shown in E″ (mock) and F″ [snap-29(RNAi)]. (E′, F′) Nomarski images corresponding to E and F, respectively. (G, H) Subcellular localization of the yolk receptor RME-2 in oocytes. In wild-type worms, RME-2-GFP localizes to cortical endosomes and the PM (G), whereas RME-2-GFP accumulates in deep cytoplasmic vesicles in the oocytes of snap-29(RNAi) animals (H). An enlarged (1.5×) image of the boxed area is shown in the inset (G and H). All images were observed in living animals. Bars, 10 μm (B, D, F″, and H) or 50 μm (F′). (I) Detection of SNAP-29 protein. Total lysates were prepared from N2 wild-type worms treated with RNAi of either control vector or snap-29. They were examined by Western blotting using anti–SNAP-29 antibody. The same membrane was also probed with anti-actin antibody.
To gain insight into the mechanisms of polarized protein transport in the epithelial cells, we looked for genes whose RNAi knockdown affected the distribution of GFP-UNC-64 and/or GFP-SYN-1. We found that the RNAi of K02D10.5 resulted in high accumulation of both GFP reporters in small cytoplasmic vesicles (Figure 1, B and D). K02D10.5 is one of three SNAP-25 family genes in the C. elegans genome (aex-4, ric-4, and K02D10.5), whereas mammals have four genes of the SNAP-25 family, SNAP-25, SNAP-23, SNAP-29, and SNAP-47 (Holt et al., 2006) (Supplemental Figure S1). The genes aex-4 and ric-4 encode the SNAP-25 homologues involved in defecation motor program and synaptic function, respectively (Mahoney et al., 2008; Wu et al., 2010). Because K02D10.5 encodes a protein most similar to SNAP-29, we named this gene snap-29. Using an antibody that we developed against worm SNAP-29, we detected a band around 35 kDa (estimated molecular weight, 31 kDa) in Western analysis (Figure 1I). The intensity of the band of SNAP-29 was greatly reduced after RNA interference (RNAi) of snap-29, confirming the specificity of the antibody (Figure 1I). RNAi of ric-4 or aex-4 did not show any apparent effects on the subcellular localization of GFP-UNC-64 or GFP-SYN-1 in the intestine, showing a specific effect of snap-29(RNAi) (Supplemental Figure S2). In the Nomarski images (Figure 1A′, arrowheads), the intestinal cells of C. elegans are normally filled with granules ∼1–2 μm in diameter, some of which are gut granules, lysosome-related organelles containing autofluorescent and birefringent materials (Hermann et al., 2005). In the intestinal cells of snap-29(RNAi) animals, there were fewer such granules, and a large area of the cytoplasm appeared to have fine vesicular materials (Figure 1B′, arrows).
We further examined whether the knockdown of snap-29 affects the secretion of the yolk protein YP170, a ligand related to the mammalian cholesterol carrier ApoB-100 (Grant and Hirsh, 1999). YP170-GFP fusion protein, like endogenous YP170, is synthesized in the intestine and secreted basolaterally into the body cavity, from which it is endocytosed by the RME-2 yolk receptors expressed in the oocytes (Grant and Hirsh, 1999). In wild-type animals, YP170-GFP was efficiently secreted from the intestinal cells, taken up by oocytes and discharged as eggs (Figure 1E). Conversely, the depletion of snap-29 resulted in a severe accumulation of YP170-GFP in small vesicles filling the cytoplasm of the intestine, demonstrating the secretion defect of snap-29(RNAi) (Figure 1, F and F″). These results suggest that, in general, SNAP-29 is required for the PM targeting of the membrane proteins and yolk secretion in the intestine. We often observed that undigested bacteria accumulated in the intestinal lumen of snap-29(RNAi) animals.
We also examined the effect of snap-29(RNAi) in nonpolarized oocytes by using RME-2-GFP. In the wild-type oocytes, RME-2-GFP is enriched on the PM and in cortical vesicles and tubules representing early and recycling endosomes (Figure 1G; Balklava et al., 2007). In the oocytes of snap-29(RNAi) animals, RME-2-GFP largely accumulated in vesicular structures deeper in the cytoplasm and around the nucleus, suggesting that the transport of RME-2-GFP to the PM and cortical endosomes was impaired (Figure 1H). Thus SNAP-29 has functions in various cell types. Consistently, snap-29(RNAi) showed a strong effect on animal growth and viability. When L1 larvae were treated with snap-29 RNAi, they did not reach adulthood and died at about the L4 stage (n = 267). When L4 larvae were treated with snap-29 RNAi, they showed abnormal oogenesis and produced a reduced number of eggs, about a half of which were unhatched dead embryos (Supplemental Table S2).
