Abstract
The relationships between autophagy and cell death are complex and still not well understood. To advance our understanding of the molecular connections between autophagy and apoptosis, we performed an RNAi-based screen of Drosophila melanogaster apoptosis-related genes for their ability to enhance or suppress starvation-induced autophagy. We discovered that six apoptosis-related genes, Dcp-1, hid, Bruce, buffy, debcl and p53 as well as Ras/ Raf/MAPK signaling pathway components play a role in autophagy regulation in Drosophila cultured cells. Our study also provides the first in vivo evidence that the effector caspase Dcp-1 and IAP protein Bruce regulate both autophagy and starvation-induced cell death at two nutrient status checkpoints, germarium and mid-oogenesis, in the Drosophila ovary. Analysis of degenerating mid-stage egg chambers in DmAtg1 and DmAtg7 mutants reveal a reduction in TUNEL staining though DNA condensation appears unaffected. Based on these and previous findings, we propose here a putative molecular pathway that might regulate the sensitivity threshold of apoptotic and autophagic responses. We also discuss multiple interpretations of the Atg mutant egg chamber TUNEL phenotype that are consistent with a possible role for autophagy in either suppressing or enhancing the efficiency of cell degradation and/or promoting cell clearance associated with the death process.
Keywords: autophagy, apoptosis, caspase, Dcp-1, Bruce
Macroautophagy (autophagy hereafter) is a lysosome-mediated catabolic process involved in the degradation and recycling of intra-cellular components. The association of autophagy with cell death has attracted considerable attention and raised many unanswered questions. To contribute to a better understanding in this area, our approach was to conduct a systematic RNAi-based screen of Drosophila apoptosis-related genes to identify potential apoptosis-related modifiers of starvation-induced autophagy. Starvation or nutrient deprivation is a well characterized inducer of autophagy in Drosophila1 and many other organisms.2,3 We developed an efficient flow cytometer-based LysoTracker Green (LTG) assay as a primary screen and coupled that to a secondary GFP-LC3 redistribution assay, both in Drosophila l(2)mbn cells, to provide readouts representing late and early stages of autophagy, respectively. As an initial validation of our screening strategy, we designed dsRNAs corresponding to autophagy genes and known autophagy regulators. Our findings show that dsRNAs corresponding to eleven Drosophila Atg homologues are able to reduce LTG levels in starved l(2)mbn cells. Knockdown of known positive and negative autophagy regulators using RNAi also produces expected alterations in LTG and GFP-LC3. We next screened twenty apoptosis-related and Ras pathway-related genes and found nine that modify LTG and GFP-LC3 levels significantly. Knockdown of Dcp-1, hid, debcl, buffy and p53 suppresses LTG and GFP-LC3 levels in starved cells, identifying these genes as positive regulators of autophagy. RNAi-mediated knockdown of Bruce, Ras, Raf and MAPK enhances LTG and GFP-LC3 levels in starved cells, identifying these genes as negative regulators of autophagy.
How might the identified gene products act to regulate or modulate the autophagic response in nutrient-deprived cells? Our data show that the proapoptotic gene, hid, but not rpr, grim, or skl, acts to regulate starvation-induced autophagy in Drosophila l(2)mbn cells. Consistent with our findings, overexpression of hid induces autophagy in various Drosophila tissues including fat body, midgut and salivary glands.4 Survival Ras/Raf/MAPK signaling specifically inhibits the proapoptotic activity of Hid,5 and our observations indicate that the Ras/Raf/MAPK pathway also plays an inhibitory role in starvation-induced autophagy. The Hid protein contains five MAPK phosphorylation consensus sites;5 thus it is possible that survival signals regulate the crosstalk between autophagy and apoptosis through different threshold levels of MAPK-mediated phosphorylation on Hid. In addition, Hid promotes polyubiquitination of DIAP1 and antagonizes its anti-apoptotic activity through proteasomal-dependent degradation.6 Surprisingly, we find that another IAP protein, Bruce, but not DIAP1 acts as a suppressor of autophagy, suggesting that Bruce, instead of DIAP1, might be the downstream target of Hid and act to antagonize Hid-mediated autophagy. Bruce and its mammalian homologue, Apollon, share sequence conservation in the BIR (baculoviral-IAP-repeat) and UBC (ubiquitin-conjugating enzyme) domains.7 Apollon ubiquitinates and promotes degradation of SMAC, the mammalian homologue of Hid.7 Perhaps Drosophila Bruce has a similar molecular function as Apollon and promotes degradation of Hid through ubiquitination, thereby acting to negatively regulate autophagy. However, Bruce is a large protein (530 kDa) and it is plausible that it could regulate autophagy through protein interactions with one of its other protein regions. Another candidate Bruce-interacting protein that we identified in our screen is the effector caspase Dcp-1. IAP family members can bind directly to caspases, and inhibit their activity.8 Thus, it is possible that Bruce suppresses Dcp-1 activity or promotes Dcp-1 degradation through its BIR and/or UBC domains. Such an interaction would be consistent with our identification of Dcp-1 as a positive regulator of autophagy. Based on our recent observations and these previous findings, we propose a hypothetical pathway for the regulation of starvation-induced autophagy in Drosophila (Fig. 1). Clearly, epistasis analyses and protein interaction studies are required to prove or disprove this model, and determine how it integrates with other components (e.g., Tor) already known to control the autophagic response to starvation.
