Abstract
Background
Somatostatin (SST) inhibits cell proliferation and negatively regulates the release of growth hormones via specific receptors (SSTR). Genetic variation in SSTR has been associated with risk of human cancers but has never been investigated in pancreatic cancer.
Methods
In this retrospective study, we sequenced the SSTR5 gene in paired tumor and blood samples from 33 pancreatic adenocarcinoma patients using the Sanger method. We analyzed 3 single nucleotide polymorphisms (SNPs) in samples from 863 patients with pancreatic ductal adenocarcinoma and 876 healthy controls using the TaqMan method. The associations between gene polymorphisms and pancreatic cancer risk and survival were analyzed by multivariate logistic regression and Cox proportional hazard models, respectively.
Results
We identified no somatic mutations but 3 nonsynonymous SSTR5 SNPs (P109S, L48M, and P335L) in pancreatic tumors. The SSTR5 P109S variant allele was associated with a 1.62-fold increased risk of pancreatic cancer (95% confidence interval [CI]: 1.08–2.43, P = 0.019). Furthermore, the SSTR5 L48M AC variant and smoking had a joint effect on pancreatic cancer risk (pinteraction = 0.035). The odds ratios (95% CIs) were 0.58 (0.34–0.97), 1.49 (1.18–1.89), and 2.27 (1.35–3.83) for the variant genotype alone, smoking alone, and both factors, respectively, compared with no factors. Finally, SSTR5 P335L CC and P109S CC combined were associated with lower overall survival durations in patients with resectable disease.
Conclusion
Our data suggest that SSTR5 genetic variants play a role in pancreatic cancer development and progression.
Keywords: Somatostatin receptor 5, single nucleotide polymorphism, pancreatic cancer risk, survival, smoking
Introduction
Pancreatic cancer is a common malignant gastrointestinal disease and is the fourth leading cause of cancer death in the United States; an estimated >35,000 people died of this disease in the United States in 2009 (1). Known risk factors for pancreatic cancer include cigarette smoking, obesity, a history of diabetes, and a family history of pancreatic cancer (2, 3). Hereditary syndromes caused by germ line mutations explain 5–10% of the pancreatic cancer cases. Genetic factors that contribute to the development of sporadic pancreatic cancer have not been well-defined.
Somatostatin (SST) is a polypeptide hormone that inhibits the proliferation of normal and neoplastic cells. Therefore, SST is thought to play a role in carcinogenesis, and SST analogues have been used as therapeutic agents for several neoplasms, including prostate cancer, breast cancer, neuroendocrine tumors, and pancreatic cancer (4–7). SST’s effects on cell growth and proliferation regulation in various organ systems are mediated via specific SST receptors (SSTRs) (8), 5 subtypes (SSTR1–5) of which have been identified and cloned in human tissue (4). SSTR subtype expression has been characterized in physiologic tissues and neoplastic breast and prostate tissues (9). SSTRs are expressed in pancreatic tumor and normal tissues, but SSTR2 and SSTR5 mRNA levels are significantly lower in tumor tissues than in adjacent normal tissues (10). In addition, acting through SSTRs, SST negatively regulates pituitary synthesis and growth hormone release, resulting in decreased synthesis of insulin-like growth factor-I (IGF-1) (11). IGF-I plays a role in cancer development by stimulating cell proliferation and inhibiting apoptosis 12). Thus, SSTRs may be directly or indirectly involved in pancreatic carcinogenesis.
The associations between genetic variants of SSTRs and cancer risk have been evaluated in prostate and breast cancer (13, 14). However, to our knowledge, no study has been performed of SSTR gene variants in pancreatic cancer. Because SSTR5 is highly expressed in pancreatic tissue and its level is lower in pancreatic tumors (10, 15, 16), we focused on the SSTR5 gene in the current study. To determine the SSTR5 gene’s role in pancreatic cancer, we performed a mutation analysis of 33 primary pancreatic adenocarcinoma samples by direct DNA sequencing. We determined the associations between SSTR5 gene variants and risk or survival of pancreatic cancer in a study of 863 pancreatic cancer patients and 876 healthy controls.
Patients and Methods
Study Population
We performed DNA sequencing in 33 pancreatic adenocarcinoma patients who had undergone tumor resection at Baylor College of Medicine (Houston, Texas). Each patient had signed an informed consent form to allow their blood sample and residual tumor sample to be used for research, and the study was approved by the institutional review board.
