Abstract
This paper combines two techniques—mass spectrometry and protein charge ladders—to examine the relationship between the surface charge and hydrophobicity of a representative globular protein (bovine carbonic anhydrase II; BCA II) and its rate of amide hydrogen-deuterium (H/D) exchange. Mass spectrometric analysis indicated that the sequential acetylation of surface lysine-ε-NH3+ groups—a type of modification that increases the net negative charge and hydrophobicity of the surface of BCA II without affecting its 2° or 3° structure—resulted in a linear decrease in the aggregate rate of amide H/D exchange at pD 7.4, 15 °C. According to analysis with MS, the acetylation of each additional lysine generated between 1.4 and 0.9 additional hydrogens that are protected from H/D exchange during the 2 h exchange experiment at 15 °C, pD 7.4. NMR spectroscopy demonstrated that none of the hydrogen atoms which became protected upon acetylation were located on the side chain of the acetylated lysine residues (i.e., lys-ε-NHCOCH3) but were instead located on amide NHCO moieties in the backbone. The decrease in rate of exchange associated with acetylation paralleled a decrease in thermostability: the most slowly exchanging rungs of the charge ladder were the least thermostable (as measured by differential scanning calorimetry). This observation—that faster rates of exchange are associated with slower rates of denaturation—is contrary to the usual assumptions in protein chemistry. The fact that the rates of H/D exchange were similar for perbutyrated BCA II (e.g., [lys-ε-NHCO(CH2)2CH3]18) and peracetylated BCA II (e.g., [lys-ε-NHCOCH3]18) suggests that the electrostatic charge is more important than the hydrophobicity of surface groups in determining the rate of H/D exchange. These electrostatic effects on the kinetics of H/D exchange could complicate (or aid) the interpretation of experiments in which H/D exchange methods are used to probe the structural effects of non-isoelectric perturbations to proteins (i.e., phosphorylation, acetylation, or the binding of the protein to an oligonucleotide or to another charged ligand or protein).
Keywords: amide H/D exchange, lysine acetylation, mass spectrometry, protein folding, carbonic anhydrase II, protein charge ladder, hydrogen/deuterium, electrostatic potential
Introduction
We wished to determine how the surface charge and hydrophobicity of a folded protein affects the rate at which it exchanges amide N-H hydrogen with buffer. To do so, we have measured the rate of hydrogen-deuterium (H/D) exchange of the rungs (successively acylated sets of proteins) of two protein charge ladders 1–5 with electro-spray ionization mass spectrometry (ESI-MS). A “protein charge ladder” is a mixture of charge isomers generated by the modification of the functional groups of a protein. The charge ladders we used were prepared by sequentially acylating all 18 lysine-ε-NH3+ at the surface of bovine carbonic anhydrase II (BCA II)6 with acetic or butyric anhydride to yield lysine-ε-NHCOCH3 and lysine-ε-NHCO(CH2)2CH3.
The isoelectric point (pI) of BCA II is ~ 5.9. Previous experiments at pH 8.4 have shown that each acetylation increases the net negative charge (Zo) of BCA II by approximately 0.9 units. The difference between ΔZ = −0.9 and the value of −1.0 that might be expected for -NH3+ ➔ -NHCOCH3 can be explained by charge regulation.7 Charge regulation in a protein is the change in the values of pKa, of functional groups (or equivalently, of local pH) in response to a change in the electrostatic potential around the protein that might result, for example, from acylating its lys-ε-NH3+ groups. The BCA II charge ladder contains 19 charge isomers or “rungs,” and therefore spans approximately 16 units of charge (Zo for BCA II = −2.9 at pH 8.4, 10 mM ionic strength; Zo for BCA-Ac(18) = −19).8 The acetylation of all 18 lysine residues (peracetylation) does not change the structure of this thermostable zinc protein (as measured previously by circular dichroism3 and X-ray crystallography9 ).
Mass spectrometry established a linear relationship between the net negative charge of folded BCA II (e.g., the number of acylations) and the number of hydrogens that do not exchange with solvent after a 2 h incubation in deuterated buffer (we say these hydrogen are protected from exchange). According to analysis with MS, the acetylation of each additional lysine generated between 1.4 and 0.9 additional hydrogen that are protected from H/D exchange during the 2 h exchange experiment at 15 °C, pD 7.4. Multi-dimensional Nuclear Magnetic Resonance (NMR) spectroscopy demonstrated that the additional protected hydrogen atoms were not located on the lysine-acetyl side chains, but were present in amide NH groups located on the backbone of the polypeptide; the side chain NHCOCH3 proteins exchanged rapidly on the time-scale of the experiments. Although the most negatively charged rungs of the ladder had the slowest rates of global10 H/D exchange, an analysis with differential scanning calorimetry showed that these rungs also had lower conformational stability than the lower (less acylated) rungs.
Hydrogen Exchange as a Tool for Studying the Structure and Folding of Proteins
The rate at which a protein exchanges its backbone amide hydrogens with tritium or deuterium in buffer has been used for nearly 60 years11,12 to study the structure 13–18, folding 19, and conformational stability20–22 of proteins. In fact, the first measurements of H/D exchange were not made with any form of spectroscopy, but rather by determining the density of H2O droplets after the addition of deuterated protein (which had been flash-frozen as a function of time in D2O, and then dried under vacuum with P2O5).12 The utility of hydrogen exchange in protein biochemistry is based on the generally observed correlation between the rate of amide hydrogen exchange and i) the rate of protein folding, ii) the local structure surrounding a backbone amide, and iii) the conformational stability of the folded protein.22,23 In spite of the historic and now widespread use of hydrogen exchange in structural biology and biochemistry—and in spite of all that is known about the processes of H/D exchange in proteins—the reasons why many amide hydrogen atoms are slow to exchange in folded polypeptides (and other types organic molecules for that matter 24–26) are still not completely understood.27
The exchange of amide hydrogens with aqueous solvent is catalyzed by both acid and base, and the minimum rate of exchange for an unstructured polypeptide occurs at ~ pH 2.5.28 Above pH 4, the primary catalyst for amide hydrogen exchange is hydroxide 29 (the pKa of the backbone amide in an unstructured polypeptide is ~ 15); below pH 4, the exchange is catalyzed by hydronium. In the case of an unstructured polypeptide, the exchange of amide hydrogen with solvent is fast: it occurs in milliseconds to seconds at pH 7 and room temperature.30 With a folded or structured protein, however, the rate of exchange can be slower by factors of 108 (at pH 7 and room temperature).31,32
A simple kinetic model, developed by Linderstrøm-Lang, has been used for decades to model the kinetics of amide hydrogen exchange in folded proteins.12,33 This model (summarized in Equation 1) involves a transition between two states: “open” and “closed”. Hydrogen exchange occurs in the “open” state and not in the “closed” state.
| (1) |
In Equation 1, kint refers to the rate constant for the exchange of an amide in an unstructured polypeptide (i.e., the intrinsic rate of exchange); kcl refers to the rate constant for a closing reaction (e.g., refolding or a change in conformation). The intrinsic rate of hydrogen exchange for all 20 amino acids have been measured (as a function of temperature and pH) using model peptides.34,35
The reaction scheme in (1) can occur at two extremes: i) kcl >> k int; that is, the closing reaction (such as folding or a change in conformation) is much faster than the intrinsic rate of exchange; and: ii) kcl <<kint. These two conditions, described by equation 2 and equation 3, are termed EX2 (e.g., kobs depends on two terms) and EX1 (kobs depends on one term). 33,36,37
| (2) |
| (3) |
The two most widely accepted theories for understanding how various amides undergo hydrogen exchange in proteins are known as “local unfolding”23,29 and “solvent penetration”.38,39 These models theorize that amide hydrogens exchange slowly in folded proteins because of hydrogen bonding (the local unfolding model) or burial in the protein and physical protection from contact with solvent and catalysts (the solvent penetration model). The local unfolding model postulates that local fluctuations in protein structure permit exchange by separating NH and CO groups (by ≥ 5Å) that are H-bonded in α-helices or β-sheets23; the solvent penetration model postulates that water or a catalyst permeate the protein (without its unfolding, per se). These two theories are not mutually exclusive; Dill has reviewed data in support of each.40
The results of studies of hydrogen exchange on small-molecule amides and model peptides have made it reasonable, in our opinion (and in the opinion of others 25,41,42 ), to suspect that amides in closed configurations (Eq. 1) are not protected from hydrogen exchange entirely because of hydrogen bonding and solvent accessibility, but that electrostatic effects can also, in some cases, greatly affect the rate of H/D exchange. Hydrogen exchange studies of model amides (i.e., N-methyllauramide and N-methylbutyramide) in the presence of cationic, neutral, and anionic micelles have suggested that the electrostatic environment of an amide affects its rate of hydrogen exchange.25 For example, the rate constant for base-catalyzed exchange (kOH) of model amides decreased by factors of 2500 in the presence of negatively charged micelles, whereas kH increased 100 fold.25 Decreases in kOH were not observed in the presence of neutral or cationic micelles (although a 30-fold decrease in kH was observed in the presence of cationic micelles). The relative rates of hydrogen exchange for diketopiperazine and 2-piperidone (the mono-amide analog of diketopiperazine) are also interesting: the kOH of diketopiperazine is ~ 740 times greater than for 2-piperidone.41
Confounding matters even further, the pioneering studies that used model amino acids and oligopeptides to determine the effect of primary structure on rates of amide hydrogen exchange were performed at high concentrations of salt (i.e., 0.5 M KCl) in order to “shield possible charge effects”.35,43 We believe, however, that understanding electrostatic effects on amide hydrogen exchange in proteins is necessary to understand them mechanistically and to understand what they reveal—and what they do not necessarily reveal—of the structure and folding of proteins.
Our lack of an understanding of amide H/D exchange in proteins is ultimately demonstrated by the known examples of amino acid residues located on the surface of proteins (i.e., lysozyme and rubredoxin) whose backbone amides are exposed to solvent and not H-bonded, but undergo H/D exchange only very slowly (e.g., a billion-fold more slowly than the rate of the corresponding model oligopeptides). These types of slow-exchanging, surface residues (i.e., Val 38 in Pyrococcus furiousus rubredoxin27) exchange as if they were in the hydrophobic core of the protein or engaged in strong H-bonds. A series of recent papers has suggested that variations in the electrostatic potential at the surface of proteins such as rubredoxin might explain why some solvent-exposed amino acids that are not hydrogen bonded exchange so slowly 27,44,45 (and why other amides exchange more rapidly than would be expected based upon their deep burial from solvent and H-bonding 46). The surface properties of a protein—especially the surface charge and hydrophobicity—have never been systematically changed, experimentally, in order to test how such properties might affect the rate of H/D exchange in folded proteins.
Using Protein Charge Ladders and Mass Spectrometry to Determine How Surface Properties Affect H/D Exchange in Proteins
There are few experimental tools available with which to study how the electrostatic environment of amides in folded proteins—especially the surface properties of the protein—might influence their rates of H/D exchange. Much of the previous work investigating electrostatic effects in the rate of H/D exchange of folded proteins has compared the rate of exchange at different values of pH 47,48 or ionic strength. 49,50 In these types of experiments, any change in the rate of exchange of amide hydrogens that occur with pH or ionic strength (e.g., kobs in Eq. 1) is compared to changes in the intrinsic rate of exchange (e.g., kint in Eq. 1); it is difficult, however, to determine the origin of effects observed in this sort of experiment. A change in pH can, inter alia, change the structure of a protein, in addition to changing its net charge51,52 (serum albumin, for example, undergoes distinct changes in conformation at pH 2.7, 4.3, 7.0, ~8 and ~10).53
A protein charge ladder provides a straightforward and internally consistent tool with which to study how the surface properties of a folded protein might affect the rate of H/D exchange, independent of the structure of the protein. By acylating lysine-ε-NH3+ groups at the surface of BCA II with different acylating agents (i.e., acetic or butyric anhydride) we can systematically change the electrostatic potential and hydrophobicity at the surface of this protein without altering its secondary or tertiary structure.2 The rates of H/D exchange of all 19 rungs can be measured simultaneously using mass spectrometry; each charge isomer is, therefore, measured under conditions of identical buffer pH, ionic strength, temperature, and deuterium concentration. BCA II has 27 positively charged residues: 18 lysine and 9 arginine residues. There are 30 negatively charged residues: 19 aspartate and 11 glutamate residues. The side chains of all 18 lysine residues are solvent exposed.2 All 18 lysine residues are equally dispersed in α-helical, β-strand and loop motifs.6 The N-terminal serine residue is acetylated (when isolated from bovine erythrocytes). Each rung of the charge ladder is probably composed of an approximately statistical mixture of regioisomers.54 Bringing about changes in net surface charge of ~ 16 units with conventional methods such as site-directed mutagenesis or changes in pH is possible alternative to experiments using charge ladders, but would require multiple rounds of mutagenesis or changes of several units in pH.
