Abstract
SHP-1 belongs to the family of non-receptor protein tyrosine phosphatases (PTPs) and generally acts as a negative regulator in a variety of cellular signaling pathways. Previously the crystal structures of the tail-truncated SHP-1 and SHP-2 revealed an autoinhibitory conformation. To understand the regulatory mechanism of SHP-1, we have determined the crystal structure of the full length SHP-1 at 3.1 Å. Although the tail was disordered in current structure, the huge conformational rearrangement of the N-SH2 domain and the incorporation of sulfate ions into the ligand-binding site of each domain indicate that the SHP-1 is in the open conformation. The N-SH2 domain in current structure is shifted away from the active site of the PTP domain to the other side of the C-SH2 domain, resulting in exposure of the active site. Meanwhile, the C-SH2 domain is twisted anticlockwise by about 110°. In addition, a set of new interactions between two SH2 domains and between the N-SH2 and the catalytic domains is identified, which could be responsible for the stabilization of SHP-1 in the open conformation. Based on the structural comparison, a model for the activation of SHP-1 is proposed.
Keywords: Protein tyrosine phosphatase, SHP-1, Conformation
Protein tyrosine phosphorylation is a post-translational modification that has important roles in cell growth, proliferation and differentiation (Fischer et al., 1991). This reversible biological process is strictly controlled in normal cells by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs). The structure and function of PTKs have been widely studied and are well understood. In contrast, much less research has focused on PTPs (Lorenz, 2009). The PTPs, which dephosphorylate the phosphorylated tyrosine in targeted proteins, can be categorized into four groups: receptor-like PTPs (RPTPs), non-receptor PTPs (non-TM PTPs), dual-specificity phosphatases (DSPs) and low molecular weight phosphatases (LMPs) (Neel and Tonks, 1997). They all contain a catalytic domain with a signature motif (H/V)C(X)5R(S/T) in the active site and have the ability to hydrolyze p-nitrophenyl phosphate (p-NPP) in vitro (Zhang, 2002). SHP-1 and SHP-2 are two members of the src homology 2 (SH2) domain-containing protein tyrosine phosphatases, which belong to non-TM PTP group. They have high sequence identity and each protein contains two SH2 domains at the N-terminus followed by a catalytic domain (PTP domain) and a C-terminal tail. However, these two enzymes have different, even opposite biological functions (Marrero et al., 1998; Kuo et al., 2010).
Unlike SHP-2, which is ubiquitously expressed in mammalian cells and mainly serves as a positive regulator (Shi et al., 2000; Salmond and Alexander, 2006), SHP-1 is primarily expressed in hematopoietic cells and is generally considered a negative regulator of a variety of signaling pathways (Marsh et al., 2003; Kamata et al., 2003; Bonaparte et al., 2006; Croker et al., 2008). Motheaten (me) mice and viable motheaten (mev) mice that have mutations in SHP-1 gene exhibit severe immunodeficiency and autoimmune diseases (Green and Shultz, 1975; Shultz et al., 1993; Tsui et al., 1993). Dysfunction of SHP-1 protein in lymphocytes has been related to the abnormal cell growth and even leukemia, lymphoma and other related diseases (Wu et al., 2003).
SHP-1 remains inactive in the resting state. Activation of SHP-1 is caused by binding of its SH2 domains to the phosphotyrosine residues of the immune receptor tyrosine-based inhibitory motifs (ITIMs) from various activators. The activated SHP-1 will then dephosphorylate the downstream targeted proteins (Mori et al., 2008). SHP-1 can also be activated by its tyrosine phosphorylated tail (Lorenz et al., 1994) and acidic phospholipids (Zhao et al., 1993; Frank et al., 1999). The activation of SHP-1 is accompanied by its conformational rearrangement.