Figure 1.
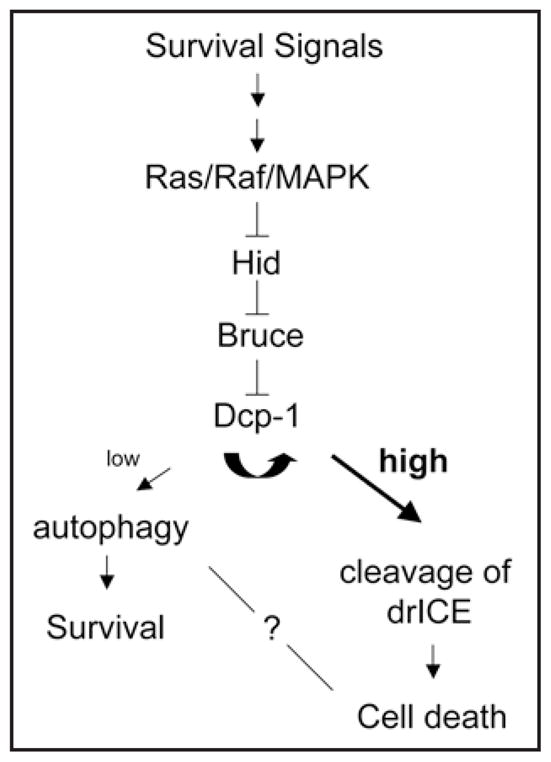
A hypothetical pathway for the regulation of sensitivity thresholds leading to autophagy or apoptosis. Based on the known apoptosis-related interactions of the gene products identified in our study, we propose a putative pathway involved in the regulation of starvation-induced autophagy in Drosophila. In this model, the effector caspase Dcp-1 plays a key role in defining the balance between autophagic and apoptotic responses.
To validate the autophagy modulating effects of some of the identified cell death-related genes in vivo, we used Drosophila melanogaster oogenesis as a model system. Nutrient deprivation triggers germline cell death at two specific stages during oogenesis, the germarium and mid-stage oogenesis.9 Using a GFP-LC3 transgenic Drosophila line10 as well as LysoTracker Red staining, we find that autophagy also occurs in response to nutrient deprivation at these two stages in oogenesis. An earlier study in Drosophila virilis similarly reports the presence of autophagic structures, as observed by TEM, in mid-stage (as well as late-stage) oogenesis.11 Dcp-1 is required for mid-stage egg chamber cell death.12 We further demonstrate that Dcp-1 is required for cell death in germaria, and is also necessary for starvation-induced autophagy in both germaria and mid-stage egg chambers. Further, overexpression of Dcp-1 was sufficient to induce autophagy at these two stages even under well-fed conditions (Fig. 2). Loss-of-function mutations in Bruce resulted in ecotopic autophagy and cell death in both stages, regardless of nutrient status, indicating that Bruce acts normally to suppress both autophagy and cell death during Drosophila oogenesis. Thus, our observations using RNAi targeting Dcp-1 and Bruce in the l(2)mbn cell line were confirmed in vivo during Drosophila melanogaster oogenesis.
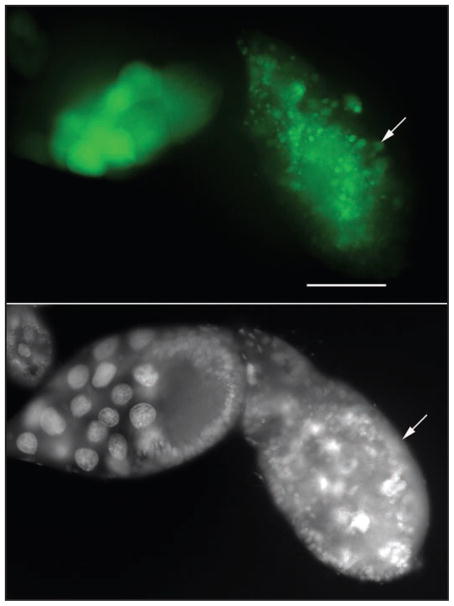
Figure 2.