Genotyping was performed in DNA samples that had been collected in a case-control study of pancreatic cancer conducted at The University of Texas MD Anderson Cancer Center (Houston, Texas) from 2000 to 2008 (17). All patients had pathologically confirmed primary pancreatic ductal adenocarcinoma. Controls were cancer-free individuals recruited from spouses, friends, and non-blood relatives of patients with non-gastrointestinal or -smoking-related cancers who had visited MD Anderson. Controls had been frequency-matched to cases by age at enrollment (5-year interval), race and sex. Demographic data and risk factor information on cigarette smoking, alcohol consumption, occupational history, medical history including diabetes, and family history of cancer had been collected by personal interview. Body mass index (BMI; kg/m2) data had been collected from study participants recruited in 2004 and later. Document-based informed consent had been obtained from each study participant for the interviews, blood sample collections, DNA extractions, and genotyping analyses. The study was approved by the institutional review board of MD Anderson. Because of the known ethnic difference in genotype distribution and the small number of minorities enrolled in this study, the current analysis was restricted to non-Hispanic white.
Clinical information was collected by reviewing patients’ medical records and included date of pathologic diagnosis, clinical tumor stage (resectable, locally advanced, metastasized, and unstaged), tumor grade, serum carbohydrate antigen 19-9 (CA19-9) values (unit/mL) at diagnosis, tumor resection date, node and margin statuses, and date of death or last follow-up. Overall survival duration was calculated from the time of pathologic diagnosis to the date of death or last follow-up. Dates of death were obtained and cross-checked usingat least 1 of the following 3 methods: Social Security Death Index, inpatient medical records, and the MD Anderson tumorregistry.
DNA Sequencing
Blood samples had been directly collected in PAXgene blood DNA tubes (PreAnalytiX, Qiagen), and DNA was isolated using the PAXgene blood DNA kit. Resected tumor specimens had been collected and stored at −80°C in a proteinase inhibitor solution (Roche Applied Science, Indianapolis, IN), and DNA was isolated using the QIAamp DNA mini kit (Qiagen, Valencia, CA) after the tissue had been washed several times in phosphate-buffered saline to remove any trace of stabilizing solution. Primer sets were designed to cover the SSTR5 exons. Fifty nanograms of each DNA sample were whole genome amplified (GenomiPhi DNA amplification kit, Amersham Biosciences), and polymerase chain reaction (PCR) was performed on 10 ng of these samples in a final reaction volume of 8 μL in a 384-well plate using the polymerase HotStar (Qiagen). Cycling parameters consisted of 40 cycles of a denaturation step at 95°C for 45 sec, followed by an annealing step at 60°C for 45 sec and an extension step at 72°C for 45 sec. The cycling process was preceded by a denaturation period at 95°C for 15 min, followed by a final extension period at 72°C for 7 min. Unconsumed deoxynucleotide triphosphates were hydrolyzed and the remaining primers were degraded using a cocktail of shrimp alkaline phosphatase and exonuclease I (ExoSAP-IT, USB). The purified PCR products were diluted and sequenced using a BigDye Terminator version 3.1 cycle sequencing kit on ABI 3700 DNA sequencers. The sequences were analyzed with SNP Detector version 3 (created by Jinhui Zhang at the National Cancer Institute) using the corresponding sequence in GenBank as the reference. To identify germline polymorphisms, the sequences were compared with the reference sequence in GenBank. Base disparities from the reference sequence identified by SNP Detector were manually verified in Consed and in Sequencher version 4.7 (Gene Codes Corp.).
Genotyping Assays
DNA was extracted from peripheral blood lymphocytes using a FlexiGene DNA kit (Qiagen, Valencia, CA) and a Maxwell16 automated system (Promega, Madison, WI), and genotyping was performed using the Taqman 5′ nuclease assay. Primers and TaqMan MGB probes were provided by TaqMan SNP Genotyping Assay Services (Applied Biosystems, Foster City, CA). PCR was performed in a 5-μL total volume consisting of TaqMan Universal PCR Master Mix, 20 ng of genomic DNA (diluted with dH2O), and TaqMan SNP genotyping assay mix. Alleles were discriminated by running end point detection using an ABI Prism 7900HT sequence detection system and SDS 2.3 software (Applied Biosystems). About 5% of samples were analyzed in duplicate, and 100% consistency was achieved.
Statistical Methods
The genotype distribution was tested for the Hardy-Weinberg equilibrium using the goodness-of-fit χ2 test. Pancreatic cancer risk was estimated with odds ratios (ORs) and 95% confidence intervals (95% CIs), calculated by logistic regression analysis with adjustments for age, sex, and known risk factors such as family history of cancer, history of diabetes, smoking status, and BMI. To detect interactions between genotypes and risk factors (exposure), ORs were calculated by logistic regression analysis for the following groups: wild-type genotype and non-exposed (reference group), at-risk genotype and non-exposed (OR10), wild-type genotype and exposed (OR01), and at-risk genotype and exposed (OR11). An OR11 that was more than the sum of OR10 + OR01 indicated an additive effect. The significance of the interaction term (Pinteraction) was obtained using the likelihood ratio test, with the full model containing the interaction term, the main genotype effect, and the exposure variable and reduced model lacking the interaction term. The associations between overall survival and each SNP were estimated using the Kaplan and Meier method and log-rank test. Hazard ratios and 95% CIs were estimated using multivariable Cox proportional regression models. All statistical tests were conducted using SPSS 17.0 and Stata 9.0 software. P values < 0.05 were considered statistically significant.