Experimental Procedures
BCA II Charge Ladders
Lyophilized bovine carbonic anhydrase II (E.C. 4.2.1.1) was purchased from Sigma, and dissolved in 100-mM HEPBS buffer (pH 9.0) for reaction with acetic anhydride or butyric anhydride. Lysine-acetyl charge ladders of BCA II were produced by allowing BCA II to react with different amounts of acetic anhydride as previously described.5 Protein charge ladders were repeatedly concentrated and were diluted in 10 mM phosphate (pH 7.4) using a Centricon centrifugal filtration device (10,000 MW; Millipore) in order to remove HEPBS buffer and acetic acid. Aliquots of acetylated BCA II (80 µM; 10 mM phosphate, pH 7.4) were flash frozen with N2 (l) for analysis with ESI-MS, capillary electrophoresis (CE) and differential scanning calorimetry (DSC). The degree of lysine acetylation was determined with ESI-MS and CE. The perbutyrated derivative of BCA II was produced and characterized using the same procedure as the peracetylated protein, with the exception of the duration of reaction. Butyric anhydride is less soluble in water than acetic anhydride, and the reaction (as a suspension) was allowed to proceed for 2 days at 4 °C.
Measuring Hydrogen-Deuterium Exchange of Protein Charge Ladders with Mass Spectrometry
H/D exchange was measured with mass spectrometry as previously described with minor modifications that are described21 in the supplemental material.
Distinguishing Backbone and Lys-ε-NHCOCH3 Amides in CAII with Multidimensional NMR
One difficulty that arises from using a Lys-NH3+ protein charge ladder to study the exchange of amide hydrogen in proteins is that the acetylation of lysine-ε-NH3+ generates an additional amide hydrogen on the lysine side chain (i.e., lys-ε-NH3+ + (CH3CO)2O ➔ lys-ε-NHCOCH3 + CH3COOH + H+). The mass spectrometric tools that we use to measure H/D exchange can not distinguish the amide hydrogen on an acetylated side chain from amide hydrogen on the backbone. We have, therefore, also used multi-dimensional NMR (which can distinguish side chain and backbone) to measure the rate of amide hydrogen exchange specifically at the acetyl side chains of acetylated lysine residues.
The NMR experiments were carried out on Bruker 600 MHz and 750 MHz spectrometers equipped with cryoprobes. Sensitivity-enhanced TROSY version55 of the HSQC experiments were used to record HSQC spectra.56 For D2O exchange experiments, a concentrated sample of HCA II (3.5 mM) in water was diluted 10-fold in deuterated buffer (pD 7.4, 10 mM phosphate). In order to obtain a “zero-time point”, an aliquot of the same concentrated sample was diluted 10 fold in buffered H2O (pH 7.4, 10 mM phosphate). A control spectrum in water was recorded under identical conditions. A detailed description of the experimental parameters of NMR experiments is included in the Supplemental section.
The H/D exchange of lys-ε-NHCOCH3 in HCA II was exclusively measured by recording the first HN plane in an HNCO experiment.57 This two dimensional experiment (referred to as 2D–HN-HNCO) is the first plane of the HNCO experiment, in which there is no evolution of the carbonyl frequency. This 2D experiment will exclusively detect the Nitrogen-Proton correlation of those amides that are directly attached to a 13C-enriched carbonyl group. This selection in the H-N plane relies on the preparation of isotopically enriched HCAII in which only the amides of the lysine side chain are attached to a 13C carbonyl. This selective enrichment is achieved by growing cells in 12C glucose media and acetylating the purified protein with acetic anhydride that is 13C enriched only at the carbonyl position. A TROSY version of the HNCO experiment where the nitrogen dimension is incremented in a semi-constant time fashion was employed to collect 2D–HN-HNCO planes. Hydrogen-deuterium exchange experiments were carried out as described above, where each H-N plane was recorded in 15 min. The data were processed with NMRPipe and the intensities of the peaks were measured using the program Sparky.58
Recombinant Expression and Purification of 15N–labeled Carbonic Anhydrase II
Human carbonic anhydrase II (HCA II) was recombinantly expressed in E. coli and purified as previously described.59 E. coli cells expressing HCA II were grown in M9 minimal media enriched with 15NH4Cl in ~90% D2O. The cells were grown to an OD600 of 0.7 at 37 °C and induced for 10–12 hrs at 30 °C with 1.5 mM IPTG. Zinc chloride (ZnCl2, 200 µM) was added before induction. The cells were lysed by sonication and centrifuged. HCA II was purified as previously described.59 In order to remove deuterium from labile sites, solutions of purified protein were heated in phosphate-buffered H2O (10 mM, pH 8.4) at 35 °C for 2 days. Proteins were then transferred to 10 mM phosphate buffer (pH 7.0) with centrifugal filtration devices and stored at 4 °C for H/D exchange experiments.
Measuring the Effects of Lysine Acetylation on the Rate of Backbone Amide H/D Exchange in Model Peptides
In order to determine how neutralizing the ε-NH3+ of lysine by acetylation affected the rate of H/D exchange of the backbone amide of lysine, we used NMR spectroscopy to compare the hydrogen exchange of N-α-acetyl-L-lysine-N-methylamide (abbreviated: Ac-Lys(ε-NH3+)-NHMe) and the analogous ε-NHCOCH3 derivative (abbreviated: Ac-Lys(ε-NHCOCH3)-NHMe). These derivatives of lysine are ‘models’ of lysine in polypeptides in that the α-NH3+ group has been acetylated (yielding a “backbone” amide; the ε-NH3+ group is also acetylated in the ε-NHCOCH3 derivative yielding a side chain amide). The α-COO− group has also been converted into a CONHCH3 (yielding a second “backbone” amide). The rate of H/D exchange of each amide was measured at pD 4.5, 5 °C, 20 mM acetate. The rate of amide hydrogen exchange was also measured at pH 7.1 using magnetization transfer 1H NMR (the rate of amide H/D exchange is too fast at neutral pH to be measured by the addition of D2O). In order to assign the resonances of “side chain” from “backbone” amide groups at each pH, we acquired homonuclear 2D–NMR spectra on a 900 MHz spectrometer (Bruker). Nuclear Overhauser effect spectroscopy (NOESY), Correlation Spectroscopy (COSY), and Total Correlation Spectroscopy (TOCSY) were performed in order to assign NMR signals unambiguously to particular amides in two relevant structures: Ac-Lys(ε-NH3+)- NHMe and Ac-Lys(ε-NHCOCH3)-NHMe. Additional experimental details can be found in the supporting information.
Differential Scanning Calorimetry (DSC)
To determine the effect of lysine acetylation on the conformational stability of BCA II, partial charge ladders were analyzed by differential scanning calorimetry (DSC). DSC was carried out on a VP-DSC instrument (MicroCal) with a scan rate of 1 °C/min. Protein samples (~25 µM; pH 7.4, 10 mM phosphate) were degassed prior to analysis. Raw DSC data was smoothed and deconvoluted using Origin 7.0 (MicroCal).
Capillary Electrophoresis (CE)
The change in surface charge for each rung of the charge ladder was confirmed by capillary electrophoresis (CE). Capillary electrophoresis was performed as previously described using a Beckman PACE instrument.60
Results and Discussion
Preparation and Characterization of Charge Ladders of Bovine Carbonic Anhydrase II
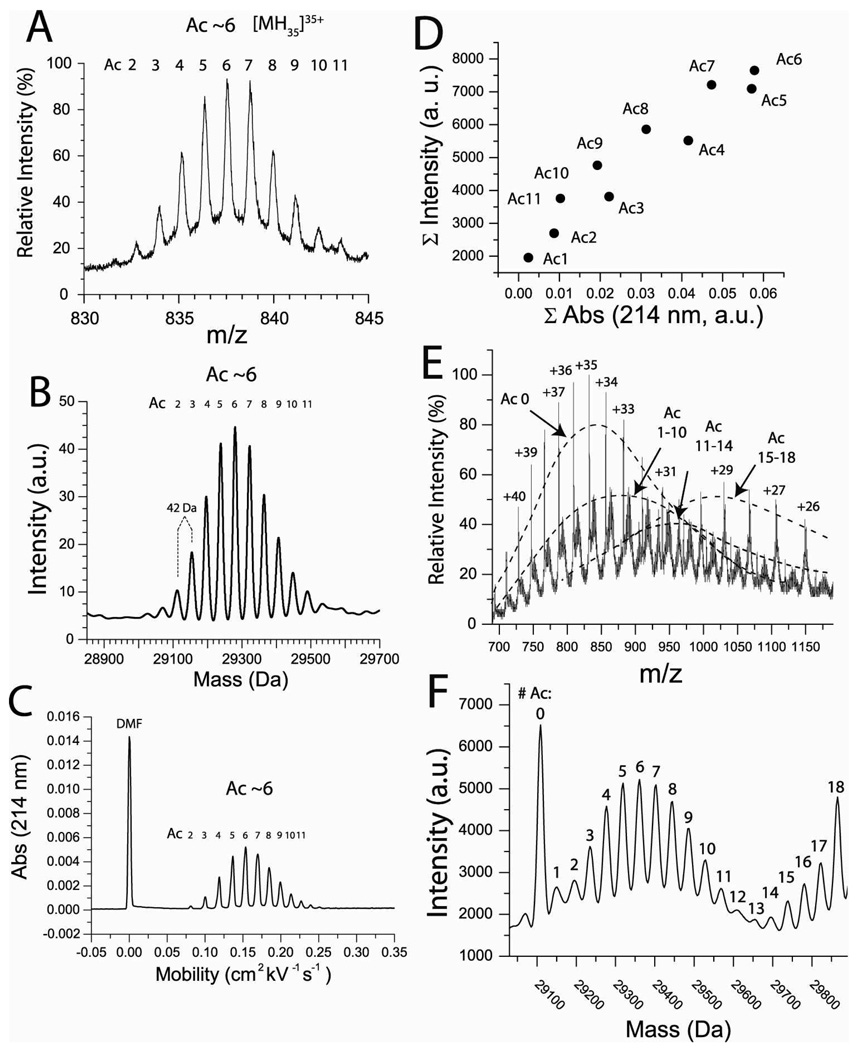
Proteins with different degrees of acetylation (as measured by ESI-MS and CE) were prepared by causing BCA II to react with different molar equivalents of acetic anhydride. For example, a solution of proteins having four to eight lys-ε-NH3+ groups acylated was prepared by reaction with eight molar equivalents of acetic anhydride (Figure 1); the most abundant species had ~ six modifications (according to mass spectrometry and capillary electrophoresis, Figure 1A–C). This partial charge ladder was denoted “BCA-Ac(~6)”. The relative abundance of each rung is similar when analyzed by either mass spectrometry or capillary electrophoresis (during CE, proteins are detected by their absorbance at 214 nm). This similarity in abundance when solutions were analyzed by CE and ESI-MS demonstrated that each rung had a similar ionization efficiency during electrospray ionization (Figure 1D).
Figure 1. Electrospray ionization mass spectrometry (ESI-MS) of a lys-ε-NHCOCH3 charge ladder of BCA II.
A) Rungs of the lys-ε-NHCOCH3 charge ladder are well resolved with electrospray ionization mass spectrometry (ESI-MS). The +35 molecular ion (A) and the mass reconstruct (B) are shown for a partial charge ladder (in H2O) having between 2 and 11 acetylated lysines (the most abundant rung is Ac(6)). C) Capillary electrophoresis of the same sample shows a similar distribution of acetylated proteins (between 2 and 11 modifications; the most abundant species had 6 modifications). D) Integrated values of intensity (from the mass spectra in B) for Ac(1)-Ac(11) plotted against integrated values of absorbance (from the electropherogram in C). The approximately linear correlation demonstrates that each rung of the charge ladder has a similar ionization efficiency during ESI-MS (although the higher rungs have lower relative values of absorbance due to their greater mobility during CE). E) The mass spectra of a full protein charge ladder was prepared by mixing partial charge ladders with unmodified and peracetylated BCA II. The charge state distribution of BCA II is shifted to higher m/z values (i.e. lower positive charge states) as the degree of acetylation increases. The predominant charge states are +34 to +37 for unmodified BCA II and +30 to +28 for peracetylated BCA II. F) Mass reconstruct of spectra of full charge ladder showing all 19 rungs (in D2O).
We analyzed the partial charge ladders denoted “BCA-Ac(~6)”, “BCA-Ac(~9)” and the peracetylated protein (with 18 lysine modifications; denoted BCA-Ac(18)) using differential scanning calorimetry (DSC), in order to determine how the acetylation of lysine affected the thermostability of folded BCA II. Combining various partial ladders with unmodified and peracetylated BCA II resulted in a full charge ladder with 19 rungs: 18 variously acetylated derivatives and the unmodified protein (Figure 1E, F).