The effector caspase Dcp-1 is sufficient for the induction of autophagy during Drosophila oogenesis. Expression of activated Dcp-1 in the germline (UASp-GFP-LC3/+; nanos-GAL4/nanos-GAL4 UASp-tDcp-1)38 resulted in nurse cell death during mid-oogenesis (arrow) and dying nurse cells had numerous GFP-LC3 puncta (green). DAPI staining of nuclei is shown in white. Scale Bar: 50 μm.
If an effector caspase is required for autophagy and apoptosis, what determines the balance between these two processes and what is the final cellular outcome? In the Drosophila ovary, the two cellular stress responses occurred together and it is possible that autophagy is part of the apoptotic response itself, an idea put forward already by Thorburn.13 Several cell death regulators have functions that are involved in the adaptation to stress.14 For example, EGL-1, a BH3-only protein, is required for metabolic stress,15 and AIF plays a role in redox stress.16 Hence, an alternative idea is that some proteins involved in stress responses, such as autophagy, also evolved roles as cell death effectors. A previous biochemical study shows that the effector caspase Dcp-1 is able to auto-cleave/auto-activate and also cleave another effector caspase, drICE.17 In contrast, drICE does not act to cleave itself.17 It is possible that the level of Dcp-1 activity could determine the sensitivity thresholds of autophagic and apoptotic responses. Starvation signals might initially induce a low level of Dcp-1 activity which promotes autophagy for cell survival, giving the cells a chance to recover and allow continued development. Prolonged starvation signals might result in a higher level of Dcp-1 activity which in turn activates drICE and triggers apoptosis. Future studies to elucidate upstream regulators and downstream substrates of Dcp-1 in cells undergoing autophagy and apoptosis will help to further establish the regulatory mechanisms governing the crosstalk between these two cellular processes.
The role of autophagy in cell death is still not well understood and appears to be context-dependent. During developmental cell death, such as embryogenesis and insect metamorphosis, it is proposed that autophagy acts to assist dead cell clearance when insufficient phagocytes are available for corpse removal.18,19 Three recent studies demonstrate that autophagy is involved in developmental cell death processes.20–22 In a mouse embryoid body cavitation model20 and in a mouse neuroepithelium model,22 autophagy is essential for the clearance of dying cells by generating engulfment signals, including lysophosphatidylcholine secretion (come-get-me signal) and phosphatidylserine exposure (eat-me signal). During Drosophila metamorphosis, autophagy genes are demonstrated to be required for complete salivary gland cell degradation.21 In the Drosophila ovary, nutrient deprivation signals trigger germarium (region 2A) and mid-oogenesis cell death to remove defective egg chambers before the investment of energy into them,9,23 and our results showed that nutrient deprivation also triggers autophagy at these two stages.
To investigate the role of autophagy during germarium and mid-oogenesis cell death, we analyzed the phenotype of DmAtg1 germline clones and DmAtg7 mutant ovaries. Although chromatin condensation appears normal, TUNEL staining, an indicator of DNA fragmentation, appears reduced in germarium cells and degenerating mid-stage egg chambers in DmAtg1 and DmAtg7 mutants. How might autophagy be involved in DNA degradation during Drosophila germarium and mid-stage nurse cell death? DNA degradation is mediated by multiple nucleases,24 including cell-autonomous and waste-management nucleases. Most cell autonomous nucleases generate TUNEL-reactive DNA fragments.25 Autophagy might positively regulate the activity of cell-autonomous nucleases and thus be involved directly in the regulation of DNA fragmentation. Alternatively, autophagy could modulate the activity of the lysosomal nuclease, DNAseII, which generates 5'-hydroxyl and 3'-phosphate ends that are unrecognizable substrates for TdT of the TUNEL assay. In this scenario, autophagy might act normally to delay or suppress DNAse II-mediated DNA degradation,25 and, thus, autophagy inhibition would result in accelerated DNAseII activity and a concomitant decrease in TUNEL-positive DNA. Electron microscopy analyses show that nurse cell debris is engulfed by surrounding follicle cells in dying mid-stage egg chambers.26 Therefore, the TUNEL-negative nurse cell nuclei in the DmAtg1 and DmAtg7 mutants could also be accumulated cell corpse DNA, which cannot be recognized by engulfing cells because of failure to display engulfment signals. However, in mouse embryoid body and retinal neuroepithelium models, dying cells in Atg gene mutants fail to display engulfment signals but still show TUNEL staining,20,22 a result that differs from our observations in the Drosophila ovary. The possible role(s) of autophagy in DNA degradation, as illustrated in Figure 3, remains to be further investigated. It is interesting to note that cell-autonomous DNA degradation is not essential for cell death but appears to affect the efficiency or extent of death at least in some systems. In contrast, removal of dead cell debris can be required for sustained organism survival.24 Thus, we propose that at least one of the functions of autophagy in the Drosophila ovary is to enhance or suppress the efficiency of cell degradation and/or promote corpse clearance associated with cell death.
