Abstract
Although DISC1 has been implicated in many psychiatric disorders, including schizophrenia, bipolar disorder, schizoaffective disorder and major depression, its biological role in these disorders is unclear. To better understand this gene and its role in psychiatric disease, we conducted transcriptional profiling and genome-wide association analysis in 1 232 pedigreed Mexican American individuals for whom we have neuroanatomic images, neurocognitive assessments and neuropsychiatric diagnoses. SOLAR was used to determine heritability, identify gene expression patterns and perform association analyses on 188 quantitative brain-related phenotypes. We found that the DISC1 transcript is highly heritable (h2=0.50; p=1.97 × 10−22), and that gene expression is strongly cis-regulated (cis-LOD=3.89) but is also influenced by trans-effects. We identified several DISC1 polymorphisms that were associated with cortical gray-matter thickness within the parietal, temporal and frontal lobes. Associated regions affiliated with memory included the entorhinal cortex (rs821639, p=4.11 × 10−5; rs2356606, p=4.71 × 10−4), cingulate cortex (rs16856322, p=2.88 × 10−4) and parahippocampal gyrus (rs821639, p=4.95 × 10−4); those affiliated with executive and other cognitive processing included the transverse temporal gyrus (rs9661837, p=5.21 × 10−4; rs17773946, p=6.23 × 10−4), anterior cingulate cortex (rs2487453, p=; 4.79 × 10−4; rs3738401, p= 5.43 × 10−4) and medial orbitofrontal cortex (rs9661837; p=7.40 × 10−4). Cognitive measures of working memory (rs2793094, p=3.38 × 10−4), as well as lifetime history of depression (rs4658966, p=4.33 × 10−4; rs12137417, p=4.93 × 10−4) and panic (rs12137417, p=7.41 × 10−4) were associated with DISC1 sequence variation. DISC1 has well-defined genetic regulation and clearly influences important phenotypes related to psychiatric disease.
Keywords: DISC1, association, neuroanatomical, neurocognitive, endophenotype, cis-regulation, depression, panic, memory, learning
Introduction
Disrupted in schizophrenia 1 (DISC1) is one of the few genes that have been consistently implicated in psychiatric illness, making it one of the most promising genes for understanding the biological mechanisms that drive such disorders. However, little is known about how DISC1 is regulated and how extensively neuroanatomical and neurocognitive measures are influenced by DISC1 genetic variation.
DISC1 was initially identified as a schizophrenia candidate gene when linkage analysis in a large Scottish family revealed co-segregation between schizophrenia and a balanced translocation (1;11)(q42.1;14.3) that disrupts the DISC1 locus (1, 2). Further analysis revealed linkage of the DISC1 locus to schizophrenia, bipolar disorder and/or schizoaffective disorder in families from Finland (3, 4), Scotland (5), and the United Kingdom/Ireland (6). It has been noted that both unaffected carriers of this transclocation and individuals affected with schizophrenia show reduced amplitude and latency of the auditory P300 event-related potential (7) and this in turn has been correlated with reduced gray matter volume in the left superior temporal gyrus, suggesting that DISC1 may mediate this important trait marker for schizophrenia risk. In addition, numerous studies have provided evidence for association of various DISC1 single nucleotide polymorphisms (SNPs) in schizophrenia (8-11), bipolar disorder (9, 10, 12, 13), depression (14) and autism and Asperger syndrome (15). Although some DISC1 variants have been associated with brain-related phenotypes, including hippocampal gray matter volume and function (16), poor concentration (in schizophrenics) (17), recall and memory (10), verbal ability and memory (10), visuospatial ability (10), psychomotor processing (10), visual working memory (10, 18, 19) and general cognitive ability (20), these studies have examined only a few selected SNPs usually based on prior evidence for potential involvement. Furthermore, DISC1 SNPs have been associated with anxiety, depression, emotional stability and neuroticism in elderly women (21).
Composed of 13 alternatively spliced exons spanning about 415kb (NG_011681.1), DISC1 has numerous isoforms and is expressed most highly during periods of neurogenesis (22). In the adult mammalian brain, expression levels are highest in the hippocampus and cerebral cortex (23, 24). However, the regulation of DISC1 expression has not been well studied, with one study finding no evidence (25), and another showing only suggestive evidence for cis-regulation (26). DISC1 interacts with a number of binding partners to regulate neuronal development (23, 27). Notably, DISC1 plays a critical role in normal microtubular dynamics (24, 27), neurite outgrowth (23, 27, 28) and neuronal migration (27).
To improve our understanding of DISC1 we used a combination of transcriptional profiling of lymphocytes and genome wide genotyping to investigate the genetic factors driving DISC1 expression in a large sample of randomly ascertained individuals. Here we show that the expression of the DISC1 gene is heritable and highly cis-regulated. Furthermore, we have identified DISC1 sequence variation that strongly influences various neuroanatomic and neurocognitive traits that commonly coincide with mental illness.
Methods
For more detailed information, please refer to online Supplementary Methods.
Population phenotypes
We utilized samples derived from individuals in the San Antonio Family Heart Study (SAFHS) (29) and San Antonio Diabetes/Gallbladder Study (SAFDGS) (30, 31), which both consist of large extended families. All study participants gave informed consent and the study was undertaken with approval by the Institutional Review Board at The University of Texas Health Science Center at San Antonio. Brain-related phenotypes were collected for up to 625 participants. Neuroanatomical images for 387 individuals have been collected using a Siemens 3T MRI scanner (Siemens, New York, NY), linearly aligned and averaged (32) and analyzed using FreeSurfer software (33). Gray-matter thickness, surface area and volume have been determined for 34 cortical regions of interest (ROI), combining left and right hemispheres and gray-matter volume has been determined for 16 subcortical regions. The South Texas Assessment of Neurocognition (STAN) neuropsychological battery (34) includes clinical and experimental tests investigating a wide range of cognitive domains, including general intellectual functioning, sensory-motor and processing speed, attention, executive functioning, working memory, long-term memory, language and social cognition. To date, 625 GOBSF individuals have received the STAN neuropsychological battery as well as the Mini-International Neuropsychiatric Interview (MINI-Plus) (35), a semi-structured interview to facilitate diagnoses of DSM-IV and ICD-10 psychiatric illnesses.
