Abstract
Inflammation of the gastrointestinal tract increases the risk of developing colon cancer especially in younger adults. Dietary compounds are not only associated with the etiology of inflammation and colon cancer, but also in their prevention. Sphingolipid metabolites have been shown to play a role in the initiation and perpetuation of inflammatory responses. In the present study, we investigated the suppression of dextran sodium sulfate-induced colitis and azoxymethane-induced colon cancer by dietary sphingomyelin in mice that lack functional PPAR-γ n intestinal epithelial and immune cells. Dietary sphingomyelin decreased disease activity and colonic inflammatory lesions in mice of both genotypes but more efficiently in mice expressing PPAR-γ. The increased survival and suppression of tumor formation in the sphingomyelin-fed mice appeared to be independent of PPAR-γ expression in immune and epithelial cells. Using a real-time PCR array, we detected an up-regulation in genes involved in Th1 (IFN-γ) and Th17 (IL-17 and IL-23) responses despite the reduced inflammation scores. However, the genes involved in Th2 (IL-4, IL-13 and IL-13ra2) and Treg (IL-10rb) anti-inflammatory responses were up-regulated in a PPAR-γ dependent manner. In line with the PPAR-γ dependency of our in vivo findings, treatment of RAW macrophages with sphingosine increased the PPAR-γ reporter activity. In conclusion, dietary sphingomyelin modulated inflammatory responses at early stages of disease by activating PPAR-γ, but its anti-carcinogenic effects followed a PPAR-γ-independent pattern.
Keywords: sphingomyelin, inflammation, macrophages, CD4+ T cells, colon cancer
Introduction
Colorectal cancer is a prevalent cancer among the older population (146,970 were diagnosed and 49,920 died of colorectal cancer in 2009) with a median age at diagnosis of 71 years [1]. The most common risk factors include genetic predisposition and environmental factors such as carcinogens and dietary patterns. Intestinal inflammation (ulcerative colitis and Crohn’s disease), however, drastically increase the risk of developing colon cancer especially at early age (< 30 years of age) [2]. The risk of developing colorectal cancer for an ulcerative colitis patient is estimated to be 2% after 10 years, 8% after 20 years, and 18% after 30 years of disease onset [3]. A prevention strategy that targets inflammation could, therefore, be successful to prevent the colon cancer in these high-risk groups.
Dietary components affect the risk of developing colon cancer [4]. In this regard, our studies have shown that dietary sphingolipids suppress early and late stages of carcinogen-induced colon cancer [5–8], and suppress tumor formation in Min mice [9,10]. Orally administered complex sphingolipids are hydrolyzed to ceramide and sphingosine throughout the intestinal tract [11–14] which are the same bioactive metabolites generated in vitro shown to regulate growth, death, differentiation, and motility of cells [15,16]. Bioactive sphingolipid metabolites have also been implicated in the generation or perpetuation of inflammatory responses in endothelial cells [17], adipose tissue [18], lung [19], and immune cells [20]. A recent report demonstrated an increase of ceramide in colonic lipid extracts from dextran sodium sulfate (DSS)-treated mice in vivo [21]. On the other hand, dietary sphingomyelin (SM) did not affect antibody formation, natural killer cell cytotoxicity, or delayed-type hypersensitivity in carcinogen-treated mice [22]. However, a recent study found that orally administered SM lowered DSS-induced inflammation in Jci:ICR mice [23]. This appears to contradict the pro-inflammatory effect of the bioactive sphingolipid metabolites summarized above.
While the exact mechanism for the elevated colon cancer risk by colonic inflammation is still unknown, recent studies have been investigating whether the marked reduction in levels of the nuclear peroxisome proliferator-activated receptor (PPAR)-γ in colons of patients with ulcerative colitis may play a role in their increased susceptibility to developing colorectal cancer [24]. PPARs belong to the superfamily of nuclear hormone receptors with 48 members identified in the human genome. There are three known PPAR isoforms; α, β or δ, and γ which differ in their tissue distribution and functional activity [25]. PPARs are endogenously controlled molecular switches that regulate inflammation, immunity and metabolism [26,27] but their main biological function is the sensing of intracellular nutrient concentrations and regulation of gene expression involved in maintaining both metabolic and immune homeostasis. Since PPAR-γ is ubiquitously expressed in immune cells and the gut, it has already been identified as a target for preventive and therapeutic efforts since its activation attenuates inflammatory responses. PPAR-γ is expressed by CD4+ T cells, and regulates their differentiation into at least four functionally distinct subsets referred to as T helper (Th) 1, Th2, Th17, and induced regulatory T cells (iTreg). Functionally, Th1 and Th17 subsets are linked to increased risk of autoimmune disorders whereas Treg cells can prevent inflammatory and immune mediated diseases [28].
In this regard, we have shown previously that conjugated linoleic acid suppressed inflammation-induced colorectal cancer in a strictly PPAR-γ dependent manner [29]. We have also demonstrated that abscisic acid can suppress experimental IBD in a manner dependent on T cell PPAR-γ [30]. In the present study, we set out to investigate the effect of dietary SM on inflammation-driven colon cancer to clarify the apparent contradiction of the immunological effects of sphingolipid metabolites, and test the dependence of an anti-inflammatory effect on PPAR-γ expression. Our studies show that dietary SM greatly inhibited the tissue damage caused by DSS-induced colitis, enhanced the survival and reduced tumor formation. Interestingly, the anti-inflammatory of SM, but not its anti-carcinogenic effects, were enhanced by PPAR-γ expression in the intestinal epithelial and immune cells.
Methods and Materials
Mice
Since whole body deletion of PPAR-γ is lethal by day E10 [31], tissue-specific PPAR-γ null mice were generated as previously described [32,33]. PPAR-γfl/fl mice carrying (PPAR-γ gene flanked with two loxP boxes on exon 2 that are recognized by a transgenic recombinase) were cross-bred with transgenic mice carrying the Cre-gene under control of the MMTV-LTR promoter which express a transgenic recombinase only in epithelial and hematopoietic cells [34]. PPAR-γ fl/fl; Cre+ (named PPAR-γ−/− throughout the manuscript) express a truncated form of mRNA transcript and do not express PPAR-γ protein in the intestinal epithelial cells, T-cells or macrophages as confirmed by real-time RT-PCR and Western Blotting. These mice are viable without any known problems and have been used in previous studies by our group [34,35]. Both female and male mice (5 each per group) were used in the present studies. All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Virginia Polytechnic Institute and State University and met requirements of the Public Health Service/National Institutes of Health and the Animal Welfare Act.
Diets and Treatments
PPAR-γ+/+ and PPAR-γ−/− mice were randomly assigned into either the control or the SM-supplemented diet (10 per group). The mice in all four groups were fed the semi-purified sphingolipid-free AIN76A diet [36] throughout the study. The sphingolipid groups received 1g/kg SM (0.1% by weight) (Avanti, Alabaster, AL) in the diet. Bovine milk SM powder (99% pure) was mixed thoroughly into small batches of diet using a mixer and stored at 4°C until use. The milk SM contains mostly saturated fatty acids [37] and is stable under these conditions. The amount of SM used does not add significant amounts of caloric value to the diet and has demonstrated to have no side effects in either of our studies or those of other groups; even 1% was tolerated well over 2 generations of rats [38]. Amounts up to 0.1% have been used in our previous studies and have suppressed carcinogen-induced colon cancer by up to 80% [7,8,37]. After 7 days on the experimental diets, the mice were injected with a single dose of azoxymethane (10 mg per kg bodyweight in bicarbonate buffer) to induce colon cancer. Inflammation was induced one week later (day 13) by adding 2.0% DSS (molecular weight 36,000 to 44,000 g/L; ICN Biomedicals, Aurora, OH) to the drinking water for 7 consecutive days. During this time the mice were weighed on a daily basis and examined by blinded observers for clinical signs of disease associated with colitis (i.e., perianal soiling, rectal bleeding, diarrhea, and piloerection). The disease activity index (DAI) consisted of a scoring for diarrhea and lethargy (0–3), whereas rectal bleeding consisted of a visual observation of blood in feces and the perianal area (0–4). On day 8 of the DSS challenge, the mice were switched to regular drinking water for the duration of the colon cancer study. DAI and weight determinations were continued weekly until the end of the study.
Determination of tumor load
After the DAI increased – bleeding indicated the existence of colon tumors- the mice were euthanized by CO2 asphyxiation 80 days after the carcinogen injection. The colons were removed, opened longitudinally, and the tumor area was measured since the colons appeared macroscopically to be mostly a solid sheet of tumor. In case of identifiable single tumors, the tumors were measured individually and their surface areas were combined. Then parts of the colons were either fixed in formalin or OCT for immunohistochemistry and determination of tumor progression by an experienced veterinarian, used for isolation of immune cells or preserved in RNAlater (Qiagen) using the same parts of the colon of each animal for these analyses.
Immune cell isolation from mesenteric lymph nodes (MLN)
MLNs were excised, crushed, and placed into 3 ml of FACS buffer (1xPBS, 5% FBS, and 0.1% Sodium Azide). After the cells were centrifuged, the supernatant was discarded and the cells were washed with 10 ml of FACS buffer and centrifuged at 1200 RPM. The supernatant was discarded and the cells resuspended at 2×106 cells/ ml. MLN cells were then immunophenotyped as described previously [29]. Briefly, MLN cells were seeded into 96-well plates, blocked with FcBlock (BD-Pharmingen), incubated with anti-F4/80-PE-Cy5, anti-CD11b-FITC (eBioscience), and anti-CCR2-PE (R&D systems, Minneapolis, MN) antibodies to assess macrophage infiltration, anti-CD4-FITC, anti-CD25-Pe-Cy5 (BD Pharmingen) and anti-FoxP3 (after permeabilization with Cytofix-Cytoperm from BD Pharmingen) to determine T regulatory cells (Tregs) infiltration. Cells were resuspended in PBS for BD LSR II flow cytometer analysis and data analysis was performed with FACS Diva software.
Histopathology
The excised colons were fixed in 10% buffered neutral formalin, embedded in paraffin, and then sectioned (5 µm) and stained with H&E stain for histological examination by RH and JBR who have more than 15 years of experience in histological/histopathological tissue analysis. Tissue slides were examined in a Nikon 80i eclipse epifluorescence microscope, equipped with DIC, color and monochromatic digital cameras. Images were digitally captured using the NIS software (Nikon) and processed in Adobe Photoshop Elements 2.0 (Adobe Systems Inc., San Jose, CA).
Transfection of RAW 264.7 Macrophages
RAW 264.7 macrophages (from ATCC) were grown in 25mm2 flasks in Dulbecco’s Modification of Eagle’s Medium (DMEM, Mediatech Inc., Herndon, VA) containing 10% FBS. Cells were then treated in replicates of 8 with sphingosine (1 and 2.5 µmol/L; Avanti, Alabaster, AL,), rosiglitazone (1 µmol/L; Cayman Chemicals, Ann Arbor, MI) as a positive control, or vehicle (DMSO) and placed in a 37°C incubator with 5% CO2 for 4 and 24 h. These concentrations are not toxic to the cells and were chosen because we have not detected the induction of apoptosis in any of our studies using dietary complex sphingolipids in mouse models for colon [7], breast or ovarian cancer (manuscripts in preparation). Reporter activity assays for PPAR-γ activity were performed as previously described [29]. Briefly, a pCMX.PPAR-γ expression plasmid and a pTK.PPRE3x luciferase reporter plasmid driven by the PPRE-containing acyl-CoA oxidase promoter (kindly provided by Dr. R.M. Evans, The Salk Institute, San Diego, CA) were transfected into RAW 264.7 cells. After 24h, the cells were seeded into 96 well dishes, treated as indicated for 4 and 24 h and luciferase activity was measured using the Dual Luciferase II reporter assay system (Promega, Madison, WI) using a Modulus 96-well luminometer (Turner Biosystems, Sunnyvale, CA). All values were normalized to control wells to calculate relative luciferase activity.