Figure 3.
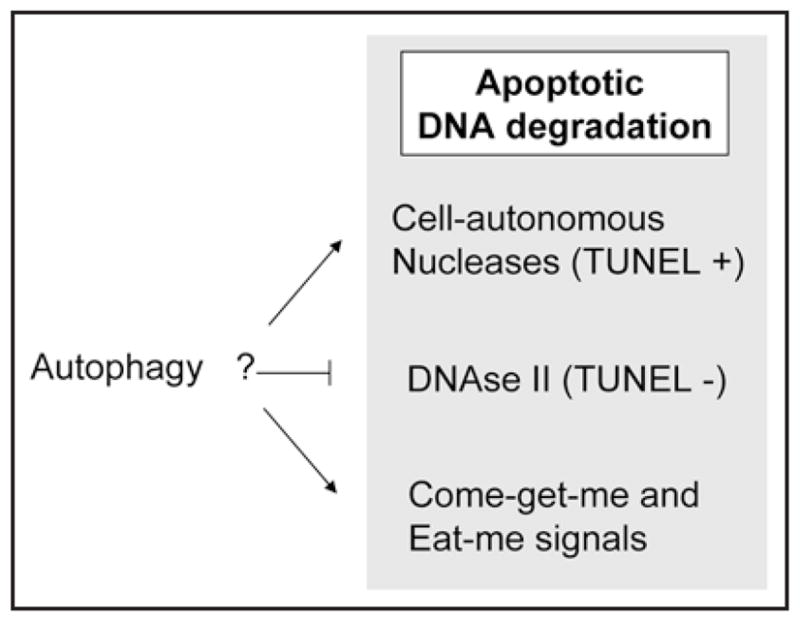
Possible relationships between autophagy and DNA degradation in Drosophila oogenesis. The diagram depicts how autophagy might play a role in the DNA degradation process based on the reduced TUNEL staining phenotype observed in dying mid-stage egg chambers of DmAtg1 and DmAtg7 mutants. Autophagy could positively regulate the activities or subcellular localization of cell-autonomous nucleases (that generate TUNEL-reactive fragments) and thereby enhance DNA degradation. Alternatively, autophagy may negatively regulate the activity of lysosomal nuclease DNAseII (that generates TUNEL non-reactive DNA ends), and thereby suppress or delay DNAseII-mediated DNA degradation. Finally, autophagy could sustain the high ATP levels that are required for display of engulfment signals, including lysophosphatidylcholine (come-get-me) and/or phosphati-dylserine (eat-me).
Mammalian homologues of Drosophila Bruce, Hid and Dcp-1 are Apollon, Smac and Caspase-3, respectively. It remains to be tested whether the autophagy-regulating functions of Bruce, Hid and Dcp-1 are conserved in these mammalian counterparts. Interestingly, overexpression of Apollon can suppress apoptosis, and recent evidence suggests that this occurs indirectly via p53.27 As noted above, Apollon can also ubiquitinate the pro-apoptotic protein Smac, as well as Caspase-9.7 Smac/DIABLO is released from the mitochondria to antagonize IAPs, namely XIAP, cIAP-1 and -2, survivin and Apollon.8 In this way, Smac promotes the activation of Caspase-3 and is proapoptotic. Perhaps it has a similar proautophagy mode of action. Low levels of Smac28 and Caspase-3,29 are associated with chemotherapy resistance, and based on our model in Figure 1, low level activation would be consistent with induction of autophagy. While genetic studies showed that autophagy may act as a tumor suppressor mechanism,30,31 autophagy can also play a protective role during chemotherapy and radiation treatment.32–36 Since Smac mimetics and suppression of IAP proteins are under active investigation as anti-cancer treatments,37 it may be worthwhile to investigate a therapeutic strategy that combines Smac mimetics with autophagy inhibition. If Smac, Apollon and Caspase-3 do function in autophagy regulation, it will be important to understand their effects in both normal and cancer cells. And given the complexity of apoptotic signaling pathways, it is likely that additional apoptosis-related genes with a link to autophagy will be discovered.