Genetic analysis
We previously generated transcriptional profiles for 1 240 individuals within the SAFHS population using Illumina® Sentrix® Whole Genome (WG-6) Version 1 BeadChips (Illumina, San Diego, CA) and utilized highly polymorphic STR markers to examine evidence of cis-linkage of DISC1 gene expression (36). To examine genetic variation that might influence DISC1 gene expression and brain-related phenotypes, we used Illumina GoldenGate technology to genotype 1 240 individuals for 125 SNPs within the 5′ UTR, 3′ UTR and DISC1 gene, 11 of which overlapped with previously implicated DISC1 SNPs (Supplementary Table 2). One 5′ UTR polymorphism (rs3738398), which could not effectively be incorporated into the OPA pool, was genotyped using a Taqman assay (Supplementary Table 2). Illumina HumanHap550k version 3.0 and Human1M version 1.0 BeadChips were used to assess 543 031 SNPs in 858 individuals to determine trans-effects on DISC1 gene expression. There were no SNPs within the DISC1 gene within this panel, making this genotyping platform independent of the analysis of DISC1 gene variation.
Statistical analysis
All statistical analyses were performed using the SOLAR software package (37). Heritability was assessed using a classical approach to deconstruct the phenotypic variance into independent genetic and environmental components and the Loki software package (38) was used to compute probabilities of multipoint identity-by-descent (IBD) allele sharing. Covariates sex, age, sex*age, age2, and sex*age2) were included in every analysis performed. Quantitative trait linkage analysis of DISC1 gene expression levels was performed using the likelihood ratio tests (37). Heritability and linkage analysis of DISC1 transcript levels is based on 1 240 individuals, while our genome-wide association of DISC1 expression is performed on a subset of 858 of these individuals. Association testing was performed by measured genotype analysis (39). All brain-related measures were performed on the same set of 625 individuals (387 for neuroanatomical measures), all were typed for each DISC1 sequence variant.
To account for multiple testing, we performed modified Bonferroni corrections adjusting for the number of effective DISC1 SNPs (n=66) for each test (required significance p=7.7×10−4.
Results
DISC1 is a cis-regulated gene
The expression of DISC1 was highly heritable (h2=0.50; p=1.97×10−22). Furthermore, quantitative trait linkage analysis performed at the genetic location of DISC1 revealed strong evidence for cis-regulation, (cis-LOD=3.89; p=1.16×10−5). This cis-acting QTL was estimated to account for a sizable 25.6% of the total phenotypic variation in DISC1 expression levels suggesting that the cis effects are large.
For the 1 240 individuals that had previously undergone transcriptional profiling, we genotyped 126 known DISC1 SNPs. A total of 1 232 individuals had complete data sets for both DISC1 transcripts levels and DISC1 SNP variation. The effective number of SNPs could be reduced to 66 based on linkage disequilibrium across the gene (Supplementary Figure 1). Global variation within the promoter region (Supplementary Figure 2) was significantly associated with expression of the DISC1 transcript (p=5.7×10−34). To determine whether particular variants were more highly associated with expression of DISC1, we performed association analysis with individual SNPs using DISC1 gene expression as a quantitative trait. Within the promoter region, 15 SNPs were significant after a Bonferroni correction for the effective number of SNPs (see Table 1, Figure 1), which is in contrast to prior studies by Hayesmoore (25) and Hennah and Porteus (26). Multivariant analysis (40) identified at least 5 independent cis-acting contributions. The best multivariant model included five SNPs (rs12042938, rs34574703, rs3738398, rs16854957, and rs16856202) and accounted for 16.6% (multivariate p=1.07×10−39) of the total phenotypic variation in DISC1 expression. This estimate of the component of heritability due to cis effects confirms the importance of local sequence variation on DISC1 regulation.
Table 1.