Cytokine Real-time PCR array
For the initial determination of changes in inflammatory responses that may be associated with the diet and genotype, PPAR-γ+/+ and PPAR-γ−/− mice (n=12 per group) were placed on the AIN76A diet with or without the 0.1% SM throughout the study (6 per group). After one week, 2% DSS was added to the drinking water of 3 animals of each group for 7 days during which they were weighed and scored daily. Then, all mice were euthanized by CO2 asphyxiation followed by secondary thoracotomy, the colons were removed, rinsed and a portion of each colon was fixed in 10% neutral buffered formaldehyde, embedded in paraffin and sectioned for H&E staining for determination of tissue architecture or placed into RNAlater. To extract RNA, the remaining colonic tissue was cut into 15–25mg pieces and rinsed with RNAse free water to remove the RNAlater. Then the tissue was snap-frozen in liquid nitrogen, homogenized, and RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and stored at −80°C until analysis.
RNA purity was assured by assessing the A260:A230 ratio and A260:A280 ratio (NanoDrop, Thermo Fisher) and the ribosomal RNA integrity using the Experion™ Nano LabChip (Biorad). In each group, 2ng of RNA per mouse (n=3 in each group) were pooled for cDNA synthesis using the RT2 First Strand Kit (SABiosciences). Real-time RT-PCR was performed in a RNA microarray format using the Inflammatory Response and Autoimmunity RT2 Profiler PCR Array (PAMM-3803 SABiosciences) according to the manufacturer’s instructions on an ABI 7900HT PCR machine (Applied Biosystems, Foster City, CA). Data analysis was performed using the SABiosciences analyzing tools provided online. Included in the arrays were controls for genomic DNA contamination, reverse transcription and positive PCR controls and 5 housekeeping genes (β-actin, GAPDH, Hsp90ab1, Hprt1, Gusb). Data presented are the fold differences between the 2^ ΔCT values for genes that were at least 2-fold up- or down-regulated in the treatment versus the appropriate control group, or the same treatment in different genotypes. Since the array was performed with pooled samples of 3 mice, no statistical analyses of the data were performed.
Statistics
Data were analyzed as 2 × 2 factorial arrangements of treatments. The statistical model was: Yijk = μ + Genotypei + Dietj + (Genotype × Diet)ij + error Aijk, in which μ was the general mean, Genotypei was the main effect of the ith level of the genotypic effect (expression of PPAR-γ by immune and epithelial cells), Dietj was the main effect of the jth level of the dietary effect (sphingomyelin vs no supplement), (Genotype × Diet)ij was the interaction effect between genotype and diet, and error A representing the random error. To determine the statistical significance of the model, analysis of variance (ANOVA) was performed using the general linear model procedure of Statistical Analysis Software (SAS Institute Inc., Cary, NC). When the model was significant, the analysis was followed by Fisher’s Least Significant Difference multiple comparisons method. Data were expressed as the means ± standard error of the mean. Statistical significance was assessed at a P value of < 0.05.
Results
Effect of dietary sphingomyelin on DSS-induced inflammation
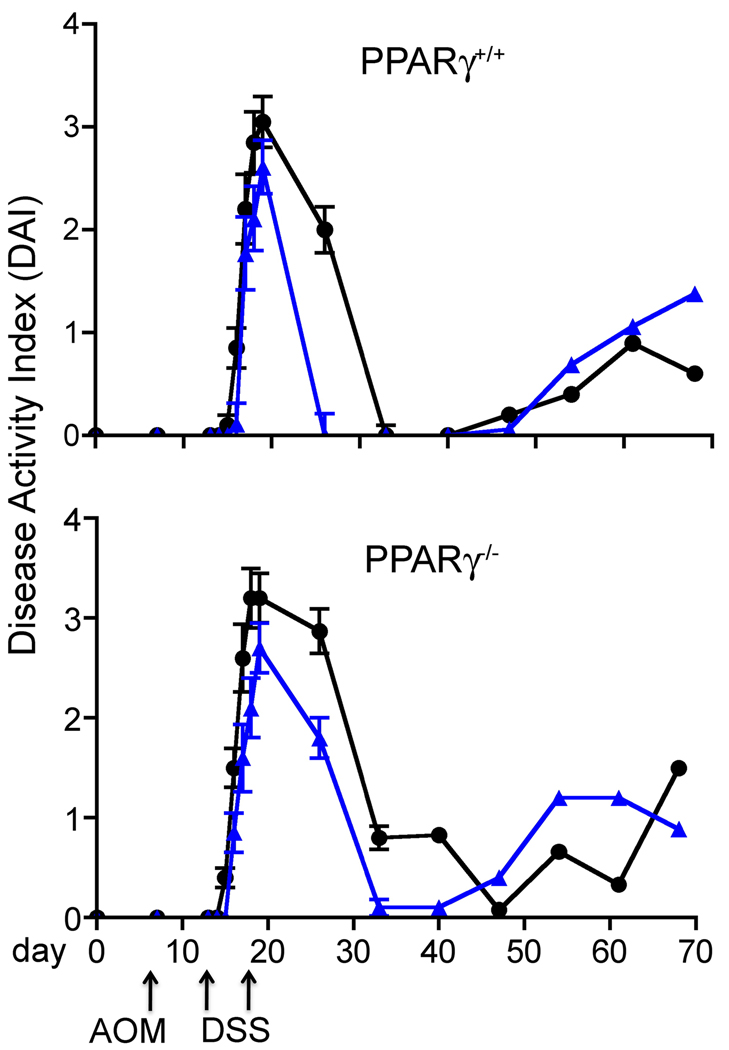
To determine if orally administered SM affect DSS-induced colitis and colitis-driven colon cancer and whether these effects are dependent on the expression of PPAR-γ, mice expressing or lacking PPAR-γ in the intestinal epithelium and immune cells were injected with a single dose of AOM after acclimatization, and treated a week later with DSS for 7 days. The DAI was determined as described under methods and materials. During the acute phase of inflammation, the mice lost weight as was expected but there was no significant difference in the weight of the control mice of either genotype during DSS treatment, recovery or throughout the rest of the study (Table 1). In both the PPAR-γ++ and PPAR-γ control groups fed the AIN76A diet without supplements, the DAI increased on day 15 of the study (after 3 days of DSS treatment) and was consistently higher in the PPAR-γ−/− group throughout the study (significant at p<0.001 on day 4 of the DSS treatment (Table 2). The recovery from the DSS challenge was significantly delayed in the PPAR-γ control group compared to the PPAR-γ+/+ group p<0.0001, p<0.0001, and p<0.007 on day 14, 21, and 28 of study, respectively) (Fig.1 and Table2). This confirms earlier studies showing an increased DAI after DSS treatment in PPAR-γ mice [34] and indicates an anti-inflammatory effect of PPAR-γ.
Table 1.
Effect of dietary supplementation with sphingomyelin (SM; 0.1 g/100 g) in PPAR-γ+/+ or PPAR-γ−/− mice on body weights prior to and following the induction of inflammation-induced colorectal cancer.
| PPAR-γ+/+ | PPAR-γ−/− | ANOVA P value | ||||||
|---|---|---|---|---|---|---|---|---|
| Day | Control | 0.1% SM |
Control | 0.1% SM |
SEM | Diet | Genotype | D × G |
| D0 | 21.66 | 20.41 | 19.41 | 20.83 | 1.203 | NS | NS | NS |
| D7 | 23.00 | 21.40 | 20.82 | 23.45 | 1.079 | NS | NS | NS |
| D13 | 23.67 | 21.50 | 21.18 | 23.90 | 1.159 | NS | NS | 0.045 |
| D14 | 23.19 | 21.96 | 21.91 | 24.00 | 1.180 | NS | NS | NS |
| D15 | 23.78 | 21.85 | 21.01 | 23.98 | 1.142 | NS | NS | 0.042 |
| D16 | 24.08 | 22.00 | 21.23 | 24.07 | 1.109 | NS | NS | 0.035 |
| D17 | 23.62 | 21.65 | 20.88 | 24.00 | 1.077 | NS | NS | 0.026 |
| D18 | 23.19 | 21.17 | 20.33 | 23.58 | 1.049 | NS | NS | 0.018 |
| D19 | 22.29 | 21.01 | 19.86 | 23.25 | 1.048 | NS | NS | 0.035 |
| D26 | 18.82ab | 18.65ab | 16.29b | 21.73a | 0.926 | 0.013 | NS | 0.008 |
| D33 | 22.55ac | 20.95bc | 17.54b | 24.60a | 0.752 | 0.006 | NS | 0.0001 |
| D40 | 24.18a | 22.525ab | 19.45b | 25.18a | 0.793 | 0.04 | NS | 0.001 |
| D47 | 24.36ab | 22.51ab | 21.38b | 25.88a | 0.836 | NS | NS | 0.005 |
| D54 | 24.98ab | 23.00ab | 21.66b | 25.98a | 0.815 | NS | NS | 0.004 |
| D61 | 24.22 | 23.70 | 21.85 | 25.79 | 0.827 | NS | NS | 0.03 |
| D68 | 25.2 | 21.88 | 20.97 | 25.79 | 0.981 | NS | NS | 0.002 |
Least squares means values (n = 10) in a row for a particular body weight (grams) with different superscripts are significantly different (P < 0.05).
On day 7, mice were challenged with azoxymethane (i.p.) and on day 13 mice were administered 2% dextran sodium sulfate in the drinking water for 7 days as described in Materials and Methods.
Data were analyzed as a 2 × 2 factorial arrangement (i.e., 2 genotypes and 2 dietary treatments).
Table 2.