Acknowledgments
We thank K. McCall for the activated Dcp-1 construct and helpful discussions. We thank L. DeVorkin and S. Chittaranjan for valuable comments on the manuscript. We gratefully acknowledge support from CIHR grant MOP-78882 to S.M.G.
References
- 1.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meléndez A, Tallóczy Z, Seaman M, Eskelinen E-L, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 4.Juhasz G, Sass M. Hid can induce, but is not required for autophagy in polyploid larval Drosophila tissues. Eur J Cell Biol. 2005;84:491–502. doi: 10.1016/j.ejcb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–41. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 6.Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–24. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 7.Hao Y, Sekine K, Kawabata A, Nakamura H, Ishioka T, Ohata H, et al. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat Cell Biol. 2004;6:849–60. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasula SM, Ashwell JD. IAPs: what’s in a name? Mol Cell. 2008;30:123–35. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–78. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- 10.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–92. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Velentzas AD, Nezis IP, Stravopodis DJ, Papassideri IS, Margaritis LH. Mechanisms of programmed cell death during oogenesis in Drosophila virilis. Cell Tissue Res. 2007;327:399–414. doi: 10.1007/s00441-006-0298-x. [DOI] [PubMed] [Google Scholar]
- 12.Laundrie B, Peterson JS, Baum JS, Chang JC, Fileppo D, Thompson SR, et al. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 2003;165:1881–8. doi: 10.1093/genetics/165.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 2008;15:1113–23. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salinas LS, Maldonado E, Navarro RE. Stress-induced germ cell apoptosis by a p53 independent pathway in Caenorhabditis elegans. Cell Death Differ. 2006;13:2129–39. doi: 10.1038/sj.cdd.4401976. [DOI] [PubMed] [Google Scholar]
- 16.Cande C, Vahsen N, Metivier D, Tourriere H, Chebli K, Garrido C, et al. Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J Cell Sci. 2004;117:4461–8. doi: 10.1242/jcs.01356. [DOI] [PubMed] [Google Scholar]
- 17.Song Z, Guan B, Bergman A, Nicholson DW, Thornberry NA, Peterson EP, et al. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid and grim. Mol Cell Biol. 2000;20:2907–14. doi: 10.1128/mcb.20.8.2907-2914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baehrecke EH. How death shapes life during development. Nat Rev Mol Cell Biol. 2002;3:779–87. doi: 10.1038/nrm931. [DOI] [PubMed] [Google Scholar]
- 19.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 21.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellen MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–90. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- 23.Buszczak M, Cooley L. Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ. 2000;7:1071–4. doi: 10.1038/sj.cdd.4400755. [DOI] [PubMed] [Google Scholar]
- 24.Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–88. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 25.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–48. [PMC free article] [PubMed] [Google Scholar]
- 26.Giorgi F, Deri P. Cell death in ovarian chambers of Drosophila melanogaster. J Embryol Exp Morphol. 1976;35:521–33. [PubMed] [Google Scholar]
- 27.Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, et al. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci USA. 2005;102:565–70. doi: 10.1073/pnas.0408744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirro E, Consoli ML, Massimino M, Manzella L, Frasca F, Sciacca L, et al. Altered expression of c-IAP1, survivin and Smac contributes to chemotherapy resistance in thyroid cancer cells. Cancer Res. 2006;66:4263–72. doi: 10.1158/0008-5472.CAN-05-3248. [DOI] [PubMed] [Google Scholar]
- 29.Philchenkov A, Zavelevich M, Kroczak TJ, Los M. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26:82–97. [PubMed] [Google Scholar]
- 30.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–10. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 33.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–58. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 35.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, et al. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem. 2008;283:19665–77. doi: 10.1074/jbc.M710169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Nikolovska-Coleska Z, Yang CY, Qian D, Lu J, Qiu S, et al. Design of small-molecule peptidic and nonpeptidic Smac mimetics. Acc Chem Res. 2008;41:1264–77. doi: 10.1021/ar8000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in drosophila oogenesis. Dev Biol. 2003;260:113–23. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]