DISC1 cis-acting SNPs
| SNP | MAF* | Gene Region | p-value |
|---|---|---|---|
|
| |||
| rs12042938 | 0.478 (C) | Intron 1 | 3.51 × 10−36 |
| rs11122319 | 0.487 (A) | Intron 1 | 1.33 × 10−33 |
| rs3738398 | 0.424 (C) | 5′ UTR | 5.98 × 10−33 |
| rs12030517 | 0.444 (C) | Intron 1 | 3.65 × 10−31 |
| rs823163 | 0.489 (C) | Intron 1 | 1.49 × 10−30 |
| rs823161 | 0.486 (T) | Intron 1 | 3.83 × 10−30 |
| rs2082552 | 0.362 (G) | Intron 1 | 7.13 × 10−27 |
| rs6541281 | 0.389 (T) | Intron 1 | 1.89 × 10−25 |
| rs1417584 | 0.399 (A) | Intron 1 | 2.12 × 10−22 |
| rs11122322 | 0.361 (G) | Intron 3 | 3.48 × 10−11 |
| rs3738401 | 0.348 (A) | Exon 2 | 3.78 × 10−10 |
| rs11122324 | 0.364 (A) | Intron 3 | 7.06 × 10−10 |
| rs2487453 | 0.379 (A) | Intron 3 | 2.58 × 10−9 |
| rs3806372 | 0.081 (T) | 5′ UTR | 5.69 × 10−6 |
| rs823162 | 0.042 (G) | Intron 1 | 1.61 × 10−4 |
Minor allele listed according to the forward strand designated by dbSNP build 131
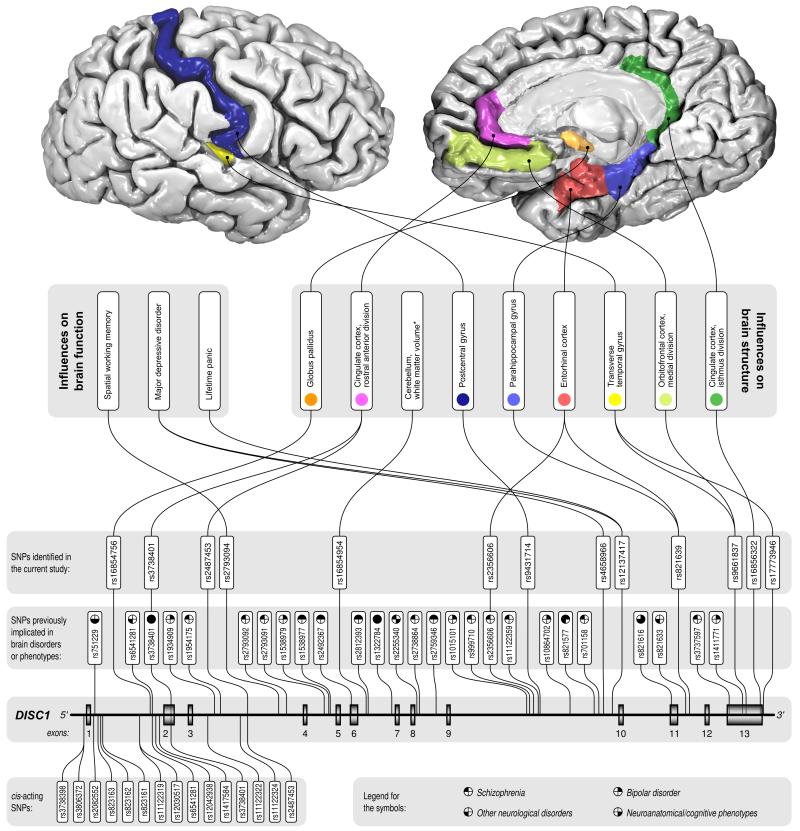
Figure 1.
Location of DISC1 SNPs and their influence on brain structure and function. Proximity to SNPs that have been previously implicated in published studies of schizophrenia, bipolar disorder, other neurological disorders, or neuroanatomical/neurocognitive phenotypes in either single SNP or haplotype analysis, is also shown.
DISC1 expression is modulated by trans-effects
We used genome-wide association analysis employing 543 031 SNPs in 857 of these individuals to identify genomic regions harboring genes that might contribute to the modulation of DISC1 expression. Eighteen SNPs were identified that were significantly associated with DISC1 expression using a false discovery rate of 0.10. Of these, 15 SNPs were cis-acting (as previously described) and the remaining three SNPs represented significant trans-effects in two genomic regions. The SNPs were within or near the NUP210 gene at 3p25 (p=5.5×10−6) and MPP6 gene at 7p15 (p=8.1×10−6). Although these genes may be directly responsible for regulating DISC1 expression it is more likely that potential upstream modifiers are located within these genomic regions. To identify potential modifiers of DISC1 expression we examined the genes within 1Mb either side of the trans-effects seen at 3p25 and 7p15 to determine whether quantitative expression of these genes was also genetically correlated (rG) with DISC1 expression. At 3p25, bivariate quantitative genetic analysis revealed several correlated genes including PPARG (rG=0.703, p=6.4×10−4), MGC2776 (rG=0.280, p=1.4×10−2), RAF1 (rG=0.252, p=2.9×10−2), RPL32 (rG=−0.293, p=4.6×10−2), NUP210 (rG=−0.219, p=3.2×10−2), HDAC11 (rG=−0.549, p=1.6×10−2), CHCHD4 (rG=0.225, p=2.4×10−2), and XPC (rG=0.252, p=1.4×10−2). At 7p15 these genes included SNX10 (rG=0.386, p=4.4×10−4), SCAP2 (rG=0.387, p=1.4×10−3), and TRA2A (rG=−0.294, p=8.8×10−3).
DISC1 is associated with brain-related endophenotypes
In those individuals, for whom we had both genotypic and brain-related phenotypic data, we examined associations between DISC1 variants and neuroanatomical and neurocognitive measures. Implementing a modified Bonferroni correction within phenotypes for the 66 effective SNPs, we identified 14 associations with neuroanatomical traits (Table 2), two associations with neurocognitive traits and two associations with a neuropsychiatric diagnosis of recurrent major depressive disorder. These associations and SNP locations, as well as proximity to previously implicated SNPs, are described in Figure 1. Notably, many associated SNPs are located within the 3′ end of the gene between exons 9 and 13, a fewer number of SNPs are located in intron 1, exon 2 and intron 3. Significant associations were detected with cortical thickness in the temporal, parietal and frontal lobes and in some cases specific DISC1 variants were associated with cortical thickness in multiple regions.
Table 2.