Effect of dietary supplementation with sphingomyelin (SM; 0.1 g/100 g) in PPAR-γ+/+ or PPAR-γ−/− mice on body weights prior to and following the induction of inflammation-induced colorectal cancer.
| PPAR-γ+/+ | PPAR-γ−/− | ANOVA P value | ||||||
|---|---|---|---|---|---|---|---|---|
| Day | Control | 0.1% SM |
Control | 0.1% SM |
SEM | Diet | Genotype | D × G |
| D0 | - | - | - | - | - | - | - | - |
| D7 | - | - | - | - | - | - | - | - |
| D13 | 0.00 | 0.00 | 0.00 | 0.00 | 0.000 | - | - | - |
| D14 | 0.00 | 0.00 | 0.00 | 0.00 | 0.000 | - | - | - |
| D15 | 0.10 | 0.00 | 0.40 | 0.00 | 0.971 | 0.01 | NS | NS |
| D16 | 0.85ab | 0.11b | 1.50a | 0.85ab | 0.196 | 0.001 | 0.001 | NS |
| D17 | 2.20 | 1.77 | 2.60 | 1.60 | 0.338 | 0.045 | NS | NS |
| D18 | 2.85 | 2.11 | 3.20 | 2.10 | 0.297 | 0.004 | NS | NS |
| D19 | 3.05 | 2.61 | 3.20 | 2.70 | 0.249 | NS | NS | NS |
| D26 | 2.00ab | 0.00c | 2.87a | 1.80b | 0.200 | 0.0001 | 0.0001 | 0.039 |
| D33 | 0.00b | 0.00b | 0.80a | 0.10b | 0.081 | 0.001 | 0.0001 | 0.001 |
| D40 | 0.00b | 0.00b | 0.83a | 0.10b | 0.125 | 0.02 | 0.007 | 0.029 |
| D47 | 0.20 | 0.06 | 0.08 | 0.40 | 0.175 | NS | NS | NS |
| D54 | 0.40 | 0.69 | 0.66 | 1.20 | 0.377 | NS | NS | NS |
| D61 | 0.90 | 1.06 | 0.33 | 1.20 | 0.318 | NS | NS | NS |
| D68 | 0.60 | 1.38 | 1.50 | 0.89 | 0.373 | NS | NS | NS |
Least squares means values (n = 10) in a row for a particular DAI with different superscripts are significantly different (P < 0.05).
On day 7, mice were challenged with azoxymethane (i.p.) and on day 13 mice were administered 2% dextran sodium sulfate in the drinking water for 7 days as described in Materials and Methods.
Data were analyzed as a 2 × 2 factorial arrangement (i.e., 2 genotypes and 2 dietary treatments).
Figure 1.
Disease Activity Index (DAI) in PPAR-γ expressing (PPAR-γ+/+) and tissue-specific knockout (PPAR-γ−/−) mice lacking PPAR-γ in intestinal epithelial cells, macrophages and T cells. All mice were injected with a single dose of AOM, treatment with 2.0% DSS for 7 days, and fed either the AIN76A diet alone (circles), or supplemented with 0.1% sphingomyelin (triangles). n=10 per group
SM supplements significantly suppressed the inflammatory response to DSS compared to the controls of both genotypes. While there was no difference in the DIA during acute inflammation between the groups fed SM, the onset of the inflammation was delayed in PPAR-γ+/+ mice and their recovery was significantly accelerated after the DSS was removed from the drinking water (p<0.039, p<0.001, and p<0.029 for diet × genotype on day 26, 33 and 40 (Fig. 1, Table 2); 7 days after the last day of DSS treatment, the DAI had already returned to zero in the SM-fed PPAR-γ+/+ mice while PPAR-γ+/+ mice still showed an average DAI of 1.80. These results suggests both PPAR-γ-dependent and independent pathways of suppression of colonic inflammation and recovery from the DSS colitis by dietary SM. After the recovery from the DSS-induced inflammation, the DAI in all groups increased over the course of the study, indicating the development of colonic tumors which is often associated with intestinal bleeding and blood in the feces. However, the number of mice in the control groups was very small at the end of the study and there were no statistically significant differences in DAI at these time points (Table 2).
Long-term effects of the combined AOM and DSS treatment
The DSS-induced intestinal inflammation is an established model for ulcerative colitis and has been used in many studies to investigate the tumor-promoting effect of intestinal inflammation. Upon removal of DSS from the drinking water mice will recover from the damage; this has been also observed in our previous studies [35]. Surprisingly, in the present study, the control mice showed an overly strong response to the treatment although the DSS concentration was similar or even lower than those used in other reports and several mice did not recover after DSS discontinuation and reached the euthanasia endpoint as defined by the animal welfare guidelines and were sacrificed when found moribund. As shown in Figure 2, only 3 of the PPAR-γ++ and 2 of the PPAR-γ control mice survived until the end of the study. In the SM-fed groups, 7 of the PPAR-γ++ and 8 of the PPAR-γ survived (p=0.075 and p=0.0051 compared to the appropriate controls, respectively). This increased survival in the SM-fed groups suggests that the suppression of the initial response to the DSS treatment may be critical for the survival of the mice. Also, the choice of basal diet may have affected the severity of the inflammatory response. AIN76A is essentially sphingolipid-free [14] while the lipid source has been changed from corn to soy oil in the AIN93 diets. Soy is a rich source of sphingolipids and even small amounts may have exerted protective effects in previous studies. However, this needs to be investigated in more detail.
Figure 2.
The survival of mice after DSS treatment (2% for 7 days) is enhanced in mice fed sphingomyelin independent of their genotype.
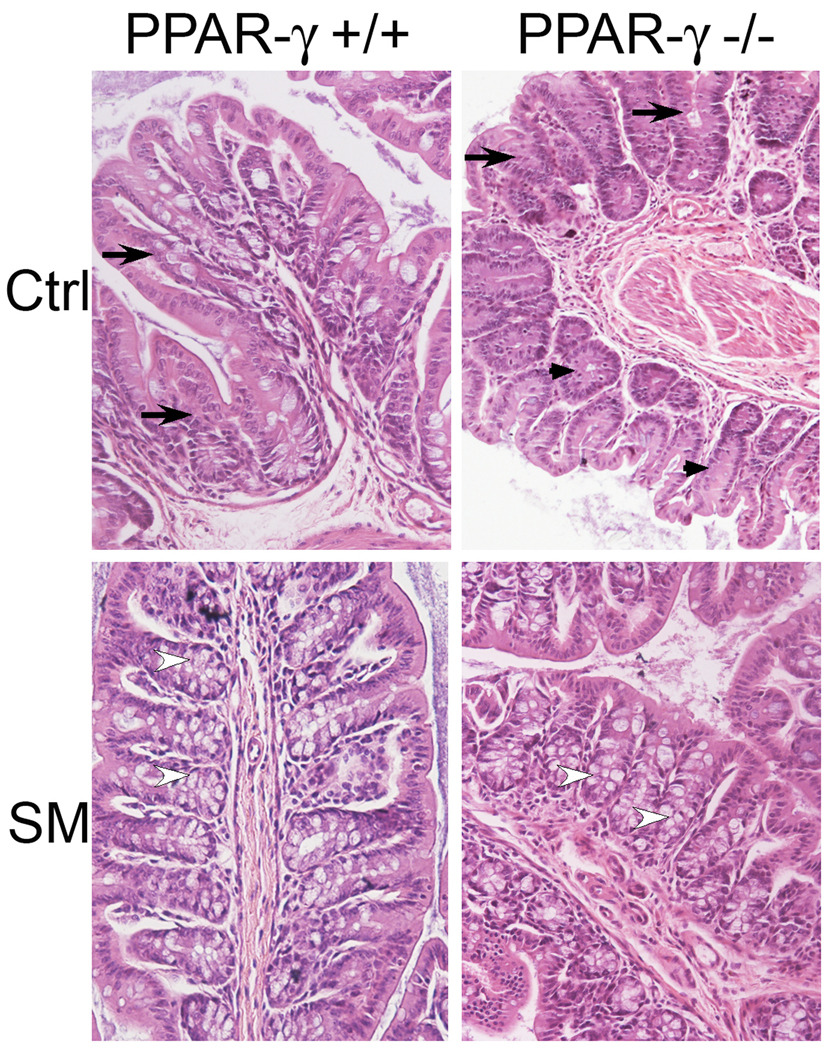
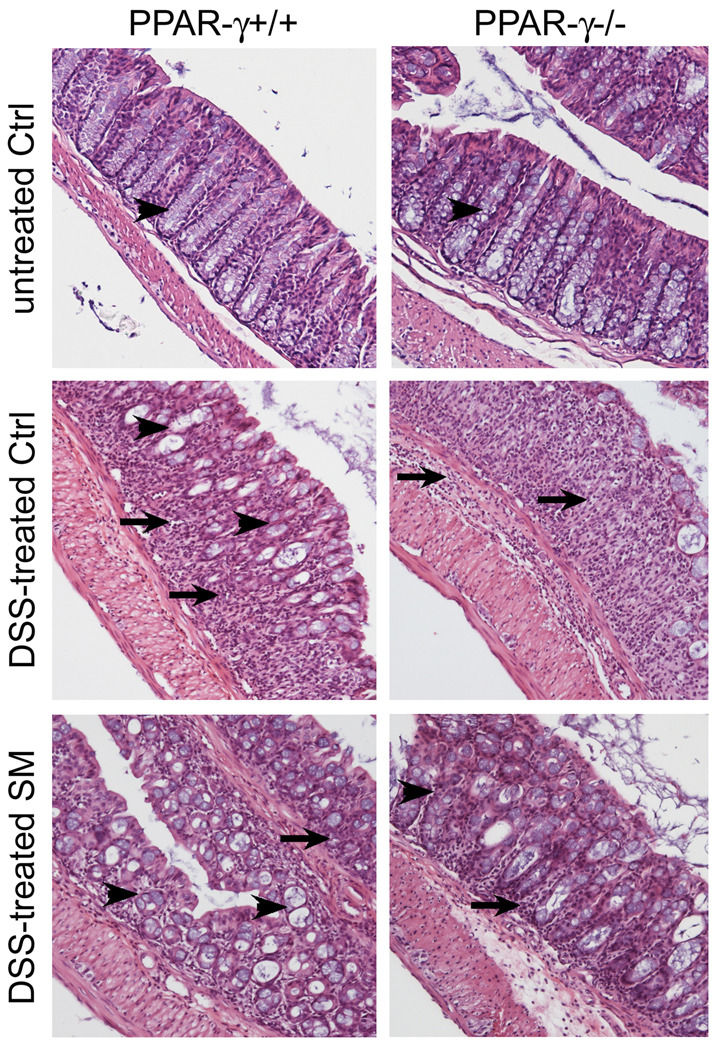
H&E stained sections of the colons of mice at the end of the study showed a distortion of the columnar shape of the colonic crypts generating a loose tissue architecture (arrows), and a lack of mucin-producing goblet cells in most areas of the colons of PPAR-γ mice (arrow heads). In PPAR-γ++ mice, the tissue architecture was better preserved, and more goblet cells were visible (Fig.3, upper panels). However, these tissues were derived from mice that survived the DSS treatment while 7/10 and 8/10 of the PPAR-γ++ and PPAR-γ mice, respectively, succumbed to the DSS-induced injuries; these colons, therefore, may represent mice with less initial damage to their tissue architecture. These deleterious changes were greatly reduced by the dietary SM in the colons of the mice of either genotype (Fig.3, lower panels). Most of the PPAR-γ+/+ mice showed the distinct colonic architecture with abundant goblet cells (open arrow heads); this was also seen in the PPAR-γ−/− mice but there were also larger areas of less tight crypts that lacked goblet cells despite the SM in the diet in this group.
Figure 3.