We estimated the false-positive report probability (FPRP) for observed statistically significant associations using methods described by Wacholder et al (18). The FPRP is the probability of no true association between a genetic variant and a phenotype given a statistically significant finding. OR values of 2.0 to 4.0 were considered likely thresholds. The prior probability used was 0.25, and the FPRP value for noteworthiness was set at 0.2.
Results
Patient Characteristics
The 33 patients in the DNA sequencing analysis were mostly white, equally divided between male and female, and predominantly 60 to 70 years old. Eighteen (54.5%) and 13 (39.4%) of the patients’ 33 tumors were resectable and metastatic, respectively (Table 1).
Table 1.
Patient Characteristics (n = 33)
| Variable | No. of patients |
|---|---|
| Ethnicity | |
| White | 25 |
| Hispanic | 3 |
| Black | 3 |
| Asian | 2 |
| Age (years) | |
| ≤50 | 1 |
| 51–60 | 7 |
| 61–70 | 16 |
| 71–80 | 7 |
| >81 | 2 |
| Sex | |
| Female | 16 |
| Male | 17 |
| Tumor stage | |
| Resectable | 18 |
| Locally advanced | 2 |
| Metastatic | 13 |
The genotyping analysis included 863 patients and 876 controls. Their demographic data and potential risk factors are shown in Table 2. Their mean ages were 62.0 ± 9.9 years and 61.5 ± 9.7 years. No significant differences were found in age or sex between cases and controls. On the other hand, a family history of cancer among first-degree relatives, a history of diabetes, smoking, and BMI were significantly associated with pancreatic cancer risk (P < 0.001).
Table 2.
Distribution of Selected Variables among Patients and Controls
| Variable | Cases, n (%) (N = 863) | Controls, n (%) (N = 876) | ORa (95% CI) | P value |
|---|---|---|---|---|
| Age at diagnosis | ||||
| <50 | 111 (12.9) | 119 (13.6) | 1.00 | |
| 51–60 | 254 (29.4) | 275 (31.4) | 0.99 (0.73–1.35) | 0.956 |
| 61–70 | 322 (37.3) | 309 (35.3) | 1.13 (0.84–1.53) | 0.427 |
| >70 | 176 (20.4) | 173 (19.7) | 1.11 (0.79–1.54) | 0.552 |
| Sex | ||||
| Female | 342 (39.6) | 355 (40.5) | 1.00 | |
| Male | 521 (60.4) | 521 (59.5) | 1.05 (0.87–1.27) | 0.611 |
| Family history of cancerb | ||||
| No | 306 (35.7) | 406 (46.6) | 1.00 | |
| Yes | 550 (64.3) | 465 (53.4) | 1.59 (1.31–1.93) | <0.001 |
| History of diabetes | ||||
| No | 663 (76.8) | 783 (89.4) | 1.00 | |
| Yes | 200 (23.2) | 93 (10.6) | 2.58 (1.97–3.36) | <0.001 |
| Smoking status | ||||
| Non-smoker | 363 (42.1) | 457 (52.2) | 1.00 | |
| Smoker | 500 (57.9) | 419 (47.8) | 1.49 (1.23–1.80) | <0.001 |
| BMI (kg/m2)c | ||||
| <25.0 | 382 (50.5) | 425 (62.4) | 1.00 | |
| 25.0–30.0 | 285 (37.6) | 219 (32.2) | 1.45 (1.16–1.81) | 0.001 |
| >30.0 | 90 (11.9) | 37 (5.4) | 2.71 (1.80–4.07) | <0.001 |
Crude odds ratio.
Missing value from 7 patients and 5 controls.
Information was available for 757 cases and 681 controls recruited after 2004.
Tumor Mutations and Polymorphisms
No somatic SSTR5 gene mutations were detected in the 33 tumors analyzed. Three nonsynonymous SNPs—P109S (rs4988487, Ex1 +325C>T), L48M (rs4988483, Ex1 +142C>A), and P335L (rs169068, Ex1 −92C>T)—were identified (Fig. 1).
Fig. 1.
