This study reveals that mRNAs are partitioned between the cytosol and endoplasmic reticulum (ER) compartments in a hierarchical manner and identifies a prominent role for the ER in global protein synthesis. Two modes of mRNA association with the ER are defined: ribosome dependent and ribosome independent.
Abstract
The mRNA transcriptome is currently thought to be partitioned between the cytosol and endoplasmic reticulum (ER) compartments by binary selection; mRNAs encoding cytosolic/nucleoplasmic proteins are translated on free ribosomes, and mRNAs encoding topogenic signal-bearing proteins are translated on ER-bound ribosomes, with ER localization being conferred by the signal-recognition particle pathway. In subgenomic and genomic analyses of subcellular mRNA partitioning, we report an overlapping subcellular distribution of cytosolic/nucleoplasmic and topogenic signal-encoding mRNAs, with mRNAs of both cohorts displaying noncanonical subcellular partitioning patterns. Unexpectedly, the topogenic signal-encoding mRNA transcriptome was observed to partition in a hierarchical, cohort-specific manner. mRNAs encoding resident proteins of the endomembrane system were clustered at high ER-enrichment values, whereas mRNAs encoding secretory pathway cargo were broadly represented on free and ER-bound ribosomes. Two distinct modes of mRNA association with the ER were identified. mRNAs encoding endomembrane-resident proteins were bound via direct, ribosome-independent interactions, whereas mRNAs encoding secretory cargo displayed predominantly ribosome-dependent modes of ER association. These data indicate that mRNAs are partitioned between the cytosol and ER compartments via a hierarchical system of intrinsic and encoded topogenic signals and identify mRNA cohort-restricted modes of mRNA association with the ER.
INTRODUCTION
On exit from the nucleus, newly synthesized mRNAs are partitioned between the two primary protein synthesis compartments of the cell: the cytosol and the endoplasmic reticulum (ER). This ubiquitous, transcriptome-wide sorting process functions to segregate mRNAs on the basis of the subcellular trafficking fates of their encoded translation products (Blobel, 1975; Blobel and Dobberstein, 1975a, 1975b; Blobel et al., 1979). Thus mRNAs encoding secretory and integral membrane proteins undergo cotranslational localization to the ER via the activity of topogenic signals encoded in the nascent polypeptide chain (Blobel, 1975; Blobel and Dobberstein, 1975a, 1975b; Blobel et al., 1979; Walter and Johnson, 1994). In contrast, mRNAs encoding cytosolic/nucleoplasmic proteins, which lack topogenic signals, are translated on cytosolic ribosomes and assume a default localization in the cytosol.
For those nascent translation products containing topogenic signals (signal sequence and/or transmembrane domain), topogenic signal recognition by the signal-recognition particle (SRP) initiates the trafficking of mRNA/ribosome/nascent chain complexes to the ER (Blobel, 1980; Lingappa and Blobel, 1980; Walter and Johnson, 1994). In directing mRNA traffic to the ER, the SRP pathway operates in an iterative manner to segregate the mRNA transcriptome between the cytosol and ER compartments (Blobel and Dobberstein, 1975a, 1975b; Walter and Blobel, 1981a, 1981b;Walter et al., 1981); Walter and Johnson, 1994). Although a primary function for the SRP pathway in mRNA localization to the ER is well established, molecular genetic and genomic studies have indicated that multiple pathways likely contribute to subcellular mRNA partitioning in the cell. Notably, molecular genetic analyses of SRP pathway function in yeast have demonstrated that SRP/SRP receptor expression is not essential for viability (Hann et al., 1989; Hann and Walter, 1991; Mutka and Walter, 2001). In yeast, the loss of SRP pathway function was accompanied by a broad physiological response yielding ER expansion and an enhanced capacity for mRNA translation on the ER, presumably through direct initiation of mRNA translation on ER-associated ribosomes (Potter and Nicchitta, 2000, 2002; Mutka and Walter, 2001). Similar to findings in yeast, siRNA-mediated silencing of SRP72 or SRP54 expression in mammalian cells had modest effects on membrane protein synthesis and trafficking, with no overt effects on cell growth and division (Ren et al., 2004). Silencing of SRP14 expression also resulted in decreased efficiencies in protein secretion efficiency and reduced cell growth rates, but was not lethal (Lakkaraju et al., 2008). Further evidence for the existence of multiple pathways for regulating mRNA partitioning to the ER was obtained in cDNA microarray studies of the population identities of cytosolic and membrane-bound mRNAs, where a substantial representation of mRNAs encoding cytosolic proteins in the ER-bound mRNA pool, was reported (Kopczynski et al., 1998; Diehn et al., 2000, 2006; de Jong et al., 2006). Currently, little is known regarding the biological rules that determine the subcellular partitioning behavior of individual mRNAs, or the biochemical mechanisms that enable distinct patterns of mRNA enrichment in the two compartments.
In this communication, we utilize cell fractionation, subgenomic and genomic analysis of mRNA population identities, and established biochemical methodologies for the study of ribosome–membrane interactions to investigate subcellular patterns and mechanisms of mRNA partitioning in mammalian cells. These studies reveal an unexpected diversity in the cellular mechanisms governing the subcellular distribution of topogenic signal-encoding mRNAs and suggest that mRNAs encoding resident proteins of the endomembrane system contain localization signals dominant to the signal peptide, which enable mRNA cohort-restricted modes of direct association with the ER.
RESULTS
Identification of cohort-specific patterns of subcellular mRNA partitioning
We examined subcellular mRNA distributions in mammalian cell lines using an established sequential detergent extraction protocol (Stephens et al., 2005, 2008; Stephens and Nicchitta, 2007). This protocol does not require cell homogenization, as used in past studies, and thus provides an important test of the contribution of fractionation methodologies to subcellular mRNA partitioning patterns (Diehn et al., 2000, 2006). To enable the quantitative assessment of subcellular mRNA distributions, mRNA levels were assayed by qPCR array using commercially available qPCR gene array sets, which were selected to include genes with products that encode prominent proteins of known function and subcellular distribution, and that represent cytosolic, nucleoplasmic, and topogenic signal-encoding mRNAs (Supplemental Table S1).
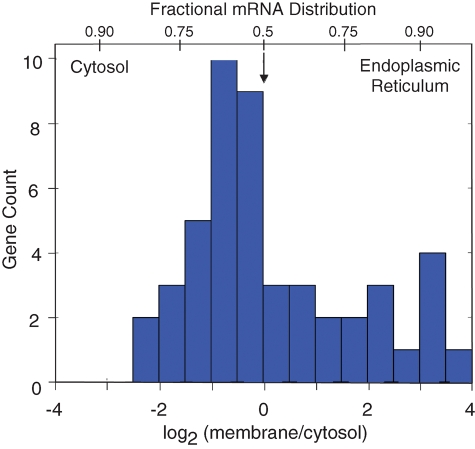
The data in Figure 1 depict the subcellular distribution values (log2 membrane/cytosol) for all gene products identified in the qPCR array analysis of J558 murine plasmacytoma cytosol and ER-bound RNA pools. The overall gene distribution profile is notable for the broad dispersion of mRNAs between the cytosol and ER compartments (∼6 log2 orders); the abundance of mRNAs present in both compartments (peak gene distribution was centered at log2 −0.5); and the differing absolute magnitudes of compartmental enrichment, ranging from −2.27 (cytosolic protein, leucine aminopeptidase) to 3.85 (secretory protein, prosaposin [Psap]). The observed mRNA distribution pattern contrasts with the bimodal pattern expected in models where topogenic signals dictate mRNA partitioning to the ER. This distribution pattern broadly mirrors the pattern previously reported in genome-scale cDNA microarray-based studies of subcellular mRNA partitioning, and demonstrates that noncanonical subcellular mRNA distribution patterns are apparent, regardless of the methodologies used for cell fractionation or for assessing mRNA distributions (Diehn et al., 2000, 2006).
FIGURE 1:
mRNAs display diverse patterns of subcellular partitioning. J558 murine plasmacytoma cells were fractionated by sequential detergent extraction and total cytosolic and ER membrane-associated RNA pools were isolated by Trizol extraction. The mRNA composition of the two pools was assayed by 96-gene qPCR arrays and compartmental enrichment values quantified using GPR software.