An auto-inhibitory conformation with the N-SH2 domain blocking the active site of the PTP domain was observed in the crystal structures of tail-truncated SHP-1 and SHP-2 proteins (Hof et al., 1998; Yang et al., 2003). In this conformation, the Nβ4-Nβ5 hairpin loop or D’E loop from N-SH2 domains inserts into the active sites of PTP domains and blocks substrate binding to the active site. The N-SH2 domain was proposed to move away from the active site and render the enzymes active upon ligand binding to the N-SH2 domain. It behaves as a molecular switch in the activation and regulation of SHP-1 and SHP-2 (Hof et al., 1998; Yang et al., 2003). This autoinhibitory mechanism has been supported by mutagenesis studies (Pei et al., 1994; Pei et al., 1996).
To understand how the autoinhibitory conformation is activated and what kinds of molecular interactions are used to stabilize the protein in the active conformation, the structure of SHP-1 in active conformation is needed. Here we report a crystal structure of full length SHP-1 (mutant C453S) in the open conformation where the N-SH2 domain is away from the active site, resulting in exposure of the active site. This open conformation is stabilized by the interactions between N-SH2 and PTP domains, and between the two SH2 domains.
MATERIAL AND METHODS
Protein expression, purification and crystallization
The procedures for protein expression and purification were modified from previous study (Liang et al., 1997). Briefly, the full length SHP-1 (residues 1–595) was cloned into vector pT7 and expressed in the E. Coli BL21 (DE3). Protein was purified by ion exchange through a Q-Sepharose fast flow column (Sigma, St. Louis, MO) and affinity chromatography through an HDBP (L-Histidyldiazobenzylphophonic acid)(Sigma, St. Louis, MO) column. Bound proteins were eluted with a linear gradient of 0 to 1M NaCl in QA buffer containing 25mM Tris-HCl, pH 7.5, 2mM β-mercaptoethanol, 1mM EDTA. The harvested protein was desalted by dialysis and concentrated to 3.5 mg/ml in buffer QA for crystallization. Crystallization experiments were performed by the hanging-drop vapor diffusion method at 4°C. Crystals were obtained from the crystallization drops by mixing 2.5 µl of protein sample with equal volume of reservoir solution, containing 1.8M ammonium sulfate, 0.1M glycine, 0.1M Tris-HCl, pH 7.0. The addition of 0.5 µl 14mM deoxy Big Chap (#8 of detergent screen I, Hampton Research) into the drops was important to obtain diffraction-quality crystals. In addition, repeated cycles of macro-seeding were required to obtain crystals of sufficient size for data collection.
Data Collection, Structural Determination and Refinement
Crystals were stabilized by addition of 25% glycerol into the reservoir solution as cryoprotectant and flash-cooled in liquid nitrogen for data collection. Diffraction data were collected with a MarCCD detector at the Gulf Coast Protein Crystallography Consortium (GCPCC) beamline of the Center for Advanced Microstructures and Devices (CAMD, Louisiana State University). Data were processed using HKL2000 software (Otwinowski and Minor, 1997). The crystals belong to the rhombohedral space group R32 with cell dimensions a=b=231.92Å and c=78.85Å. There is one molecule per asymmetric unit. The best dataset was taken in the resolution range of 116-3.1 Å.
The structure was determined by molecular replacement with the program Phaser (McCoy et al., 2007). The C-terminal tail-truncated SHP-1 structure (PDB code: 2B3O) was used as the search model. The search was based on domains with linkers between domains removed. The large PTP domain was first searched for, followed by locating the two smaller SH2 domains. Structural refinement was done with Refmac (Murshudov et al., 1997), and graphical modeling was performed with COOT (Emsley et al., 2010). The grouped B factor refinement strategy was used in the B factor refinement as in CNS (Brünger et al., 1998). The linker regions and missing amino acid residues, as well as water molecules and sulfate ions were built on difference electron density maps. The final model is composed of residues of 1–529, five sulfate ions and 20 water molecules. The statistics for data collection and structural refinement are listed in Table 1.
Table 1.