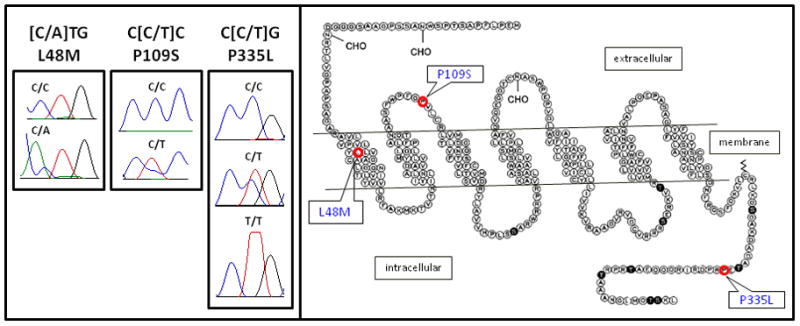
Typical Sanger sequencing chromatograms of the SSTR5 P109S, L48M, and P335L gene sequence variants (left panel). The SNPs’ locations in the protein are indicated in this topographic arrangement of the SSTR5 amino acid sequence and the N-glycosylation (CHO), phosphorylation (S, T, and ●), and palmitoylation (ΛΛ) sites (right panel). The SSTR5 topographic amino acid arrangement was published in Molecular Endocrinology, 13:82–90, 1999 and modified with permission of The Endocrine Society.
SSTR5 Genetic Variation and Pancreatic Cancer Risk
The genotype distributions were consistent with the Hardy-Weinberg equilibrium among cases and controls (χ2 = 0.107–2.53, P = 0.112–0.744). All 3 SSTR5 SNPs were in linkage disequilibrium, with |D′|> 0.85. The minor allele frequencies in controls were 0.04, 0.06, and 0.41 for the P109S, L48M, and P335L SNPs, respectively, which is similar to the reported frequencies of 0.04, 0.05, and 0.43 in the general population. No homozygous P109S or L48M variants were detected in cases or controls. The SSTR5 P109S CT genotype was significantly associated with pancreatic cancer risk (OR, 1.62 [95% CI, 1.08–2.43]; P = 0.019) (Table 3). The FPRP was 0.11, indicating noteworthiness. The SSTR5 P335L and L48M genotypes were not associated with pancreatic cancer risk. A haplotype analysis revealed that the CCT haplotype of P335L/L48M/P109S was associated with a significantly higher pancreatic cancer risk than was the most common TCC haplotype (OR, 1.54; 95% CI, 1.03–2.33; P = 0.038, Table 4).
Table 3.
Genotype Distribution and Association with Pancreatic Cancer Risk
| SSTR5 Genotype | Cases n=863 (%) | Controls n=876 (%) | OR (95% CI)a | OR (95% CI)b | P valueb |
|---|---|---|---|---|---|
| P335L (rs4988487) | |||||
| TT | 243 (28) | 267 (31) | 1.00 (reference) | 1.00 (reference) | |
| CT | 434 (51) | 417 (49) | 1.15 (0.92–1.44) | 1.02 (0.75–1.39) | 0.902 |
| CC | 176 (21) | 175 (20) | 1.08 (0.81–1.42) | 1.13 (0.88–1.46) | 0.332 |
| L48M (rs4988483) | |||||
| CC | 749 (90) | 764 (90) | 1.00 (reference) | 1.00 (reference) | |
| AC | 87 (10) | 8 (10) | 0.99 (0.72–1.38) | 0.94 (0.66–1.34) | 0.739 |
| P109S (rs169068) | |||||
| CC | 764 (90) | 798 (92) | 1.00 (reference) | 1.00 (reference) | |
| CT | 84 (10) | 65 (8) | 1.39 (0.98–1.97) | 1.62 (1.08–2.43) | 0.019 |
OR (95% CI) was adjusted for age, sex, family history of cancer, history of diabetes, and smoking.
OR was further adjusted for BMI.
Table 4.
Haplotype Frequency and Risk of Pancreatic Cancer
| Haplotype | Frequency | * OR (95% CI) | P |
|---|---|---|---|
| SSTR5 P335L/L48M/P109S | |||
| TCC | 0.55 | Reference | |
| CCC | 0.36 | 0.99 (0.84–1.17) | .898 |
| CAC | 0.05 | 0.99 (0.69–1.42) | .963 |
| CCT | 0.04 | 1.54 (1.03–2.33) | .038 |
| others | <0.01 | 0.41 (0.08–2.18) | .293 |
OR was adjusted for age, sex, family history of cancer, history of diabetes, smoking status, and BMI.