Higher (More Highly Charged) Rungs of the BCA II Charge Ladder have Lower Conformational Stability
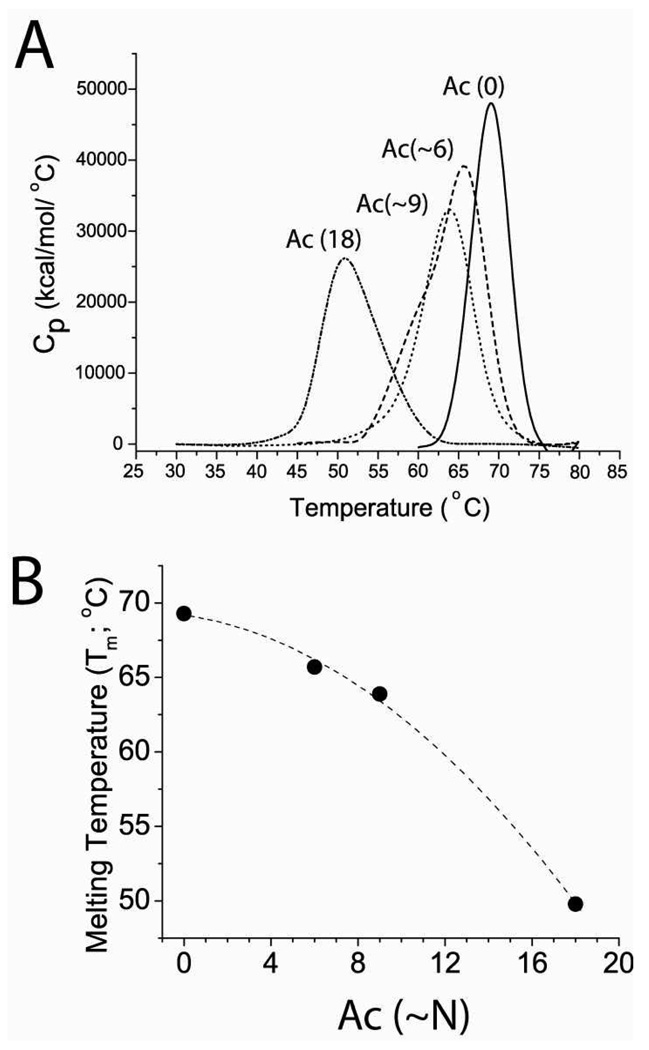
The thermal denaturation of unmodified BCA II (denoted BCA-Ac(0)), peracetylated BCA II (BCA-Ac(18)), and partial BCA II charge ladders produced well-defined changes in heat capacity. The endothermic transitions shown in Figure 2A were generated by deconvoluting the raw data (using Origin 7.0). Integration of each endotherm yielded temperatures of the melting transition (Tm). For BCA II-Ac(0), Tm = 69.3 °C; for BCA-Ac(~6), Tm = 65.7 °C; for BCA-Ac(~9), Tm = 63.9 °C; for BCA-Ac(18), Tm = 49.8 °C. We observed a non-linear relationship between the number of acetyl modifications to BCA II and its thermostability; the first ~9 modifications lower the Tm by 5.4 °C; the next 9 modifications, however, lower the Tm by 14.1 °C (Figure 2B).
Figure 2. Lysine acetylation decreases the thermostability of BCA II.
A) Thermal denaturation of unacetylated BCA II (denoted Ac(0)), partially acetylated BCA II (Ac(~6) and Ac(~9)) and peracetylated BCA II (Ac(18)) measured by differential scanning calorimetry (DSC). Integration of peaks produced melting temperature (Tm) values of 69.3 °C (Ac(0)), 65.7 °C (Ac(~6)), 63.9 °C (Ac(~9)), and 49.8 °C (Ac(18)). B) Plot of Tm of Ac(0), Ac(~6), Ac(~9), Ac(18) versus the average number of modifications.
Measuring Amide H/D Exchange in a BCA II Charge Ladder with Electrospray Ionization-Mass Spectrometry (ESI-MS)
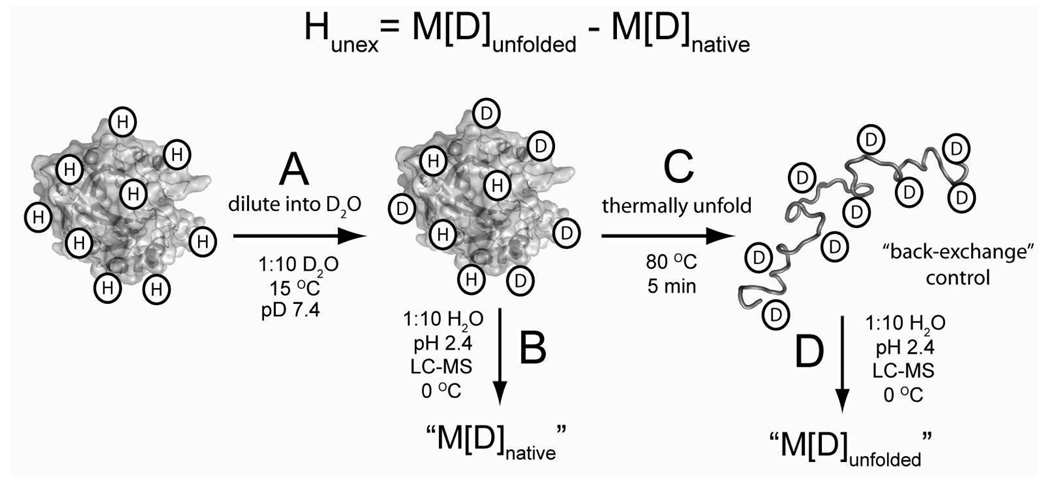
The mass spectrometric method that we use to measure H/D exchange (illustrated in Figure 3) will measure the global exchange of hydrogen in BCA II, but can not distinguish individual residues.61 We expressed the kinetics of H/D exchange for each rung of the charge ladder in terms of its number of unexchanged hydrogens (as opposed to the number of exchanged hydrogen or incorporated deuterons) because the number of unexchanged hydrogens provides a more accurate description of the overall structure of a protein than does the number of exchanged hydrogens or incorporated deuterium.62 The number of unexchanged hydrogens (denoted Hunex in Equation 4) for each rung is calculated by subtracting the measured mass of each rung throughout the H/D exchange experiment (denoted M[D]native; illustrated in Figure 3B) from the measured mass of each perdeuterated rung (denoted M[D]unfolded; Figure 3C).
Figure 3. Measuring the amide hydrogen-deuterium exchange of proteins using liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS).
A) H/D exchange was initiated by diluting concentrated protein solutions (1:10 v/v) from buffered H2O into buffered D2O. B) The mass of the protein was measured as a function of time by quenching the isotopic exchange of an aliquot with low pH buffer (pH 2.4, 100 mM PO43−) and injection onto an LC-ESI-MS apparatus that was equilibrated at 0 °C (he ionization solvent used in LC-ESI-MS is 0.3 % formic acid, 49.85 % acetonitrile and 49.85 % H2O). We refer to this measured mass as M[D]native. C) The perdeuterated protein is prepared by thermally unfolding an aliquot from B and measuring the mass. We refer to this measured mass as M[D]unfolded. Deuterons on side chain functionalities (OH, -COOH, -NH3+, -C(NH2)2+) rapidly exchanged with H2O during the analysis with HPLC-MS (e.g., during steps “B” and “D”) and cannot be measured with this method. The number of unexchanged amide hydrogen (Hunex) at any given time during the experiment is calculated as Hunex = M[D]unfolded – M[D]native.
| (4) |
Hydrogen/deuterium exchange is initiated and measured as previously described.63 Briefly, a concentrated solution of protein charge ladder was diluted ten-fold from buffered H2O (20 mg/mL protein, 15 °C, 10 mM PO4 3−, pH 7.4) into buffered D2O (15 °C, 10 mM PO4 3−, pD 7.4; Figure 3A; see supplemental information for additional experimental details). Aliquots were removed over time, and isotopic exchange was immediately quenched by diluting aliquots again (1:10) into ice-chilled, acidic, aqueous buffer (0 °C, 100 mM PO4 3−, pH 2.4). Solutions were then injected onto a short HPLC column (in order to remove salts that suppress ESI) that was chilled on ice and coupled to the ESI-MS (Figure 3B). During quenching and analysis with LC-ESI-MS, deuterons on side chain functionalities such as carboxylic acid, indole, guanidinium or alcohol groups will typically undergo back-exchange with water.64,65 The LC-ESI-MS methods we use will therefore measure the exchange of hydrogen primarily at the amide nitrogen, and not hydrogen on rapidly exchanging groups. 64,66 A substantial number of amide deuterons, however, will also undergo “back-exchange” with solvent during quenching and LC-ESI-MS (Figure 3B). Consequently, the number of deuterons that are incorporated into BCA II during the in-exchange experiment (Figure 3A) will be underestimated unless this back-exchange is taken into account. The percent of deuterons that undergo back-exchange (% BE) is calculated as the difference between the measured mass of each perdeuterated rung and the theoretical mass of each perdeuterated rung; the perdeuterated ladder is prepared by thermally unfolding the proteins (Figure 3C). We found that approximately 27 % of amide deuterons had undergone back-exchange with solvent during quenching and analysis with LC-ESI-MS (Supplemental Table 1). This value is consistent with reported values that involved similar LC-ESI-MS methods.63
We emphasize that the acetylation of lysine results in an additional amide (lys-ε-NH3+ + (CH3CO)2O ➔ lys-ε-NHCOCH3 + CH3COOH +H+). Consequently, we expect the maximum number of deuterons that can be incorporated into amide sites of unfolded BCA II to increase (in 90 % D2O) by ~1 with each additional modification. This result is in fact what we observed by thermally unfolding the charge ladder in deuterated buffer and measuring the mass of each rung (Supplemental Table 1). The number of deuterons incorporated into unfolded BCA II increased by ~1 with each additional acetylation (Supplemental Table 1, Supplemental Figure 1C). The incorporation of deuterons into the acetyl amide of lys-ε-NHCOCH3 can, therefore, be measured with ESI-MS, and are retained during liquid chromatography and mass spectrometric analysis.
H/D Exchange of the BCA II Charge Ladder
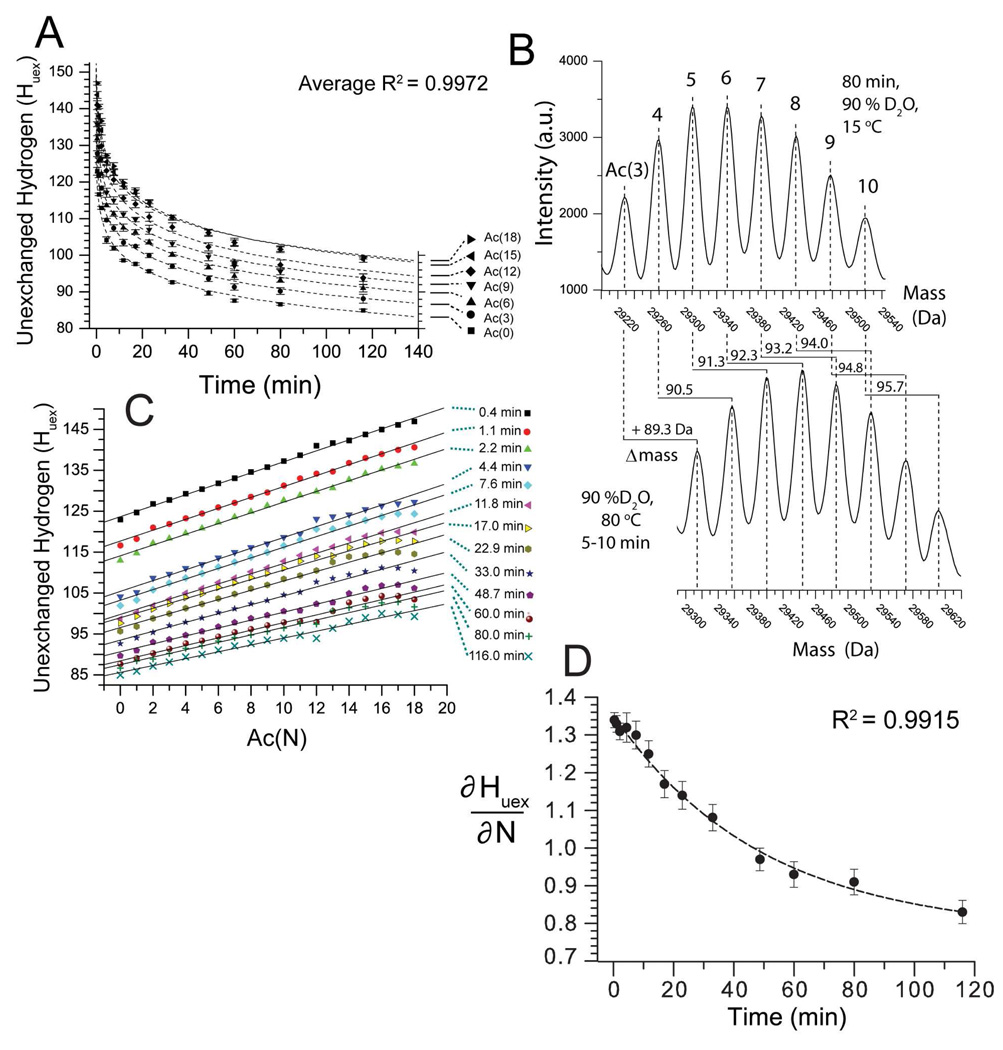
Figure 4A shows the kinetics of H/D exchange of the charge ladder monitored by mass spectrometry. In the case of the first rung, BCA-Ac(0), there are 37 hydrogen atoms that exchange with solvent before the first time point (typically ~20 s). These hydrogens exchange rapidly with solvent because, presumably, they are solvent accessible and/or located in loosely structured regions. Approximately 85 hydrogens in BCA-Ac(0) remain unexchanged with solvent after 100 minutes (90 % D2O, pD 7.4, 15 °C). These 85 hydrogen exchange slowly because, we assume, they are hydrogen bonded and/or are buried from solvent (according to the ‘solvent penetration’ or ‘local unfolding’ models of H/D exchange).40
Figure 4. Lysine acetylation decreases the rate of H/D exchange of BCA II as measured by ESI-MS.