Brain-related phenotypes associated with DISC1 variation
| Phenotype | Hemisphere | SNP | MAF* | p-value |
|---|---|---|---|---|
| Temporal Lobe, Medial Aspect, Entorhinal Cortex; thickness |
- overall - left - left |
rs821639 rs821639 rs2356606 |
0.277 (C) 0.136 (C) |
4.11 × 10−5 1.11 × 10−4 4.71 × 10−4 |
| Left Pallidum; volume | rs16854756 | 0.010 (C) | 2.01 × 10−4 | |
| Cingulate Cortex, Retrosplenial Cortex; thickness |
rs16856322 | 0.271 (T) | 2.88 × 10−4 | |
| Left Cerebellum White Matter; volume | rs16854954 | 0.168 (C) | 4.31 × 10−4 | |
| Temporal Lobe, Medial Aspect, Parahippocampal Gyrus; thickness |
- left - overall |
rs821639 rs821639 |
0.277 (C) | 4.68 × 10−4 4.95 × 10−4 |
| Cingulate Cortex, Rostral Anterior Division; thickness |
- overall - overall |
rs2487453 rs3738401 |
0.379 (A) 0.348 (A) |
4.79 × 10−4 5.43 × 10−4 |
| Temporal Lobe, Lateral Aspect, Transverse Temporal Cortex; thickness |
- overall - right |
rs9661837 rs17773946 |
0.009 (G) 0.012 (G) |
5.21 × 10−4 6.23 × 10−4 |
| Parietal Lobe, Postcentral Gyrus; thickness |
- left | rs9431714 | 0.308 (G) | 7.22 × 10−4 |
| Frontal Lobe, Medial Division, Orbitofrontal Cortex; thickness |
- right | rs9661837 | 0.009 (G) | 7.40 × 10−4 |
Minor allele listed according to the forward strand designated by dbSNP build 131
Additionally, associations were detected with measures of spatial working memory (rs2793094, p=3.38×10−4), recurrent major depressive disorder (MDD) (rs4658966, p=4.33×10−4; rs12137417, p=4.93×10−4) and lifetime measures of panic (rs12137417, p=7.41×10−4).
All observed associations are obligately independent of sex and age effects which were utilized as covariates in each analysis. Additionally, there was no evidence for any correlation between age or sex and genotype for any DISC1 sequence variant.
Discussion
The DISC1 gene is a strong candidate gene for a number of psychiatric illnesses. Using a combination of transcriptional profiling of lymphocytes, targeted genotyping and genome wide association analysis, we show here that DISC1 expression is heritable, highly regulated by variation within its promoter region and to a lesser extent influenced by other genomic regions (3p25, 7p15). Furthermore, variation within DISC1 is associated with a number of neuroanatomical and neurocognitive phenotypes, most significantly cortical thickness.
One obvious caveat regarding our findings is that while we correct on a phenotype by phenotype basis for the multiple testing of DISC1 sequence variants, we do not perform such correction across all phenotypes examined. We anticipate that replication of these observed associations for specific phenotypes will be needed to confirm our findings.
Lymphocyte-derived or lymphoblastoid-derived DISC1 gene expression has previously shown variability in psychiatric diseases, such as bipolar disorder (41), schizophrenia and schizoaffective disorder (42), and in unaffected family members carrying the DISC1 t(1;11) translocation (43). Further, this variability has been correlated with markers for schizophrenia risk (7). This current study supplements the increasing body of evidence that suggests transcriptional profiling of lymphocytes is a good surrogate model for diseases in which the tissue of interest is difficult to obtain (36, 44). Moreover, they suggest that DISC1 gene expression in lymphocytes may provide a suitable marker for psychiatric disease risk.
We determined DISC1 to be strongly cis-regulated. Using allelic expression analysis of a single common SNP (rs3738401) within exon 2 of DISC1 in 148 unrelated individuals, Hayesmoore and colleagues failed to detect cis-regulation of DISC1 (25). Using a significantly larger population size, we found moderate evidence for cis-regulation at this same SNP (p=3.78×10−10), although the strongest cis-effects were seen in intron 1 (p=1.61×10−4-3.51×10−36) and near the 5′ end of the gene (5.69×10−6-5.98×10−33). Hennah, et al, probed existing databases for evidence of cis-regulation at 754 SNPs, also for a small number of samples (210 cell lines derived from the four HapMap population cohorts). They found only suggestive evidence for cis-regulation in three of the four populations for six SNPs, four of which were located about 4-30kb upstream of our region of significance (26). The remaining two SNPs were located at the 5′ end of our region of significance, including one (rs3738398) that we also found to be significant (26). It is possible that ethnic heterogeneity and low sample numbers contributed to an underpowered study, which was unable to detect significant cis-effects. Given that we investigated cis-effects in 1 240 ethnically similar individuals, we believe that our study was more highly powered to identify strong cis-regulation of the DISC1 gene.
The DISC1 protein-protein “interactome” is a highly connected network consisting of 127 proteins and 158 interactions (45), in which DISC1 acts as a “hub” protein functioning in cAMP signaling, axon elongation and neuronal migration. We have identified two genomic regions, which may comprise genes that regulate DISC1 gene expression. Although there are no obvious candidate genes within the 3p25 or 7p15 regions, it is possible that one or more genes within these regions may act to regulate DISC1 expression through protein modification (e.g. HDAC11), cell-specific splicing (e.g. TRA2A), or DNA repair (e.g. XPC). Given the importance of DISC1 as a “hub” protein within many neurological processes, it is important to identify upstream modifiers of DISC1 gene expression, which could potentially be targeted for therapeutic benefit. Other factors regulating DISC1 gene or protein expression that are outside the context of this study should also be considered, such as epigenetic modifications. Recently, it was shown that enriched histone methylation occurs proximal to transcription start sites of annotated genes and that reorganization of the neuronal epigenome is correlated with age, suggesting an important role for epigenetics in the regulation of genes involved in neuronal development (46).