Joint Effect of SSTR5 Genotype and Smoking
We determined the joint effect of the SSTR5 genotype and known pancreatic cancer risk factors, including cigarette smoking, diabetes, obesity, and heavy alcohol consumption and found a significant association between the L48M SNP and cigarette smoking (Pinteraction = 0.035); the adjusted ORs (95% CI) were 0.58 (0.34–0.97), 1.49 (1.18–1.89), and 2.27 (1.35–3.83) for non-smokers with the variant AC genotype (OR10), smokers with the CC genotype (OR01), and smokers with the AC genotype (OR11), respectively, compared with non-smokers with the CC genotype (Table 5). No significant interaction was found between genotype and diabetes, BMI, and alcohol intake (data not shown).
Table 5.
Joint Effect of SSTR5 Genotype and Smoking on Pancreatic Cancer Risk
| Genotype | Smoking status | Cases/controls n/n | OR (95% CI)a | Pinteraction |
|---|---|---|---|---|
| P335L | 0.230 | |||
| TT | Never | 104/123 | 1.00 (reference) | |
| CC and CT | Never | 256/323 | 0.92 (0.65–1.31) | |
| TT | Ever | 139/144 | 1.34 (0.90–2.00) | |
| CC and CT | Ever | 356/269 | 1.71 (1.21–2.42) | |
| L48M | 0.035 | |||
| CC | Never | 319/385 | 1.00 (reference) | |
| AC | Never | 32/54 | 0.58 (0.34–0.97) | |
| CC | Ever | 430/379 | 1.49 (1.18–1.89) | |
| AC | Ever | 55/34 | 2.27 (1.35–3.83) | |
| P109S | 0.345 | |||
| CC | Never | 323/411 | 1.00 (reference) | |
| CT | Never | 34/36 | 1.34 (0.75–2.37) | |
| CC | Ever | 441/385 | 1.61 (1.28–2.03) | |
| CT | Ever | 50/28 | 3.17 (1.77–5.66) |
OR (95% CI) was adjusted for age, sex, family history of cancer, history of diabetes, and BMI.
Association between Clinical Predictors and Genotype and Overall Survival
Tumor characteristics and clinical predictors of survival are shown in Table 6. Tumor stage, tumor resection, tumor grade, and serum CA19-9 level at diagnosis were significantly associated with survival duration in all patients. Margin and node status were additional predictors in patients with resected tumors. By the end of the follow-up in May 2010, 661 (77%) patients had died. The median survival duration of the overall study population was 14.7 months (95% CI, 13.5–15.9 months). SSTR P335L CC genotype compared to the TT or CT genotype and P109S CC genotype compared to the CT genotype had shorter overall survival duration in patients with resectable tumors (Table 7) but not in those with advanced disease (data not shown). When the two SNPs were analyzed in combination (Fig. 2), patients with the P335L variant CC genotype and P109S CC genotype (CC-CC) had significantly shorter survival duration than did those with the P335L TT/CT and P109S CC genotypes (TT/CT-CC) or those with P335L any genotype and P109S CT genotype (Table 7), which translate into a hazard ratio of 1.57 and 95% CI of 1.06–2.34 using the TT/CT-CC genotype carriers as the referent group.
Table 6.
Patient Characteristics and Overall Survival (n = 863)
| Variable | No. of patients | No. of deaths (%) | MST (months) | P (log-rank) |
|---|---|---|---|---|
| Age (years) | 0.292 | |||
| ≤50 | 111 | 84 (76) | 15.4 | |
| 51–60 | 254 | 185 (73) | 13.9 | |
| 61–70 | 322 | 219 (68) | 14.7 | |
| >70 | 176 | 123 (69) | 17.1 | |
| Sex | 0.845 | |||
| Female | 342 | 250 (73) | 15.5 | |
| Male | 521 | 361 (69) | 14.8 | |
| Tumor stage | <0.001 | |||
| NED | 20 | 10 (50) | 46.7 | |
| Resectable | 250 | 143 (57) | 30.6 | |
| Locally advanced | 201 | 156 (78) | 14.4 | |
| Metastatic | 373 | 289 (77) | 9.5 | |
| Unstaged | 19 | 13 (68) | 19.3 | |
| Tumor resection | <0.001 | |||
| Yes | 284 | 155 (55) | 39.1 | |
| No | 579 | 456 (79) | 10.8 | |
| Margin statusa | 0.005 | |||
| Negative | 235 | 121 (51) | 40.0 | |
| Positive | 49 | 456 (79) | 24.9 | |
| Lymph node statusa | <0.001 | |||
| Negative | 122 | 50 (41) | 67.7 | |
| Positive | 162 | 105 (65) | 28.2 | |
| CA19-9 (units/mL)b | <0.001 | |||
| ≤47 | 192 | 107 (58) | 26.4 | |
| 48–500 | 326 | 232 (71) | 17.2 | |
| 501–1000 | 92 | 67 (73) | 13.4 | |
| >1000 | 248 | 202 (81) | 9.6 | |
| Tumor grade | <0.001 | |||
| Well | 33 | 19 (58) | 26.5 | |
| Moderate | 333 | 220 (66) | 21.4 | |
| Poor | 154 | 115 (75) | 12.4 | |
| Unknown | 343 | 257 (74) | 12.2 |
MST, median survival time; NED, no evidence of disease.