A) H/D exchange kinetics of the BCA II charge ladder (90 % D2O, pD 7.4, 15 °C). For clarity only BCA-Ac(0), (3), (6), (9), (12), (15) and (18) are shown. BCA-Ac(0) retained ~85 unexchanged hydrogens after 100 minutes in D2O. The higher rungs are more protected from H/D exchange than the lower rungs. BCA-Ac(3) retained ~ 87 unexchanged hydrogens after 100 minutes; BCA-Ac(6), 90; BCA-Ac(9), 93 and BCA-Ac(12), 97. The last four rungs, BCA-Ac(15) through BCA-Ac(18) have nearly superimposable exchange profiles and retain ~ 100 unexchanged hydrogens after 100 min. Error bars represent the standard deviation of average mass values calculated from seven charge states for each rung. B) Mass reconstruct showing BCA-Ac(3) through BCA-Ac(10) after 80 minutes in 90% D2O, pD 7.4, 15 °C (top) and after the same sample is heated and the proteins are unfolded (bottom). Each higher rung incorporates (after 80 minutes) approximately one additional deuteron upon thermal unfolding as observed by an increase of 0.8–1.0 Da in the mass for each rung. C) Plot of number of unexchanged hydrogen (Huex) for each rung of the charge ladder (N) after various periods of time in D2O, ranging from 0.4 min to 116 min (e.g., ∂Huex/∂N). D) The slope of each line in Figure 4C (denoted ∂Huex/∂N) is plotted as a function of time (t) and fit with the single exponential function y = yo + Ae-kx. The ∂Huex/∂N value is decreasing exponentially with time with a half life t1/2= 34.1 min. The quantity ∂2Huex/∂N∂t expresses the magnitude by which the acetylation of lys-ε-NH3+reduces the aggregate rate of H/D exchange in BCA II. The error bars represent the standard deviation that was calculated from the linear fit of each data set in Figure 4C.
For visual clarity, Figure 4A shows the kinetic profile for only seven rungs of the charge ladder (e.g., BCA-Ac(0), (3), (6), (9), (12), (15) and (18); data for all 19 rungs are included in supporting information). Figure 4A shows that the number of unexchanged hydrogens (after 100 min, pD 7.4, 15 °C) in the charge ladder increases from ~85, for the first rung (BCA-Ac(0)), by approximately one hydrogen for each additional rung. The sixteenth rung (BCA-Ac(15)), for example, has 101 unexchanged hydrogens after 100 min in D2O (Figure 4A, see also Supplemental Table 1).
We also fitted the kinetic plots in Figure 4A with triexponential functions in order to extract kinetic parameters for “fast,” “medium” and “slow” exchanging hydrogens, as previously described63 (we found that a triexponential function fit the plots in Figure 4A better than a biexponential function; the average R2 value for the triexponential fit was 0.9972 (Figure 4A) and the average R2 = 0.9841 for biexponential fits). This analysis is described in the supplementary material, and the results for all 19 rungs of the charge ladder are listed in Supplemental Table 2. From this kinetic analysis we can estimate the reduction in the rate of H/D exchange that occurs from acetylation, if we assume (for the moment—the matter is solved experimentally below; see Figures 5 and 6) that i) the additionally protected hydrogens are located on the backbone of BCA II and not on the side chain of acetylated lysine residues, and ii) that the amide hydrogens that become protected by acetylation were not protected from exchange before acetylation. Our kinetic analysis of the plots in Figure 4A and Supplemental Figure 1 indicates that unmodified BCA II has approximately 36.2 ± 3.6 amide hydrogens that exchange, with an average rate constant k = 5.8 ± 3.6 · min−1; there are 24.3 ± 2.9 amide hydrogens that exchange more slowly with an average rate constant k = 1.9 ± 0.5 · 10−1 min−1; and 96.4 ± 2.7 amide hydrogens that exchange slowly with an average rate constant k = 1.9 ± 0.5 · 10−3 min−1. For peracetylated BCA II (the kinetic results of the other 17 rungs are listed in Supplemental Table 2), 34.8 ± 3.1 hydrogens exchange with a rate constant k = 4.3 ± 1.1 · min−1; 29.1 ± 2.9 hydrogens with a rate constant k = 0.9 ± 0.2 · 10−1 min−1; and 112.5 ± 3.6 hydrogens with a rate constant k = 1.1 ± 0.3 · 10−3 min−1. The hydrogens in peracetylated BCA II that become protected by acetylation exchange, therefore, with a rate constant k = 1.1 ± 0.3 · 10−3 min−1. We do not know the rate constant at which these hydrogens underwent exchange prior to acetylation, but if we assume that the rate constant is between 5.8 ± 3.6 · min−1 and 1.9 ± 0.5 · 10−1 min−1 (e.g., the rate constants for “fast” and “medium” exchanging hydrogens in unmodified BCA II) then we can make a zeroth order approximation that each acetylation reduces the rate of H/D exchange of amides hydrogens in BCA II by at least two or three orders of magnitude.
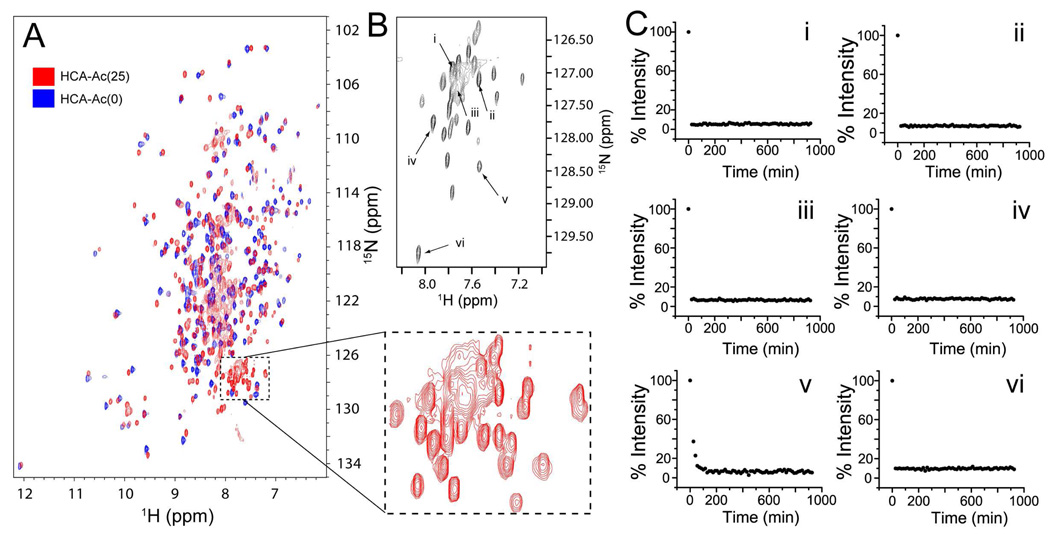
Figure 5. Hydrogen-deuterium exchange of peracetylated HCA II measured by multidimensional NMR.
A) Overlay of HSQC 1H-15N NMR spectra for HCA II (blue) and peracetylated HCA II (red; denoted HCA-Ac(25)). HCA II was acetylated with 13C-labelled acetic anhydride ((CH313 CO)2O). HCA II was expressed by E. coli in media enriched with 15N, 12C and 2H. In order to remove deuterons from non-alkyl groups, HCA II proteins were heated at 35 °C in H2O for 48 hours prior to analysis with NMR. The dashed box highlights, for HCA-Ac(25), a set of 22 resolved peaks and a broad set of overlapping peaks (containing 4–6 peaks) that are not observed in HCA-Ac(0). B) The 2D-HN-HNCO of a TROSY-HNCO experiment; each signal represents a correlated 1H-15N that is also correlated to 13C=O. C) Plots of the intensity of signals from the TROSY-HNCO(2D-HN-HNCO) experiment as a function of time (in deuterated buffer; 15 °C, pD 7.4, 10 mM PO43−). The H/D exchange of only six different amides (i–vi) are shown here (the remaining are shown in Supplemental Figure 3).
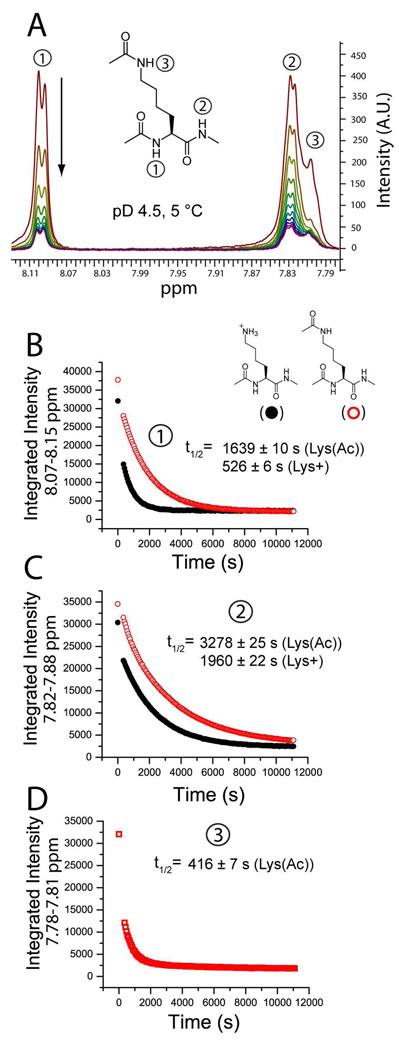
Figure 6. Neutralizing the ε-NH3+ group of Ac-Lys(ε-NH3+)-NHMe reduces the rate of H/D exchange at the backbone amide of the amino acid.
The rate of backbone amide exchange for model lysine compounds Ac-Lys(ε-NH3+)-NHMe and Ac-Lys(ε-NHCOCH3)-NHMe were measured with 1H NMR spectroscopy at pD 4.5, 5°C. A) The exchange of the amide hydrogen of the α-nitrogen (e.g., the ‘left-handed’ amide) was monitored in both compounds by the disappearance of the signal at ~ 8.10 ppm and the ‘right-handed’ amide by the signal at ~ 7.83 ppm. The ε-amide hydrogen (ε-NHCOCH3) of Ac-Lys(ε-NHCOCH3)-NHMe appears at ~ 7.80 ppm. B–D). The amide H/D exchange of both compounds expressed as a function of the integrated signal and time. Solid black circles represent Ac-Lys(ε-NH3+)-NHMe and open red circles represent the neutral compound Ac-Lys(ε-NHCOCH3)-NHMe. The function y = yo + Ae(-x/k) was fit to each plot. B) For the left handed amide (denoted “1”), the half-life of exchange (t1/2) = 526 ± 6 s for Ac-Lys(ε-NH3+)-NHMe (R2 = 0.9897) and 1639 ± 10 s for Ac-Lys(ε-NHCOCH3)-NHMe (R2 =0.9985). The ratio of half lives = 3.1. C) The neutralization of ε-NH3+ decreased the rate of exchange of the right handed amide (denoted “2”) by a lesser degree than the left handed amide. For Ac-Lys(ε-NH3+)-NHMe t1/2 = 1960 ± 22 s (R2 = 0.9958) and for Ac-Lys(ε-NHCOCH3)-NHMe t1/2 = 3278 ± 25 s (R2 = 0.9989); the ratio of half lives = 1.7. D) The ε-amide of Ac-Lys(ε-NHCOCH3)-NHMe (denoted “3”) exchanges faster than any of the backbone amide hydrogen: t1/2 = 416 ± 7 s.
Figure 4B shows that the mass distribution of each rung of the charge ladder remained unimodal and shifted gradually to higher values of mass as a result of deuteration—as opposed to being bimodal, with a lower mass peak (protonated protein) decreasing in intensity and a higher mass peak (deuterated protein) increasing in intensity. The unimodal distribution that we observed suggests that the exchange of most hydrogen of each rung occurs by a predominantly EX2 mechanism. Further support for an EX2 mechanism of exchange for the slowest exchanging hydrogen in both acetylated and unmodified BCA II is shown in Supplemental Figure 1B: the number of unexchanged hydrogen after > 5 min for both peracetylated BCA II and unmodified BCA II is similarily dependent upon the pD of solvent.