Organization of the Golgi and endosomes in the C. elegans intestinal cells
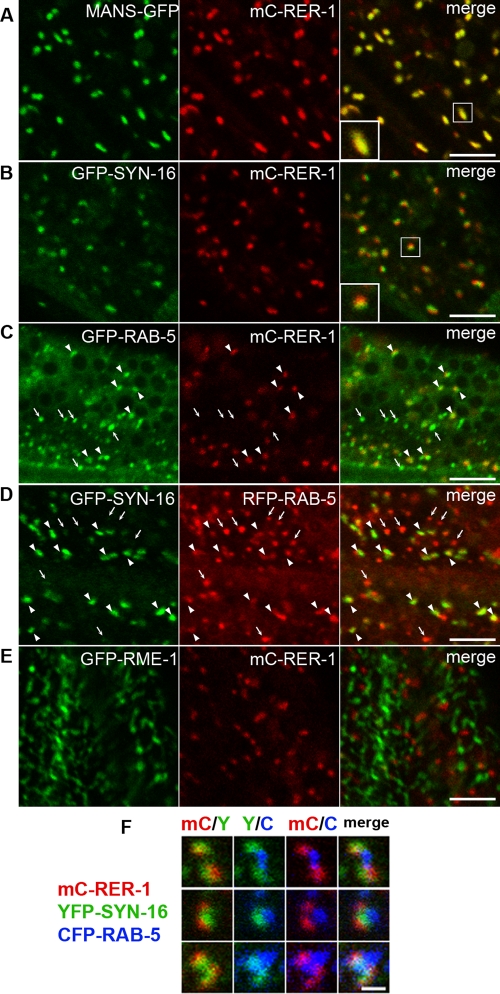
We next sought to determine subcellular localization of SNAP-29. In cultured mammalian cells, SNAP-29 localization partially overlaps with the Golgi, endosomal, and lysosomal markers (Steegmaier et al., 1998). We first examined the spatial organization of the Golgi and endosomes in the intestinal cells of living animals. Unlike vertebrate cells, which contain one large juxtanuclear Golgi stack, C. elegans cells instead contain many Golgi ministacks scattered throughout the cytoplasm like most invertebrate and plant cells (Chen et al., 2006). To visualize early-Golgi compartments, we created a transgenic animal that expressed mCherry-RER-1 (mC-RER-1) under the control of the intestine-specific act-5 promoter. RER-1 (F46C5.8) is the C. elegans homologue of Rer1p, which functions as a retrieval receptor for endoplasmic reticulum (ER) membrane proteins and localizes to the early-Golgi compartment in yeast (Sato et al., 2001). mC-RER-1 localized to the punctate structures, and almost all puncta of mC-RER-1 (95%) extensively colocalized with mannosidase (MANS)-GFP, another Golgi marker (Figure 2A; Chen et al., 2006). To visualize the late-Golgi compartments, we used SYN-16, the closest C. elegans homologue of mammalian syntaxin-16 that localizes to the trans-Golgi network (TGN) (Simonsen et al., 1998; Jantsch-Plunger and Glotzer, 1999; Gengyo-Ando et al., 2007). GFP-tagged SYN-16 (GFP-SYN-16) mainly localized to punctate structures, although a weak GFP signal was also detected on cytoplasmic tubulovesicular structures. We observed that 99% of mC-RER-1–positive puncta were tightly attached with GFP-SYN-16–positive structures (Figure 2B), suggesting that C. elegans Golgi ministacks are composed of at least the early and late compartments. Time-lapse images of intestinal cells coexpressing mC-RER-1 and GFP-SYN-16 revealed that C. elegans Golgi is highly dynamic structures (Supplemental Movie). We observed that tubular extensions frequently formed from the GFP-SYN-16–labeled membranes, and these extensions sometimes seemed to contact with distinct Golgi puncta. We also found that the Golgi structures are frequently associated with endosomes as reported previously (Shi et al., 2009). Greater than 90% of mC-RER-1–positive or GFP-SYN-16–positive puncta were adjacent to puncta labeled with GFP- or RFP-tagged RAB-5, a marker of endosomes (Figure 2, C and D, arrowheads; Chen et al., 2006). This close organization of the Golgi and endosomes was further confirmed in animals expressing mC-RER-1, YFP-SYN-16, and CFP-RAB-5 simultaneously (Figure 2F). We often but not always observed a directed arrangement of mC-RER-1, YFP-SYN-16, and CFP-RAB-5. Conversely, about a half of the RAB-5–positive puncta did not associate with mC-RER-1, suggesting that a Golgi-independent population of endosomes also exists (Figure 2, C and D, arrows). We also examined the colocalization of mC-RER-1 with the recycling endosomal marker RME-1 (Grant et al., 2001). GFP-RME-1 is highly enriched on tubulovesicular basolateral recycling endosomes. The puncta of mC-RER-1 did not overlap with GFP-RME-1 but often juxtaposed to the tips of GFP-RME-1–labeled tubules (Figure 2E). These observations suggest that the Golgi and endosomes are organized very closely in the C. elegans intestinal epithelial cells.
FIGURE 2:
Organization of the Golgi and endosomes in C. elegans intestinal cells. GFP- and mCherry-tagged proteins were coexpressed under the intestine-specific promoters and observed in intact living animals. (A) MANS-GFP and mC-RER-1 colocalize in punctate Golgi structures. (B) Puncta of GFP-SYN-16 and mC-RER-1 closely associate with each other. (C) Most mC-RER-1 closely associate with GFP-RAB-5 (arrowheads), but small punctate structures labeled only with GFP-RAB-5 are also observed (arrows). (D) Puncta of GFP-SYN-16 closely associate with RFP-RAB-5 (arrowheads), but small punctate structures labeled only with RFP-RAB-5 are also observed (arrows). (E) GFP-RME-1 and mC-RER-1 do not overlap with each other. An enlarged (2×) image of the boxed area is shown in the inset (A and B). Bars, 5 μm. (F) Spatial organization of the Golgi and endosomes was observed using animals expressing mC-RER-1, YFP-SYN-16, and CFP-RAB-5 simultaneously. Bar, 1 μm.