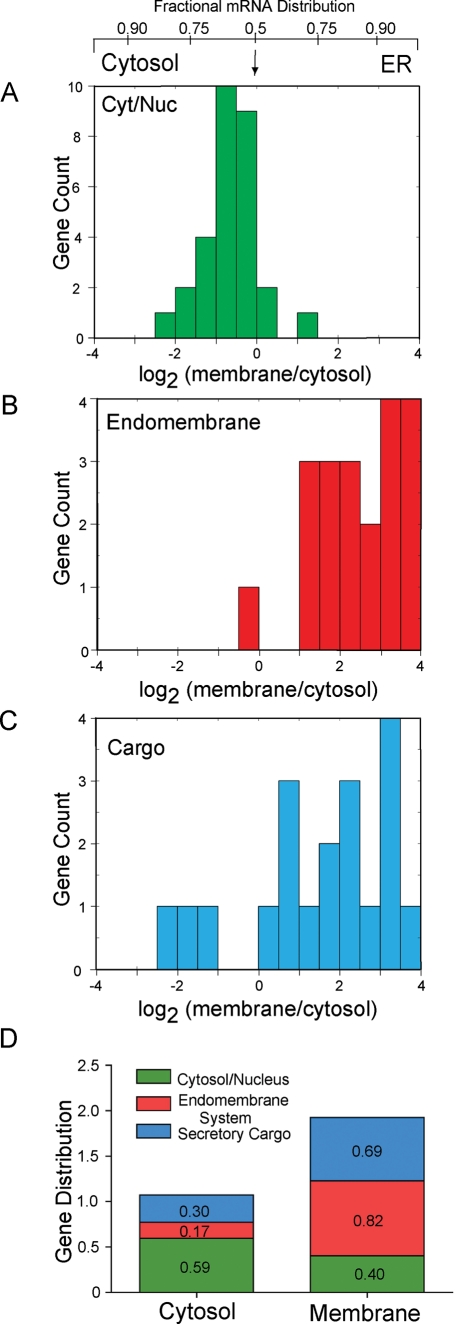
To gain insight into the biological origins of the overlapping compartmental mRNA distribution, the gene list was first sorted on the basis of the presence or absence of an encoded topogenic signal. Similar to past reports, this analysis demonstrated a substantial cross-representation of topogenic signal-encoding mRNAs in the cytosol and ER compartments, suggesting that topogenic signals alone are incomplete determinants of subcellular mRNA distribution patterns (Diehn et al., 2000, 2006). When the topogenic signal-encoding mRNAs were sorted on the basis of the subcellular trafficking fate of the encoded translation product, however, an mRNA cohort-specific subcellular distribution pattern was revealed (Figure 2A–D). In this analysis, three mRNA cohorts were defined: mRNAs encoding cytosolic/nucleoplasmic proteins, predicted to be enriched in the cytosolic mRNA pool; mRNAs encoding secretory pathway cargo, which encode proteins bearing topogenic signals and are thus predicted to be enriched in the ER; and mRNAs encoding resident proteins of the endomembrane system (ER, Golgi apparatus, lysosomes, endosomes), which encode proteins bearing topogenic signals and intrinsic organellar localization signals and which are predicted to be enriched in the ER (Table S2). As shown in Figure 2A, mRNAs encoding cytosolic/nucleoplasmic proteins displayed a peak gene enrichment at log2 −0.5, with a range of approximately −2.271 (Lap3) to 1.322 (CaMKIIα; Table S2). In addition to displaying an overall fractional enrichment in the cytosol, this mRNA cohort includes mRNAs that undergo noncanonical enrichment in the ER (Table S2). The biological mechanism and rationale for such noncanonical enrichments is unknown and is under separate investigation. In contrast to the cytosolic/nucleoplasmic mRNA cohort, mRNAs encoding endomembrane-resident proteins were highly enriched on the ER, with log2 distribution values of −0.1 (Lamp1) to 3.56 (Lgmn, a lysosomal, cysteinyl protease; Figure 2B and Table S2). The combined subcellular distribution patterns of mRNAs encoding cytosolic/nucleoplasmic and endomembrane-resident proteins approximate the predicted bimodal distribution predicted in a topogenic signal-based mRNA sorting model, though biased to an ER-associated distribution. This distribution pattern was lost when the analysis was expanded to include the topogenic signal-encoding secretory pathway cargo cohort (Figure 2C and Table S2). The latter cohort was distinguished by an unexpectedly broad spectrum of log2 partitioning values, ranging from −2.27 (FcRγ-IIb) to 3.85 (Psap). The broad distribution pattern for this cohort may in part reflect signal peptide functionality; the critical signal peptide hydrophobic region (H region) of the FcRγ-IIb (log2 −2.27) signal peptide is relatively short (seven amino acids), whereas the H region of Psap (log2 3.85) is substantially longer (12 amino acids). Indeed, differences in signal peptide functionality have previously been demonstrated to influence protein translocation efficiencies and the subcellular distribution of the translation products (Kim et al., 2001, 2002; Kim and Hegde, 2002; Shaffer et al., 2005; Hegde and Bernstein, 2006). Interestingly, in the current annotation, Psap is reported as a dual secretory/integral membrane protein, and thus is assigned to the secretory pathway cargo cohort. However, the primary subcellular trafficking destination of Psap is the lysosome, where saposins A, B, C, and D function as lysosomal glycosphingolipid hydrolases (O'Orien and Kishimoto, 1991; Kishimoto et al., 1992). The high ER partitioning value for Psap mRNAs likely reflects a resident lysosomal (endomembrane) function for the encoded products.
FIGURE 2:
Subcellular mRNA distributions are marked by cohort-specific enrichment patterns. Gene product distributions (Figure 1) were grouped on the basis of predicted subcellular localization, and cohort-specific subcellular distributions patterns were analyzed. Predicted subcellular distributions were determined by manual interrogation of existing databases. (A) mRNAs encoding cytosolic and nucleoplasmic proteins (Cyt/Nuc). (B) mRNAs encoding resident proteins of the endomembrane system. (C) mRNAs encoding secretory pathway cargo. (D) Cohort-specific population distributions of mRNAs in the cytosol and ER membrane RNA pools. Total gene distributions were determined, and the overall and relative pool compositions were determined as a function of a three-cohort model.
Summary gene distribution patterns for the cytosolic and ER mRNA pools are depicted in Figure 2D. The overall gene product distributions favor the ER by approximately twofold, suggesting that the ER is the predominant site of mRNA translation in the cell. The cytosolic mRNA pool was comprised predominantly (∼55%) of mRNAs encoding cytosolic/nucleoplasmic proteins, with the remaining fraction comprising noncanonically enriched secretory pathway cargo-encoding and endomembrane organelle protein-encoding mRNAs. The ER fraction was highly enriched in mRNAs encoding secretory pathway cargo (36%) and endomembrane organelle proteins (43%), with the remaining fraction (21%) comprising mRNAs encoding cytosolic/nucleoplasmic proteins that undergo noncanonical localization to the ER.
The data presented in Figures 1 and 2 suggest that subcellular mRNA distribution patterns are determined by multiple localization elements. Importantly, topogenic signals, which are essential for protein translocation and sufficient to direct mRNA localization to the ER in vitro, appear to represent one of potentially many such signals. These findings, observed at a subgenomic level, are consistent with the emerging view that a large fraction of the mRNA transcriptome undergoes localization to serve fundamental roles in conferring cellular organization and function, and that signal sequences can encode diverse protein localization and localization information (Hegde and Bernstein, 2006; Lecuyer et al., 2007, 2009).