Changes in the colonic architecture, determined by H&E staining in colons from mice treated with a single dose of AOM, 2% DSS for 7 days and maintained on control AIN76A diet alone (upper panel) or supplemented with 0.1% sphingomyelin (80 days after begin of study; n=2/17 PPAR-γ+/+ ctrl/SM fed, n=3/18 PPAR-γ−/− ctrl/SM)
Tumor formation
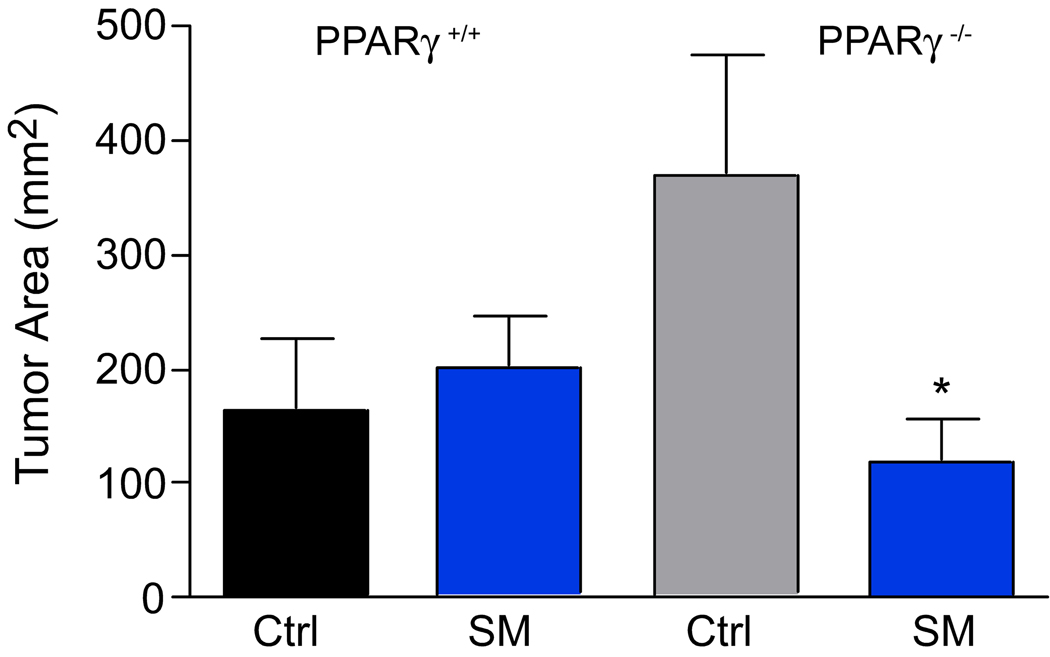
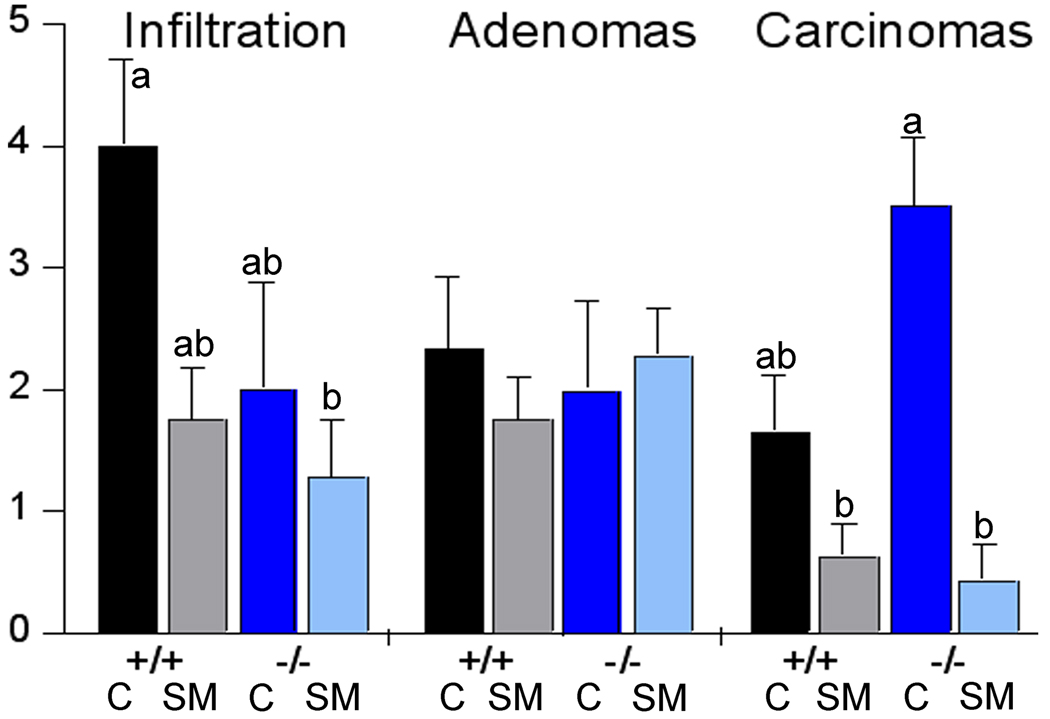
Mice of both genotypes were injected with a single dose of azoxymethane to induce colon tumors. After 80 days, the colons of control mice macroscopically appeared as a solid tumor sheet without discernible single tumors. Therefore, the tumor area rather than tumor number was determined. As shown in Figure 4, the tumor area in the PPAR-γ mice was larger than in the PPAR-γ++. There was no effect of the SM supplement in the PPAR-γ++ mice but the tumor area in the PPAR-γ−− mice was significantly smaller in the SM-fed mice (p=0.0251). This confirms our observations that by dietary SM suppressed tumor formation in other models; however, since the number of mice in this study was small, these results need to be confirmed. Next, we determined the infiltration of immune cells into the colonic tissue and the progression of the colonic tumors microscopically in H&E stained colonic sections. As shown in Figure 5, there was a substantial lymphoplasmacytic infiltrate in the colonic tissue of control-fed mice of either genotype as determined by histological examination of the H&E stained colonic sections; this was reduced by dietary SM. There was no difference in the number of adenomas per section but fewer areas that had progressed to adenocarcinomas in the PPAR-γ++ mice. Dietary SM had no effect on the number of adenomas but SM-fed mice exhibited fewer adenocarcinomas, confirming the results of our earlier studies in carcinogen-induced colon cancer models and suggesting that the suppression of tumor progression by dietary SM is PPAR-γ independent.
Figure 4.
Tumor area PPAR-γ+/+ and PPAR-γ−/− mice 80 days after a single AOM injection.
*p<0.05
Figure 5.
Immune cell infiltration and progression of colon cancer in PPAR-γ+/+ and PPAR-γ−/− mice fed AIN76A diet alone or with sphingomyelin supplements. Means without common letter are different (p<0.05).
SM changes immune cell population in mesenteric lymph nodes (MLN)
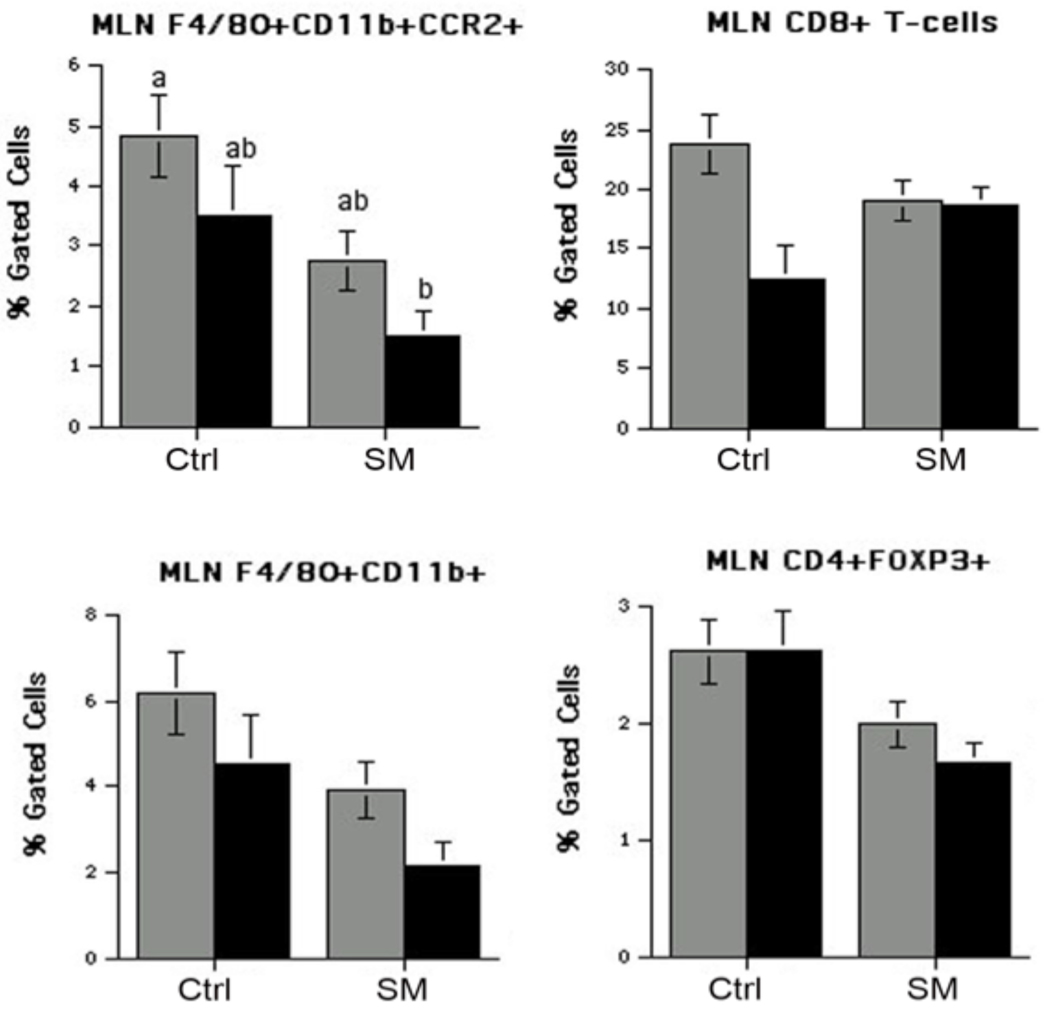
MLN and spleen were harvested to determine if the effect of SM is restricted locally to the intestinal mucosa and inductive mucosal sites (MLN) or if there is a systemic change in immune cells populating the spleen. While there was no effect on immune cell population in the spleen (data not shown), there was a reduction in the F4/80+ macrophage population in MLN in the SM-fed groups of either genotype, and a trend to reduced number of CD4+ T cells (Fig. 6). These numbers, however, need to be confirmed in a larger group, but suggest that dietary SM can affect immune cell populations in the mesentery lymph nodes.
Figure 6.
Changes in macrophage (left panels) and T-cell (right panels) populations in mesentery lymph nodes (MLN) of PPAR-γ+/+ mice (black bars) and PPAR-γ−/− mice (grey bars) 80 days after AOM injection. Mice were fed AIN76A diet alone (control) or supplemented with 0.1% sphingomyelin.