SNAP-29 localizes to the late-Golgi, endosomes, and the PM
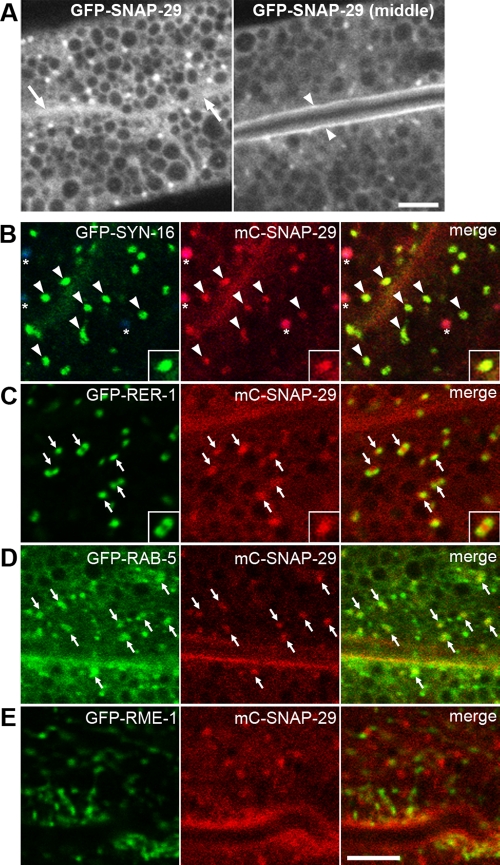
To examine the subcellular localization of SNAP-29, GFP-SNAP-29 or mCherry-SNAP-29 (mC-SNAP-29) was expressed in the intestine under the control of the intestine-specific act-5 promoter and the universal let-858 terminator. Expression of either tagged protein in the intestine rescued the growth defect caused by the RNAi using the 3′ untranslated region (UTR) of the snap-29 gene, confirming that the tagged SNAP-29 proteins are functional (Supplemental Table S3). Both GFP-SNAP-29 and mC-SNAP-29 mainly localized to intracellular punctate structures and the apical and basolateral PMs, but a significant portion of the proteins also seemed to be cytosolic in nature (Figure 3A). Most puncta of mC-SNAP-29 extensively colocalized with GFP-SYN-16 and closely attached with GFP-RER-1, suggesting the late-Golgi localization of mC-SNAP-29 (Figure 3, B and C). Because mC-SNAP-29 was also associated with GFP-RAB-5, mC-SNAP-29 could distribute to the Golgi-associated endosomes (Figure 3D). mC-SNAP-29–positive structures were often juxtaposed directly to RME-1–labeled recycling endosomes (Figure 3E). These results suggest that a large part of SNAP-29 localizes to the late-Golgi, endosomes, and the PM.
FIGURE 3:
Subcellular localization of SNAP-29. (A) Subcellular localization of GFP-SNAP-29 in the intestine. GFP-SNAP-29 localizes to punctate structures in the cytoplasm and on the basolateral (arrows) and apical (arrowheads) PMs. GFP-SNAP-29 signal is also detected in the cytosol. (B–D) mC-SNAP-29 and GFP-tagged markers were coexpressed in the intestine. mC-SNAP-29 colocalizes with GFP-SYN-16 on punctate structures (B). Asterisks indicate autofluorescence of gut granules because they appear in green, red, and blue channels. Puncta of mC-SNAP-29 closely associate both with GFP-RER-1 (C) and GFP-RAB-5 (D). Puncta of mC-SNAP-29 are adjacent to GFP-RME-1, but colocalization is observed only in the minor population (E). An enlarged (2×) image is shown in the inset (B and C). Note: All images were observed in living animals. Bars, 5 μm.
Loss of SNAP-29 causes aberrant morphology of the Golgi and endosomes
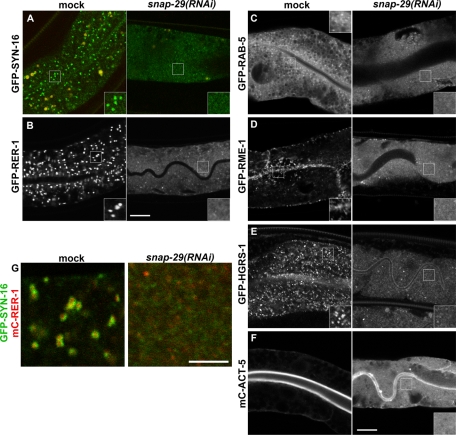
To better understand the function of SNAP-29 in the exocytic pathway, we investigated the effect of SNAP-29 depletion on organelle morphology. Because mC-SNAP-29 localizes to the Golgi, we observed the morphology of the Golgi by using the early- and late-Golgi–resident transmembrane markers. A depletion of SNAP-29 by RNAi resulted in the redistribution of both GFP-SYN-16 and GFP-RER-1 from the punctate Golgi structures to the fine membrane structures dispersed in the cytoplasm (Figure 4, A and B). We further analyzed this Golgi vesiculation phenotype by using animals coexpressing GFP-SYN-16 and mC-RER-1. Of interest, even in the snap-29(RNAi) animals, the early- and late-Golgi markers appeared to reside on distinct vesicles rather than be mixed (Figure 4G).