Genome-scale analysis of cohort-specific patterns of mRNA partitioning
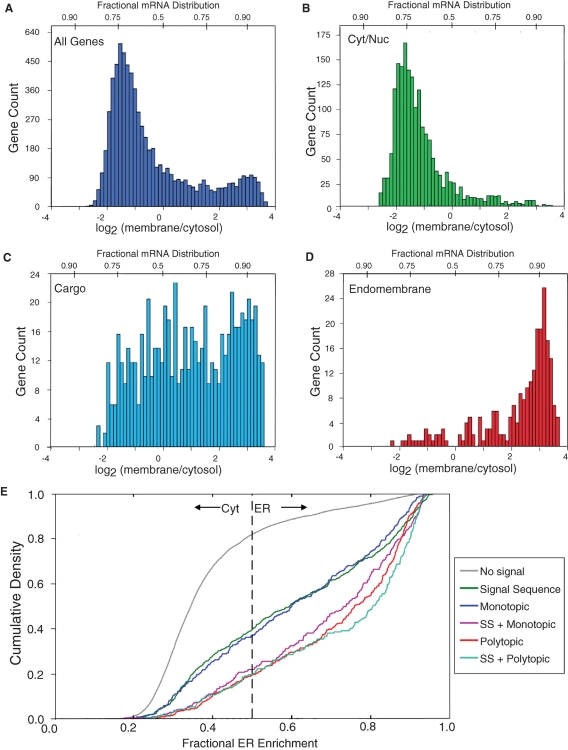
The cohort-specific subcellular mRNA distribution patterns described above were derived from a limited gene set; of the 96 genes examined in the qPCR arrays, significant qPCR signals were obtained for 60 genes, or ∼0.3% of the murine genome. To determine whether the cohort-specific subcellular enrichment patterns noted above were recapitulated at the genome scale, publicly available genome-scale subcellular mRNA partitioning data (Stanford Microarray Database, http://smd.stanford.edu/resources/databases.shtml) were analyzed using the cytosolic/nucleoplasmic/secretory cargo/resident endomembrane protein distribution model described in the preceding section. In this analysis, the mRNA subcellular distribution data set from K-562 leukemia cells, described in Diehn et al. (2006), was examined. As shown in Figure 3A, peak K-562 gene enrichment was centered at log2 –1.6, with maximal cytosol and ER enrichments extending from log2 –2.6 to 4.5, respectively (Table S3). The overall K-562 subcellular mRNA distribution pattern was similar to that determined in the J558 cell line and was recapitulated in numerous cell lines (Supplemental Figure S1). As in the mRNA distributions determined in J558 cells, the overall pattern of subcellular mRNA partitioning in K-562 cells is notable for the broad distribution of mRNAs between the cytosol and ER compartments, as well the differences in the maximal degree of enrichment in the ER relative to the cytosol (Figure 3A). The distribution pattern of mRNAs encoding cytosolic/nucleoplasmic proteins is shown in Figure 3B. This gene set was identified by selecting genes via the gene ontology (GO) category “cytoplasm” (GO: 0005737) and computationally filtered to remove any topogenic domain-encoding mRNAs that were included through inaccurate GO annotation. The K-562 cytoplasm cohort (1814 genes) displayed a peak at −1.8 log2 (membrane/cytosol). A sizable fraction of mRNAs encoding cytosolic proteins was represented in the ER mRNA pool, with a small subset of the cytosolic protein-encoding mRNAs displaying relative ER enrichment levels that approximate those seen with topogenic domain-encoding mRNAs. The latter cohort was distinguished by its enrichment in mRNAs encoding proteins with a nuclear function as well as proteins functioning in cytoskeletal organization (Table S3). To identify genes encoding resident proteins of the endomembrane system, a custom GO category, “endomembrane system,” was defined to include proteins that reside in the rough ER and/or rough ER lumen (defined as bearing a C-terminal ER retention/retrieval motif), or are resident membrane proteins of the Golgi apparatus or lysosomes. This cohort (245 genes) displayed a peak log2 enrichment of ∼3.4, and ranged from −1.8 to 4.5 (Figure 3D). The subcohort of mRNAs displaying the lowest ER partitioning values was highly enriched in genes encoding enzymes functioning in lipid biosynthesis (Table S3). In contrast, the subcohort displaying the highest ER partitioning values was enriched in genes encoding resident molecular chaperones of the ER lumen (i.e., GRP94 and calreticulin), proteins functioning in protein translocation/modification (β-subunit of the SRP receptor, ribophorin I), resident Golgi proteins functioning in oligosaccharide modification, and resident lysosomal proteins (Table S3). Similar to the observations obtained in J558 cell fractionations, the genome-scale mRNA enrichment patterns of the cytosolic/nucleoplasmic and endomembrane system mRNA cohorts’ genes displayed a bimodal distribution pattern with a clear bias to the ER compartment.
FIGURE 3:
Genome-scale analysis of subcellular mRNA distribution reveals topogenic signal-independent partitioning patterns. Publicly available subcellular gene production distribution data were analyzed by a three-cohort model. The genome database for the K-562 (human myelogenous leukemia) cell line was used. Gene product cohorts were identified by algorithmic sorting, using gene ontology (GO) criteria. (A) All genes. (B) Genes encoding cytosolic and nucleoplasmic proteins, selected via the GO category “cytoplasm” (GO: 0005737) and filtered to remove topogenic signal-encoding genes. (C) Secretory pathway cargo. The K-562 gene set was sorted using the GO categories “extracellular” (GO: 0005615) and “plasma membrane” (GO: 0005886). (D) Endomembrane system. The K-562 gene set was sorted using a custom GO category “endomembrane” to include genes whose translation products reside in the ER membrane, the ER lumen, or the Golgi apparatus or lysosomes. (E) Subcellular mRNA distributions were analyzed by cumulative density distribution, using a six-cohort model: no topogenic signal, signal sequence-encoding, single transmembrane domain-encoding (monotopic), multiple transmembrane domain-encoding (polytopic), and single/multi-transmembrane domain plus signal sequence.
As observed in the subgenomic analysis, the bimodal distribution pattern displayed by the K-562 cytosolic/nucleoplasmic and endomembrane system mRNA cohorts was compromised when mRNAs encoding secretory pathway cargo (668 genes) were included in the analysis (Figure 3C and Table S3). This category was selected by filtering the K-562 gene set by the GO categories “extracellular” (GO:0005615) and “plasma membrane” (GO:0005886) and displayed log2 (membrane/cytosol) enrichment values ranging from −1.9 to 4.2. At the genome scale, no clear correlation between signal peptide H region length and subcellular distribution was discernible, with both the high and low relative ER membrane enrichment cohorts displaying strong signal peptide H region SignalP (www.cbs.dtu.dk/services/SignalP/) scores (unpublished data; Bendtsen et al., 2004). As observed in the qPCR array studies (Figures 1 and 2), the secretory pathway cargo-encoding mRNA cohort displayed a broad subcellular distribution pattern, with a substantial number of topogenic domain-encoding mRNAs partitioning to the cytosol.
To further investigate the role of topogenic signals in the regulation of subcellular mRNA partitioning, log2 (membrane/cytosol) K-562 gene distributions were evaluated with respect to encoded topogenic signals (signal peptide, monotopic membrane protein, polytopic membrane protein). Using the programs SignalP and TMHMM (www.cbs.dtu.dk/services/TMHMM), genes were classified into six categories: lacking topogenic signal, signal peptide-encoding, single transmembrane domain-encoding, multiple transmembrane domain-encoding, single transmembrane domain and signal peptide-encoding, and lastly, multiple transmembrane domain and signal peptide-encoding (Krogh et al., 2001; Bendtsen et al., 2004). Cumulative density functions were then generated for each cohort (Figure 3E). In the case of mRNAs lacking an encoded topogenic signal (i.e., cytosolic/nucleoplasmic), the gene density function is curvilinear, with a substantial gene representation at log2 = 0 and a cohort of genes displaying noncanonical enrichment in the ER. Plots depicting topogenic signal-encoding genes are also curvilinear and include a substantial fraction of gene products that are cross-represented in the cytosol and ER compartments. For those genes encoding mono- and polytopic transmembrane domain-bearing proteins, a more pronounced bias to ER enrichment was observed, suggesting that transmembrane domain topogenic signals are dominant to signal peptides (Figure 3E). Interestingly, overall ER-enrichment values were most enhanced for those mRNAs encoding both transmembrane domains and signal peptides (Figure 3E). The extended fractional enrichments in the cumulative density plots suggest that multiple processes contribute to individual subcellular mRNA distribution patterns, a conclusion that is supported by the divergence between the observed cumulative density distributions and the distributions predicted by a positive selection mechanism of mRNA partitioning to the ER.
In summary, analysis of genome-scale subcellular mRNA partitioning data via the three-cohort model described above (Figure 2) supports the view that mRNA partitioning is under hierarchical regulation and suggests that mRNAs encoding resident proteins of the endomembrane system contain localization information that is dominant to that expressed by topogenic protein-encoded signals. When sorted with respect to topogenic signals, the data also demonstrate that transmembrane domain topogenic signals are dominant to signal peptides in conferring an ER-enriched subcellular distribution pattern.
Given the substantial variations in the subcellular partitioning patterns of mRNA cohorts that share the property of an encoded topogenic domain (secretory pathway cargo and endomembrane system), we postulated that a hierarchical mechanism of mRNA partitioning to the ER would include cohort-specific modes of biochemical association with the ER membrane. This hypothesis was examined in the experiments described below.
mRNAs associate with the ER by distinct biochemical mechanisms
Expanding on the qPCR and cDNA microarray studies described above (Figures 2 and 3), where the endomembrane system-encoding mRNA cohort was distinguished from the secretory pathway cargo-encoding cohort via log2 (membrane/cytosol) enrichment values, we sought to determine whether endomembrane system-encoding and secretory pathway cargo-encoding mRNAs displayed similar or distinct modes of interaction with the ER membrane. To this end, mRNA–ER membrane interactions were examined by biochemical fractionation of equilibrium density gradient–purified rough microsomes (RM) (Stephens et al., 2005, 2008; Stephens and Nicchitta, 2008).