Changes in cytokine and chemokine expression levels by dietary SM
To direct our mechanistic studies of how dietary SM reduced DSS colitis, changes in mRNA expression levels of common pro- and anti-inflammatory and regulatory genes were determined. This is especially important since endogenous sphingolipids have been associated with increased inflammation while dietary SM reduced the severity of DSS-induced inflammation and significantly increased the recovery in an at least partially PPAR-γ dependent manner as shown above. Therefore, mice of both genotypes (n=3 per group) were either treated with DSS or plain water and fed the AIN76A diet alone or supplemented with SM as described under methods. To confirm inflammation after 7 days of treatment, colonic H&E stained sections were evaluated. At the peak of inflammation, all three PPAR-γ+/+ mice (Fig. 7, middle panels) showed a greatly compromised colonic tissue architecture when compared to the colons of mice not treated with DSS (upper panels). There was a large influx of immune cells (arrows) and the typical columnar shape of the colonic crypts (arrowheads) was apparent in less than 40% of the observed tissue. In PPAR-γ−/− mice, the colonic tissue architecture was completely destroyed; there was a massive influx of immune cells and intact colonic crypts were not found. These changes were attenuated in the SM-fed mice (Fig.7, lower panel). The colons of 2 of 3 PPAR-γ+/+ mice had recognizable crypt-like colonic architecture in more than 80% of the observed area while this was less in the colons of PPAR-γ−/− mice. This confirms our observations that dietary SM reduced the DSS-induced damage to the colonic tissue, and suggests that retaining of some tissue architecture may have been critical for the survival of the SM-fed mice in the study described above. However, a large influx of immune cells, albeit less than in the controls, was also apparent in the SM-fed mice.
Figure 7.
Representative images of H&E stained colonic sections from untreated PPAR-γ+/+ or PPAR-γ−/− mice (upper panels), at the peak of intestinal inflammation induced by 2.0% DSS. Images from mice fed the control AIN76A diet are depicted in the middle panels while the lower panels show representative images from mice fed SM supplements. Arrows: immune cell infiltration; arrowheads: typical colonic columnar tissue architecture.
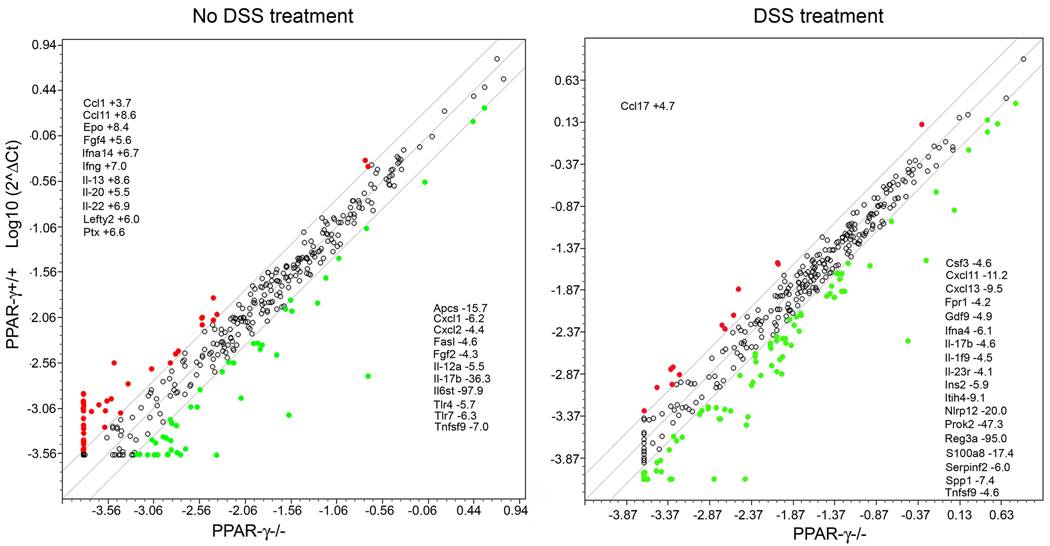
Since the composition of the inflammatory cells may have been critical for the survival of the SM-fed mice after the DSS challenge, we next began to investigate the pro- and anti-inflammatory gene expression levels in the colons using a real-time RT-PCR array that contained 372 genes. These genes are involved in inflammatory response, autoimmunity, tissue regeneration but some have also been associated with differentiation, proliferation and carcinogenesis and angiogenesis, allowing for the identification of critical signaling events or pathways that are targeted by SM to direct our future mechanistic studies. We first compared gene expression levels in the colons of untreated mice of both genotypes to identify genes specifically regulated by dietary SM and not inherent to the genotype. There were few genes differentially expressed in PPAR-γ+/+ versus PPAR-γ mice (Fig.8, left panel). Most of the genes that were more than 2-fold higher expressed in PPAR-γ+/+ than in PPAR-γ−/− mice were cytokines and chemokines; however, these were also the categories of genes with lower expression levels in PPAR-γ+/+ mice. Cytokine and chemokine expression levels were elevated in both genotypes after DSS treatment (29 genes in PPAR-γ+/+ and 66 in PPAR-γ−/− mice). Most of the inflammatory genes up-regulated in PPAR-γ+/+ mice were also up-regulated in PPAR-γ−/− mice; only a small number of genes were down-regulated by DSS treatment (20 in PPAR-γ+/+ and 11 in PPAR-γ−/− with very little overlap). Comparing the expression levels of these genes after DSS treatment between the groups, only one gene was higher expressed in the acute inflamed colons of PPAR-γ+/+ mice by more than 4-fold (Ccl17) while several cyto- and chemokines (Csf3, Cxcl11, Cxcl13, IFNa4) and inflammatory response genes (Prok2, Reg3a, S100a8) showed lower expression levels (Fig.8, right panel). These results demonstrate that PPAR-γ expression in the intestinal epithelium and immune cells greatly affects the inflammatory response to DSS treatment but has a less profound effect in untreated animals.
Figure 8.
Comparison of pro-and anti-inflammatory gene expression levels between PPAR-γ+/+ and PPAR-γ−/− mice (left panel), and after treatment with 2% DSS in the drinking water for 7 days (right panel) as determined by a real-time PCR array.
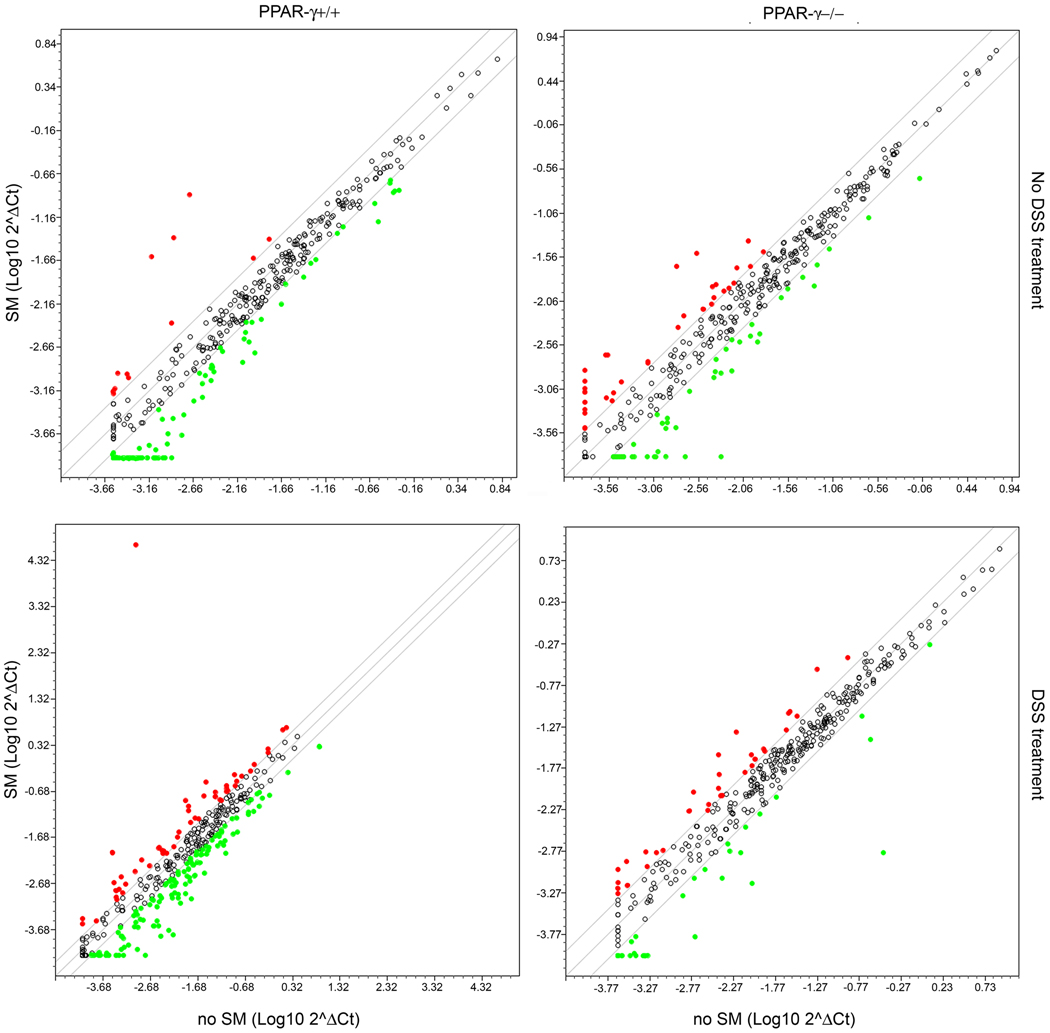
We next determined which inflammatory genes are targeted by dietary SM and if this is dependent on PPAR-γ expression. Changes in gene expression levels in response to SM supplements were compared in colons from mice either treated with DSS or vehicle alone. As shown in Fig. 9 (upper panels), SM supplements increased several cyto- and chemokines by more than 2-fold in both genotypes. However, more cytokines, cytokine receptors and genes involved in the inflammatory response were down-regulated in PPAR-γ+/+ mice (genes listed in Table 3A). While the categories of genes were similar and several genes were regulated in the same direction as in PPAR-γ+/+ mice (Table 3, underlined), several genes were changed in the opposite direction in PPAR-γ−/− mice (Table 3A, in bold). In DSS-treated mice, the response to SM was more pronounced compared to untreated mice (Fig. 9, lower panels; gene list in Table 3B). The patterns of genes up-regulated by dietary SM during DSS colitis consisted essentially of: 1) chemokines and chemokine receptors expressed in epithelial cells and involved in cell trafficking to the gut (Ccl19, Ccl11, Ccl20, Cxcl9, Cxcl11), 2) genes involved in CD4+ T cell differentiation and fate, including Th1 (IFN-γ), Th17 (IL-17 and IL-23), Th2 (IL-4, IL-13 and IL-13R) and Treg (IL-10R). CD4+ naïve T helper cells respond to antigenic stimulation by differentiating into one of, at least, four known fates: Th1, Th2, Th17, and regulatory T cell (Treg). Each of these phenotypes is characterized by their function as well as the cytokines being secreted. The majority of genes and pathways upregulated by SM (Th1, Th17, gut homing chemokines and chemokine receptors) are immunostimulatory and pro-inflammatory which is in agreement with results from previous studies demonstrating the pro-inflammatory effect of sphingolipid metabolites. However, the up-regulation of genes in the Th2 differentiation pathway (IL-4, IL-13 and IL-13R) and Treg-related genes such as IL-10R is in line with an anti-inflammatory effect and are modulated by SM in a PPAR-γ-dependent manner. Furthermore, dietary SM suppressed the expression of other regulatory molecules such as members of the TGF superfamily that mediate apoptosis (Fasl), inflammatory signaling intermediates such as Myd88, DOCK2, and transcriptional regulators such as Nfe2l1, NfrκB and Stat3 also in a PPAR-γ dependent manner, thereby favoring an anti-inflammatory environment.
Figure 9.