An analysis of the partial derivatives (∂Huex/∂N) of the H/D exchange data from Figure 4A revealed a linear relationship to exist between the number of acetylated lysine residues in BCA II and the number of unexchanged hydrogens retained by BCA II during the 2 h experiment in D2O. Figure 4C shows a plot of the partial derivatives (∂Huex/∂N) of the H/D exchange data from Figure 4A. Each set of data in Figure 4C expresses the number of unexchanged hydrogens for each rung of the charge ladder, after various periods of time in D2O. Each separate plot is labeled with the time of exposure of BCA II to D2O (ranging from 0.4 to 116 minutes). The number of unexchanged hydrogens in BCA II increased linearly—at all points in time during the 2 h experiment—with the number of acetylated lysine residues (denoted N); Figure 4C). Although the 13 plots in Figure 4C appear to be parallel, and the slopes of each fitted line (∂Huex/∂N) appear to be equal, the value ∂Huex/∂N is actually decreasing exponentially with time. Figure 4D plots the value ∂Huex/∂N as a function of the time (t) at which each set of data was collected. The value ∂Huex/∂N decreased exponentially from 1.35 Da·N−1 (at 0.4 min) to 0.86 Da·N−1 (at 116 min). Fitting the plot in Figure 4D with the function y = yo + Ae−kx yielded an R2 = 0.9915 and a time constant k = 34.1 min. The slope of the plot in Figure 4D (∂2Huex/∂N∂t) represents the extent by which The value ∂2Huex/∂N∂t can be expressed as function of ΔG‡ for the amide H/D exchange reaction, N(H) → N(D):
| (5) |
| (6) |
The quantity ∂2Huex/∂N∂t expresses the magnitude by which the acetylation of lys-ε-NH3+ reduces the aggregate rate of H/D exchange in BCA II. The exponential decrease in ∂Huex/∂N demonstrates that the protective effect of acetylation of lys-ε-NH3+against H/D exchange is diminishing exponentially with time. The negative slope of ∂2Huex/∂N∂t also suggests—but does not prove—that the acetylation of lysine residues has a greater effect on the fastest exchanging amide hydrogens in BCA II (e.g., surface hydrogens that exchange first during the two hour experiment) than on the slowest exchanging amide hydrogens (e.g., those that might be in the hydrophobic core).
It is important to remember that each rung of the charge ladder represents, to an extent that depends on the number of acetylations, a mixture of regioisomers. The kinetics of H/D exchange measured for each rung (Figure 4 Supplemental Table 1 and 2) are, therefore, a population-weighted average value that represents the H/D exchange of all regioisomers within the rung. Mass spectrometry is the only tool that can measure the rate of H/D exchange of an entire protein charge ladder, but it has one significant drawback: the results of MS do not tell us if the rate of exchange of a specific amide, or a set of amides, is changed as a result of acetylation. Although the increase in the number of unexchanged hydrogens for each higher rung is small (e.g., < 2)—and this value strongly suggests that the rate of a only a few amides are affected by acetylation—these values are determined from a measured change in mass (e.g., M[D]unfolded and M[D]native; see Eq 4, and Supp. Table 1). It is possible, therefore, that the acetylation of a single lysine residue resulted in a reduction in the rate of exchange of > 2 amides and it is the net reduction in rate—as measured with ESI-MS—that appears as an aggregate value.
We also point out that the results shown in Figure 4 cannot be the result of an unexpectedly fast back-exchange of the side chain amide hydrogen on lysine-ε-NHCOCH3 during the LC-MS analysis at pH 2.4 (a plausible artifact). The H/D exchange of the side chain amide groups can be measured by our LC-MS apparatus: the number of deuterons that can be incorporated into thermally denatured BCA II increases by approximately 1 per each acylation, from 160.6 for Ac(1), to 176.4 for Ac(18) (see Supplemental Table 1). A complete list of the masses of the perdeuterated forms of each rung of the charge ladder (denoted “M[D]Unfolded ”) as well as the number of deuterons that can be incorporated into the thermally denatured protein (“M[D]Unfolded – M[H]”) are listed in Supplemental Table 1. A plot of these values can be found in Supplemental Figure 1C.
Are the Additional Protected Hydrogens Located on the Protein Backbone or Side Chain of Lys-ε-NHCOCH3?
A partial explanation for the effects of acetylation on the rate of H/D exchange within the charge ladder is that the additional hydrogen that were protected in each rung include those amides introduced onto the side chain of lysine via acetylation. All 18 lysine in BCA II are, however, located on the surface of the protein and X-ray crystallography of peracetylated BCA II revealed that the lys-ε-NHCOCH3 groups are neither buried nor inaccessible to solvent.9 A few of these amide groups were, however, engaged in intramolecular hydrogen bonds, for example, with aspartate-β-COO− functionalities.9 Nevertheless, the rate of exchange of lys-ε-NHCOCH3 are difficult to predict and not necessarily slower than backbone –CONH hydrogen.
In order to determine if the additional hydrogens that were protected in the charge ladder included those amide hydrogen on lys-ε-NHCOCH3 (and not backbone amide hydrogen), we measured the rate of H/D exchange of lys-ε- 15 NH 13 COCH3 on peracetylated CA II using multidimensional NMR. For these experiments, we recombinantly expressed, and purified, carbonic anhydrase II proteins that were labeled with 15 N and 12C, and then acetylated these proteins, after purification, with 13 C-labelled acetic anhydride ((CH313 CO)2O). We used the human variant of carbonic anhydrase II (HCA II) for NMR experiments because an expression system was readily available. The human variant has 24 lysines and, when expressed in the prokaryotic expression system, is not acetylated at its N-terminus (there are, therefore, 25 R-NH3+ rather than 18 for BCA II). HCA II is similar to BCA II in its sequence and structure 67,68 (e.g., the two proteins have 79 % sequence homology); the net negative charge of HCA II at pH 8.4 was measured with CE to be −2.3; the net charge of BCA II was measured to be −3.3 under similar conditions.49
Using a TROSY (Transverse Relaxation Optimized Spectroscopy) version of the HSQC (Heteronuclear Single Quantum Coherence) NMR experiment, we have demonstrated that amide hydrogens on lys-ε-NHCOCH3 exchange rapidly with solvent (t1/2 << 30 min, 15 °C, pD 7.4) and are not included in the set of hydrogens that became protected as a result of acetylation (Figure 5). This result demonstrates that the additional hydrogen that were protected in each rung of the charge ladder are main-chain amides located somewhere on the backbone of the protein (Figure 5). An explanation of the results of NMR experiments that led us to this conclusion is as follows.
An overlay of the HSQC NMR spectra of HCA II (blue) and peracetylated HCA II (red) is shown in Figure 5A. A HSQC experiment correlates the amide nitrogen with the amide hydrogen; a cross peak is observed for each amide N-H pair. Each peak, therefore, represents an amide N-H that is in a unique chemical environment. Other N-H signals (i.e., δ-guanidino, β- and γ-CONH2, ε-NH3+, δ-NH, β-imidazole and β-indole) are typically not observed because they exchange too rapidly to be detected, or because they are suppressed in these types of experiments. 69 Many peaks in the HSQC spectra of HCA II and peracetylated HCA II do not overlap (this difference is not surprising, considering that the chemical environment of many residues will be altered by the neutralization of charge in the conversion of lys-ε-NH3+ to lys-ε-NHCOCH3). The majority of peaks in the spectra of both proteins were dispersed and well resolved, indicating that both proteins were folded.69,70
The HSQC spectrum of HCA II included approximately 260 observable peaks; this observation indicates that approximately 260 amide N-H species in HCA II were in unique chemical environments. This number is approximately equal to the number of amides in the HCA II polypeptide: HCA II has 259 residues; 17 are proline residues (which do not have backbone NH groups and are not observable in this type of HSQC spectrum). There are, therefore, 242 backbone amides in HCA II. This difference of 18 peaks is plausibly due to amide residues that exist in more than one sterioisomeric form. The HSQC spectrum of peracetylated HCA II contained 310 peaks (Figure 5A, red spectrum). The additional 50 peaks in the spectrum of peracetylated HCA II are due, in part, to the 25 additional amides introduced by acetylation. The additional ~ 25 peaks, beyond those that can be attributed to acetylation, are likely to represent amides in different HCA II conformers (these could be amides located on the backbone of HCA II, or the lysine-ε-NHCOCH3, or both).
The HSQC spectrum of peracetylated HCA II shows a group of peaks (at 7–8 ppm, 126–129 ppm; Figure 5A) that includes 22 resolved peaks and a broad set of overlapping peaks that appear as a cluster of approximately 6 peaks. This entire group is conspicuous because it is not present in the HSQC spectrum of unmodified HCA II. In order to determine if these amides were coupled to 13C and therefore represented the side chain amides of lysine-ε-NHCOCH3 (and possibly the acetylated N-terminus), we performed a TROSY version of an HNCO NMR experiment. An HNCO experiment is a triple resonance NMR experiment that correlates the nuclear spin of the 1H and 15N of the amide to the 13C=O that is attached directly to the nitrogen. The H-N plane of an HNCO experiment will, therefore, reveal only the side chain N-H groups of lys-ε-15NH13COCH3 (because the protein was expressed in a media containing 12C glucose, and acetylated with (CH313CO)2O). The triple resonance spectrum shows that each N-H signal in this conspicuous group is coupled to 13C, and therefore represents the N-H from lysine-ε-NHCOCH3 (Figure 5B). Twenty-two resolved peaks were observed in the triple resonance spectrum in addition to a broad set of overlapping peaks (Figure 5B). No other signals were observed in the N-H plane of this triple resonance spectrum (Supplemental Figure 2).
The rate of H/D exchange of the acetyl amides was measured by monitoring the disappearance of each peak in D2O. Concentrated solutions of peracetylated HCA II (3.5 mM) were diluted 1:10 (v/v) into deuterated buffer (pD 7.4, 15 °C, 10 mM PO43−). An aliquot from the same concentrated solution was diluted 1:10 into buffered H2O (pH 7.4, 15 °C, 10 mM PO43−) in order to obtain a “zero” time point. The decrease in intensity of six of the peaks are shown with respect to time in D2O (Figure 5C). Plots of the remaining 22 peaks are shown in Supplemental Figure 3. Twenty one of the peaks representing amides from lysine-ε-NHCOCH3 (and α-NHCOCH3) disappeared before the first time point could be measured (e.g., in less than 27 minutes: 12 min was required to dilute the concentrated HCA II protein into deuterated buffer, shim the magnet, tune the probe, and begin scanning; each spectrum is a collection of scans collected over 15 minutes).
In order to illustrate the persistence of an NMR signal that represents a backbone amide in peracetylated HCA II that is protected from H/D exchange, we chose three N-H systems (that are not correlated with 13C and are presumably backbone amides) from the HSQC spectrum of peracetylated HCA II. The intensity of each of these peaks is plotted as a function of time in D2O (Supplemental Figure 4). These three N-H systems exchange at different rates (and were chosen based on their different rates of exchange).
Together, the data presented in Figure 5 demonstrates that the amides from lysine-ε-NHCOCH3 exchange too rapidly to be included among those that are protected upon acetyltion. We conclude the amide N-H hydrogens that are protected from exchange as a result of acetylation are, therefore, located on the backbone of BCA II.
Acetylating the ε-NH3+ Group of a Derivative of Lysine Decreases the Rate of H/D Exchange of its Backbone Amide
In order to determine specifically how the neutralization of ε-NH3+ affects the rate of H/D exchange at the backbone amide of a derivative of lysine that models lysine in polypeptides, we compared the rates of exchange of a model of lysine (N-α-acetyl-L-lysine-N-methylamide; abbreviated: Ac-Lys(ε-NH3+)-NHMe) with the acylated derivative (abbreviated: Ac-Lys(ε-NHCOCH3)- NHMe) using 1H NMR spectroscopy. The rate of H/D exchange was monitored by the disappearance of signal for the α-nitrogen amide at ~ 8.10 ppm (e.g., amide 1 in Figure 6) and the β-nitrogen amide (e.g., amide 2 in Figure 6) which appears at ~ 7.83 ppm. These experiments were performed at pD 4.5, instead of pD 7.4 (protein samples were measured at pD 7.4) because pD 4.5 is the value where H/D exchange is both slow enough to be measured with NMR by the addition of D2O, and predominantly base-catalyzed and therefore, mechanistically comparable to H/D exchange at pD 7.4.