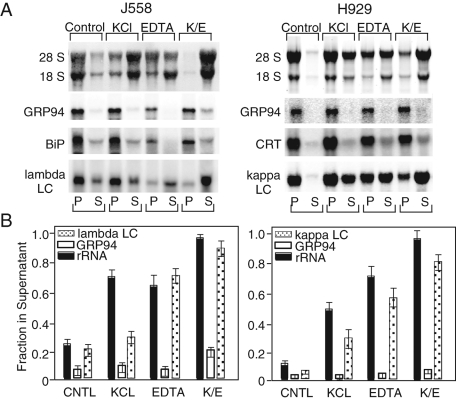
Past studies have established that exposure of RM to high salt concentrations can elicit 80S ribosome release from the ER membrane, whereas divalent cation chelators (e.g., EDTA) elicit ribosomal subunit dissociation and efficient 40S and partial 60S ribosomal subunit release from the ER membrane (Sabatini et al., 1966; Adelman et al., 1973). To assay for ribosome-dependent mRNA–ER interactions, RM were prepared from J558 plasmacytoma and H929 myeloma cells; adjusted to 0.5M KCl, 20 mM EDTA, or 0.5M KCl/20 mM EDTA; and subjected to ultracentrifugation to separate the membrane-associated and released fractions. The two fractions were assayed via analysis of 28S/18S rRNA for ribosome content and via Northern blot for mRNA. As shown in Figure 4, A and B, when RM suspensions were diluted into physiological salt solutions and subjected to ultracentrifugation, ribosomes and mRNAs encoding the resident ER chaperones GRP94 and BiP (J558 cells), or GRP94 and calreticulin (H929 cells), were efficiently recovered in the membrane (P) fraction. Also recovered in the membrane fraction were mRNAs encoding the secretory pathway cargo proteins λ light chain (J558 cells) and κ light chain (H929 cells). Following dilution of the RM fractions with 0.5 M KCl solutions, a partial release of ribosomes was observed for both J558 and H929 RM, with modest effects on mRNA distributions. As noted in past studies, ribosomes released under such conditions include vacant 80S monosomes (Stephens et al., 2005). Under experimental conditions, where ribosomes were dissociated into their component subunits (addition of EDTA), the membrane association behavior of the two mRNA species could be clearly distinguished. In the presence of 20 mM EDTA, mRNAs encoding the secretory pathway cargo proteins λ and κ light chain were recovered in the supernatant (S) fraction, whereas mRNAs encoding the resident ER proteins GRP94, BiP, and calreticulin were recovered in the membrane fraction (Figure 4, A and B). These differences were further distinguished by extraction of RM in high salt/EDTA-supplemented buffers. Under these conditions, mRNAs encoding ER-resident proteins remained tightly associated with the ER membrane, whereas secretory pathway-encoding proteins and ribosomes were efficiently released into the supernatant fraction (Figure 4, A and B). These data identify two biochemically distinguishable modes of mRNA association with the ER. In one mode, displayed by mRNAs encoding the secretory pathway cargo proteins λ and κ light chain, mRNAs displayed membrane-binding properties similar to those established for ribosomes and were released upon addition of high salt/EDTA-containing buffers. In contrast, mRNAs encoding resident ER proteins remained predominantly membrane-associated under experimental conditions that elicited ribosome dissociation and release. These data suggest a model where mRNAs can be localized to the ER through distinct, though not necessarily exclusive, modes via their functional association with membrane-bound ribosomes and/or via salt and divalent cation-insensitive, ribosome-independent interactions with components of the ER membrane.
FIGURE 4:
mRNAs display cohort-specific modes of interaction with the ER membrane. RM were purified from J558 murine plasmacytoma and H929 human myeloma cells by equilibrium density gradient centrifugation and mRNA–ER membrane interactions were determined by biochemical fractionation. RM suspensions were diluted in physiological salts buffer (Control), 0.5 M KCl (KCl), 20 mM EDTA (EDTA), or 0.5 M KCl/20 mM EDTA (K/E) and incubated on ice. Membrane-bound (P) and released (S) fractions were separated by ultracentrifugation and total RNA was isolated. (A) rRNA (28S, 18S) distributions were determined by dye staining, and mRNAs encoding the ER-resident proteins GRP94 (J558, H929), BiP (J558), calreticulin (CRT)(H929), λ light chain (J558), and κ light (chain (H929) were determined by Northern blot analysis. (B) Digital images for rRNA and mRNA distributions are depicted. rRNA and mRNA distributions were quantified by ImageJ analysis of SYBR Safe stained RNA gels or phosphorimager scans of Northern blots. Data represent mean ± SD of seven (J558) or three (H929) individual experiments.
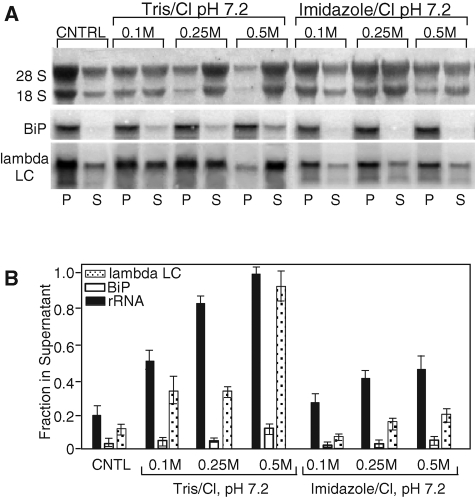
As a further test of this model, an alternative biochemical method for disrupting peripheral protein–membrane interactions was examined. Previous studies have demonstrated that high concentrations of protonated amines (e.g., neutral Tris solution) efficiently extract the peripheral membrane protein fraction of coated vesicles (Keen et al., 1979). This experimental approach was adapted to the analysis of ribosome–mRNA interactions in experiments where RM were treated with increasing concentrations of protonated amines, and ER membrane–mRNA interactions were examined by the centrifugation assay detailed above (Figure 4). The results of these studies are depicted in Figure 5, A and B. When J558 RM were treated with increasing concentrations of Tris/Cl (pKa = 8.3), a progressive release of ribosomes was observed (Figure 5, A and B). Ribosome release was discernibly less efficient in the presence of increasing concentrations of imidazole/Cl (pKa = 5.9), an observation consistent with the requirement for the protonated form of the base in the disruption of peripheral membrane interactions (Keen et al., 1979). At Tris/Cl or imidazole/Cl concentrations up to 0.25 M, BiP- and λ light chain–encoding mRNAs were retained on the ER. However, in the presence of 0.5 M Tris/Cl, BiP-encoding mRNAs were retained on the ER, whereas λ light chain mRNAs were efficiently released.
FIGURE 5:
mRNA–ER binding interactions of mRNAs are distinguished by protonated amine extraction. RM were purified from J558 murine plasmacytoma cells by equilibrium density gradient centrifugation and mRNA–ER membrane interactions were examined by extraction with the protonated amine buffers (neutral Tris or neutral imidazole). J558 RM suspensions were diluted in physiological salts buffer (Control), neutral Tris/HCl (0.1, 0.25, 0.5 M), or neutral imidazole/HCl (0.1, 0.25, 0.5 M), and incubated on ice, and the membrane-associated and -released fractions were isolated by ultracentrifugation. Samples were processed as described in the Figure 4 caption. (A) Digital images of rRNA gels and phosphorimager data (BiP, λ light chain). (B) Data from three independent experiments are summarized, with mean ± SD values indicated.
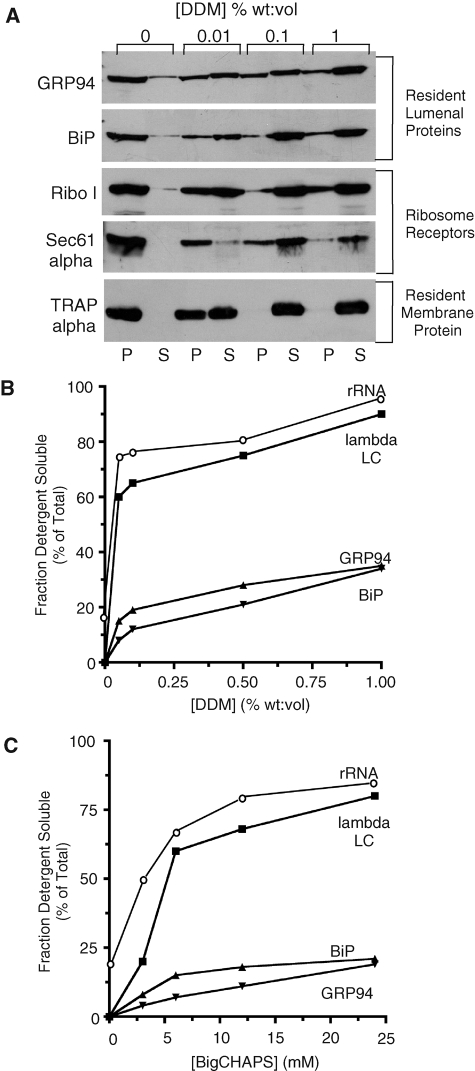
As an additional independent means of distinguishing mRNA–ER binding interactions, detergent extraction studies were performed to examine the membrane interaction profiles of the ER-associated mRNA populations. In these experiments, we postulated the detergent solubilization profile for a given cohort of mRNAs associated with the ER membrane primarily via a functional association with ER-bound ribosomes would mirror the profile of ribosomes and ribosome receptor proteins (e.g., Sec61α). Alternatively, if mRNAs engage in multiple interactions with components of the ER membrane, the detergent solubilization profiles would reflect the composite solubilization behavior of ribosome receptor proteins and (putative) ER-membrane mRNA/RNP-binding proteins. As a test of this hypothesis, RM were prepared from J558 cells and the detergent solubilization profiles of both resident ER proteins (GRP94, BiP) and λ light chain–encoding mRNAs were determined. Two detergents were compared, BigCHAPS and dodecylmaltoside (DDM). BigCHAPS and DDM differ in structure, mechanism of membrane solubilization, and relative efficacy in solubilizing biological membranes (Lin and Guidotti, 2009). In these experiments, J558 RM suspensions were adjusted to 0.5M KCl, detergent stocks were added to the indicated concentrations, and the detergent-soluble and detergent-resistant fractions were separated by ultracentrifugation. The data in Figure 6A depict the DDM solubilization behavior of resident ER lumenal proteins (GRP94, BiP), ribosome-interacting membrane proteins (ribophorin I, Sec61α), and a resident ER membrane protein (TRAPα). Whereas ER-resident lumenal proteins were predominantly released at 0.01% DDM, efficient solubilization of resident ER membrane proteins required DDM concentrations of 0.05–0.1%, with modest increases in solubilization efficiencies as DDM concentrations were increased to 1% (Figure 6A). The solubilization behavior of bound ribosomes was similar to that of the ribosome-interacting proteins ribophorin I and Sec61α; in the presence of increasing DDM concentrations membrane-associated ribosomes were recovered in the supernatant fraction, in parallel with ribophorin I and Sec61α (Figure 6B).