Data only available for 284 patients with resected disease.
Missing value in 5 patients.
Table 7.
SSTR5 Genotype and Overall Survival of Patients with Resectable Pancreatic Cancer
| Genotype | No. of patients | No. of deaths | MST | P (log- rank) | HR (95% CI)a |
|---|---|---|---|---|---|
| P335L | |||||
| TT | 70 | 38 | 28.2 | Ref | 1.0 |
| CT | 123 | 65 | 35.9 | 0.655 | 0.87 (0.57–1.31) |
| CC | 53 | 36 | 26.0 | 0.235 | 1.35 (0.85–2.13) |
| L48M | |||||
| CC | 238 | 134 | 30.7 | Ref | 1.0 |
| CA | 34 | 17 | 40.0 | 0.583 | 1.04 (0.58–1.87) |
| P109S | |||||
| CC | 224 | 132 | 29.2 | Ref | 1.0 |
| CT | 20 | 7 | - | 0.037 | 0.48 (0.22–1.03) |
| P335L-P109S | |||||
| TT/CT-CC | 177 | 97 | 31.3 | Ref | 1.0 |
| CC-CC | 46 | 34 | 20.2 | 0.036 | 1.57 (1.06–2.34) |
| Any-CT | 19 | 6 | - | 0.050 | 0.46 (0.20–1.05) |
HR, hazard ratio; MST, median survival time.
Hazard ratio was adjusted for resection status and CA19-9 level.
Fig. 2.
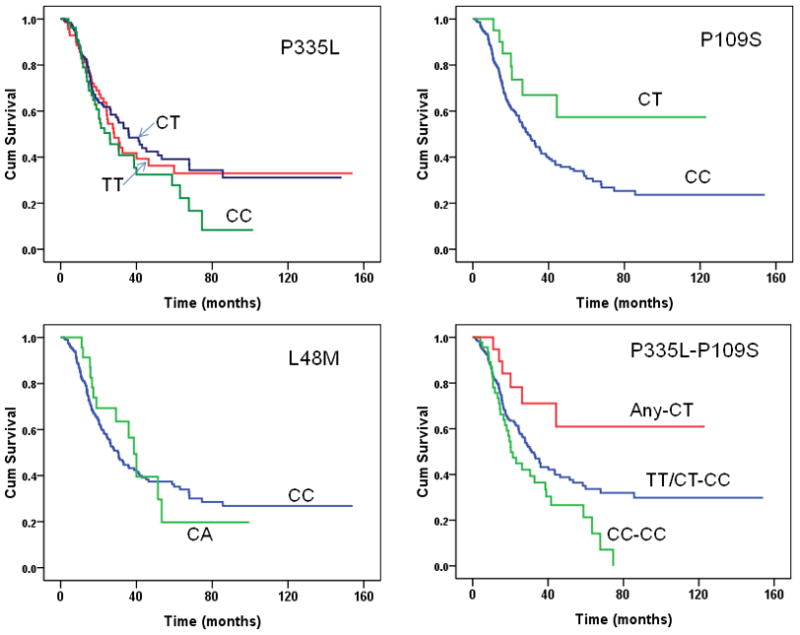
Overall survival curves of patients with resectable pancreatic cancer by SSTR5 genotype. In the combined “P335L-P109S” panel, “Any-CT” indicate P335L any genotype and P109S CT genotype; “TT/CT-CC” indicates P335L TT/CT and P109S CC genotype; “CC-CC” indicates CC genotype for both SNPs.
Discussion
In the current study, we detected no somatic mutations but found 3 nonsynonymous SNPs—P109S, L48M, and P335L of the SSTR5 gene—in 33 primary pancreatic adenocarcinomas. A case-control analysis revealed that both the P109S variant T allele alone and the L48M variant A allele and smoking were significantly associated with increased pancreatic cancer risk. The P335L CC and P109S CC genotypes were associated with reduced overall survival duration in patients with resectable disease. To our knowledge, this is the first study to demonstrate an association between SSTR5 gene variants and pancreatic cancer.