FIGURE 4:
Knockdown of snap-29 affects a wide range of organelle morphology. (A–F) Subcellular localizations of various GFP- and mCherry-tagged marker proteins were observed in the intestine of mock or snap-29(RNAi) animals. In the intestine of snap-29(RNAi) animals, all marker proteins are diffused in the cytoplasm or mislocalized to cytoplasmic fine vesicles. An enlarged (2×) image of the boxed area is shown in the inset. GFP-SYN-16 and mC-RER-1, the Golgi; GFP-RAB-5, endosomes; GFP-RME-1, recycling endosomes; GFP-HGRS-1, multivesicular endosomes; mC-ACT-5, microvillus actin. (G) Animals coexpressing GFP-SYN-16 and mC-RER-1 were treated with snap-29(RNAi). In snap-29(RNAi) animals, GFP-SYN-16 and mC-RER-1 disperse in cytoplasmic vesicles. Bars, 10 μm (A–F) or 5 μm (G).
We further investigated whether the morphologies of endocytic organelles are affected by SNAP-29 depletion. In snap-29(RNAi) animals, both the GFP-RAB-5 on early endosomes and GFP-RME-1 on recycling endosomes were mislocalized to the cytoplasm or cytoplasmic fine vesicles (Figure 4, C and D). Similar mislocalization was observed for GFP-HRGS-1, which is the C. elegans homologue of Hrs, the late endosomes/mutivesicular endosomes protein (Figure 4E; Shi et al., 2007). After snap-29(RNAi), fine cytoplasmic vesicles were labeled by at least some of these endosomal markers, although the cytosolic population of these markers may also have increased (Figure 4, C–E). These results reveal a quite wide-ranging impact of snap-29(RNAi) on the morphologies of exocytic and endocytic organelles and suggest that multiple pathways of membrane trafficking are impaired by snap-29(RNAi).
SNAP-29 depletion impairs organization of the endomemebrane system and microvillus formation
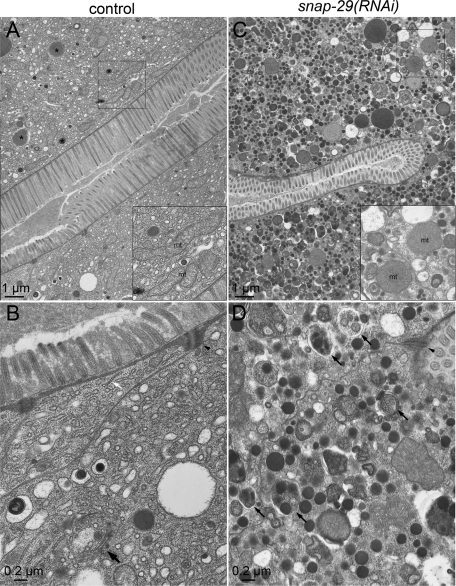
We further sought to define the defect in the intestine of snap-29(RNAi) animals by using an electron microscope (EM). In the intestinal cells of wild-type animals, we observed a tubular ER network coated with electron-dense ribosomes, a stack of cisternae assumed to be the Golgi apparatus, and a number of vesicles and granules (Figure 5, A and B). Large granules filled with electron-dense materials are likely to be granules containing yolk components to be exocytosed (Figure 5A, asterisks). Conversely, in the snap-29(RNAi) animals, we were unable to find the typical Golgi-like cisternae, whereas many small, electron-dense vesicles and multivesicular membrane structures accumulated in the cytoplasm (Figure 5, C and D). This observation demonstrates again that the organization of the endomembrane system is severely impaired by the depletion of SNAP-29. It is striking that the length of the microvilli projecting from the apical surface was apparently shortened in the snap-29(RNAi) animals compared with that of the control animals (Figure 5, A and C). Electron-dense adherens junctions were still visible in the snap-29(RNAi) animal (Figure 5, B and D, arrowheads). To confirm the effect of SNAP-29 depletion on the microvillus morphology, we analyzed the distribution of an mCherry-fusion protein with ACT-5 (mC-ACT-5), which is one of the actin isoforms that constitute the intestinal microvilli in C. elegans (MacQueen et al., 2005). As expected, mC-ACT-5 extensively localized to the apical PM in the wild-type intestine (Figure 4F). Consistent with the EM observation, a large population of mC-ACT-5 was mislocalized from the apical PM to the cytoplasm in the snap-29(RNAi) animals (Figure 4F). These results suggest that proper membrane trafficking is important for microvillus formation and/or maintenance. In the intestine of snap-29(RNAi) animals, we frequently observed autophagosome- and autolysosome-like structures (Figure 5D, small arrows). Furthermore, a number of smaller, rounder mitochondria were observed in the snap-29(RNAi) (Figure 5C, inset), whereas many elongated mitochondria were found in the control animal (Figure 5A, inset), suggesting that mitochondrial network formation is also affected directly or indirectly by the loss of the SNAP-29 function.