FIGURE 6:
mRNAs encoding endomembrane-resident proteins are bound to the ER via ribosome-independent interactions. The ribosome dependence of mRNA–ER interactions for mRNAs encoding secretory pathway cargo (λ light chain) and endomembrane-resident proteins (GRP94, BiP) was determined by detergent solubilization profiling. (A) J558 plasmacytoma RM were diluted into buffers containing 0.3 M KCl and the indicated concentrations of the detergent DDM. The detergent-soluble and -resistant fractions were separated by ultracentrifugation, and the protein composition of the two fractions was determined by immunoblot analysis of the ER-resident lumenal proteins GRP94 and BiP, the ribosome-interacting proteins ribophorin I and Sec61α, and the ER-resident membrane protein TRAPα. The detergent solubilization profiles for ribosomes (rRNA), mRNAs encoding the secretory pathway cargo protein λ light chain (λ LC), and the resident endomembrane (ER) proteins BiP and GRP94 are depicted in (B and C), for the detergents DDM and BigCHAPS, respectively. rRNA levels were determined by SYBR Green Supermix staining of denaturing RNA gels; mRNA levels were determined by Northern blot and phosphorimager analysis.
Analyses of detergent-mediated mRNA solubilization revealed substantially different solubilization profiles for mRNAs encoding the secretory pathway cargo protein λ light chain and the ER lumenal chaperones GRP94 and BiP (Figure 6, B and C). In experiments conducted with DDM, detergent concentrations of ≥ 0.05% yielded efficient solubilization of membrane-bound ribosomes and λ light chain–encoding mRNAs, with ≥ 80% of both the membrane-bound ribosomes and λ light chain–encoding mRNAs being recovered in the detergent-soluble fraction at DDM concentrations of 1% (Figure 6B). In contrast, mRNAs encoding BiP and GRP94 were relatively insensitive to DDM-mediated solubilization and were predominantly recovered in the detergent-resistant fraction. At the highest DDM concentration examined (1%), ∼65% of this mRNA cohort remained in the detergent-resistant fraction. Such differences in the relative efficacy of detergent-mediated mRNA solubilization were also evident in experiments conducted with BigCHAPS (Figure 6C). Here the majority (∼80%) of the mRNAs encoding BiP and GRP94 were recovered in the detergent-resistant fraction, although ribosome solubilization efficiencies approached 90% (Figure 6C). mRNAs encoding λ light chain were recovered in the detergent-soluble supernatant fraction, with the release profile mirroring that of the bound ribosomes (Figure 6C). These data demonstrate that mRNAs encoding resident proteins of the ER, and which are essential for ER function and biogenesis, are bound via mechanisms largely independent of bound ribosomes. In the case of mRNAs encoding secretory pathway cargo, the primary mechanism of ER association is via ribosome association. These findings raise the possibility that mRNAs whose translation products are essential for secretory pathway function are localized to the ER by mechanisms distinct from that operating in translation-dependent mRNA localization (i.e., the SRP pathway). This hypothesis was further examined in subgenomic studies of ER–mRNA interaction.
Sub-genomic analysis of mRNA–ER association patterns
To give insight into the composition of the mRNA pools comprising the biochemically distinct modes of ER membrane–mRNA interaction identified above, qPCR array analyses were performed on the membrane-associated and -released fractions derived from high salt/EDTA-extracted RM (Figure 4). In these experiments, multiple, independent RM preparations were generated from J558 cells, extracted in 0.5M KCl/20 mM EDTA, and centrifuged to separate the membrane-associated and released mRNA pools, and the mRNA composition of the membrane-associated and -released mRNA pools was assessed by qPCR array. Two commercially available 96-gene qPCR array sets used in combination provided a broad representation of prominent gene products functioning in the cytosol/nucleoplasm, serving as secretory pathway cargo, or resident in the endomembrane system.
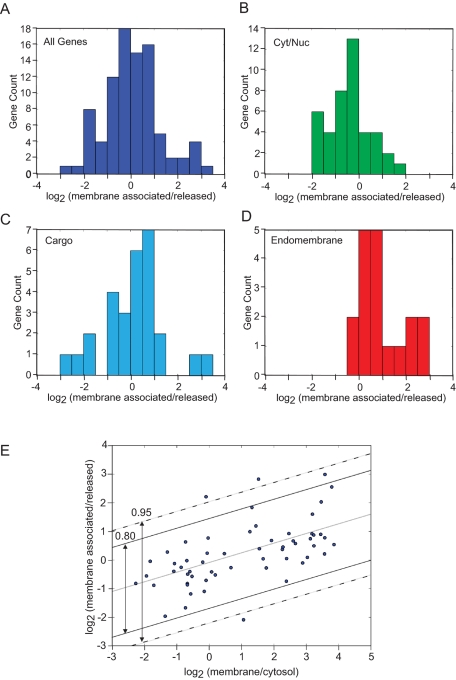
The results of these studies are summarized in Figure 7, A–E. For all mRNA species (i.e., those copurifying with the RM fraction), a broad range (∼6 log2 orders) of membrane-associated/released values was observed, ranging from −2.9 (secretory cargo proteins thrombopoietin and β2-microglobulin) to 2.9 (endomembrane-resident proteins Tap1 and Lamp1; Figure 7A and Table S6). These data mirror the results depicted in Figure 4 regarding the secretory cargo proteins λ and κ light chain and the ER-resident proteins GRP94 and BiP. Notably, however, the majority of the gene products displayed membrane-associated versus -released ratios centered near 0, suggesting that individual mRNAs undergo heterogeneous modes of interaction with components of the ER membrane. For ER-associated gene products encoding cytosolic/nucleoplasmic proteins (n = 42), the distribution was centered at log2 −0.5, with a range of −2.00 to 1.84 (Figure 7B). Within this cohort, distinct gene product subsets could be identified. For example, mRNAs encoding cytosolic proteins with a nuclear function (e.g., transcription factors Taf4a, Rfxa5, Fos) were enriched in the membrane-associated fraction, whereas gene products encoding proteins functioning in the cytosol (e.g., caspase-8, c-srk tyrosine kinase, Birc5, inhibitor of apoptosis) were enriched in the released fraction (Table S4). Intriguingly, within this cohort, the relative distributions of mRNAs between the membrane-associated and -released fractions mirrored their subcellular log2 (membrane/cytosol) distribution patterns (Figure 2 and Table S2). For example, mRNAs encoding Lap3, Tcp1, and Psme1 displayed cytosol-enriched subcellular distribution patterns and were distributed predominantly in the released fraction (Table S4). Conversely, mRNAs encoding the cytosolic proteins CaMK2α and protein kinse Cδ displayed membrane-enriched subcellular distribution patterns and were enriched in the ER-associated fraction of the RM extraction experiments. Thus the capacity to undergo noncanonical localization to the ER correlated with a multimodal mechanism of ER association.