Changes in gene expression levels in the colons of PPAR-γ+/+ (left panels) and PPAR-γ−/− mice (right panels) in response to dietary SM. The mice were not treated (upper panels) or treated with 2% DSS for 7 days (lower panels).
| A | ||||
|---|---|---|---|---|
| No DSS treatment |
||||
| PPAR-γ+/+ | PPAR-γ−/− | |||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| Cytokines | Gdf9, Epo, Il11, Il12, Il17, Muc4 | Cd70, Cer1, Cmtm1,2, Csf3, Ctf2, Epo, Fasl, Fgf3, Fgf4, Fgf5, Fgf6, Fgf8, Gdf2, Gdf3, Gdf6, Grem1, Ifna11, Ifna14, IIfna2, Ifna4, Ifna9, Ifnab, Ifnb1, Ifne, Ifng, Il1f10, Il1f5, Il1f6, Il1f8, Il1f9, Il12, Il13, Il16, Il17a, Il17c, Il17d, Il17f, Il19, Il1b, Il20, Il21, Il22, Il23a, Il31, Il4, Il5, Il6, Il9, Inhba, Lefty2, Ltb, Nodal, Nrg1, Prl, Slco1a4, Spp1, Thpo, Tnfsf15, Tnfsf9 | Cd27, Ebi1, Fgf3, Gdf6, Ifna4, Ifne, Ifng, Il10, Il17d, Il1f10, Il24, Lefty2, Muc4, Spp1 | Areg, Clcf1, Csf3, Fasl, Fgf2, Fgf5, Gdf2, Gdf5, Ifna2, Ifna9, Il11, Il5, Il17a, Il1a, Ltb, Slurp1, Thpo, Tnf, Tnfsf11, Tnfsf15, Tnfsf8 |
| Cytokine receptors | Il23r, Il6st | Epor, Ifgr1, Il13ra2, Il18r1, Il18rap, Il12rb1, Il12rb, Il1rapl2, Il1rl1, Il1rl2, Il20ra, Il2rb, Il21r, Il3ra, Il5ra, Il7r, Il8ra, Il8rb, Mpl, | Gfra1, Il2ra, Prlr | Il20ra, Il21r, Il21ra, Il22ra2, Il23r |
| Chemokines | Ccl12, Cxcl2 | Ccl28, Ccl3, Ccl4, Ccl8, Cxcl15, Cxcl16 | Ccl12, Ccl3, Cxcl9, Pxmp2 | Ccl17, Cxcl1, Cxcl5 |
| Chemokine rec. | Ccr2, Ccr3, Ccr5, Ccr9, Ccrl2, Cxcr4 | Ccrl1, Ccr3, Ccrl2, Cxcr6 | Ccr8, Ccr9, Cxcr5, | |
| Inflamm. response | Apol7a, Reg3g, Tlr7 | Ahsg, Apol8, Bmp7, Fos, Fpr1, Kng1, Mmp25, Nfam1, Nos2, Olr1, Prg2, Prg3, Prok2, Ptgs2, Ptx3 | Apol8, C3ar1 | Adora1, Ahsg, Cd180, Fos, Ltb4r1, Mmp25, Siglec1, Tnfaip6, |
| Acute phase | Crp, Fn1,Ins1, Ins2, Itih4, Reg3a, Sele, Serpinf2 | Itih4, Reg3a, Reg3g, Saa4 | Apcs, Crp, F2, Serpina1a | |
| Cytokine metab. | Nlrp12, Sftpd | Csf2ra, Irf4 | ||
| B | ||||
|---|---|---|---|---|
| DSS treatment |
||||
| PPAR-γ+/+ | PPAR-γ−/− | |||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | |
| Cytokines | Figf, Gdf6, Gdf9, Ifng, Il1f6, Il1f10, Il1f6, Il1f9, Il17b, Il1a, Il1b, Il23b, Il27, Il4, Il7, Mif, Nrg1, Pglyrp1, Tnfsf14,Tnfsf 9, Trap1 | Bmp2, Bmp3, Cast, Clcf1, Csf1, Csf3, Ctf1, Erbb2, Fasl, Fgf3, Flt3l, Gdf1,Gdf5, Gdf7, Glmn, Grem1, 2, Grn, Ifna2, Ifna9, Ifnk, Il11, Il15, Il16, Il17a, Il7c, Il17d, Il17f, Il19, Il24, Il3, Inha, Inhba, Inhbb, Pdgfb, Pdgfc, Ptn, Scg2, Slco1a4, Spred1, Srgap1, Tnfsf13, Tnfsf13b, Tnfsf4, Tymp, Vegfa, Vgefb, Yars | Cd70, Fgf7, Ifna2, Ifnk, Il1a, Il1b, Il1f9, Il10, Il6, Inhba, Lefty2, Osm, Slurp1, Tnf, Tnfsf4, Tnfsf11, Tnfsf13b | Cer1, Cmtm1, Crp, Il17a, Il17b, Il19, Epo, Fgf3, Fgf4,Fgf5, Fgf6, Fgf8, Gdf3, Gdf9, Infa11, Ifna14, Ifna4, Ifna9, Ifnb1, Ifne, Il12b, Il13, Il16, Il17a, Il17b, Il19, Il1f5, Il1f6, Il1f8, Il20, Il21, Il3, Il31, Il5, Il9, Mstn, Nodal, Prl, Slco1a4, Tnfsf15, Tnfsf18, Tymp |
| Cytokine receptors | Il10rb, Il1r2, Il31ra, Il2ra | Cfra2, Cntfr, Csf2ra, Epor, Ifngr2, Il11ra1, Il12rb1, Il15ra, Il1rl2, Il21r, Il2rb, Il5ra, Il17r, Il17rb, Il18r1, Il18rap, Il1rapl2, Il20ra, Il21r, Il22ra1, Il28ra, Il7r, Il9r, Lifr, Ttn | Il1rl1, Il2ra, Il31ra, Il5ra | Il6st, Il1rapl2, Il23r, Il8ra, Mpl |
| Chemokines | Ccl19, Ccl3, Ccl4, Ccl8, Cxcl10,Cxcl11 Cxcl3, Cxcl2, Cxcl5, Cxcl9 | Ccl1, Ccl17, Ccl22, Ccl24, Ccl25, Cyp26b, Pf4, Ppbp, Pxmp2 | Ccl3, Ccl4, Cxcl11, Cxcl2 | Ccl24, Cxcl13, Cxcl15, |
| Chemokine rec. | Cxcr3 | Ccr10, Ccr3, Ccr4, Ccr8, Ccr9, Ccrl1, Cx3cr1, Cxcr4, Cxcr5, Cxcr6 | Ccr7, Ccrl2 | Ccr8, Ccrl1 |
| Inflamm. response | Aif1, Cd14, Cd40, Itgb2, Ly86, Nmi, Pla2g7,Reg3g, S100a8, Spp1, Tacr1 | Adora1, C3ar1, Dock2,F11r, Gpr68, Hdac 4,5,9, Hrh1, Lta, Ltb, Ltb4r1, Mmp25, Myd88, Nfam1, Nfatc4, Nfe2l1, Nfrkb,Nfx1, Nos2, Prdx5, Ptafr, Reg3a, Syk, Tlr2, Tlr5, Vsp45 | Ptx3, Sele, Tnfaip6 | Ahsg, Apol8, Kng1, Prg2, Prg3, Ptpra, Reg3a, Spaca3 |
| Acute phase | F2, Fn1, Lbp, Serpina1a, Serpinf2, Stat3 | F8, Saa4 | Ins1, Ins2, Itih4, Reg3a, Serpinf2 | |
| Cytokine metab. | Cd28, Irf4 | Cd27, Irf4, Sfpd | ||
Several SM-regulated genes have also been associated with tissue protection or regeneration (Fasl [39], Serpina3n, Spp1 [40], Trap1[41]) or have a potential effect on colon cancer (ErbB2, Slurp1 [42], Lta [43], Ltb [44], NfrkB [45], Serpina1 [46], Tlr5 [47]), suggesting that in addition to modifying the pro-and anti-inflammatory balance towards an anti-inflammatory environment, SM also may affect tumor initiation, and promotion.
Effect of sphingolipid metabolite sphingosine on PPAR-γ transcriptional activity of acrophages
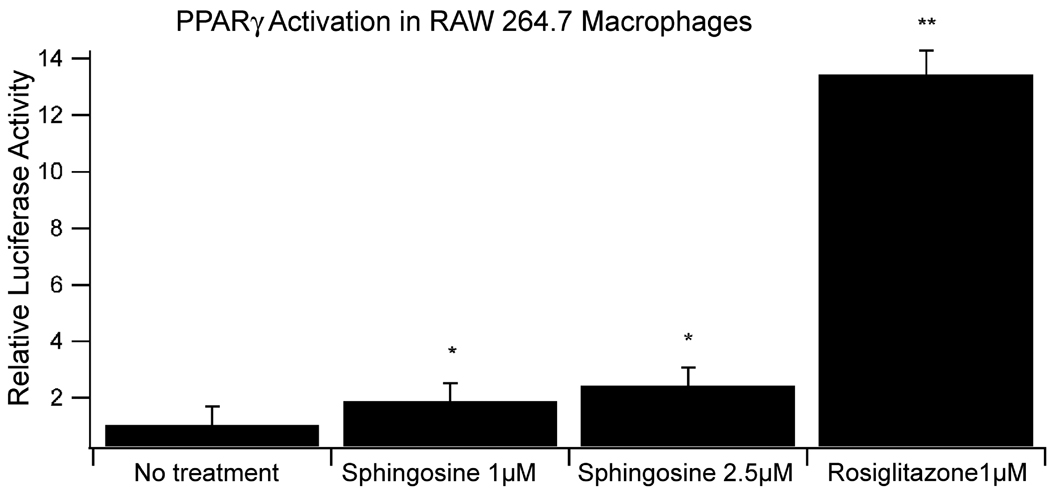
To support our findings that SM affects anti-inflammatory events in a PPAR-γ dependent manner, we determined whether sphingolipid metabolites activate PPAR-γ in immune cells we treated RAW 264.7 macrophages with sphingosine, the metabolite also generated in the intestinal tract after oral administration of complex sphingolipids and presumably responsible for the suppression of carcinogen-induced colon cancer [5]. As shown in Figure 10, sphingosine modestly but significantly activated PPAR-γ transcriptional activity in a concentration-dependent manner.
Figure 10.
Non-toxic concentrations of sphingosine activate PPAR-γ transcriptional activity in macrophages, determined by luciferase promoter activity assay, compared to the PPAR-γ agonist rosilitazone.
*p<0.05; **p<0.01
Discussion
Endogenous sphingolipid metabolites have been implicated in the induction of inflammatory responses both in vitro and in vivo. The present studies show that dietary SM reduced DSS-induced inflammation, accelerated disease recovery, and reduced AOM-induced colon tumors. These findings were associated with an improved preservation of the colonic microscopic architecture during acute inflammation in both PPAR-γ+/+ and PPAR-γ−/− mice, and an increased survival of the mice. Interestingly, the expression of PPAR-γ in immune and epithelial cells enhanced the anti-inflammatory effects of dietary SM while its anti-carcinogenic actions appear to be mostly independent of the mouse genotype.