FIGURE 5:
Electron microscope analysis of wild-type and snap-29(RNAi) worms. Electron micrographs of cross sections of the intestine of wild-type and snap-29(RNAi) animals are shown. (A, B) Control RNAi animals display a normal intestinal organelle distribution, electron-dense adherens junctions (arrowheads), and microvilli. The tubular network of the ER, vesicles, granules, and elongated mitochondria (A, inset) are also observed. Large granules filled with electron-dense materials are likely to be granules containing yolk components to be exocytosed (A, asterisks). A stack of cisternae similar to the Golgi apparatus is also found (B, large arrow). In the intestine of the control RNAi animals, 0.76 Golgi, 0.41 multivesicular structures, and 5.0 electron-dense structures were observed in 10 μm2. (C, D) In snap-29(RNAi) animals, many electron-dense small vesicles containing yolk and multivesicular structures have accumulated. The typical branching tubular network of the ER is difficult to detect. The snap-29(RNAi) animals frequently exhibited shortened microvilli on the apical surface, whereas the adherens junctions still exist. It should be noted that many rounder mitochondria (B, inset) and autophagosome- and autolysosome-like structures (D, arrows) are observed in the snap-29(RNAi) animals. In the intestine of the snap-29(RNAi) animals, 15.3 multivesicular structures and 45.0 electron-dense structures were observed in 10 μm2.
SNAP-29 interacts with various worm syntaxins
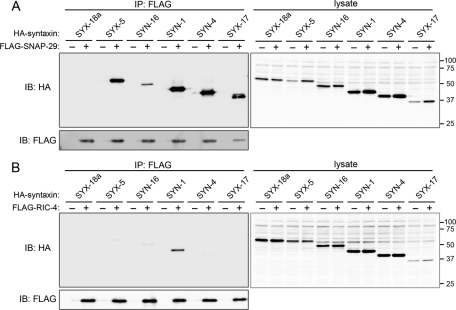
Because depletion of SNAP-29 affected a wide range of endomembrane organelles, we determined whether SNAP-29 binds to syntaxins present on distinct internal membranes. Hemagglutinin (HA)-tagged C. elegans SYX-18a (ER syntaxin homologue), SYX-5 (cis-Golgi syntaxin homologue), SYN-16 (trans-Golgi syntaxin homologue), SYN-1(PM syntaxin homologue), SYN-4 (PM syntaxin homologue), and SYX-17 (endosome/lysosome syntaxin homologue) were coexpressed with FLAG-tagged SNAP-29 or RIC-4 in mammalian cultured cells and subjected to coimmunoprecipitation experiments (Figure 6). We found that SNAP-29 bound to SYX-5, SYN-16, SYN-1, SYN-4, and SYX-17 (Figure 6A), whereas RIC-4 preferentially bound to SYN-1 (Figure 6B). These results suggest that unlike RIC-4, SNAP-29 is able to form binary t-SNARE complex with various syntaxins on the distinct compartments in C. elegans.
FIGURE 6:
SNAP-29 binds to various syntaxins in vivo. HEK293T cells were transiently transfected with pCI-3xHA-syn-18a, syx-5, syn-16, syn-1, syn-4, or syx-17 in the presence (+) or absence (−) of CMV10-snap-29 (A) or CMV10-ric-4 (B). After 36 h, cells were subjected to immunoprecipitation (IP) using FLAG M2 agarose beads. The resulting precipitates and total cell lysates were examined by immunoblotting (IB) using anti-FLAG (to SNAP-29 or RIC-4) and anti- HA (to syntaxin families) antibodies.
DISCUSSION
In this study, we identified C. elegans SNAP-29 as an essential regulator of membrane trafficking in the intestinal cells of living animals. We showed that SNAP-29 depletion blocks yolk secretion and results in the accumulation of small vesicles containing yolk. This phenotype strongly suggests that SNAP-29 is required for anterograde transport through the secretory pathway and controls the fusion of the transport intermediates with the target membrane. PM targeting of apical and basolateral membrane proteins is also inhibited by SNAP-29 depletion, suggesting a general requirement of SNAP-29 for the apical- and basolateral-directed transport pathways in polarized intestinal cells. Given the apical and basolateral PM localization of functional GFP-SNAP-29 and mC-SNAP-29, SNAP-29 could directly mediate the fusion of post-Golgi carriers with the PM. In humans, a mutation in the SNAP-29 gene results in CEDNIK syndrome (Sprecher et al., 2005). Consistent with our observations, the secretion of lamellar granules, Golgi-derived vesicular structures containing proteases and lipids, is impaired in the epidermis of these patients. Recently, it was also reported that a knockdown of SNAP-29 results in an exocytosis defect of an artificial model cargo in HeLa-M cells (Gordon et al., 2010).