The neutralization of the ε -NH3+ in this derivative of lysine resulted in a significant decrease in the rate of exchange of both the “backbone” amides 1 and 2 (Figure 6B and 6C). The data in Figure 6 were fit to the exponential function y = yo + Ae(-x/k); the time constants (k or t1/2) for amide 1 (Figure 6) were 526 ± 6 s for Ac-Lys(ε-NH3+)-NHMe (R2 = 0.9897) and 1639 ± 10 s for Ac-Lys(ε-NHCOCH3)-NHMe (R2 = 0.9985). The acetylation of the ε-NH3+ of model lysine, therefore, increases the half-life (t1/2) of exchange of its backbone amide hydrogen by a factor of approximately 3. The neutralization of ε-NH3+ decreased the rate of exchange of amide 2 (Figure 6) to a smaller extent. The time constants for the right handed amide were 1960 ± 22 s for Ac-Lys(ε-NH3+)-NHMe (R2 = 0.9958) and 3278 ± 25 s for Ac-Lys(ε-NHCOCH3)-NHMe (R2 = 0.9989); the neutralization of ε-NH3+, therefore, increased the half-life of exchange by a factor of 1.7.
The decrease in the rate of exchange of the backbone amide hydrogens in Ac-Lys(ε-NH3+)-NHMe that resulted from the acetylation of ε-NH3+ (e.g., a 2–3 fold decrease for each of the two amides—a 4–6 fold decrease in total; Figure 6) is significantly less than the decrease that we estimate to occur in BCA II as a result of acetylation of lysine (e.g., a 100–1000 fold decrease; Figure 4, Supplemental Table 2). One plausible explanation for this disparity is that the small-molecule experiments were performed at a different value of pD (i.e., pD 4.5) rather than at pD 7.4, where experiments on BCA II were done. Previous work has shown that lysine-ε-NH3+ derivatives (similar to those studied here) have a minimum rate of H/D exchange near pH 4.0.34,35 At pD 4.5, the exchange of amide hydrogen with deuterium is, therefore, catalyzed by D3O+ to a greater degree than at pD 7.4. Neutralization of ε-NH3+ will most likely accelerate the rate of acid (D3O+)-catalyzed exchange (although the neutralization of ε-NH3+ is expected to reduce the rate of base-catalyzed exchange).
Measuring the rate of amide hydrogen exchange of lysine-ε-NHCOCH3 and lysine-ε-NH3+ derivatives at pH 7.1 with magnetization transfer 1H NMR
Although the acetylation of Ac-Lys(ε-NH3+)-NHMe decreased the rate of H/D exchange by less than 3-fold, at pD 4.5, it is possible that acetylation would result in even larger decreases in the rate of amide H/D exchange at neutral pH. Such differences might be large enough to account for the decreased rate of H/D exchange that is estimated to occur throughout the protein charge ladder (e.g., 100–1000-fold). Amide hydrogen exchange in small molecules is, however, typically too rapid at neutral pH to be measured with NMR via the addition of D2O.
In order to test the hypothesis that the acetylation of lysine will have a much greater effect on the rate of amide H/D exchange at pD 7.4, than at pD 4.5, we used magnetization transfer NMR methods71 to approximate the rate of exchange of water protons, at pH 7.1, to amides of Ac-Lys(ε-NHCOCH3)-NHMe and Ac-Lys(ε-NH3+)-NHMe. This experiment is important because it will test whether the decreased rates of H/D exchange of each higher rung of the BCA II charge ladder are simply due to differences in kint of lysine-ε-NH3+ and lysine-ε-NHCOCH3. The details of these experiments are discussed in the supplemental information. Briefly, we found that the neutralization of the positive charge of Ac-Lys(ε-NH3+)-NHMe resulted in a 3.0-fold reduction in the rate of hydrogen exchange of amide 1 at pH 7.1, and a 2.9-fold reduction of amide 2 at pH 7.1 (Supplemental Figure 5). From comparing these data to those at pD 4.5, where neutralization caused a 3.1-fold reduction in the rate of H/D exchange of amide 1, and a 1.7-fold reduction in the rate of H/D exchange of amide 2 at pD 4.5, we conclude that acetylating Ac-Lys(ε-NH3+)-NHMe affects the rate of hydrogen exchange by similar magnitudes at pD 7.4 and pD 4.5. The different rates of H/D exchange of the protein charge ladder can not be explained, therefore, by large differences in kint between lysine-NH3+and lysine-ε-NHCOCH3.
Surface Properties of Proteins and H/D Exchange
We hypothesized that the correlation between the net negative charge of BCA II and its rate of hydrogen exchange is—at least in part—a manifestation of charge regulation at the surface of BCA II. Charge regulation in proteins is the change in local pH or local pKa of functional groups in response to a change in the local electrostatic potential (for example, acylation). These differences between pHlocal and pHsolv affect the ionization of residues that have values of pKa within ± 3 units of pHsolv.7 The change in local pH, and in values of pKa’s, will be in a direction that will tend to decrease the change in charge on the protein.
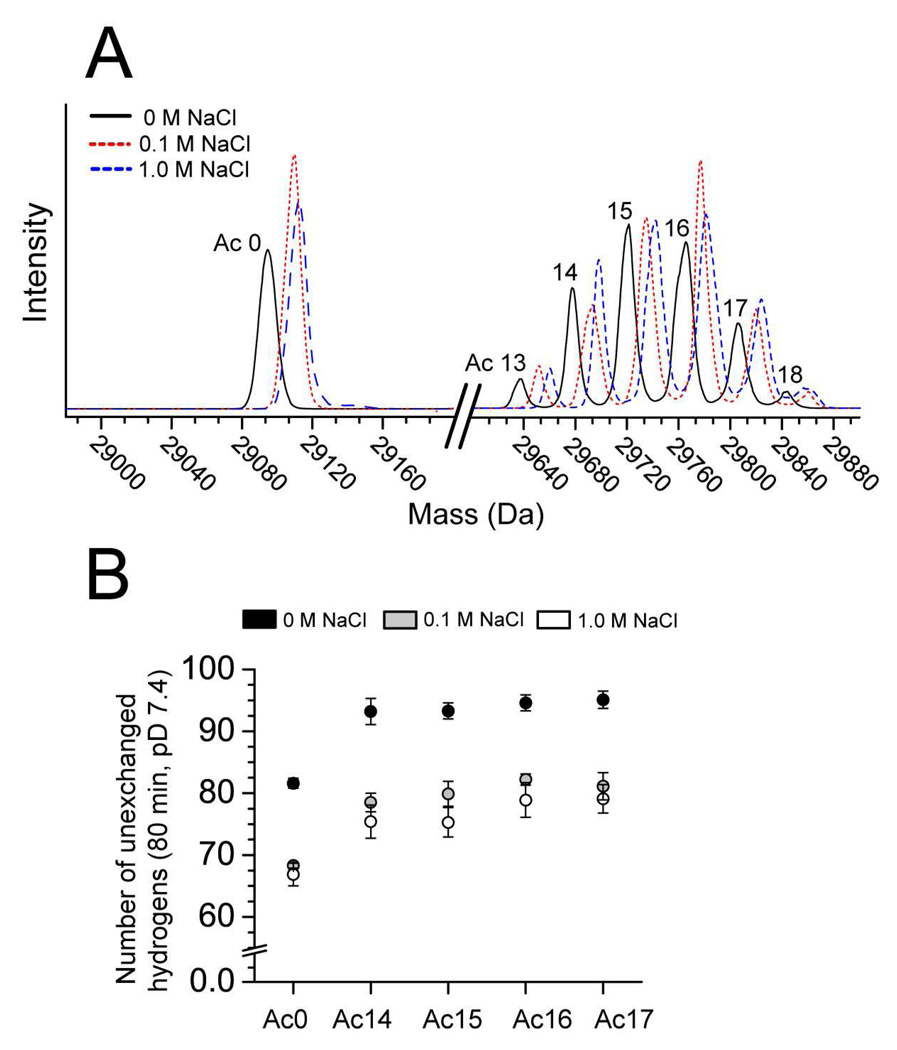
In order to begin to quantify how changes in electrostatics at the surface of BCA II (i.e., changes in local pH and the reorganization of solvent ions) that result from acetylation might affect the rate of H/D exchange, we measured the H/D exchange of unmodified and acetylated BCA II in 0.1 M and 1.0 M sodium chloride. Sodium and chloride ions will effectively screen electrostatic interactions between ions in solution and charged groups at the surface of the BCA II protein, for example between OH¯ and lys-ε-NH3+. For these experiments, we used a partial charge ladder of BCA II that consisted only of the higher rungs (i.e., rungs Ac(14)-Ac-(17)). Working with a partial ladder consisting of two or three abundant rungs, rather than a full ladder consisting of 19 rungs is convenient for mass spectrometric experiments that involve high concentrations of NaCl.72 We found that the number of unexchanged hydrogens decreased by 15–19 in both acetylated and unmodified BCA II (Figure 7) when the concentration of added NaCl was increased from 0 M to 1 M; that is, sodium chloride led to an overall increase in the rate of H/D exchange for both acetylated and unmodified proteins. For unmodified BCA II, the number of unexchanged hydrogens retained after 80 min decreased by 14.7 ± 2.1 as the concentration of sodium chloride was increased from 0 M to 1 M. For rungs Ac(13)-(18), the number of unexchanged hydrogens decreased by 19.1 ± 4.0, 17.9 ± 3.4, 18.0 ± 2.7, 15.7 ± 3.1, 16.0 ± 2.7 and 14.1 ± 3.4. Interestingly, the number of unexchanged hydrogens in rungs Ac(14)-Ac(17) is, at 1.0 M NaCl, equivalent (within error) to the number of unexchanged hydrogens in unmodified BCA II at 0 M NaCl (Figure 7). When the acetylated and unmodified proteins are both at 1.0 M NaCl, however, the acetylated derivatives still have more unexchanged hydrogen than the unmodified protein after 80 min in deuterated buffer. The observation that the rate of exchange of the higher rungs of the charge ladder is still slower than the unmodified protein, when both sets of proteins are in 1 M NaCl, suggests that 1 M NaCl does not entirely screen all of the inter- or intramolecular electrostatic interactions that occur in BCA II.
Figure 7. Sodium chloride increases the rate of H/D exchange in both acetylated and unmodified BCA II.
A) Mass spectra of unmodified BCA II and acetylated BCA II (e.g., rungs Ac(13)-Ac(18)) after 80 min in D2O, 10 mM PO43−, pD 7.4, 0–1 M NaCl. The mass of each BCA II species increases with the concentration of NaCl (indicating an increase in the incorporation of deuterium). B) Plot showing the number of unexchanged hydrogens in unmodified and acetylated BCA II after 80 min in D2O, pD 7.4, 0–1 M NaCl. The number of unexchanged hydrogens in each protein decreases with increasing sodium chloride. Unmodified BCA II in 0 M NaCl and acetylated BCA II in 1 M NaCl have a similar number of unexchanged hydrogens after 80 min in D2O (note: the rungs corresponding to Ac(13) and Ac(18) are omitted from the bar graph because these rungs are low in intensity and did not yield precise values of mass).
The dissimilarity between the rates of H/D exchange for each rung at 1.0 M NaCl does not, however, prove that electrostatic effects are not causing the different rates of H/D exchange throughout the protein charge ladder. Previous work by Englander34,35 has shown that the presence of 0.1 M and 0.5 M KCl does not completely eliminate what appear to be charge effects in the rates of H/D exchange of model peptides: the protonation the imidazole group of histidine peptides, for example, increases the rate constant of base-catalyzed exchange by nearly an order of magnitude, even in the presence of 0.5 M KCl (similar results were obtained for the protonated and deprotonated forms of glutamate and aspartate peptides). Furthermore, in the presence of 0.1 M KCl, the rate constant of base-catalyzed exchange of the amide in CH3(=O)NHCH3 is kOH = 2.51 × 108; the cationic analog, +H3NCH2C(=O)NHCH3, has the rate constant: kOH = 2.94 × 1010. Nevertheless, the results shown in Figure 7 also suggest that some other characteristic of the acetylated lysine residue—something other than the change in its charge on acetylation—(i.e., perhaps hydrophobicity or activity as an acid catalyst, or the local surface electrostatic potential of lysine in the folded protein) might contribute to the decreased rates of H/D exchange observed throughout the protein charge ladder.
Increasing Surface Hydrophobicity Does Not Affect the Rate of H/D Exchange of BCA II
In addition to neutralizing positive charges at the surface of BCA II, the acetylation of lysine also increases the average (and local) hydrophobicity of its surface. The Hansch π-parameter (log P) for –NH3+ groups is log P = −2.12; and for – NHCOCH3 groups, log P = −1.21.73,74 To test if the differences in the hydrogen exchange of each rung of the charge ladder were due (entirely or in part) to an increase in surface hydrophobicity, we prepared a perbutyrated derivative of BCA II (e.g., lys-ε-NHCO(CH2)2CH3) by acylating all 18 lys-ε-NH3+ groups with butyric anhydride. The perbutyrated protein has 36 more –CH2– groups on its surface than peracetylated BCA II; the perbutyrated protein will have a net charge similar to peracetylated BCA II, but a greater surface hydrophobicity (the Hansch parameter for NHCO(CH2)2CH3 is not available, however, log P = 0.5 for –CH3 and log P = 1.5 for –(CH2)2CH373).