Studies of SSTR gene polymorphisms and cancer risk are limited and have been performed mainly in hormone-related cancers. One study showed that SSTR2 gene polymorphisms are significantly but weakly associated with breast cancer risk (14). Another showed no association between SSTR gene polymorphisms and prostate cancer risk (13). Thus, the relationship between SSTR genetic variants and cancer risk is controversial and may depend on cancer type. In the current study, we found a positive association between SSTR5 genotype and pancreatic cancer risk. Although the functional significance of the SSTR5 P109S variant has not been demonstrated experimentally, it was predicted to be damaging or deleterious using a bioinformatics approach (19). Considering that SST’s role in inhibiting cell growth and proliferation is mediated via SSTR (8) and the SSTR5 expression level is decreased in pancreatic cancer tissue (10), the variant genotypes may result in decreased expression or impaired function, which promotes cell proliferation and cancer development in the pancreas.
The SSTR5 receptor has a high affinity for SST, a multifunctional neuropeptide that is widely distributed throughout the central nervous system and acts in the anterior pituitary gland to inhibit growth hormone secretion (20). Therefore, SSTR5 negatively regulates IGF-1 levels through the growth hormone (GH)-IGF-1 axis (11). Circulating IGF-1 in the blood has been correlated with breast and prostate cancer risk (21) but not with pancreatic cancer risk (22). However, IGF-1 and IGF-1 receptors are highly expressed in pancreatic cancer cell lines (23). Notably, the SSTR5 L48M variant allele has been associated with lower circulating IGF-1 and IGFBP3 levels (13, 14, 24). In the current study, the L48M variant had no significant main effect but had a differential effect on the risk of pancreatic cancer by cigarette smoking status, i.e. the L48M variant was associated with reduced risk among non-smokers but increased risk among smokers. A similar but insignificant interaction between the P109S variant and smoking was also observed. A previous study has observed that smokers had a lower serum IGF1:IGFBP3 molar ratio and IGF1 level than non-smokers among African Americans but not among whites (25). Thus the increased risk of pancreatic cancer in smokers by L48M variant allele could not be explained by its impact on IGF1 level. We can only speculate that the reduced risk associated with the valiant allele in non-smokers was related to a lower level of IGF1 while the increased risk in smokers could be related to impaired inhibition of cell growth and proliferation conferred by the variant allele. Further investigation is required to confirm these observations and to illustrate the mechanisms underlying such associations. Because of the extremely low frequency of the SNPs’ homozygous variants and the relatively small sample size, the effect of the homozygote on pancreatic cancer could not be assessed in this study.
The SSTR5 P335L SNP has a much higher minor allele frequency than do the P109S and L48M SNPs but little effect on pancreatic cancer risk. Nevertheless, patients with resectable tumors who have the P335L and P109S CC genotypes had significantly shorter overall survival durations than did patients with the common P335L TT/CT and P109S CC genotypes. The underlying mechanism of this association remains unknown. A recent study showed that P335L T allele overexpression in pancreatic cancer cells leads to increased cell proliferation and PDX-1 expression, whereas C allele overexpression enhances the SSTR5 agonist’s inhibitory effect on cell proliferation and insulin secretion (manuscript in preparation). SSTR5’s role in pancreatic cancer development and its complex interactions with PDX1 or IGF-1 and progression require further investigation. The P109S variant CT genotype’s protective effect on patient survival could indicate that its association with pancreatic cancer risk is confounded by a survival bias. However, it was not associated with survival in the most patients with advanced disease.
Overall, we observed weak associations between 3 nonsynonymous SSTR5 gene SNPs and pancreatic cancer risk and survival. Because the minor allele frequencies of 2 of these gene variants (L48M and P109S) were low (≤6%), these observations need to be confirmed in much larger studies. If confirmed, this genetic information will be useful for estimating pancreatic cancer risk and survival.