FIGURE 7:
mRNAs display cohort-restricted modes of interaction with the ER membrane. Three independent J558 RM preparations were isolated and extracted with 0.5 M KCl/20 mM EDTA, as described in the Figure 4 caption. Total RNA was isolated from all fractions by Trizol extraction, and the mRNA composition was analyzed by qPCR array analysis, using Lonza's StellARray qPCR Array. Two Lonza gene arrays were examined: 1) murine antigen-processing and presentation, and 2) growth and development. Together these provide a diverse representation of genes encoding prominent cytosolic, nucleoplasmic, secretory pathway cargo, and resident endomembrane proteins. qPCR data were analyzed as described in the Figure 1 caption. (A) All genes. (B) Genes encoding cytosolic and nucleoplasmic proteins. (C) Genes encoding secretory pathway proteins. (D) Genes encoding resident proteins of the endomembrane system. In (E), the gene product distribution data for genes common to the experiments depicted in Figures 1 and 2 and (A–D) were examined for correlation. Statistical analysis was performed by Spearman's correlation. The 80% and 95% confidence intervals for the data correlation analysis are depicted by the solid and dashed lines, respectively.
The data describing the membrane association behavior of mRNAs encoding resident proteins of the endomembrane system are consistent with the conclusion that a high log2 (membrane/cytosol) subcellular distribution coefficient was predictive of a salt/EDTA-insensitive mode of mRNA association with the ER. Of the 19 arrayed genes comprising this cohort, which include the genes encoding the resident ER proteins GRP94, BiP, PDI, and calreticulin, and the resident lysosomal proteins Lamp1 and cathepsin D, all were substantially enriched in the ER membrane fraction in the subcellular partitioning experiments reported in Figures 2 and 3 and in the membrane-associated fraction of the RM extraction data depicted in Figure 7D. Again, similar to what was observed in the subcellular mRNA partitioning studies, where population comparisons of the cytosol- and ER-resident protein-encoding cohorts yielded a clear bimodal mRNA distribution pattern, population comparisons of the cytosol- and ER-resident protein-encoding cohorts from the RM extraction experiments exhibited a bimodal mRNA distribution pattern, and this bimodal pattern was compromised when the analysis was expanded to include the secretory pathway cargo protein-encoding mRNA cohort (Figure 7C). The secretory pathway cargo protein-encoding mRNA cohort displayed a wide distribution range in the RM fractionation experiments, ranging from highly enriched in the membrane fraction (ca. log2 3; insulin-like growth factor binding protein) to strongly released (ca. log2 −3, Thpo). Thus the subcellular mRNA partitioning patterns of the secretory pathway cargo-encoding mRNA cohort mirrored the subcellular distribution patterns obtained in the cellular partitioning experiments. To examine the statistical relationship between subcellular mRNA distribution and mode of membrane association, the correlation between the two data sets was evaluated. Analysis by Spearman's r demonstrates a statistically significant relationship between log2 (membrane/cytosol) and log2 (membrane-associated/released), ρ = 0.64 overall, with ρ = 0.8 for those mRNAs in the 95th percentile confidence interval, p < 0.001 (Figure 7E). This analysis provides statistical support for the conclusion that mRNAs encoding resident proteins of the endomembrane system undergo directed, likely autonomous, localization to the ER.
Given the likelihood that the binding interactions between individual mRNA/ribonucleoprotein complexes and components of the ER reflect avidity-based association phenomena, we tested the hypothesis that KCl/EDTA-resistant binding is correlated with mRNA length. For mRNAs enriched in the membrane pellet fraction (KCl/EDTA resistant), the average message size was 2639 bases, with a range of 997–4738 bases. For mRNAs enriched in the supernatant fraction, the average message size was 2387 bases, with a range of 786–5735 bases. Analysis of the two data sets by Pearson's r yielded an r value of 0.18, indicating that the mode of mRNA binding to the ER was not correlated with message size. We have also investigated the hypothesis that the biochemical mechanism of mRNA–ER association reflects functional links to mRNA degradation processes, with KCl/EDTA-resistant binding conferring enhanced mRNA stability. The correlation between mRNA stability and KCl/EDTA-resistant binding to the ER membrane was examined using the mRNA stability database of Sharova et al. (2009). Analysis of the RM extraction data by Pearson r indicated the biochemical mode of mRNA association with the ER membrane was not statistically correlated with mRNA stability (r = 0.28).
In summary, the included data demonstrate that steady-state subcellular mRNA distributions are governed by a hierarchical system of ER-directed mRNA localization processes. A primary function of this mRNA localization system appears to be the segregation of mRNAs encoding proteins essential for the biogenesis of the endomembrane system into mRNA localization pathways distinct from those utilized by mRNAs whose translation products likely function in cell nonautonomous processes. The mRNA cohort-specific patterns of mRNA localization and ER membrane association were readily discernible at the subgenome and genome scales and thus likely reflect a ubiquitous characteristic of eukaryotic cells.
DISCUSSION
The hierarchical, cohort-specific patterns of cellular mRNA partitioning reported here identify a previously unappreciated role for the ER in global protein synthesis, demonstrate that mRNAs utilize both canonical (topogenic signal-dependent) and noncanonical (topogenic signal-independent) ER localization pathways, and reveal that mRNAs encoding the resident proteins of the endomembrane system directly associate with components of the ER membrane. These findings provide a conceptual framework to decode the complex patterns of subcellular mRNA partitioning characteristic of eukaryotic cells and illuminate long-standing controversies regarding the role of direct mRNA–ER interactions in the compartmentalization of protein synthesis to the ER.
The role of topogenic signals in directing mRNA localization to the ER
The SRP pathway provides a well-established, signal peptide- and translation-dependent mechanism functioning in mRNA localization to the ER (Blobel, 1980, 2000; Walter and Blobel, 1981a, 1981b; Walter et al., 1981; Walter and Johnson, 1994). Though it is generally accepted that the SRP pathway directs mRNA localization to the ER, past studies of mRNA partitioning between the cytosol and ER have indicated that multiple pathways likely function in mRNA localization to the ER (Mechler and Rabbitts, 1981; Mueckler and Pitot, 1981; Kopczynski et al., 1998; Diehn et al., 2000, 2006; Lerner et al., 2003). We have examined subcellular mRNA distribution patterns, and consistent with past studies, we have also observed broadly overlapping distributions of mRNAs between the cytosol and ER compartments. Through analysis of the subcellular partitioning of defined mRNA cohorts, the studies described here now demonstrate that topogenic signals comprise one of likely many mRNA localization elements functioning in mRNA localization to the ER. Importantly, in such a model, topogenic signals are not uniquely predictive of an ER-enriched mRNA distribution. Topogenic signals are, however, essential for protein translocation across the ER membrane, and we propose that topogenic signal function in mRNA localization to the ER is distinct from that operating in protein translocation. This hypothesis is supported by findings in yeast and mammalian cells demonstrating that the SRP pathway is not essential for viability. However, the loss of SRP pathway function can negatively impact the efficiency of protein translocation, and clearly presents a significant, though surmountable, physiological challenge to cells (Hann et al., 1989; Hann and Walter, 1991; Mutka and Walter, 2001; Ren et al., 2004; Lakkaraju et al., 2008). The observations reported here and in a past report (Pyhtila et al. 2008) provide some insight into why this might be. For example, those mRNAs for which ER localization can proceed in the absence of SRP necessarily would be insensitive to the loss of SRP function. Also, and as discussed further below, the observations that ER-bound ribosomes can initiate mRNA translation, and that secretory proteins whose synthesis is initiated on ER-bound ribosomes are translocated in the absence of SRP receptor function, suggest additional mechanisms whereby loss of SRP function can be tolerated (Potter and Nicchitta, 2000; Potter et al., 2001).
A role for the ER in global protein synthesis
We observed that the cytosolic/nucleoplasmic protein-encoding mRNA pool was well represented on the ER, a finding that suggests a broad role for the ER in global protein synthesis. The mechanism or mechanisms enabling the translation of cytosolic/nucleoplasmic protein-encoding mRNAs on the ER remain to be determined. It is possible that this phenomenon reflects a stochastic distribution of mRNAs between free and membrane-bound ribosomes. For example, if newly exported mRNAs are accessible to translation by membrane-bound and cytosolic ribosomes alike, mRNAs that lack specific compartmental enrichment information would be predicted to partition between both ribosome pools, with steady-state mRNA subcellular distribution profiles determined by both ribosome distribution and the elongation-coupled ribosome release pathway previously proposed (Potter and Nicchitta, 2000; Potter et al., 2001). Given the physical and functional continuity between the outer nuclear envelope and the ER, such a mechanism may favor an ER locale for the translation of newly exported mRNAs. In models where the initiation of translation is restricted to the cytoplasm, ER-bound ribosomes would necessarily need to be refractory to initiation. However, where this has been experimentally examined, it was observed that ER-bound ribosomes were capable of de novo initiation of mRNAs encoding cytosolic and secretory proteins alike (Potter and Nicchitta, 2000; Potter et al., 2001; Nicchitta, 2002; Nicchitta et al., 2005). Regardless of the mechanisms governing the subcellular site of initiation, the fact that mRNAs encoding cytosolic/nucleoplasmic proteins are abundantly represented on ER-bound ribosomes indicates multiple pathways function to establish subcellular mRNA distributions.