Complex sphingolipids are digested to the bioactive metabolites ceramide and sphingosine [11,12,14,48]. These metabolites are likely the mediators of the observed tumor suppression since all complex sphingolipids tested (SM, glucosylceramide, lactosylceramide, GM1, GD3) [7,49] showed the same effect independent of the headgroup; additional effects of other molecular structures such as the fatty acid and the choline headgroup, however, cannot be ruled out. Many recent studies have focused on the association of ceramide and the initiation or perpetuation of inflammation. In macrophages, an increase in ceramide levels was associated with a higher Cox-2 expression [20] via NF-κB activation [18]. Exogenous ceramide mimicked these events in murine macrophages and it is thought that a ceramide-mediated suppression of PPAR-γ prevents suppression of NF-κB activity (see recent review [50]). Other studies suggest a key role of ceramide in the increased secretion of pro-inflammatory cytokines via NF-κB activation in adipose tissue-associated macrophages [18], and Cox-2, cPLA2 and 12-LOX in primary astrocytes [51]. Another sphingolipid metabolite, sphingosine 1-phosphate (S1P), generated by sphingosine kinase-1, is a regulator of cell growth and a potent immune stimulator. The immune suppressor FTY720 activates specifically S1P receptor1 and mediates sequestration of circulating lymphocytes within secondary lymphoid tissues, thereby decreasing CD4+ T-cells in the colonic lamina propria, and homing of circulating lymphocytes into Peyer’s patches and MLN [52]. In DSS-induced colitis, FTY720 attenuated infiltration of CD4+ T-cells into the colonic lamina propria, and the disease severity [53].
Based on our gene expression analyses, we observed that SM induced a pro-inflammatory response that was paralleled by an anti-inflammatory or counter-regulatory immune response characterized by up-regulation of genes associated with anti-inflammatory phenotypes of CD4+ T cells (i.e., Th2 and Treg) as well as suppression of inflammatory chemokines and their receptors. Recent evidence supports the existence of cross-talk between these pathways and demonstrates specificity and plasticity in CD4+ T cell fate determination [54,55]. Indeed, our data provide evidence that dietary SM supplements trigger the expression of pro-inflammatory and immunostimulatory genes consistent with Th1 and Th17 effector and pro-inflammatory responses while at the same time up-regulates genes involved in a counter-regulatory immune response that may inhibit pro-inflammatory genes. More specifically, dietary SM up-regulated the expression of genes involved in the differentiation of CD4+ T cells toward Th1 (IFN-γ) and Th17 (IL-17 and IL-23) effector phenotypes involved in cellular responses against pathogens and autommunity such as inflammatory bowel disease (IBD), rheumatoid arthritis, and multiple sclerosis [55,56]. This predominant Th1 and Th17-related gene expression pattern observed in SM-fed mice was consistent with increased expression of chemokines involved in leukocyte recruitment into the colonic mucosa (i.e., Ccl19, Ccl11, Ccl20, Cxcl9, Cxcl11). Surprisingly, this inflammatory gene expression pattern was associated with decreased colonic inflammatory lesions, increased survival and improved clinical disease. This apparent contradiction may be explained by the existence of a regulatory response that may have paralleled the effector/pro-inflammatory gene expression pattern described above. The parallel induction of effector and regulatory responses is typical in mucosal sites during inflammation or infection [57,58]. More specifically, SM-fed mice had increased expression of Th2 genes involved in humoral immune responses (IL-4, IL-13 and IL-13R) and a Treg-related gene (IL-10R) involved anti-inflammatory responses. Interestingly, the up-regulation of Th2 and Treg genes was modulated by dietary SM in a PPAR-γ-dependent manner. The better understanding of the potential regulatory actions of dietary components of CD4+ T cell differentiation in vivo is important and further investigations of the effects of sphingolipids in CD4+ T cell differentiation networks and immune function are currently ongoing in our laboratory.
Dietary SM administration decreased the number of macrophages in the MLN and lymphoplasmacytic infiltration in colons of mice with inflammation-induced colorectal cancer. These immunological changes were associated with a faster recovery from DSS-induced inflammation PPAR-γ+/+ but not in PPAR-γ−/− mice. Macrophages, T cells and intestinal epithelial cells are the primary contributors to intestinal inflammation during inflammatory bowel disease and express PPAR-γ. We hypothesized that the anti-inflammatory effects of SM were mediated through a PPAR-γ-dependent mechanism. We provide in vitro evidence demonstrating that SM increased PPAR-γ reporter activity in RAW 264.7 macrophages. These in vitro findings are in line with the PPAR-γ-dependent induction of Th2- and Treg-related genes in colons of SM-fed mice with DSS colitis. This pattern is likely to be associated with an increased presence of M2 alternatively activated macrophages, a cell type that is highly responsive to M2 fate determination following activation of PPAR-γ and δ [59–61]. In this regard, we found a decrease in F4/80CD11bCCR2+ macrophages in MLN of SM-fed mice, a cell phenotype that is pro-inflammatory but also responsive to PPAR-γ [62].
The model of inflammation-induced colorectal cancer utilized followed a biphasic response pattern in which the first peak corresponded to acute inflammation and a second peak that coincided with colonic tumorigenesis. Even though the effect of dietary SM on the first inflammatory peak follows a pattern that depends partly on PPAR-γ expression in epithelial and immune cells, the improvement in tumor area as well as adenoma and carcinoma formation observed in SM-fed mice at the end of the study appears to be independent of PPAR-γ expression in immune and epithelial cells. However, several genes that may promote cancer (ErbB2, Ltb, NfrkB) or exert anti-tumor effects (Gdf9; [63]) were down- or up-regulated, respectively, by dietary SM only in PPAR-γ expressing mice, suggesting that PPAR-γ may also play a protective role in the suppression of colon cancer by dietary SM. Our results also suggest that in addition to the immune cells, epithelial cells and, perhaps, their interactions may be targets of dietary SM in the suppression of inflammation. These findings will direct our further mechanistic studies.
In conclusion, the presented data demonstrate the ability of dietary SM to suppress colonic inflammation and inflammation-driven colorectal cancer. Further time-course studies are needed to dissect the stage-specific PPAR-γ-dependent and independent effects of this compound and to further characterize the immune modulatory actions of dietary sphingolipids in CD4+ T cells and macrophages.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supported by NIH R01 CA118866 (EMS), NIH R01AT004308 (JBR), and funds of the Nutritional Immunology and Molecular Medicine Laboratory (JBR)
References
- 1.Horner MJRL, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK, editors. Bethesda, md: Seer cancer statistics review, 1975–2006, national cancer institute. 2009 http://seer.Cancer.Gov/csr/1975_2006/, based on november 2008 seer data submission, posted to the seer web site.
- 2.Bleyer AOLM, Barr R, Ries LAG, editors. Cancer epidemiology in older adolescents and young adults 15 to 29 years of age, including seer incidence and survival: 1975–2000. Bethesda, MD: National Cancer Institute; 2006. NIH Pub. No. 06-5767. [Google Scholar]
- 3.Lakatos PL, Lakatos L. Risk for colorectal cancer in ulcerative colitis: Changes, causes and management strategies. World J Gastroenterol. 2008;14:3937–3947. doi: 10.3748/wjg.14.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WCRF/AICR. Nutrition, physical activity and the prevention of cancer: A global perspective. Washington, DC: AICR; 2007. [Google Scholar]
- 5.Schmelz EM, Bushnev AS, Dillehay DL, Liotta DC, Merrill AH., Jr Suppression of aberrant colonic crypt foci by synthetic sphingomyelins with saturated or unsaturated sphingoid base backbones. Nutr Cancer. 1997;28:81–85. doi: 10.1080/01635589709514556. [DOI] [PubMed] [Google Scholar]
- 6.Schmelz EM, Bushnev AS, Dillehay DL, Sullards MC, Liotta DC, Merrill AH., Jr Ceramide-beta-d-glucuronide: Synthesis, digestion, and suppression of early markers of colon carcinogenesis. Cancer Res. 1999;59:5768–5772. [PubMed] [Google Scholar]
- 7.Schmelz EM, Sullards MC, Dillehay DL, Merrill AH., Jr Colonic cell proliferation and aberrant crypt foci formation are inhibited by dairy glycosphingolipids in 1, 2-dimethylhydrazine-treated cf1 mice. J Nutr. 2000;130:522–527. doi: 10.1093/jn/130.3.522. [DOI] [PubMed] [Google Scholar]
- 8.Lemonnier LA, Dillehay DL, Vespremi MJ, Abrams J, Brody E, Schmelz EM. Sphingomyelin in the suppression of colon tumors: Prevention versus intervention. Arch Biochem Biophys. 2003;419:129–138. doi: 10.1016/j.abb.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Symolon H, Schmelz EM, Dillehay DL, Merrill AH., Jr Dietary soy sphingolipids suppress tumorigenesis and gene expression in 1,2-dimethylhydrazine-treated cf1 mice and apcmin/+ mice. J Nutr. 2004;134:1157–1161. doi: 10.1093/jn/134.5.1157. [DOI] [PubMed] [Google Scholar]
- 10.Schmelz EM, Roberts PC, Kustin EM, et al. Modulation of intracellular beta-catenin localization and intestinal tumorigenesis in vivo and in vitro by sphingolipids. Cancer Res. 2001;61:6723–6729. [PubMed] [Google Scholar]
- 11.Nilsson A. Metabolism of sphingomyelin in the intestinal tract of the rat. Biochim Biophys Acta. 1968;164:575–584. doi: 10.1016/0005-2760(68)90187-2. [DOI] [PubMed] [Google Scholar]
- 12.Nilsson A. The presence of spingomyelin- and ceramide-cleaving enzymes in the small intestinal tract. Biochim Biophys Acta. 1969;176:339–347. doi: 10.1016/0005-2760(69)90192-1. [DOI] [PubMed] [Google Scholar]
- 13.Ohlsson L, Hertervig E, Jonsson BA, et al. Sphingolipids in human ileostomy content after meals containing milk sphingomyelin. Am J Clin Nutr. 2010 doi: 10.3945/ajcn.2009.28311. [DOI] [PubMed] [Google Scholar]
- 14.Schmelz EM, Crall KJ, Larocque R, Dillehay DL, Merrill AH., Jr Uptake and metabolism of sphingolipids in isolated intestinal loops of mice. J Nutr. 1994;124:702–712. doi: 10.1093/jn/124.5.702. [DOI] [PubMed] [Google Scholar]
- 15.Bartke N, Hannun YA. Bioactive sphingolipids: Metabolism and function. J Lipid Res. 2009;50 Suppl:S91–S96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan RD, Nilsson A. Metabolism of sphingolipids in the gut and its relation to inflammation and cancer development. Prog Lipid Res. 2009;48:62–72. doi: 10.1016/j.plipres.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Lin CI, Chen CN, Lin PW, Lee H. Sphingosine 1-phosphate regulates inflammation-related genes in human endothelial cells through s1p1 and s1p3. Biochem Biophys Res Commun. 2007;355:895–901. doi: 10.1016/j.bbrc.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 18.Wu D, Ren Z, Pae M, et al. Aging up-regulates expression of inflammatory mediators in mouse adipose tissue. J Immunol. 2007;179:4829–4839. doi: 10.4049/jimmunol.179.7.4829. [DOI] [PubMed] [Google Scholar]