Acknowledgments
Grant support: Supported by National Institutes of Health (NIH) R01 grant CA098380 (D.L.), the Effie and Wolford Cain Foundation (M.C.G.), and NIH R01 grant CDK046441 (F.C.B.).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Li D, Morris JS, Liu J, et al. Body mass index and risk, age of onset, and survival in patients with pancreatic cancer. JAMA. 2009;301:2553–62. doi: 10.1001/jama.2009.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 4.Hejna M, Schmidinger M, Raderer M. The clinical role of somatostatin analogues as antineoplastic agents: much ado about nothing? Ann Oncol. 2002;13:653–68. doi: 10.1093/annonc/mdf142. [DOI] [PubMed] [Google Scholar]
- 5.Butturini G, Bettini R, Missiaglia E, et al. Predictive factors of efficacy of the somatostatin analogue octreotide as first-line therapy for advanced pancreatic endocrine carcinoma. Endocrine-Related Cancer. 2006;13:1213–21. doi: 10.1677/erc.1.01200. [DOI] [PubMed] [Google Scholar]
- 6.Canobbio L, Cannata D, Miglietta L, et al. Somatuline (BIM 23014) and tamoxifen treatment of postmenopausal breast cancer patients: clinical activity and effect on insulin-like growth factor-I (IGF-I) levels. Anticancer Res. 1995;15:2687–90. [PubMed] [Google Scholar]
- 7.Raderer M, Hamilton G, Kurtaran A, et al. Treatment of advanced pancreatic cancer with the long-acting somatostatin analogue lanreotide: in vitro and in vivo results. Br J Cancer. 1999;79:535–7. doi: 10.1038/sj.bjc.6690084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollak MN, Schally AV. Mechanisms of antineoplastic action of somatostatin analogs. Proc Soc Exp Biol Med. 1998;217:143–52. doi: 10.3181/00379727-217-44216. [DOI] [PubMed] [Google Scholar]
- 9.Reubi JC, Waser B, Schaer JC, Laissue JA. Somatostatin receptor sst1 sst5 expression in normal and neoplastic human tissues using receptor autoradiography with subtype-selective ligands. Eur J Nucl Med. 2001;28:836–46. doi: 10.1007/s002590100541. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Li W, Kim HJ, Yao Q, Chen C, Fisher WE. Characterization of somatostatin receptor expression in human pancreatic cancer using real-time RT-PCR. J Surg Res. 2004;119:130–7. doi: 10.1016/j.jss.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Butler AA, Yakar S, LeRoith D. Insulin-like growth factor-I: compartmentalization within the somatotropic axis? News Physiol Sci. 2002;17:82–5. doi: 10.1152/nips.01351.2001. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst. 2000;92:1472–89. doi: 10.1093/jnci/92.18.1472. [DOI] [PubMed] [Google Scholar]
- 13.Johansson M, McKay JD, Wiklund F, et al. Genetic variation in the SST gene and its receptors in relation to circulating levels of insulin-like growth factor-I, IGFBP3, and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:1644–50. doi: 10.1158/1055-9965.EPI-08-0893. [DOI] [PubMed] [Google Scholar]
- 14.Canzian F, McKay JD, Cleveland RJ, et al. Genetic variation in the growth hormone synthesis pathway in relation to circulating insulin-like growth factor-I, insulin-like growth factor binding protein-3, and breast cancer risk: results from the European prospective investigation into cancer and nutrition study. Cancer Epidemiol Biomarkers Prev. 2005;14:2316–25. doi: 10.1158/1055-9965.EPI-04-0874. [DOI] [PubMed] [Google Scholar]
- 15.Taniyama Y, Suzuki T, Mikami Y, Moriya T, Satomi S, Sasano H. Systemic distribution of somatostatin receptor subtypes in human: an immunohistochemical study. Endocr J. 2005;52:605–11. doi: 10.1507/endocrj.52.605. [DOI] [PubMed] [Google Scholar]
- 16.Szepeshazi K, Schally AV, Halmos G, et al. Targeting of cytotoxic somatostatin analog AN-238 to somatostatin receptor subtypes 5 and/or 3 in experimental pancreatic cancers. Clin Cancer Res. 2001;7:2854–61. [PubMed] [Google Scholar]
- 17.Hassan MM, Bondy ML, Wolff RA, et al. Risk factors for pancreatic cancer: case-control study. Am J Gastroenterol. 2007;102:2696–707. doi: 10.1111/j.1572-0241.2007.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–24. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel YC, Greenwood MT, Panetta R, Demchyshyn L, Niznik H, Srikant CB. The somatostatin receptor family. Life Sci. 1995;57:1249–65. doi: 10.1016/0024-3205(95)02082-t. [DOI] [PubMed] [Google Scholar]
- 21.Renehan AG, Zwahlen M, Minder C, O’Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–53. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 22.Wolpin BM, Michaud DS, Giovannucci EL, et al. Circulating insulin-like growth factor axis and the risk of pancreatic cancer in four prospective cohorts. Br J Cancer. 2007;97:98–104. doi: 10.1038/sj.bjc.6603826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmann U, Funatomi H, Yokoyama M, Beger HG, Korc M. Insulin-like growth factor I overexpression in human pancreatic cancer: evidence for autocrine and paracrine roles. Cancer Res. 1995;55:2007–11. [PubMed] [Google Scholar]
- 24.Filopanti M, Ronchi C, Ballare E, et al. Analysis of somatostatin receptors 2 and 5 polymorphisms in patients with acromegaly. J Clin Endocrinol Metab. 2005;90:4824–8. doi: 10.1210/jc.2005-0132. [DOI] [PubMed] [Google Scholar]
- 25.Hoyo C, Grubber J, Demark-Wahnefried W, et al. Predictors of variation in serum IGF1 and IGFBP3 levels in healthy African American and white men. J Natl Med Assoc. 2009;101:711–6. doi: 10.1016/s0027-9684(15)30981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