There is little overlap between cis-regulated SNPs and those that influence brain-related phenotypes, suggesting that genetic variation influencing lymphocyte-derived gene expression and functional outcome are largely independent. The 5′ region of DISC1 contains 15 cis-acting SNPs, of which 11 are located within the 5′UTR or intron 1, where the strongest signal is seen. In contrast, most SNPs influencing brain structure and function are located toward the 3′ end of the DISC1 gene, and might be more likely to influence post-transcriptional or post-translational modifications. Only two SNPs, rs3738401 and 2487453, were found to be cis-acting and influence brain structure, specifically thickness of the cingulate cortex. Of these, rs3738401 is part of a haplotype block (HEP3) that has previously been implicated in schizophrenia, schizoaffective disorder, bipolar disorder and MDD as well as related phenotypes of delusions/hallucinations, manic and depressive thoughts and visual memory (10, 18, 47). Findings from our study, in addition to previous studies, suggest this SNP may be extremely important in not only regulating DISC1 gene expression, but also in eliciting a functional response the predisposes to various psychiatric disorders.
Only one other previously implicated SNP, rs2356606, was also implicated within this study and was found to be associated with thickness of the entorhinal cortex and postcentral gyrus. This SNP is within a haplotype block that has previously been associated with schizophrenia in females (48). By analyzing a large number (n=115) of previously unstudied SNPs, we have identified a number of novel associations with brain-related phenotypes, which need to be verified by follow-up studies. It is important to note that a number of SNPs that were found to be associated with brain-related phenotypes in our study are in close proximity to previously implicated SNPs (seen in Figure 1). It is therefore possible that they are in linkage disequilibrium with previously implicated SNPs, and the causal variant would likely lie somewhere within this region.
Many of the associated SNPs identified in this study are within intronic regions of the DISC1 gene, for which we have no current evidence of a functional effect on the DISC1 protein. It is possible that these SNPs may play a role in regulation of or targeting by miRNAs, or in RNA stability or splicing efficiency, but it is perhaps more likely that associated intronic SNPs are in linkage disequilibrium with other functional SNPs. Few studies have identified functional variants of the DISC1 gene and most have looked at the effect of the Ser704Cys variant (rs821616) on psychiatric disease. Variation at this locus has been associated with schizophrenia brain morphology (14, 49). Despite its proximity to a number of SNPs for which we have strong evidence of association with brain morphology, we have found no such association with the Ser704Cys variant. Given that we identified significant brain-related associations within multiple regions of the DISC1 gene, we expect that additional functional analyses will likely reveal a number of polymorphisms that directly or indirectly act to influence brain structure and phenotypes associated with cognitive functioning.
Reduction in cortical thickness is well known to be associated with schizophrenia (50-52) and has been implicated in bipolar disorder (53), depression (54), posttraumatic stress disorder (55), autism spectrum disorder (56) and Alzheimers disease (57). In this study, we identified a number of brain regions in which cortical thickness is associated with DISC1 genetic variation, including regions of the temporal, parietal and frontal lobes and the cingulate cortex. DISC1 gene variation was associated with cortical thickness of brain regions involved in memory (entorhinal cortex, cingulate cortex, parahippocampal gyrus) (58, 59) and emotion- and reward-based cognitive processing and learning (anterior cingulate cortex, medial orbitofrontal cortex) (60, 61). Given the inherent functions of these brain regions, it is not surprising that variation in cortical thickness of such regions has already been identified in patients with schizophrenia (parahippocampal gyrus, anterior cingulate cortex, entorhinal cortex, medial orbitofrontal cortex) (50-52); bipolar I disorder (left cingulate gyrus) (53); depression (postcentral gyrus, anterior cingulate cortex, medial orbitofrontal cortex) (54); posttraumatic stress disorder (parahippocampal gyrus) (55); autism spectrum disorder (left anterior cingulate gyrus, left parahippocampal gyrus, left medial orbitofrontal gyrus) (56, 62); and Alzheimer’s disease or mild cognitive impairment (parahippocampal gyrus, entorhinal cortex, retrosplenial cortex, anterior cingulate cortex) (57, 63, 64). Furthermore, individuals at ultra high risk for psychosis and relatives of individuals affected with schizophrenia also show reduction in cortical thickness in certain brain regions (50, 65-67), suggesting a strong genetic component to regional cortical thinning, which may be progressive in nature.
The spatial working memory measure, a delayed response task, significantly associated with DISC1 was previously shown to be sensitive to genetic liability for schizophrenia (68) and psychotic bipolar disorder (69). Furthermore, brain regions associated with DISC1 (e.g. entorhinal cortex, cingulate cortex and parahippocampal gyrus) were engaged when healthy subjects (70) and first episode patients with schizophrenia (71) performed this task during functional MRI. Although SNPs associated with cortical thickness of regions associated with memory are located in the 3′ region of the DISC1 gene (between exons 9 and 13), the SNP associated with working memory is located closer to the 5′ end of the gene (intron 3). It is therefore likely that the true functional SNP(s) that play a role in cortical thickness are different from those involved in spatial working memory and that multiple DISC1 SNPs may contribute to memory functioning. Measures of lifetime panic and depression were also associated with variation in DISC1. Interestingly, SNPs associated with MDD and lifetime panic were located in close proximity, within intron 9 of the DISC1 gene (Figure 1). Variation associated with the orbitofrontal cortex, previously implicated in panic disorder and MDD (72-74), was located nearby in exon 13. However, variation influencing the rostral cingulate cortex, a brain region associated with affective and anxiety disorders, was located in the 5′ region of the gene (exon 2, intron 3).
The DISC1 gene, although well implicated in psychiatric disease, is not well understood. We have attempted here to determine factors that influence DISC1 expression and identify DISC1 sequence variation associated with neuroanatomical and neurocognitive traits. We found significant associations in regions of the brain associated with memory and cognitive processing, which was also reflected in associations with cognitive measures of memory, depression and panic. The results presented here suggest that variation in DISC1 sequence and gene expression acts coordinately to affect both brain structure and cognition, specifically within phenotypes that are implicated in psychiatric disease.