- 19.Teichgraber V, Ulrich M, Endlich N, et al. Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat Med. 2008;14:382–391. doi: 10.1038/nm1748. [DOI] [PubMed] [Google Scholar]
- 20.Claycombe KJ, Wu D, Nikolova-Karakashian M, et al. Ceramide mediates age-associated increase in macrophage cyclooxygenase-2 expression. J Biol Chem. 2002;277:30784–30791. doi: 10.1074/jbc.M204463200. [DOI] [PubMed] [Google Scholar]
- 21.Bauer J, Liebisch G, Hofmann C, et al. Lipid alterations in experimental murine colitis: Role of ceramide and imipramine for matrix metalloproteinase-1 expression. PLoS One. 2009;4:e7197. doi: 10.1371/journal.pone.0007197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exon JH, South EH. Effects of sphingomyelin on aberrant colonic crypt foci development, colon crypt cell proliferation and immune function in an aging rat tumor model. Food Chem Toxicol. 2003;41:471–476. doi: 10.1016/s0278-6915(02)00297-1. [DOI] [PubMed] [Google Scholar]
- 23.Furuya H, Ohkawara S, Nagashima K, Asanuma N, Hino T. Dietary sphingomyelin alleviates experimental inflammatory bowel disease in mice. Int J Vitam Nutr Res. 2008;78:41–49. doi: 10.1024/0300-9831.78.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Desreumaux P, Ghosh S. Review article: Mode of action and delivery of 5-aminosalicylic acid - new evidence. Aliment Pharmacol Ther. 2006;24 Suppl 1:2–9. doi: 10.1111/j.1365-2036.2006.03069.x. [DOI] [PubMed] [Google Scholar]
- 25.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassaganya-Riera J, Guri A, King J, Hontecillas R. Peroxisome proliferator-activated receptors: The nutritionally controlled molecular networks that integrate inflammation, immunity and metabolism. Current Nutrition & Food Science. 2005;1:179–187. [Google Scholar]
- 27.Guri AJ, Hontecillas R, Bassaganya-Riera J. Peroxisome proliferator-activated receptors: Bridging metabolic syndrome with molecular nutrition. Clin Nutr. 2006;25:871–885. doi: 10.1016/j.clnu.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Hontecillas R, Bassaganya-Riera J. Peroxisome proliferator-activated receptor gamma is required for regulatory cd4+ t cell-mediated protection against colitis. J Immunol. 2007;178:2940–2949. doi: 10.4049/jimmunol.178.5.2940. [DOI] [PubMed] [Google Scholar]
- 29.Evans NP, Misyak SA, Schmelz EM, Guri AJ, Hontecillas R, Bassaganya-Riera J. Conjugated linoleic acid ameliorates inflammation-induced colorectal cancer in mice through activation of ppar{gamma} J Nutr. 2010 doi: 10.3945/jn.109.115642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guri AJ, Evans NP, Hontecillas R, Bassaganya-Riera J. T cell ppar gamma is required for the anti-inflammatory efficacy of abscisic acid against experimental ibd. Journal of Nutritional Biochemistry. 2010 doi: 10.1016/j.jnutbio.2010.06.011. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barak Y, Nelson MC, Ong ES, et al. Ppar gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 32.Akiyama TE, Sakai S, Lambert G, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of abca1, abcg1, and apoe in macrophages and reduced cholesterol efflux. Mol Cell Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Y, Miyoshi K, Claudio E, et al. Loss of the peroxisome proliferation-activated receptor gamma (ppargamma) does not affect mammary development and propensity for tumor formation but leads to reduced fertility. J Biol Chem. 2002;277:17830–17835. doi: 10.1074/jbc.M200186200. [DOI] [PubMed] [Google Scholar]
- 34.Bassaganya-Riera J, Reynolds K, Martino-Catt S, et al. Activation of ppar gamma and delta by conjugated linoleic acid mediates protection from experimental inflammatory bowel disease. Gastroenterology. 2004;127:777–791. doi: 10.1053/j.gastro.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 35.Guri AJ, Hontecillas R, Ferrer G, et al. Loss of ppar gamma in immune cells impairs the ability of abscisic acid to improve insulin sensitivity by suppressing monocyte chemoattractant protein-1 expression and macrophage infiltration into white adipose tissue. J Nutr Biochem. 2008;19:216–228. doi: 10.1016/j.jnutbio.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Nutrition AIo. Report of the american institute of nutrition ad hoc committee on standards for nutritional studies. J Nutr. 1977;107:1340–1348. doi: 10.1093/jn/107.7.1340. [DOI] [PubMed] [Google Scholar]
- 37.Schmelz EM, Dillehay DL, Webb SK, Reiter A, Adams J, Merrill AH., Jr Sphingomyelin consumption suppresses aberrant colonic crypt foci and increases the proportion of adenomas versus adenocarcinomas in cf1 mice treated with 1,2-dimethylhydrazine: Implications for dietary sphingolipids and colon carcinogenesis. Cancer Res. 1996;56:4936–4941. [PubMed] [Google Scholar]
- 38.Kobayashi T, Shimizugawa T, Osakabe T, Watanabe S, Okuyama H. A long-term feeding of sphingolipids affected the levels of plasma cholesterol and hepatic triacylglycerol but not tissue phospholpids and sphingolipids. Nutr Res. 1997;17 [Google Scholar]
- 39.Vetuschi A, Latella G, Sferra R, Caprilli R, Gaudio E. Increased proliferation and apoptosis of colonic epithelial cells in dextran sulfate sodium-induced colitis in rats. Dig Dis Sci. 2002;47:1447–1457. doi: 10.1023/a:1015931128583. [DOI] [PubMed] [Google Scholar]
- 40.Zhong J, Eckhardt ER, Oz HS, Bruemmer D, de Villiers WJ. Osteopontin deficiency protects mice from dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2006;12:790–796. doi: 10.1097/00054725-200608000-00015. [DOI] [PubMed] [Google Scholar]
- 41.Costantino E, Maddalena F, Calise S, et al. Trap1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 42.Pettersson A, Nylund G, Khorram-Manesh A, Nordgren S, Delbro DS. Nicotine induced modulation of slurp-1 expression in human colon cancer cells. Auton Neurosci. 2009;148:97–100. doi: 10.1016/j.autneu.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Jeong CH, Bode AM, Pugliese A, et al. [6]-gingerol suppresses colon cancer growth by targeting leukotriene a4 hydrolase. Cancer Res. 2009;69:5584–5591. doi: 10.1158/0008-5472.CAN-09-0491. [DOI] [PubMed] [Google Scholar]
- 44.Ihara A, Wada K, Yoneda M, Fujisawa N, Takahashi H, Nakajima A. Blockade of leukotriene b4 signaling pathway induces apoptosis and suppresses cell proliferation in colon cancer. J Pharmacol Sci. 2007;103:24–32. doi: 10.1254/jphs.fp0060651. [DOI] [PubMed] [Google Scholar]
- 45.Joshi N, Johnson LL, Wei WQ, et al. Gene expression differences in normal esophageal mucosa associated with regression and progression of mild and moderate squamous dysplasia in a high-risk chinese population. Cancer Res. 2006;66:6851–6860. doi: 10.1158/0008-5472.CAN-06-0662. [DOI] [PubMed] [Google Scholar]
- 46.Kloth JN, Gorter A, Fleuren GJ, et al. Elevated expression of serpina1 and serpina3 in hla-positive cervical carcinoma. J Pathol. 2008;215:222–230. doi: 10.1002/path.2347. [DOI] [PubMed] [Google Scholar]
- 47.Niedzielska I, Niedzielski Z, Tkacz M, et al. Toll-like receptors and the tendency of normal mucous membrane to transform to polyp or colorectal cancer. J Physiol Pharmacol. 2009;60 Suppl 1:65–71. [PubMed] [Google Scholar]
- 48.Ohlsson L, Burling H, Nilsson A. Long term effects on human plasma lipoproteins of a formulation enriched in butter milk polar lipid. Lipids Health Dis. 2009;8:44. doi: 10.1186/1476-511X-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dillehay DL, Webb SK, Schmelz EM, Merrill AH., Jr Dietary sphingomyelin inhibits 1,2-dimethylhydrazine-induced colon cancer in cf1 mice. J Nutr. 1994;124:615–620. doi: 10.1093/jn/124.5.615. [DOI] [PubMed] [Google Scholar]
- 50.Meydani SN, Wu D. Nutrition and age-associated inflammation: Implications for disease prevention. JPEN J Parenter Enteral Nutr. 2008;32:626–629. doi: 10.1177/0148607108325179. [DOI] [PubMed] [Google Scholar]
- 51.Prasad VV, Nithipatikom K, Harder DR. Ceramide elevates 12-hydroxyeicosatetraenoic acid levels and upregulates 12-lipoxygenase in rat primary hippocampal cell cultures containing predominantly astrocytes. Neurochem Int. 2008;53:220–229. doi: 10.1016/j.neuint.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Mizushima T, Ito T, Kishi D, et al. Therapeutic effects of a new lymphocyte homing reagent fty720 in interleukin-10 gene-deficient mice with colitis. Inflamm Bowel Dis. 2004;10:182–192. doi: 10.1097/00054725-200405000-00002. [DOI] [PubMed] [Google Scholar]
- 53.Deguchi Y, Andoh A, Yagi Y, et al. The s1p receptor modulator fty720 prevents the development of experimental colitis in mice. Oncol Rep. 2006;16:699–703. [PubMed] [Google Scholar]
- 54.Wei G, Wei L, Zhu J, et al. Global mapping of h3k4me3 and h3k27me3 reveals specificity and plasticity in lineage fate determination of differentiating cd4+ t cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarra M, Pallone F, Macdonald TT, Monteleone G. Il-23/il-17 axis in ibd. Inflamm Bowel Dis. doi: 10.1002/ibd.21248. [DOI] [PubMed] [Google Scholar]
- 56.Reynolds JM, Pappu BP, Peng J, et al. Toll-like receptor 2 signaling in cd4(+) t lymphocytes promotes t helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity. 32:692–702. doi: 10.1016/j.immuni.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hontecillas R, Bassaganya-Riera J, Wilson J, Hutto DL, Wannemuehler MJ. Cd4+ t-cell responses and distribution at the colonic mucosa during brachyspira hyodysenteriae-induced colitis in pigs. Immunology. 2005;115:127–135. doi: 10.1111/j.1365-2567.2005.02124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wendelsdorf K, Bassaganya-Riera J, Hontecillas R, Eubank S. Model of colonic inflammation: Immune modulatory mechanisms in inflammatory bowel disease. J Theor Biol. 264:1225–1239. doi: 10.1016/j.jtbi.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, et al. Macrophage-specific ppargamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, et al. Alternative m2 activation of kupffer cells by ppardelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang K, Reilly SM, Karabacak V, et al. Adipocyte-derived th2 cytokines and myeloid ppardelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassaganya-Riera J, Misyak S, Guri AJ, Hontecillas R. Ppar gamma is highly expressed in f4/80(hi) adipose tissue macrophages and dampens adipose-tissue inflammation. Cell Immunol. 2009;258:138–146. doi: 10.1016/j.cellimm.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. The role of growth differentiation factor-9 (gdf-9) and its analog, gdf-9b/bmp-15, in human breast cancer. Ann Surg Oncol. 2007;14:2159–2166. doi: 10.1245/s10434-007-9397-5. [DOI] [PubMed] [Google Scholar]