It is striking that the morphology of a broad range of organelles comprising the exocytic and even endocytic pathways is totally affected by SNAP-29 depletion. In agreement with this phenotypic analysis, we found that tagged SNAP-29 broadly localizes to the late-Golgi and possibly the endosomes as well as the PM. In addition, worm SNAP-29 has an ability to bind various syntaxins. Thus SNAP-29 could be the common regulator involved in multiple transport pathways rather than a specific regulator of exocytosis. SNAP-29 could potentially participate in any transport directed to the Golgi (anterograde transport and retrograde transport from the endosomes) and to the endosomes in addition to exocytosis. Consistent with this possibility, it has been reported that mammalian SNAP-29 binds in vitro to various syntaxins localizing to the PM (syntaxins 1A, 3, 4), TGN (syntaxin 6), early/late endosomes (syntaxins 7, 13), and ER (syntaxin 17) (Steegmaier et al., 1998). It is also possible that some of the observed phenotypes of SNAP-29 depletion on the organellar morphology are indirect effects of the inhibition of the earlier transport step.
A characteristic of SNAP-29 is that it lacks cysteine residues for palmitoylation that are conserved among the other SNAP family proteins. Nevertheless we observed that SNAP-29 localizes to various membrane compartments, suggesting a palmitoylation-independent membrane-targeting mechanism. It has been also reported that the mutant SNAP-25 protein that lacks all cysteines required for palmitoylation still possesses a partial membrane localization activity and that co-overexpression of syntaxin 1A facilitates membrane localization of the mutant SNAP-25 (Gonelle-Gispert et al., 2000). Given that C. elegans SNAP-29 interacts with various syntaxins, it is reasonable to speculate that SNAP-29 localizes to the membrane through binding to syntaxin on each compartment.
We demonstrate that after SNAP-29 depletion, the Golgi and likely the endosomes are dispersed into smaller vesicular structures and are no longer able to maintain their typical morphology and size. This suggests that these organelles are not static structures but dynamic ones that are maintained by the balance between budding and fusion of the membrane. One possible explanation is that the budding of transport intermediates continues to take place, but fusion events with target membranes are blocked in the snap-29(RNAi) animals, resulting in the consumption of the organellar membranes.
We observed that autophagy is significantly induced in the intestine of snap-29(RNAi) animals, suggesting that these intestinal cells are starved. SNAP-29 depletion may impair the secretion of digestive enzymes and the apical targeting of permeases and transporters, resulting in starvation and induction of autophagy. An abnormal morphology of mitochondria might also result from such change of cellular nutritional condition. Of interest, the depletion of SNAP-29 caused the mislocalization of ACT-5 (the actin essential for intestinal microvillus formation) from the apical membrane to the cytoplasm, resulting in disorganized and shorter microvilli in the intestine of snap-29(RNAi) animals (Figure 5). These results imply that proper membrane trafficking to the apical surface is a prerequisite for microvillus formation and maintenance in epithelial cells.
MATERIALS AND METHODS
General methods and strains
Methods for the handling and culturing of C. elegans were essentially those as described previously (Brenner, 1974). Strains expressing GFP- or mCherry-tagged proteins were grown at 20°C. The wild-type parent for all strains was C. elegans var Bristol strain N2. The unc-119(ed3) mutant (Maduro and Pilgrim, 1995) was obtained from the Caenorhabditis Genetic Center. The transgenic strains used are listed in Supplemental Table S1.
Plasmids and transgenic C. elegans
Genomic fragments containing the coding regions of rer-1, unc-64, syn-1, snap-29, or act-5 and cDNAs of snap-29, ric-4, syn-16, syx-18a, syx-5, syn-16, syn-1, syn-4, syx-17, and rab-5 were amplified by PCR and cloned into the entry vector pDONR221 by using Gateway recombination cloning technology (Invitrogen, Tokyo, Japan). These fragments of rer-1, snap-29, and syn-16 were cloned into the destination vector pKS18 or pMS14.1. Vectors pKS18 and pMS14.1 use act-5 and opt-2 5′ UTRs, respectively, to express an N-terminal GFP-tagged fusion protein in the intestine. The genomic fragments of rer-1 and snap-29 were also cloned into the destination vector pMS6.2 or pMS16.1, which drive the expression of an N-terminal mCherry-tagged fusion protein under the control of act-5 or opt-2 5′ UTRs, respectively. The genomic fragment of act-5 was cloned into a destination vector pMS6.3, which drives the expression of an N-terminal mCherry-HA (HA epitope)–tagged fusion protein under the act-5 5′ UTR control (Sato et al., 2008b). To express YFP- or CFP-tagged proteins, a region corresponding to GFP in pKS18 was replaced with a fragment encoding EYFP (pMS8.1) or Cerulean (pMS9.1). cDNAs of syn-16 and rab-5 were then cloned into these vectors. All destination vectors use the universal let-858 3′ UTR. The vectors pMS14.1, pMS6.2, pMS6.3, pMS8.1, and pMS9.1 contain a genomic fragment of Caenorhabditis briggsae unc-119 to select transgenic animals. Transgenic lines were created using the microparticle bombardment method as described previously (Praitis et al., 2001). For pKS18-based plasmids, the wild-type unc-119 fragment was introduced simultaneously. cDNAs of snap-29 and ric-4 were prepared from expressed sequence tag clones provided by Yuji Kohara (National Institute of Genetics, Shizuoka, Japan) and subcloned into L4440. A snap-29 3′ UTR fragment (926 base pairs) and a genomic fragment of aex-4 were amplified from the N2 genome and cloned into L4440. RNAi using the snap-29 3′ UTR fragment showed similar but slightly milder phenotypes compared with the RNAi using snap-29 cDNA. As a negative control of the RNAi experiments, L4440 alone or L4440 harboring cDNA of human transferrin receptor was used (Sato et al., 2006). To generate a mammalian expression vector for the expression of FLAG and HA fusions in mammalian cells, the two mammalian expression vectors p3XFLAG-CMV-10 (Sigma-Aldrich, Tokyo, Japan) and pCIneo (Promega, Tokyo, Japan) together with HA-epitope tag (Itakura et al., 2008) were used to incorporate the Gateway cassettes rfB of the Gateway conversion system (Invitrogen). To express FLAG fusion proteins in mammalian cells, cDNA fragments of snap-29 and ric-4 were cloned into a destination vector (CMV10-rfB) by Gateway recombination cloning. To express HA fusion proteins in mammalian cells, cDNA fragments of snap-29, ric-4, syn-16, syx-18a, syx-5, syn-16, syn-1, syn-4, and syx-17 were cloned into a destination vector (pCIneo-3xHA-rfB) by Gateway recombination cloning.