Statistics for data collection and refinement. Highest resolution shell statistics given in parentheses.
| Data collection | |
| Beamline | GCPCC |
| Wavelength (Å) | 1.3807 |
| Space group | R32 |
| Cell constants | a=b=231.92Å c=78.85Å |
| Resolution (Å) | 116-3.1 (3.21-3.10) |
| Total reflections | 632873 |
| Unique reflections | 14036 |
| I/σ (I) | 18.1 (1.5) |
| Completeness (%) | 99.9 (98.5) |
| Redundancy | 12.4 (8.5) |
| Rmerge(%) | 8.6(59.6) |
| Structural Refinement | |
| Resolution (Å) | 116-3.1 |
| R factor | 0.225 |
| Rfree factor | 0.276 |
| RMS (Bond length, Å) | 0.005 |
| (Bond angle, °) | 0.855 |
| Mean B factor (Å2) | 83.0 |
| Ramachandran statistics | |
| Favored (%) | 92.0 |
| Allowed (%) | 5.3 |
| Disallowed (%) | 2.7 |
RESULTS
Overall structure of SHP-1
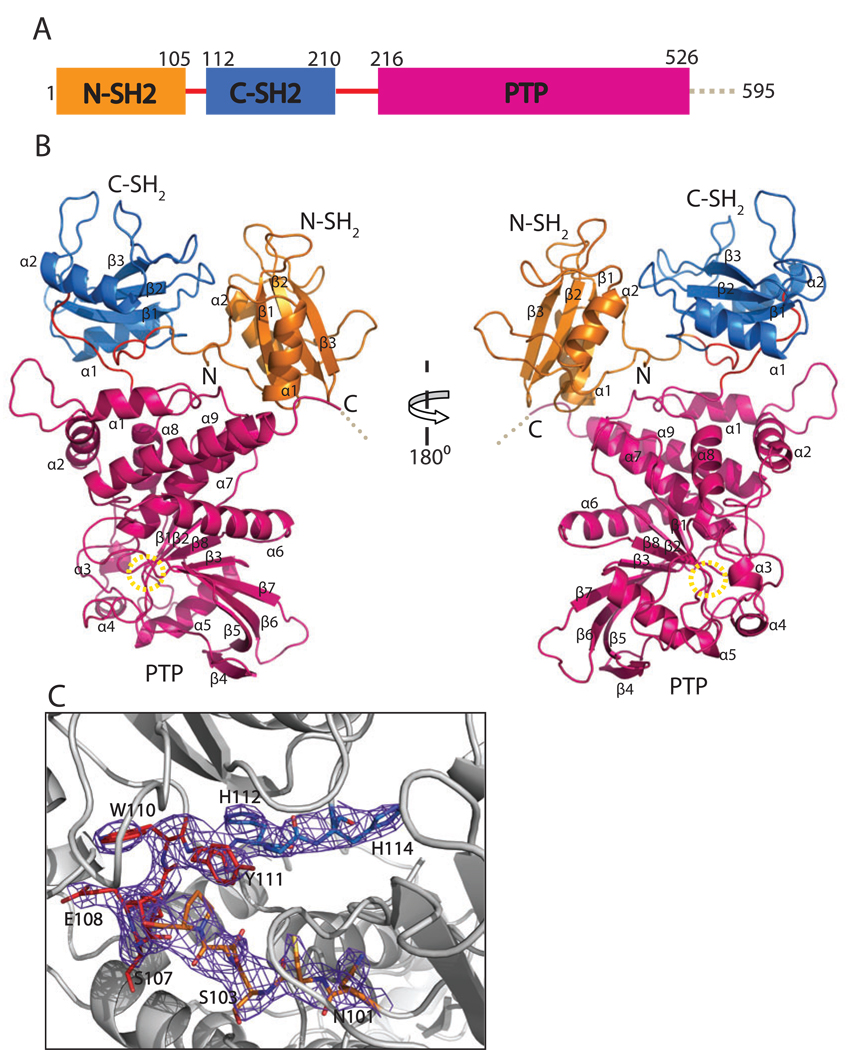
To understand the regulatory mechanism of SHP-1 we determined the crystal structure of the catalytically inactive full-length SHP-1 mutant C453S (residues 1–595) by the molecular replacement method at 3.1Å resolution. The structure was refined to an R-factor of 22.5%. This SHP-1 was crystallized as a monomer in the asymmetric unit. Residues 1–529 were built into the electron density map. The C-terminal tail was disordered and missing from the final model (Fig. 1A–B).