In both subgenomic and genomic analyses, mRNAs encoding a subset of soluble protein kinases and transcription factors displayed noncanonical enrichments on the ER. The biological function or functions enabled through such noncanonical localization events remain to be determined. In all likelihood, such noncanonical localization patterns have direct biological functions, as proposed by Krause and colleagues (Lecuyer et al., 2007, 2009). In one prominent mechanism, displayed by the stress-response transcription factors Hac1 (yeast) and XBP1 (higher eukaryotes), mRNA-intrinsic localization information (Hac1) and/or nascent chain-encoded translational regulatory elements (XBP1) direct noncanonical localization to the ER as an essential regulatory step in the mRNA-processing reactions that yield the activation of transcription factor function (Aragón et al., 2009; Yanagitani et al., 2011).
Specific enrichment of endomembrane protein-encoding mRNAs on the ER
Examinations of the composition of the highly ER-enriched mRNA pool revealed an unexpected finding: this mRNA cohort is predominantly comprised of mRNAs encoding resident proteins of the endomembrane system. In contrast, mRNAs encoding secretory/integral membrane protein secretory pathway cargo, although they share N-terminal signal peptides and/or transmembrane domain topogenic signals with the endomembrane-resident proteins, were broadly distributed between the cytosol and ER pools. These mRNA distribution patterns were evident in multiple cell lines fractionated by different techniques (homogenization vs. sequential detergent extraction), with mRNA distributions determined by distinct approaches (cDNA microarray vs. qPCR array). These data provide key experimental support for the proposal that mRNA localization to the ER is subject to hierarchical regulation. If, as current data suggest, endomembrane-resident protein-encoding mRNAs undergo direct, autonomous localization to the ER, this process would provide the ER with the information necessary for the biogenesis of the organelles of the endomembrane system. Such a phenomenon may be of particular importance during cell division, when the endomembrane system is distributed between daughter cells (Warren and Wickner, 1996; Lowe and Barr, 2007). mRNA localization to the ER might thus be viewed as contributing a critical functionality to the self-organization processes that enable the biogenesis of the endomembrane system (Misteli, 2001; Snapp, 2004). It may thus be informative to view mRNA localization to the ER in the context of cell-autonomous (i.e., biogenesis of cell architecture) and cell-nonautonomous (i.e., endocrine/exocrine signaling) processes. In this scenario, a primary role of the SRP pathway may be to provide reserve ER-directed mRNA localization function, as might be needed when cell activation and/or differentiation is associated with the transcriptional activation of topogenic signal-encoding genes. The B-cell differentiation program serves as a relevant example, where mitogen or antigen activation yields a dramatic induction of immunoglobulin (Ig) gene expression and a concomitant demand for secretory pathway capacity.
The subcellular mRNA distribution patterns of secretory pathway cargo and endomembrane-resident protein-encoding mRNA cohorts were mirrored in the mechanism(s) of mRNA association with the ER membrane. In general terms, mRNAs encoding secretory pathway cargo displayed ribosome-dependent interactions with the ER membrane, and mRNAs encoding resident proteins of the endomembrane system displayed ribosome-independent ER membrane-binding interactions. These findings are consistent with past studies, where it was reported that mRNAs encoding ER-resident lumenal chaperones bind to the ER primarily via ribosome-independent interactions and can efficiently localize to the ER in the absence of an encoded signal peptide or when mRNA-intrinsic translation is blocked in cis (Pyhtila et al., 2008). Our data demonstrating the endomembrane-resident protein-encoding mRNA fraction was enriched in detergent-resistant membrane domains, whereas mRNAs encoding secretory pathway cargo displayed detergent solubilization properties mirroring known ribosome-interacting proteins, agrees with these past data and suggests the existence of ER-resident mRNA and/or RNA-binding protein receptors that function to segregate this mRNA cohort to the ER.
Reconciling past observations on mRNA–ER interactions
The question of whether mRNAs participate in direct binding interactions with components of the ER membrane has existed for decades (Rosbash and Penman, 1971a, 1971b; Milcarek and Penman, 1974). In early studies, mRNA–ER membrane interactions were examined in tissue culture cell-derived microsomal fractions, and it was demonstrated that addition of EDTA yielded the partial release of membrane-associated mRNAs. The remaining mRNA fraction was released following treatment with detergent admixtures, leading these authors to suggest that “detergent-sensitive proteins” may function in mRNA binding to the ER (Milcarek and Penman, 1974). The view that mRNAs can participate in direct interactions with the ER membrane received further experimental support in studies demonstrating that mRNA–ER interactions in tissue culture cell-derived microsomes were stable to both high salt/EDTA or high salt/puromycin addition (Lande et al., 1975; Adesnik et al., 1976). In subsequent studies, conducted in rat liver-derived RM, it was concluded that mRNAs do not participate in direct interactions with the ER (Kruppa and Sabatini, 1977; Freidlin and Patterson, 1980); these results have been challenged. Nonetheless, and in an effort to reconcile their conflicting data, Kruppa and Sabatini suggested that different classes of mRNAs may display ribosome-dependent versus ribosome-independent binding interactions with the ER membrane, with the relative contributions of either mode of membrane interaction varying in a cell type–dependent manner (Kruppa and Sabatini, 1977). In the discovery that mRNAs can display cohort-specific patterns of partitioning between the cytosol and the ER, and that such patterns are reflected in distinct biochemical mechanisms of mRNA–ER interaction, the data reported here provide a biological rationale to resolve this long-standing controversy.
MATERIALS AND METHODS
Cell culture and fractionation
J558 murine plasmacytoma and H929 human myeloma cells were obtained from ATCC (http://www.atcc.org) and cultured in accordance with ATCC recommendations. Cell fractionation was performed using an established sequential detergent extraction protocol (Stephens et al., 2005, 2008; Stephens and Nicchitta, 2007). Where indicated, RM were prepared from J558 cells, as previously described. In brief, cells were collected, resuspended in hypotonic media, and homogenized; the homogenate was supplemented with a broad spectrum RNase inhibitor (aurin tricarboxylic acid), a postnuclear fraction was obtained by centrifugation, and the RM were subsequently purified by equilibrium density-gradient centrifugation, using a membrane flotation protocol (Stephens et al., 2005, 2008).
Protein and RNA analysis
Immunoblots of protein distribution were performed as described previously (Lerner et al., 2003; Stephens et al., 2005, 2008). Total RNA was obtained from cell fractions by Trizol extraction, the concentration was determined by UV spectrometry, and mRNA composition was analyzed by either Northern blot analysis or by qPCR array (Lerner et al., 2003; Stephens et al., 2005, 2008). For qPCR array experiments, three biological replicates for each of the samples were generated and a control spike-in RNA (Alien qRT-PCR inhibitor alert; Agilent, Santa Clara, CA) was added to all samples per the manufacturer's instructions. Samples were treated with Turbo DNAse (Ambion, Austin, TX) and quality-assessed by Agilent Bioanalyzer. RNA integrity number (RIN) scores were typically in the range of 9.0–9.8. Samples prepared from salt/EDTA extraction of J558 RM necessarily had skewed rRNA levels, and RIN scoring was thus not informative. For these samples, RNA quality was assessed by agarose gel electrophoresis and Northern blot.
cDNA synthesis, RT-PCR, and qPCR arrays
cDNA synthesis was conducted using M-MLV reverse transcriptase and random hexamer priming. Real-time PCR (RT-PCR) was performed on a 7900HT Sequence Detection System (Applied Biosystems, Bedford, MA) using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). StellARray qPCR Array (Lonza, Allendale, NJ) was performed according to the manufacturer's protocols. Threshold cycle was manually set to identical values for all qPCR arrays. Data were analyzed using the Global Pattern Recognition (GPR) program (Bar Harbor Technology, Trenton, ME). Primer sequences used for individual qPCR reactions were as follows:
MMU_GRP94 (NM011631)
F: TGGGTCAAGCAGAAAGGAG
R: TCTCTGTTGCTTCCCGACTT
MMU_Hspa5 (NM_022310)
F: ACCCACCAAGAAGTCTCAGATCTT
R: CGTTCACCTTCATAGACCTTGATTG
18s rRNA
F: CACGGGAAACCTCACCCGGC
R: CGGGTGGCTGAACGCCACTT
λ light chain
F: CAGGCTGTTGTGACTCAGGAA
R: CTTGTCTCCAATCAGGGAGC
κ light chain
F: CGCTGGCCTTCACACAGAA
R: CGCCATGACAACAGACACA
Genomic analysis of RNA partitioning
Microarray data from Diehn et al. (2006) were downloaded from the Stanford Microarray Database (http://smd.stanford.edu) as net intensity values. The data set was intensity filtered with a cutoff of 2.5-fold over background. Gene categories were generated using modified GO. Secretory pathway cargo was generated by combining “extracellular region” GO (GO:0005576) and “integral to plasma membrane” GO (GO:0005587), filtered to include only proteins containing transmembrane domains as predicted by TMHMM. The resident endomembrane category was defined as the “endoplasmic reticulum” GO (GO:0005783), filtered to include either a TMHMM-predicted transmembrane domain or an ER retention sequence; the “lysosome” (GO:0005764); or the “Golgi apparatus” (GO:0005794). The cytoplasm category uses the “cytoplasm” GO (GO:0005737), filtered to exclude our custom ER cargo and endomembrane categories, along with TMHMM-predicted transmembrane proteins. Miscategorized entries were eliminated by manual curation using GeneCards (www.genecards.org), Swiss-Prot (www.expasy.org/sprot/), and Mouse Genome Informatics (www.informatics.jax.org) as references. Plots were generated with the Python graphing utility Matplotlib (http://matplotlib.sourceforge.net).