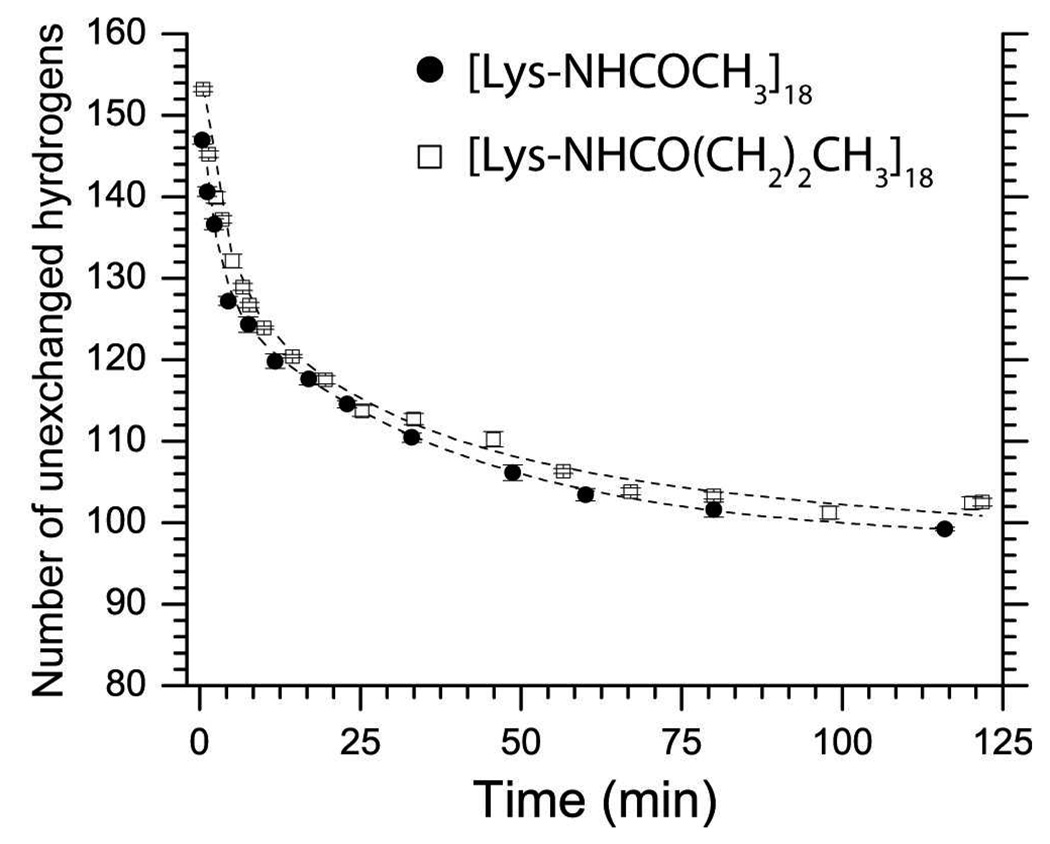
We measured the hydrogen exchange of perbutyrated BCA II with LC-ESI-MS, and compared its exchange profile with peracetylated BCA II (Figure 8). The global hydrogen exchange kinetics of these two (we believe isostructural) derivatives of the same protein were indistinguishable (Figure 8). Each set of data was fit with equation S1 (see supplemental material) and the resulting curves were superimposable (Figure 8). The similarity in the rates of exchange of BCA-(NHCOCH3)18 and BCA-(NHCOC3H7)18 demonstrates that surface hydrophobicity does not affect the kinetics of amide hydrogen exchange in BCA II; we conclude that increased surface hydrophobicity does not explain the different rates of H/D exchange throughout the charge ladder.
Figure 8. Increasing the surface hydrophobicity of BCA II has no effect upon the kinetics of hydrogen-deuterium exchange as measured by ESI-MS.
The surface hydrophobicity of BCA II was varied, but the net charge was maintained, by attaching either 18 acetyl groups or 18 butanoyl groups. Both proteins exhibited similar protection from hydrogen exchange and nearly superimposable exchange profiles. Hydrogen exchange was monitored at 15 °C, pD 7.4, 10 mM PO43−. Error bars represent the standard deviation of average mass values calculated from seven charge states for each protein.
Acetylation of Lysine Decreases the Thermostability of BCA II in Spite of Increasing its Protection from H/D Exchange
The rates of amide hydrogen exchange in folded proteins (and their chemical or genetic variants) should correlate inversely with the conformational stability as measured by unfolding with heat or chaotropic agent. This correlation has been observed on several occasions among homologous proteins, and also among sets of protein variants prepared by site directed mutagenesis.21,22,75 Equation 7 expresses the free energy of the transition from a closed to an open state (under EX2 conditions) as a function of the observed rate of H/D exchange.
| (7) |
The rate of hydrogen exchange has, therefore, been used to measure the conformational stability of folded proteins and the free energy of folding for individual residues in proteins.20,76,77 We found, however, that the least thermally stable (and most highly charged) rungs of the charge ladder were more protected from H/D exchange than the more stable (and less charged) rungs. Our finding suggests that electrostatic factors such as net charge, or perturbations in the electrostatic environment of proteins that accompany post-translational modification, amino acid substitution, or ligand binding, can complicate the estimation of the conformational stability of a folded protein by measuring the rate of amide H/D exchange.
Conclusion
We have used protein charge ladders and mass spectrometry to quantify the effects of structural and electrostatic changes produced by conversion of lys-ε-NH3+ to lys-ε-NHCOCH3 on the rate of H/D exchange in BCA II. Eliminating cationic sites on the surface of this protein by acetylation of its surface lysine-ε-NH3+ groups resulted in a decrease in the rate of backbone amide H/D exchange that was linear in the number of lysine-ε-NHCOCH3 groups formed (Figure 4C). The neutralization of all 18 lysine residues by acetylation does not result in any significant changes in the 2° or 3° structure of BCA II (but did diminish the thermostability of the protein).
The rate of H/D exchange of amides in the backbone of folded polypeptides is almost universally explained by the hydrogen bonding and solvent accessibility of the amide NH. There is an increasing number of reports, however, that suggest the rate of base-catalyzed H/D exchange of surface amides in proteins can be, at least in some cases, a better indicator of the local electrostatic potential of the amide, than the structural environment (e.g., the conformational flexibility, H-bonding, or solvent accessibility) of the amide.27,44,45 These previous studies 27,44,45 arrived at this conclusion by comparing the rates of base-catalyzed H/D exchange of individual amides at the surface of proteins, with the electrostatic solvation free energies of the peptide anions (calculated with Poisson-Boltzmann methods) that form during the base-catalyzed H/D exchange of the amide NH. Our current study has shown, using protein charge ladders, that systematic variations in the electrostatic potential of the surface of a protein can affect the rate of amide H/D exchange by a magnitude that is large enough to be detected with mass spectrometry (and also, we assume, with HSQC).
The electrostatic environment of an amide must therefore be considered along with H-bonding and solvent accessibility when interpreting the meaning of hydrogen exchange kinetics of folded proteins. The type of kinetic electrostatic effect—not structural effect—that we report in this paper might also arise, for example, from non-isoelectric perturbations such as amino acid substitution78, post-translational modification79 (especially the acetylation of lysine or N-terminal –NH3+ 80,81) and the binding of charged small molecules60,82–85, oligonucleotides 86, or metal cofactors87,88 to proteins. For example, a recent paper has used mass spectrometry and H/D exchange to study how the acetylation of the N-terminus affects the structure of polypeptide chains that comprise the ribosomal stalk complex of Escherichia coli.75 This paper reported that acetylation resulted in small increases in the number of amide hydrogens (i.e., between one and three) that were protected from exchange with buffer after 10 min in 90 % D2O. The authors concluded that this decrease in the rate of H/D exchange was caused by a change in the structure of the protein (and of the protein complex as a whole), but we hypothesize that the decreased rate of H/D exchange might have occurred—in whole or in part—because of the same kinetic electrostatic effect that we observe with the protein charge ladder, and not because of a change in the structure or flexibility of the ribosomal stalk complex upon acetylation.
We hypothesize that the kinetic electrostatic effect that we observe arises from decreases in pHlocal at the surface of the protein, and/or changes in the electrostatic environment near lysine residues that result in a reduction of the pKa of the lysine amide, or backbone amides proximal to lysine. A definite, unambiguous identification of the specific residues that were protected from H/D exchange by acetylation in BCA II (which is necessary in order to define the operative mechanisms completely) is prevented by the lack of NMR peak assignments for BCA II. In addition to charge regulation, there are other plausible mechanistic explanations for why the acylation of ε-NH3+ might decrease the rate of exchange in BCA II. The number of covalent bonds separating the ε-NH3+ group and backbone amide of lysine (e.g., six) is too great for the ε-NH3+ to exert a through-bond inductive effect on the backbone amide NH (which would reduce the pKa of the amide NH); the ε-NH3+ group might, however, stabilize an anionic amide intermediate (e.g., –CON¯ –) that formed during base-catalyzed exchange of the amide hydrogen. We reiterate that the electrostatic effect that we observe can not be entirely abolished (at least in the case of BCA II) by the addition of NaCl to the buffer; the rates of H/D exchange of acetylated and non-acetylated BCA II are still different in the presence of 1 M NaCl (Figure 7). The future use of charge ladders of other stable proteins whose NMR structures have been determined (i.e., rubredoxin or superoxide dismutase-1) should clarify the mechanism by which surface electrostatics affect the hydrogen exchange of folded proteins.
This study has, nevertheless, shown that protein charge ladders offer a unique tool to use in understanding the structural and electrostatic factors that govern the rate of hydrogen exchange in folded proteins (and that have, so far, been intractably difficult to explore experimentally, and hence largely ignored). We hope that a more complete theory of hydrogen exchange in folded polypeptides might explain some of the surprising and sometimes confusing results of H/D exchange experiments that have accumulated for over 50 years in biochemistry and in organic and polymer chemistry.24–27,40–42,89,90
Supplementary Material
Acknowledgment
The authors acknowledge NIH GM 51559 for financial support. The authors acknowledge Drs. Jiong Yu and Eric T. Mack for technical assistance with mass spectrometry and protein purification. The authors also acknowledge Dr. Charisse Crenshaw for technical assistance with magnetization transfer NMR experiments. The authors also gratefully acknowledge Debby Pheasant of the Biophysical Instrumentation Facility (Massachusetts Institute of Technology) for technical assistance operating the DSC instrument. BFS thanks a NIH Ruth Kirchstein National Research Service Award (GM081055) for post-doctoral support.
Footnotes
Supporting Information Available: Additional experimental details, including mass spectrometric experiments and kinetic analysis of mass spectrometric data, and additional data, including tabulated values of mass and kinetic parameters, NMR spectra, and kinetic exchange profiles for all 19 rungs of the BCA II charge ladder. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gao J, Mammen M, Whitesides GM. Science. 1996;272:535–537. doi: 10.1126/science.272.5261.535. [DOI] [PubMed] [Google Scholar]
- 2.Gitlin I, Carbeck JD, Whitesides GM. Angew. Chem. Int. Ed. Engl. 2006;45:3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- 3.Gitlin I, Gudiksen KL, Whitesides GM. J. Phys. Chem. B. 2006;110:2372–2377. doi: 10.1021/jp055699f. [DOI] [PubMed] [Google Scholar]
- 4.Negin RS, Carbeck JD. J. Am. Chem. Soc. 2002;124:2911–2916. doi: 10.1021/ja0169567. [DOI] [PubMed] [Google Scholar]
- 5.Gudiksen KL, Gitlin I, Moustakas DT, Whitesides GM. Biophys. J. 2006;91:298–310. doi: 10.1529/biophysj.106.081547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem. Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menon MK, Zydney AL. Anal. Chem. 2000;72:5714–5717. doi: 10.1021/ac000752b. [DOI] [PubMed] [Google Scholar]
- 8.Gudiksen KL, Gitlin I, Yang J, Urbach AR, Moustakas DT, Whitesides GM. J. Am. Chem. Soc. 2005;127:4707–4714. doi: 10.1021/ja043804d. [DOI] [PubMed] [Google Scholar]
- 9.Snyder PW. Unpublished observation. [Google Scholar]
- 10.The mass spectrometric methods that we used in this research did not involve fragmenting the protein with proteolysis prior to analysis with mass spectrometry. We, therefore, measured the H/D exchange of the entire polypeptide and refer to this exchange as ‘global’.