Figure 1.
Overall structure of SHP-1. The N-SH2 domain is in orange, the C-SH2 domain is in marine, the PTP domain is in hot pink, and the linkers between them are in red. A. The domain organization of SHP-1. The dashed line represents the C-terminal tail which is disordered in the structure. B. The overall structure of SHP-1. Two SH2 domains are arranged like horns of the PTP domain. The yellow dashed circle shows the position of the active site. C. Electron density map around the linker of two SH2 domains. The 2Fo-Fc map is contoured at 1σ level.
The three domains and the connected linkers were well defined from the electron density map. Figure 1C shows a sample electron density map in which the linker region between two SH2 domains is clearly visible. The PTP domain and two SH2 domains closely resemble the published phosphatase and SH2 domain structures, respectively (Hof et al., 1998; Yang et al., 2003; Stuckey et al., 1994; Barford et al., 1994; Eck et al., 1996; Yang et al., 1998). The PTP domain is composed of 9 α-helices and 8 β-strands. The highly twisted eight-stranded β-sheet is flanked by α helices on both sides. Two SH2 domains each contain three-stranded antiparallel β-sheet flanked by an α helix on either side. The SH2 domains are close and located on one side of the PTP domain, looking like its two horns in the overall view (Fig. 1B). The C-terminal tail is disordered and in the solvent channel.
SHP-1 is in the open conformation as reflected by the movement of the N-SH2 domain and the sulfate binding to the ligand binding site of each domain
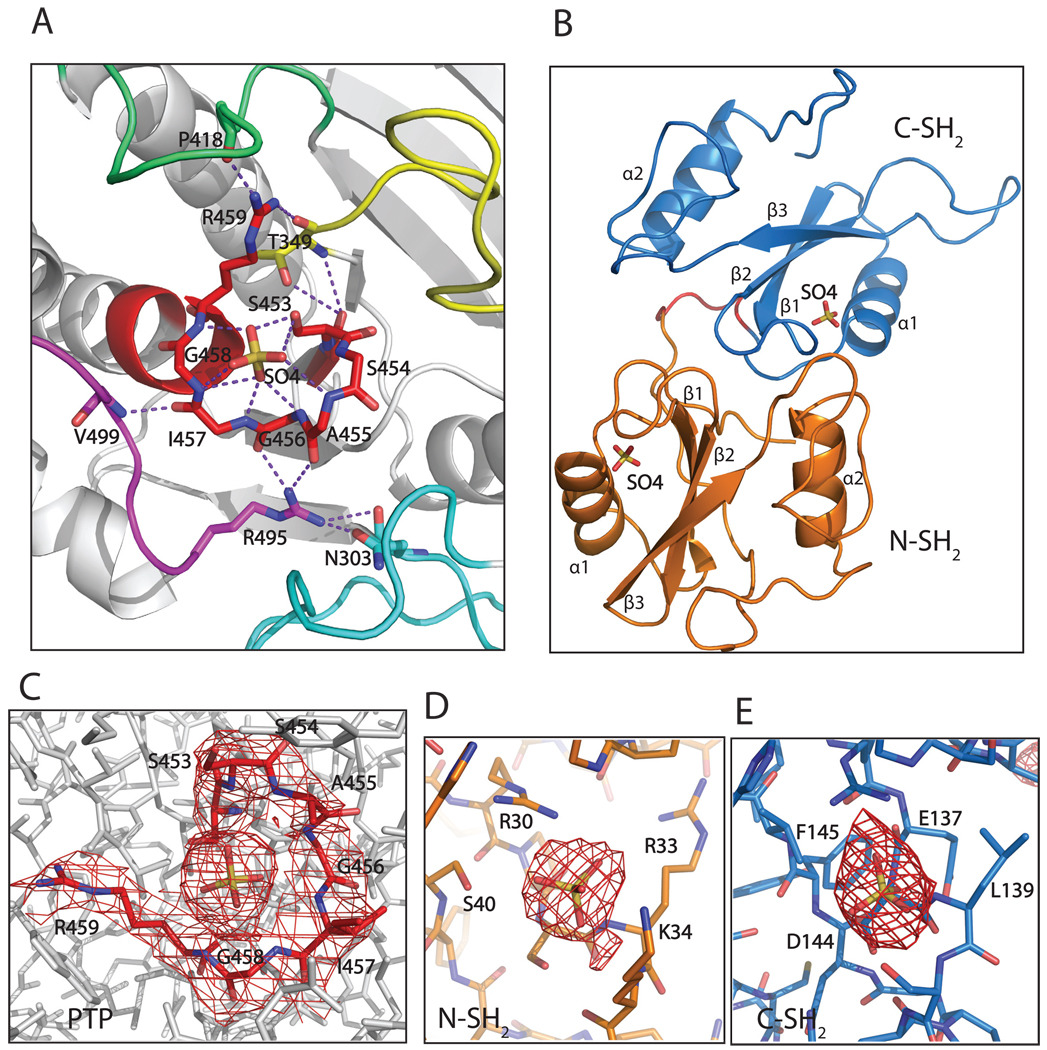