Supplementary Material
Acknowledgments
Financial support
Financial support for this study was provided by NIMH grants MH0708143 (PI: DC Glahn), MH078111 (PI: J Blangero), and MH083824 (PI: DC Glahn). SOLAR is supported by NIMH grant MH59490 (PI: J Blangero). The collection of the family data and blood samples in the San Antonio Family Heart Study was supported by NHLBI program project grant P01 HL045522 (PI: J Blangero). We would also like to thank the Azar and Shepperd families for their support of the transcriptional profiling. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant Number C06 RR017515 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990 Jul 7;336(8706):13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 2.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000 May 22;9(9):1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 3.Ekelund J, Hovatta I, Parker A, Paunio T, Varilo T, Martin R, et al. Chromosome 1 loci in Finnish schizophrenia families. Hum Mol Genet. 2001 Jul 15;10(15):1611–1617. doi: 10.1093/hmg/10.15.1611. [DOI] [PubMed] [Google Scholar]
- 4.Ekelund J, Hennah W, Hiekkalinna T, Parker A, Meyer J, Lonnqvist J, et al. Replication of 1q42 linkage in Finnish schizophrenia pedigrees. Mol Psychiatry. 2004 Nov;9(11):1037–1041. doi: 10.1038/sj.mp.4001536. [DOI] [PubMed] [Google Scholar]
- 5.Macgregor S, Visscher PM, Knott SA, Thomson P, Porteous DJ, Millar JK, et al. A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry. 2004 Dec;9(12):1083–1090. doi: 10.1038/sj.mp.4001544. [DOI] [PubMed] [Google Scholar]
- 6.Hamshere ML, Bennett P, Williams N, Segurado R, Cardno A, Norton N, et al. Genomewide linkage scan in schizoaffective disorder: significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. Arch Gen Psychiatry. 2005 Oct;62(10):1081–1088. doi: 10.1001/archpsyc.62.10.1081. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001 Aug;69(2):428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saetre P, Agartz I, De Franciscis A, Lundmark P, Djurovic S, Kahler A, et al. Association between a disrupted-in-schizophrenia 1 (DISC1) single nucleotide polymorphism and schizophrenia in a combined Scandinavian case-control sample. Schizophr Res. 2008 Dec;106(2-3):237–241. doi: 10.1016/j.schres.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Hennah W, Thomson P, McQuillin A, Bass N, Loukola A, Anjorin A, et al. DISC1 association, heterogeneity and interplay in schizophrenia and bipolar disorder. Mol Psychiatry. 2008 Mar 4; doi: 10.1038/mp.2008.22. [DOI] [PubMed] [Google Scholar]
- 10.Palo OM, Antila M, Silander K, Hennah W, Kilpinen H, Soronen P, et al. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum Mol Genet. 2007 Oct 15;16(20):2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- 11.Liu YL, Fann CS, Liu CM, Chen WJ, Wu JY, Hung SI, et al. A single nucleotide polymorphism fine mapping study of chromosome 1q42.1 reveals the vulnerability genes for schizophrenia, GNPAT and DISC1: Association with impairment of sustained attention. Biol Psychiatry. 2006 Sep 15;60(6):554–562. doi: 10.1016/j.biopsych.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 12.Perlis RH, Purcell S, Fagerness J, Kirby A, Petryshen TL, Fan J, et al. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch Gen Psychiatry. 2008 Jan;65(1):53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- 13.Thomson PA, Wray NR, Millar JK, Evans KL, Hellard SL, Condie A, et al. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry. 2005 Jul;10(7):657–668. 616. doi: 10.1038/sj.mp.4001669. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, Ishimoto T, et al. Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet. 2006 Oct 15;15(20):3024–3033. doi: 10.1093/hmg/ddl244. [DOI] [PubMed] [Google Scholar]
- 15.Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry. 2008 Feb;13(2):187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- 16.Di Giorgio A, Blasi G, Sambataro F, Rampino A, Papazacharias A, Gambi F, et al. Association of the SerCys DISC1 polymorphism with human hippocampal formation gray matter and function during memory encoding. Eur J Neurosci. 2008 Nov;28(10):2129–2136. doi: 10.1111/j.1460-9568.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HJ, Park HJ, Jung KH, Ban JY, Ra J, Kim JW, et al. Association study of polymorphisms between DISC1 and schizophrenia in a Korean population. Neurosci Lett. 2008 Jan 3;430(1):60–63. doi: 10.1016/j.neulet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Hennah W, Tuulio-Henriksson A, Paunio T, Ekelund J, Varilo T, Partonen T, et al. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005 Dec;10(12):1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- 19.Burdick KE, Hodgkinson CA, Szeszko PR, Lencz T, Ekholm JM, Kane JM, et al. DISC1 and neurocognitive function in schizophrenia. Neuroreport. 2005 Aug 22;16(12):1399–1402. doi: 10.1097/01.wnr.0000175248.25535.f6. [DOI] [PubMed] [Google Scholar]
- 20.Thomson PA, Harris SE, Starr JM, Whalley LJ, Porteous DJ, Deary IJ. Association between genotype at an exonic SNP in DISC1 and normal cognitive aging. Neurosci Lett. 2005 Nov 25;389(1):41–45. doi: 10.1016/j.neulet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Harris SE, Hennah W, Thomson PA, Luciano M, Starr JM, Porteous DJ, et al. Variation in DISC1 is associated with anxiety, depression and emotional stability in elderly women. Mol Psychiatry. 2010 Mar;15(3):232–234. doi: 10.1038/mp.2009.88. [DOI] [PubMed] [Google Scholar]
- 22.Lipska BK, Peters T, Hyde TM, Halim N, Horowitz C, Mitkus S, et al. Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum Mol Genet. 2006 Apr 15;15(8):1245–1258. doi: 10.1093/hmg/ddl040. [DOI] [PubMed] [Google Scholar]