RNA-mediated interference
RNAi experiments in this study were performed using the feeding method (Timmons et al., 2001). L1 larvae were placed on plates containing nematode growth medium agar with 5 mM isopropyl β-d-thiogalactopyranoside (IPTG) and bacteria strain HT115(DE3) carrying double-stranded RNA expression constructs and allowed to grow to the L4 larvae stage at 20°C. For the EM, L3 larvae were incubated on RNAi plates at 20°C for 48 h and allowed to grow to L4 larvae or young adult animals.
Antibodies and Western blotting
The DNA fragment of the snap-29 ORF was cloned into the 6xHis expression vector pDEST17 (Invitrogen). The His-tagged recombinant protein was expressed in Escherichia coli strain Rosetta (Merck, Tokyo, Japan) at 37°C for 2 h in the presence of 1 mM IPTG and purified with a nickel Sepharose 6 Fast Flow column under denaturing conditions according to the manufacturer's instructions (GE Healthcare Biosciences, Uppsala, Sweden). The purified proteins were used for antibody production in rabbits at TK Craft (Gunma, Japan). To examine the amount of endogenous SNAP-29, total lysates were prepared from 30 adult hermaphrodites and subjected to Western blotting as described previously (Sato et al., 2006). An anti-actin monoclonal antibody (C4) was purchased from Millipore (Tokyo, Japan). A mouse monoclonal anti-FLAG (M2) antibody and M2 agarose beads were purchased from Sigma-Aldrich. A rat monoclonal anti-HA antibody (3F10) and a rat monoclonal anti-HA-peroxidase–conjugated antibody (3F10) were purchased from Roche Diagnostics (Tokyo, Japan).
Cell culture and transfection
HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum and 50 μg/ml penicillin and streptomycin (complete medium) in a 5% CO2 incubator. Fugene HD (Roche Diagnostics) was used for transfection.
Immunoprecipitation
Cell lysates were prepared in a regular lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, and a protease inhibitor cocktail [Roche Diagnostics]). The lysates were clarified by centrifugation at 15,000 rpm for 15 min and then subjected to immunoprecipitation using a FLAG M2 agarose beads (Sigma-Aldrich) as described previously (Hara et al., 2008).
Microscopy
To observe live worms expressing transgenes, worms were mounted on agarose pads with 20 mM levamisole in M9 buffer (Sato et al., 2008a). Images were obtained using an Olympus confocal microscope system FV1000 (Olympus, Tokyo, Japan). Time-lapse images were taken using a DeltaVision Deconvolution microscope system (Applied Precision, Issaquah, WA).
Electron microscopy
N2 and snap-29(RNAi) animals were washed in an H2O and cacodylate buffer (0.1 M sodium cacodylate [pH 7.4]) and then fixed in Karnovsky fixative (2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer [pH 7.4]) for 10 min at room temperature. The animals were then transferred onto a depression glass slide, cut immediately at the level of the pharynx to facilitate penetration of fixatives, and transferred to a 1.7-ml presiliconized tube. After 2 h of fixation on ice, the fixed animals were incubated in 10% sucrose/0.1 M cacodylate buffer (pH 7.4) for 5 min and then in 1% OsO4/0.1 M cacodylate buffer for 1 h on ice. These were further incubated in 0.5% uranyl acetate/H2O overnight at room temperature. After fixation and dehydration, embedding in Epon (Queto 812) and staining were performed as previously described (Harada et al., 1990, 2002). Electron micrographs were taken using JEOL1010 (JEOL, Tokyo, Japan).
Supplementary Material
Acknowledgments
We thank Yuji Kohara for supplying the important reagents, Shiori Inoue for technical assistance, and the Caenorhabditis Genetic Center for supplying the many strains used in this study. This research was supported by the Ministry of Education, Culture, Sports, Science and Technology, Grant-in-Aid for Scientific Research on Priority Areas 2010 (Protein Community) and Scientific Research (C), 2008. K.S. and M.S. were supported by the Uehara Memorial Foundation and Astellas Foundation for Research on Metabolic Disorders. M.S., A.H., and K.S. also received support from grants-in-aid and the Global Center of Excellence Program from the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work was supported by the joint research program of the Institute for Molecular and Cellular Regulation, Gunma University.