Previous structures of tail-truncated SHP-1 and SHP-2 have revealed an auto-inhibited conformation in which the association between the N-SH2 domain and the PTP domain blocks the active site of the catalytic domain (Hof et al., 1998; Yang et al., 2003). In the present structure, the N-SH2 domain is away from the active site of the PTP domain, resulting in exposure of the active site (Fig. 2A, Fig. 5A). Interestingly, a sulfate ion, an analog of the phosphate, was found in the active site (Fig. 2A, C). The phosphatase active site is located in a positively charged deep pocket, which is surrounded and stabilized by several residues from other surface loops: T349 from the loop between strands β3 and β4; P418 from the WPD loop between helix α3 and strand β7; R495 and V499 from the loop between helices α5 and α6; R495 also forms a hydrogen bond with N303 from the loop between helix α1 and strand β1. These loops form the wall of the active site pocket (Fig. 2A). The nucleophile C453S is located in the center of the PTP signature motif (Fig. 2A). In addition, a sulfate ion was also modeled in the ligand-binding pocket of each SH2 domain (Fig. 2B, D, and E). The movement of the N-SH2 domain away from the PTP domain and the sulfates in the binding pockets of all three domains indicate that the structure is in the open conformation.
Figure 2.
The ligand-binding site of each domain has a bound sulfate ion. A. The active site (in red) has a sulfate ion, which is stabilized by surface loops: the loop between strands β3 and β4 (yellow); the WPD loop between helix α3 and strand β7 (lime green); the loop between helices α5 and α6 (magenta), and indirectly by the loop between helix α1 and strand β1 (cyan). Residues N303, T349, P418, R495 and V499 from these loops contribute to the stabilization of the active site. B. A sulfate ion was observed in the binding site of each SH2 domain as well. C–E. Electron density maps around the active site and the sulfates in the binding pocket of PTP, and around the sulfates in the binding sites of N-SH2 and C-SH2 domains. The 2Fo-Fc maps are contoured at 1σ level.