Supplementary Material
Acknowledgments
The authors are grateful for the critical and insightful comments of Shelton Bradrick and the members of the Duke Center for RNA Biology. C.N. is supported by grants GM077382 and GM077382-OS31 from the National Institutes of Health.
Abbreviations used:
- DDM
dodecylmaltoside
- ER
endoplasmic reticulum
- GO
gene ontology
- GPR
Global Pattern Recognition
- H region
hydrophobic region
- Ig
immunoglobulin
- Psap
prosaposin
- RIN
RNA integrity number
- RM
rough microsomes
- RT-PCR
real-time PCR
- SRP
signal-recognition particle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0239) on May 25, 2011.
REFERENCES
- Adelman MR, Sabatini DD, Blobel G. Ribosome-membrane interaction: non-destructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973;56:206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesnik M, Lande M, Martin T, Sabatini DD. Retention of mRNA on the endoplasmic reticulum membranes after in vivo disassembly of polysomes by an inhibitor of initiation. J Cell Biol. 1976;71:307–313. doi: 10.1083/jcb.71.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Protein targeting (Nobel Lecture) ChemBioChem. 2000;1:86–102. doi: 10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975a;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975b;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Walter P, Chang CN, Goldman BM, Erickson AH, Lingappa VR. Translocation of proteins across membranes: the signal hypothesis and beyond. Symp Soc Exp Biol. 1979;33:9–36. [PubMed] [Google Scholar]
- de Jong M, van Breukelen B, Wittink FR, Menke FL, Weisbeek PJ, Van Den Ackerveken G. Membrane-associated transcripts in Arabidopsis; their isolation and characterization by DNA microarray analysis and bioinformatics. Plant J. 2006;46:708–721. doi: 10.1111/j.1365-313X.2006.02724.x. [DOI] [PubMed] [Google Scholar]
- Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- Freidlin PJ, Patterson RJ. Heparin releases monosomes and polysomes from rough endoplasmic reticulum. Biochem Biophys Res Commun. 1980;93:521–527. doi: 10.1016/0006-291x(80)91108-0. [DOI] [PubMed] [Google Scholar]
- Hann BC, Poritz MA, Walter P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kD subunit of the signal recognition particle that in Scerevisiae is essential for growth. J Cell Biol. 1989;109:3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Keen JH, Willingham MC, Pastan IH. Clathrin-coated vesicles: isolation, dissociation and factor-dependent reassociation of clathrin baskets. Cell. 1979;16:303–312. doi: 10.1016/0092-8674(79)90007-2. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Hegde RS. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol Biol Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Rahbar R, Hegde RS. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem. 2001;276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- Kishimoto Y, Hiraiwa M, O'Orien JS. Saposins: structure, function, distribution, and molecular genetics. J Lipid Res. 1992;33:1255–1267. [PubMed] [Google Scholar]
- Kopczynski CC, Noordermeer JN, Serano TL, Chen WY, Pendleton JD, Lewis S, Goodman CS, Rubin GM. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc Natl Acad Sci USA. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kruppa J, Sabatini DD. Release of poly A(+) messenger RNA from rat liver rough microsomes upon disassembly of bound polysomes. J Cell Biol. 1977;74:414–427. doi: 10.1083/jcb.74.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AKK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande MA, Adesnik M, Sumida M, Tashiro Y, Sabatini DD. Direct association of messenger RNA with microsomal membranes in human diploid fibroblasts. J Cell Biol. 1975;65:513–528. doi: 10.1083/jcb.65.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Krause HM. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol. 2009;21:409–415. doi: 10.1016/j.ceb.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Guidotti G. Purification of membrane proteins. Methods Enzymol. 2009;463:619–629. doi: 10.1016/S0076-6879(09)63035-4. [DOI] [PubMed] [Google Scholar]
- Lingappa VR, Blobel G. Early events in the biosynthesis of secretory and membrane proteins: the signal hypothesis. Recent Prog Horm Res. 1980;36:451–475. doi: 10.1016/b978-0-12-571136-4.50018-8. [DOI] [PubMed] [Google Scholar]
- Lowe M, Barr FA. Inheritance and biogenesis of organelles in the secretory pathway. Nat Rev Mol Cell Biol. 2007;8:429–439. doi: 10.1038/nrm2179. [DOI] [PubMed] [Google Scholar]
- Mechler B, Rabbitts TH. Membrane-bound ribosomes of myeloma cells. IV. mRNA complexity of free and membrane-bound polysomes. J Cell Biol. 1981;88:29–36. doi: 10.1083/jcb.88.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek C, Penman S. Membrane-bound polyribosomes in HeLa cells: association of polyadenylic acid with membranes. J Mol Biol. 1974;89:327–338. doi: 10.1016/0022-2836(74)90522-1. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler MM, Pitot HC. Structure and function of rat liver polysome populations. I. Complexity, frequency distribution, and degree of uniqueness of free and membrane-bound polysomal polyadenylate-containing RNA populations. J Cell Biol. 1981;90:495–506. doi: 10.1083/jcb.90.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol Biol Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV. A platform for compartmentalized protein synthesis: protein translation and translocation in the endoplasmic reticulum. Curr Opin Cell Biol. 2002;14:412–416. doi: 10.1016/s0955-0674(02)00353-8. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem Cell Biol. 2005;83:687–695. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- O'Orien JS, Kishimoto Y. Saposin proteins: structure, function, and role in human lysosomal storage disorders. FASEB J. 1991;5:301–308. doi: 10.1096/fasebj.5.3.2001789. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J Biol Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Endoplasmic reticulum ribosomes reside in stable association with the translocon following termination of protein synthesis. J Biol Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- Potter MD, Seiser RM, Nicchitta CV. Ribosome exchange revisited: a mechanism for translation-coupled ribosome detachment from the ER membrane. Trends Cell Biol. 2001;11:112–115. doi: 10.1016/s0962-8924(00)01905-x. [DOI] [PubMed] [Google Scholar]
- Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell. 2004;15:5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M, Penman S. Membrane-associated protein synthesis of mammalian cells. I. The two classes of membrane-associated ribosomes. J Mol Biol. 1971a;59:227–241. doi: 10.1016/0022-2836(71)90048-9. [DOI] [PubMed] [Google Scholar]
- Rosbash M, Penman S. Membrane-associated protein synthesis of mammalian cells. II. Isopycnic separation of membrane-bound polyribosomes. J Mol Biol. 1971b;59:243–253. doi: 10.1016/0022-2836(71)90049-0. [DOI] [PubMed] [Google Scholar]
- Sabatini DD, Tashiro Y, Palade GE. On the attachment of ribosomes to microsomal membranes. J Mol Biol. 1966;19:503–524. doi: 10.1016/s0022-2836(66)80019-0. [DOI] [PubMed] [Google Scholar]
- Shaffer KL, Sharma A, Snapp EL, Hegde RS. Regulation of protein compartmentalization expands the diversity of protein function. Dev Cell. 2005;9:545–554. doi: 10.1016/j.devcel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik E, Ko MSH. Database for mRNA half-life of 19,977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–48. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. Endoplasmic reticulum biogenesis: proliferation and differentiation. In: Mulins C., editor. The Biogenesis of Cellular Organelles. New York: Kluwer Academic/Plenum; 2004. pp. 1–34. [Google Scholar]
- Stephens SB, Dodd RD, Brewer JW, Lager PJ, Keene JD, Nicchitta CV. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol Biol Cell. 2005;16:5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SB, Dodd RD, Lerner RS, Pyhtila BM, Nicchitta CV. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol Biol. 2008;419:197–214. doi: 10.1007/978-1-59745-033-1_14. [DOI] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum. Methods Enzymol. 2007;431:47–60. doi: 10.1016/S0076-6879(07)31004-5. [DOI] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell. 2008;19:623–632. doi: 10.1091/mbc.E07-07-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981a;91:551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981b;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory proteins. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.