- 23.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T, et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003 Jul;8(7):685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 24.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayesmoore JB, Bray NJ, Owen MJ, O’Donovan MC. DISC1 mRNA expression is not influenced by common Cis-acting regulatory polymorphisms or imprinting. Am J Med Genet B Neuropsychiatr Genet. 2008 Oct 5;147B(7):1065–1069. doi: 10.1002/ajmg.b.30715. [DOI] [PubMed] [Google Scholar]
- 26.Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS ONE. 2009;4(3):e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005 Dec;7(12):1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 28.Hattori T, Baba K, Matsuzaki S, Honda A, Miyoshi K, Inoue K, et al. A novel DISC1-interacting partner DISC1-Binding Zinc-finger protein: implication in the modulation of DISC1-dependent neurite outgrowth. Mol Psychiatry. 2007 Apr;12(4):398–407. doi: 10.1038/sj.mp.4001945. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996 Nov 1;94(9):2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 30.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, et al. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet. 1999 Apr;64(4):1127–1140. doi: 10.1086/302316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duggirala R, Mitchell BD, Blangero J, Stern MP. Genetic determinants of variation in gallbladder disease in the Mexican-American population. Genet Epidemiol. 1999;16(2):191–204. doi: 10.1002/(SICI)1098-2272(1999)16:2<191::AID-GEPI6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Kochunov P, Lancaster JL, Glahn DC, Purdy D, Laird AR, Gao F, et al. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum Brain Mapp. 2006 Dec;27(12):957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999 Feb;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 34.Glahn DC, Almasy L, Barguil M, Hare E, Peralta JM, Kent JW, Jr., et al. Neurocognitive endophenotypes for bipolar disorder identified in multiplex multigenerational families. Arch Gen Psychiatry. 2010 Feb;67(2):168–177. doi: 10.1001/archgenpsychiatry.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33. quiz 34-57. [PubMed] [Google Scholar]
- 36.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007 Oct;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 37.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998 May;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997 Sep;61(3):748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986 May;50(Pt 2):181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 40.Blangero J, Goring HH, Kent JW, Jr., Williams JT, Peterson CP, Almasy L, et al. Quantitative trait nucleotide analysis using Bayesian model selection. Hum Biol. 2005 Oct;77(5):541–559. doi: 10.1353/hub.2006.0003. [DOI] [PubMed] [Google Scholar]
- 41.Maeda K, Nwulia E, Chang J, Balkissoon R, Ishizuka K, Chen H, et al. Differential expression of disrupted-in-schizophrenia (DISC1) in bipolar disorder. Biol Psychiatry. 2006 Nov 1;60(9):929–935. doi: 10.1016/j.biopsych.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 42.Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry. 2005 Aug;10(8):758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- 43.Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005 Nov 18;310(5751):1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 44.Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005 Feb 5;133B(1):1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- 45.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007 Jan;12(1):74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 46.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, Weng Z, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010 May 11;107(19):8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, Haukka J, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003 Dec 1;12(23):3151–3159. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- 48.Schumacher J, Laje G, Abou Jamra R, Becker T, Muhleisen TW, Vasilescu C, et al. The DISC locus and schizophrenia: evidence from an association study in a central European sample and from a meta-analysis across different European populations. Hum Mol Genet. 2009 Jul 15;18(14):2719–2727. doi: 10.1093/hmg/ddp204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi T, Suzuki M, Tsunoda M, Maeno N, Kawasaki Y, Zhou SY, et al. The Disrupted-in-Schizophrenia-1 Ser704Cys polymorphism and brain morphology in schizophrenia. Psychiatry Res. 2009 May 15;172(2):128–135. doi: 10.1016/j.pscychresns.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Jung WH, Kim JS, Jang JH, Choi JS, Jung MH, Park JY, et al. Cortical Thickness Reduction in Individuals at Ultra-High-Risk for Psychosis. Schizophr Bull. 2009 Dec 21; doi: 10.1093/schbul/sbp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkatasubramanian G, Jayakumar PN, Gangadhar BN, Keshavan MS. Automated MRI parcellation study of regional volume and thickness of prefrontal cortex (PFC) in antipsychotic-naive schizophrenia. Acta Psychiatr Scand. 2008 Jun;117(6):420–431. doi: 10.1111/j.1600-0447.2008.01198.x. [DOI] [PubMed] [Google Scholar]
- 52.Baiano M, Perlini C, Rambaldelli G, Cerini R, Dusi N, Bellani M, et al. Decreased entorhinal cortex volumes in schizophrenia. Schizophr Res. 2008 Jul;102(1-3):171–180. doi: 10.1016/j.schres.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 53.Lyoo IK, Sung YH, Dager SR, Friedman SD, Lee JY, Kim SJ, et al. Regional cerebral cortical thinning in bipolar disorder. Bipolar Disord. 2006 Feb;8(1):65–74. doi: 10.1111/j.1399-5618.2006.00284.x. [DOI] [PubMed] [Google Scholar]