Abbreviations used:
- ER
endoplasmic reticulum
- PM
plasma membrane
- RNAi
RNA interference
- SNAP-29
synaptosomal-associated protein 29
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0279) on May 25, 2011.
REFERENCES
- Balklava Z, Pant S, Fares H, Grant B. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD. RAB-10 is required for endocytic recycling in the C. elegans intestine. Mol Biol Cell. 2006 doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Kuroyanagi H, Kobayashi T, Murate M, Fujimoto K, Okabe S, Mitani S. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–157. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonelle-Gispert C, Molinete M, Halban PA, Sadoul K. Membrane localization and biological activity of SNAP-25 cysteine mutants in insulin-secreting cells. J Cell Sci. 2000;113:3197–3205. doi: 10.1242/jcs.113.18.3197. [DOI] [PubMed] [Google Scholar]
- Gordon DE, Bond LM, Sahlender DA, Peden AA. A targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic. 2010;11:1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Sobue K, Hirokawa N. Developmental changes of synapsin I subcellular localization in rat cerebellar neurons. Cell Struct Funct. 1990;15:329–342. doi: 10.1247/csf.15.329. [DOI] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Schroeder LK, Hieb CA, Kershner AM, Rabbitts BM, Fonarev P, Grant BD, Priess JR. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol Biol Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt M, Varoqueaux F, Wiederhold K, Takamori S, Urlaub H, Fasshauer D, Jahn R. Identification of SNAP-47, a novel Qbc-SNARE with ubiquitous expression. J Biol Chem. 2006;281:17076–17083. doi: 10.1074/jbc.M513838200. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Glotzer M. Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr Biol. 1999;9:738–745. doi: 10.1016/s0960-9822(99)80333-9. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Baggett JJ, Perumov N, Bauer RA, Januszewski T, Schriefer L, Waddle JA. ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation. Mol Biol Cell. 2005;16:3247–3259. doi: 10.1091/mbc.E04-12-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney TR, Luo S, Round EK, Brauner M, Gottschalk A, Thomas JH, Nonet ML. Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:16350–16355. doi: 10.1073/pnas.0803617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Lugassy Y, Sprecher E, Horowitz M. Loss of SNAP29 impairs endocytic recycling and cell motility. PLoS One. 2010;5:e9759. doi: 10.1371/journal.pone.0009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem-Yehudar R, Galperin E, Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]
- Saifee O, Wei L, Nonet ML. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17:3085–3094. doi: 10.1091/mbc.E06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Grant BD, Harada A, Sato K. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J Cell Sci. 2008a;121:3177–3186. doi: 10.1242/jcs.034678. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008b;27:1183–1196. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardt A, Brinkmann BG, Mitkovski M, Sereda MW, Werner HB, Nave KA. The SNARE protein SNAP-29 interacts with the GTPase Rab3A: implications for membrane trafficking in myelinating glia. J Neurosci Res. 2009;87:3465–3479. doi: 10.1002/jnr.22005. [DOI] [PubMed] [Google Scholar]
- Shi A, Pant S, Balklava Z, Chen CC, Figueroa V, Grant BD. A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Sun L, Banerjee R, Tobin M, Zhang Y, Grant BD. Regulation of endosomal clathrin and retromer-mediated endosome to Golgi retrograde transport by the J-domain protein RME-8. EMBO J. 2009;28:3290–3302. doi: 10.1038/emboj.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Bremnes B, Ronning E, Aasland R, Stenmark H. Syntaxin-16, a putative Golgi t-SNARE. Eur J Cell Biol. 1998;75:223–231. doi: 10.1016/S0171-9335(98)80116-7. [DOI] [PubMed] [Google Scholar]
- Sprecher E, et al. A mutation in SNAP29, coding for a SNARE protein involved in intracellular trafficking, causes a novel neurocutaneous syndrome characterized by cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma. Am J Hum Genet. 2005;77:242–251. doi: 10.1086/432556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem. 1998;273:34171–34179. doi: 10.1074/jbc.273.51.34171. [DOI] [PubMed] [Google Scholar]
- Su Q, Mochida S, Tian JH, Mehta R, Sheng ZH. SNAP-29: a general SNARE protein that inhibits SNARE disassembly and is implicated in synaptic transmission. Proc Natl Acad Sci USA. 2001;98:14038–14043. doi: 10.1073/pnas.251532398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- Wu QL, Rui Q, He KW, Shen LL, Wang DY. UNC-64 and RIC-4, the plasma membrane-associated SNAREs syntaxin and SNAP-25, regulate fat storage in nematode Caenorhabditis elegans. Neurosci Bull. 2010;26:104–116. doi: 10.1007/s12264-010-9182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Iwasaki K, Doi M. The non-neuronal syntaxin SYN-1 regulates defecation behavior and neural activity in C. elegans through interaction with the Munc13-like protein AEX-1. Biochem Biophys Res Commun. 2009;378:404–408. doi: 10.1016/j.bbrc.2008.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.