- 11.Englander SW, Krishna MM. Nat. Struct. Biol. 2001;8:741–742. doi: 10.1038/nsb0901-741. [DOI] [PubMed] [Google Scholar]
- 12.Hvidt A, Linderstrom-Lang K. Biochim. Biophys. Acta. 1954;14:574–575. doi: 10.1016/0006-3002(54)90241-3. [DOI] [PubMed] [Google Scholar]
- 13.Wagner G, Wuthrich K. J. Mol. Biol. 1979;134:75–94. doi: 10.1016/0022-2836(79)90414-5. [DOI] [PubMed] [Google Scholar]
- 14.Wagner G, Wuthrich K. Nature. 1978;275:247–248. doi: 10.1038/275247a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Garcia-Ordonez RD, Pascal BD, Burris TP, Dodge JA, Griffin PR. Structure. 2010;18:1332–1341. doi: 10.1016/j.str.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng X, Liu G, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcsisin SR, Engen JR. Anal. Bioanal. Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carulla N, Zhou M, Giralt E, Robinson CV, Dobson CM. Acc. Chem. Res. 2010;43:1072–1079. doi: 10.1021/ar9002784. [DOI] [PubMed] [Google Scholar]
- 19.Dobson CM. Semin. Cell Dev. Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Huyghues-Despointes BM, Langhorst U, Steyaert J, Pace CN, Scholtz JM. Biochemistry. 1999;38:16481–16490. doi: 10.1021/bi9919450. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez JA, Shaw BF, Durazo A, Sohn SH, Doucette PA, Nersissian AM, Faull KF, Eggers DK, Tiwari A, Hayward LJ, Valentine JS. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner G, Wuthrich K. J. Mol. Biol. 1979;130:31–37. doi: 10.1016/0022-2836(79)90550-3. [DOI] [PubMed] [Google Scholar]
- 23.Englander SW. Annu. Rev. Biophys. Biomol. Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 24.Hvidt A, Corret R. J. Am. Chem. Soc. 1970;92:5546–5550. [Google Scholar]
- 25.Perrin C, Chen J, Ohta B. J. Am. Chem. Soc. 1999;121:2448–2455. [Google Scholar]
- 26.Scarpa J, Mueller D, Klotz I. J. Am. Chem. Soc. 1967;89:6024–6030. [Google Scholar]
- 27.Anderson JS, Hernandez G, Lemaster DM. Biochemistry. 2008;47:6178–6188. doi: 10.1021/bi800284y. [DOI] [PubMed] [Google Scholar]
- 28.Bai Y. Chem. Rev. 2006;106:1757–1768. doi: 10.1021/cr040432i. [DOI] [PubMed] [Google Scholar]
- 29.Maity H, Lim WK, Rumbley JN, Englander SW. Protein Sci. 2003;12:153–160. doi: 10.1110/ps.0225803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connelly GP, Bai Y, Jeng MF, Englander SW. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 31.Bai Y, Sosnick TR, Mayne L, Englander SW. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.One example of this type of slow exchange is the amide hydrogen on Leucine 98 in native cytochrome c; this hydrogen exchanges on a timescale of ~100 years at neutral pH.
- 33.Hvidt A, Nielsen SO. Adv. Protein. Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 34.Molday RS, Englander SW, Kallen RG. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Milne JS, Mayne L, Englander SW. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner G. Biochem. Biophys. Res. Commun. 1980;97:614–620. doi: 10.1016/0006-291x(80)90308-3. [DOI] [PubMed] [Google Scholar]
- 37.Roder H, Wagner G, Wuthrich K. Biochemistry. 1985;24:7396–7407. doi: 10.1021/bi00346a055. [DOI] [PubMed] [Google Scholar]
- 38.Kim KS, Woodward C. Biochemistry. 1993;32:9609–9613. doi: 10.1021/bi00088a013. [DOI] [PubMed] [Google Scholar]
- 39.Woodward C, Barbar E, Carulla N, Battiste J, Barany G. J. Mol. Graph. Model. 2001;19:94–101. doi: 10.1016/s1093-3263(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 40.Miller DW, Dill KA. Protein Sci. 1995;4:1860–1873. doi: 10.1002/pro.5560040921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsyth W, Robertson A. J. Am. Chem. Soc. 1996;118:2694–2698. [Google Scholar]
- 42.Radkiewicz JL, Zipse H, Clarke S, Houk KN. J. Am. Chem. Soc. 2001;123:3499–3506. doi: 10.1021/ja0026814. [DOI] [PubMed] [Google Scholar]
- 43.High ionic strength (> 100 mM) is nevertheless more physiologically relevant than low ionic strength (< 100 mM).
- 44.Hernandez G, Anderson JS, LeMaster DM. Biochemistry. 2009;48:6482–6494. doi: 10.1021/bi900526z. [DOI] [PubMed] [Google Scholar]
- 45.Anderson JS, Hernandez G, LeMaster DM. Biophys. Chem. 2009;141:124–130. doi: 10.1016/j.bpc.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez G, Anderson JS, LeMaster DM. ChemBioChem. 2008;9:768–778. doi: 10.1002/cbic.200700465. [DOI] [PubMed] [Google Scholar]
- 47.Dempsey CE. Biochemistry. 1986;25:3904–3911. doi: 10.1021/bi00361a025. [DOI] [PubMed] [Google Scholar]
- 48.Matthew JB, Richards FM. J. Biol. Chem. 1983;258:3039–3044. [PubMed] [Google Scholar]
- 49.Christoffersen M, Bolvig S, Tuchsen E. Biochemistry. 1996;35:2309–2315. doi: 10.1021/bi951711q. [DOI] [PubMed] [Google Scholar]
- 50.Kim PS, Baldwin RL. Biochemistry. 1982;21:1–5. doi: 10.1021/bi00530a001. [DOI] [PubMed] [Google Scholar]
- 51.Goldfarb NE, Lam MT, Bose AK, Patel AM, Duckworth AJ, Dunn BM. Biochemistry. 2005;44:15725–15733. doi: 10.1021/bi0511686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leikina E, Ramos C, Markovic I, Zimmerberg J, Chernomordik LV. EMBO J. 2002;21:5701–5710. doi: 10.1093/emboj/cdf559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw AK, Pal SK. J. Photochem. Photobiol. B. 2008;90:187–197. doi: 10.1016/j.jphotobiol.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 54.Shaw BF, Schneider GF, Bilgicer B, Kaufman GK, Neveu JM, Lane WS, Whitelegge JP, Whitesides GM. Protein Sci. 2008;17:1446–1455. doi: 10.1110/ps.035154.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pervushin K, Riek R, Wider G, Wuthrich K. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruben DJ, Bodenhausen G. Chem. Phys. Lett. 1980;69:185–189. [Google Scholar]
- 57.Bax A, Ikura M, Kay LE, Barbato G, Spera S. Ciba. Found. Symp. 1991;161:108–119. doi: 10.1002/9780470514146.ch8. discussion 119–35. [DOI] [PubMed] [Google Scholar]
- 58.Goddard T, Kneller D. SPARKY 3-NMR Assignment and Integration Software. San Francisco: University of California; 2006. 2006. [Google Scholar]
- 59.Burton RE, Hunt JA, Fierke CA, Oas TG. Protein Sci. 2000;9:776–785. doi: 10.1110/ps.9.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider GF, Shaw BF, Lee A, Carillho E, Whitesides GM. J. Am. Chem. Soc. 2008;130:17384–17393. doi: 10.1021/ja804736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Proteolysis of BCA II under conditions where H/D exchange is quenched could provide low resolution information about the rate of specific amino acids in CA II; the resolution provided by these methods is limited by the size of the proteolytic fragment that is generated by reactions with pepsin.
- 62.To illustrate this point, consider for example a 100 residue protein and a 150 residue protein that both incorporate 75 amide deuterons after 80 min in D2O. Although both proteins have identical rates of deuterium incorporation, the 150 residue protein is, in fact, more protected from hydrogen exchange, or more “structured” than the 100 residue protein because the 150 residue protein has 75 residues that have not exchanged with solvent (the 100 kDa protein has only 25).
- 63.Shaw BF, Durazo A, Nersissian AM, Whitelegge JP, Faull KF, Valentine JS. J. Biol. Chem. 2006;281:18167–18176. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- 64.Jonasson P, Kjellsson A, Sethson I, Jonsson BH. FEBS Lett. 1999;445:361–365. doi: 10.1016/s0014-5793(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 65.Wang L, Smith DL. Curr. Protoc. Protein Sci. 2002 doi: 10.1002/0471140864.ps1706s28. Chapter 17, Unit 176. [DOI] [PubMed] [Google Scholar]
- 66.The indole-H of deeply buried tryptophan residues in BCA II may not exchange rapidly with solvent. An analysis of the hydrogen exchange properties of tryptophan residues in human CA II, with NMR, revealed that several solvent- exposed Trp residues (i.e., Trp5, 16, and 245) undergo H/D exchange rapidly (e.g., t1/2<< 20 min) while buried residues (i.e., Trp97) did not exchange with solvent (e.g., t1/2>> 20 min) unless the protein was unfolded with guanidinium hydrochloride. Some tryptophan residues, therefore, might be mistaken as amide hydrogen when using mass spectrometric methods to measure H/D exchange of BCA II.
- 67.Colton IJ, Anderson JR, Gao JM, Chapman RG, Isaacs L, Whitesides GM. J. Am. Chem. Soc. 1997;119:12701–12709. [Google Scholar]
- 68.An alignment of amino acid sequences of human and bovine CA II (using Clusta1X software) revealed a sequence homology of 79 %. The pI of HCA II is 7.6 (the pI of BCA II =5.9). Previous analysis of HCA II and BCA II with X-ray crystallography show that the two proteins have the same over-all fold and nearly identical structures. Approximately 1715 atoms could be aligned from each crystal structure of HCA II and BCA II (HCA II contains 4083 atoms; BCA II contains 4048) and the root mean squared deviation (RMS) for these 1715 aligned atoms was 0.448 angstrom.
- 69.Cavanagh J, Fairbrother WJ, Palmer AG, III, France A, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic Press; 2007. [Google Scholar]
- 70.Wuthrich K. J. Biol. Chem. 1990;265:22059–22062. [PubMed] [Google Scholar]
- 71.Gueron M, Leroy JL. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 72.The solutions of charge ladder must be diluted considerably (i.e., 100 fold) when 1 M NaCl is present before injection into the mass spectrometer, in addition to the initial 10-fold dilution into D2O from H2O. Achieving an appropriate final concentration of protein for analysis with ESI-MS is more convenient with a partial ladder that contains only 3–4 abundant rungs rather than a full charge ladder that contains 19.
- 73.Hansch C, Coats E. J. Pharm. Sci. 1970;59:731–743. doi: 10.1002/jps.2600590602. [DOI] [PubMed] [Google Scholar]
- 74.Hansch C, Steward AR. J. Med. Chem. 1964;7:691–694. doi: 10.1021/jm00336a001. [DOI] [PubMed] [Google Scholar]
- 75.Wagner G, Wuthrich K. J. Mol. Biol. 1982;160:343–361. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- 76.Huyghues-Despointes BM, Scholtz JM, Pace CN. Nat. Struct. Biol. 1999;6:910–912. doi: 10.1038/13273. [DOI] [PubMed] [Google Scholar]
- 77.Mullins LS, Pace CN, Raushel FM. Protein Sci. 1997;6:1387–1395. doi: 10.1002/pro.5560060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chitta RK, Rempel DL, Grayson MA, Remsen EE, Gross ML. J. Am. Soc. Mass Spectrom. 2006;17:1526–1534. doi: 10.1016/j.jasms.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Jones DD, Stott KM, Howard MJ, Perham RN. Biochemistry. 2000;39:8448–8459. doi: 10.1021/bi992978i. [DOI] [PubMed] [Google Scholar]
- 80.Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. J. Mol. Biol. 2008;380:404–414. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- 81.Szewczuk Z, Konishi Y, Goto Y. Biochemistry. 2001;40:9623–9630. doi: 10.1021/bi002762c. [DOI] [PubMed] [Google Scholar]
- 82.Wildes D, Marqusee S. Protein Sci. 2005;14:81–88. doi: 10.1110/ps.04990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marques MR, Vaso A, Neto JR, Fossey MA, Oliveira JS, Basso LA, dos Santos DS, de Azevedo WF, Junior, Palma MS. Biochemistry. 2008;47:7509–7522. doi: 10.1021/bi800134y. [DOI] [PubMed] [Google Scholar]
- 84.Das R, Esposito V, Abu-Abed M, Anand GS, Taylor SS, Melacini G. Proc. Natl. Acad. Sci. U.S.A. 2007;104:93–98. doi: 10.1073/pnas.0609033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rey M, Man P, Clemencon B, Trezeguet V, Brandolin G, Forest E, Pelosi L. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.146209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sperry JB, Wilcox JM, Gross ML. J. Am. Soc. Mass Spectrom. 2008;19:887–890. doi: 10.1016/j.jasms.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, Sohn SH, Durazo A, Faull KF, Gralla EB, Nersissian AM, Valentine JS. J. Am. Chem. Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 88.Ferguson PL, Pan J, Wilson DJ, Dempsey B, Lajoie G, Shilton B, Konermann L. Anal. Chem. 2007;79:153–160. doi: 10.1021/ac061261f. [DOI] [PubMed] [Google Scholar]
- 89.Brorsson AC, Lundqvist M, Sethson I, Jonsson BH. J. Mol. Biol. 2006;357:1634–1646. doi: 10.1016/j.jmb.2006.01.090. [DOI] [PubMed] [Google Scholar]
- 90.Perrin C. Acc. Chem. Res. 1989;22:268–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.