- 54.Peterson BS, Warner V, Bansal R, Zhu H, Hao X, Liu J, et al. Cortical thinning in persons at increased familial risk for major depression. Proc Natl Acad Sci U S A. 2009 Apr 14;106(15):6273–6278. doi: 10.1073/pnas.0805311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodward SH, Schaer M, Kaloupek DG, Cediel L, Eliez S. Smaller global and regional cortical volume in combat-related posttraumatic stress disorder. Arch Gen Psychiatry. 2009 Dec;66(12):1373–1382. doi: 10.1001/archgenpsychiatry.2009.160. [DOI] [PubMed] [Google Scholar]
- 56.Jiao Y, Chen R, Ke X, Chu K, Lu Z, Herskovits EH. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage. 2010 Apr 1;50(2):589–599. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Julkunen V, Niskanen E, Muehlboeck S, Pihlajamaki M, Kononen M, Hallikainen M, et al. Cortical thickness analysis to detect progressive mild cognitive impairment: a reference to Alzheimer’s disease. Dement Geriatr Cogn Disord. 2009;28(5):404–412. doi: 10.1159/000256274. [DOI] [PubMed] [Google Scholar]
- 58.Zola-Morgan S, Squire LR, Ramus SJ. Severity of memory impairment in monkeys as a function of locus and extent of damage within the medial temporal lobe memory system. Hippocampus. 1994 Aug;4(4):483–495. doi: 10.1002/hipo.450040410. [DOI] [PubMed] [Google Scholar]
- 59.Sutherland RJ, Whishaw IQ, Kolb B. Contributions of cingulate cortex to two forms of spatial learning and memory. J Neurosci. 1988 Jun;8(6):1863–1872. doi: 10.1523/JNEUROSCI.08-06-01863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bush G, Vogt BA, Holmes J, Dale AM, Greve D, Jenike MA, et al. Dorsal anterior cingulate cortex: a role in reward-based decision making. Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):523–528. doi: 10.1073/pnas.012470999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coricelli G, Critchley HD, Joffily M, O’Doherty JP, Sirigu A, Dolan RJ. Regret and its avoidance: a neuroimaging study of choice behavior. Nat Neurosci. 2005 Sep;8(9):1255–1262. doi: 10.1038/nn1514. [DOI] [PubMed] [Google Scholar]
- 62.Meguid N, Fahim C, Yoon U, Nashaat NH, Ibrahim AS, Mancini-Marie A, et al. Brain Morphology in Autism and Fragile X Syndrome Correlates With Social IQ: First Report From the Canadian-Swiss-Egyptian Neurodevelopmental Study. J Child Neurol. 2010 Jan 27; doi: 10.1177/0883073809341670. [DOI] [PubMed] [Google Scholar]
- 63.Oliveira PP, Jr., Nitrini R, Busatto G, Buchpiguel C, Sato JR, Amaro E., Jr. Use of SVM methods with surface-based cortical and volumetric subcortical measurements to detect Alzheimer’s disease. J Alzheimers Dis. 2010 Jan;19(4):1263–1272. doi: 10.3233/JAD-2010-1322. [DOI] [PubMed] [Google Scholar]
- 64.Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. Focal decline of cortical thickness in Alzheimer’s disease identified by computational neuroanatomy. Cereb Cortex. 2005 Jul;15(7):995–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- 65.Harms MP, Wang L, Campanella C, Aldridge K, Moffitt AJ, Kuelper J, et al. Structural abnormalities in gyri of the prefrontal cortex in individuals with schizophrenia and their unaffected siblings. Br J Psychiatry. 2010 Feb;196(2):150–157. doi: 10.1192/bjp.bp.109.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prasad KM, Goradia D, Eack S, Rajagopalan M, Nutche J, Magge T, et al. Cortical surface characteristics among offspring of schizophrenia subjects. Schizophr Res. 2010 Feb;116(2-3):143–151. doi: 10.1016/j.schres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goghari VM, Rehm K, Carter CS, MacDonald AW., 3rd. Regionally specific cortical thinning and gray matter abnormalities in the healthy relatives of schizophrenia patients. Cereb Cortex. 2007 Feb;17(2):415–424. doi: 10.1093/cercor/bhj158. [DOI] [PubMed] [Google Scholar]
- 68.Glahn DC, Therman S, Manninen M, Huttunen M, Kaprio J, Lonnqvist J, et al. Spatial working memory as an endophenotype for schizophrenia. Biol Psychiatry. 2003 Apr 1;53(7):624–626. doi: 10.1016/s0006-3223(02)01641-4. [DOI] [PubMed] [Google Scholar]
- 69.Glahn DC, Bearden CE, Barguil M, Barrett J, Reichenberg A, Bowden CL, et al. The neurocognitive signature of psychotic bipolar disorder. Biol Psychiatry. 2007 Oct 15;62(8):910–916. doi: 10.1016/j.biopsych.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 70.Glahn DC, Kim J, Cohen MS, Poutanen VP, Therman S, Bava S, et al. Maintenance and manipulation in spatial working memory: dissociations in the prefrontal cortex. Neuroimage. 2002 Sep;17(1):201–213. doi: 10.1006/nimg.2002.1161. [DOI] [PubMed] [Google Scholar]
- 71.Cannon TD, Glahn DC, Kim J, Van Erp TG, Karlsgodt K, Cohen MS, et al. Dorsolateral prefrontal cortex activity during maintenance and manipulation of information in working memory in patients with schizophrenia. Arch Gen Psychiatry. 2005 Oct;62(10):1071–1080. doi: 10.1001/archpsyc.62.10.1071. [DOI] [PubMed] [Google Scholar]
- 72.Kent JM, Coplan JD, Mawlawi O, Martinez JM, Browne ST, Slifstein M, et al. Prediction of panic response to a respiratory stimulant by reduced orbitofrontal cerebral blood flow in panic disorder. Am J Psychiatry. 2005 Jul;162(7):1379–1381. doi: 10.1176/appi.ajp.162.7.1379. [DOI] [PubMed] [Google Scholar]
- 73.Sobanski T, Wagner G, Peikert G, Gruhn U, Schluttig K, Sauer H, et al. Temporal and right frontal lobe alterations in panic disorder: a quantitative volumetric and voxel-based morphometric MRI study. Psychol Med. 2010 Jan 8;:1–8. doi: 10.1017/S0033291709991930. [DOI] [PubMed] [Google Scholar]
- 74.Smoski MJ, Felder J, Bizzell J, Green SR, Ernst M, Lynch TR, et al. fMRI of alterations in reward selection, anticipation, and feedback in major depressive disorder. J Affect Disord. 2009 Nov;118(1-3):69–78. doi: 10.1016/j.jad.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.