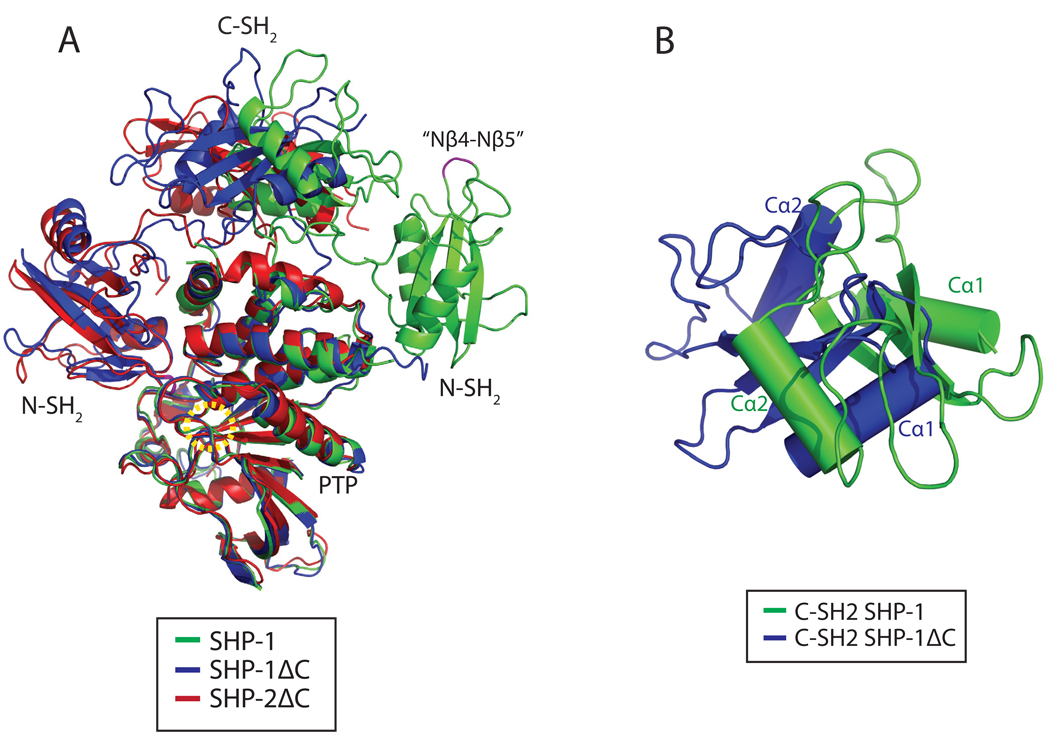
Figure 5.
Structural comparisons of SHP-1 with the tail-truncated SHP-1 and SHP-2. A. Structural comparison of SHP-1 (green) with tail-truncated SHP-1 (blue) and SHP-2 (red). The yellow dashed circle shows the position of the active site. The Nβ4–Nβ5 hairpin loops are colored magenta. The Nβ4–Nβ5 loop in SHP-1 is quoted because we use this name from tail-truncated SHP-1 regardless of the differences between their secondary structures. The N-SH2 domains of the tail-truncated SHP-1 and SHP-2 are located on the left side of the PTP domain, but shifted to the right side in the current SHP-1 structure, resulting in the exposure of the active site. B. The conformational change between the C-SH2 domains in SHP-1 (green) and tail-truncated SHP-1 (blue) structures. The C-SH2 domain of the intact protein is rotated anticlockwise by about 110° relative to the truncated protein as revealed from the superposition of their PTP domains.
The shifted N-SH2 domain is stabilized by inter-domain interactions
The displacement of the N-SH2 domain away from the active site results in reorganization of the interactions between the domains. In the published tail-truncated structures, there is no obvious interaction between two SH2 domains, other than the linkage between them (Yang et al., 2003). In the current structure, residue M1 from N-SH2 domain forms a hydrogen bond with H114 from the C-SH2 domain and R7 from N-SH2 domain forms salt bridges with E137 and D144 (Fig. 3A(a)).
Figure 3.
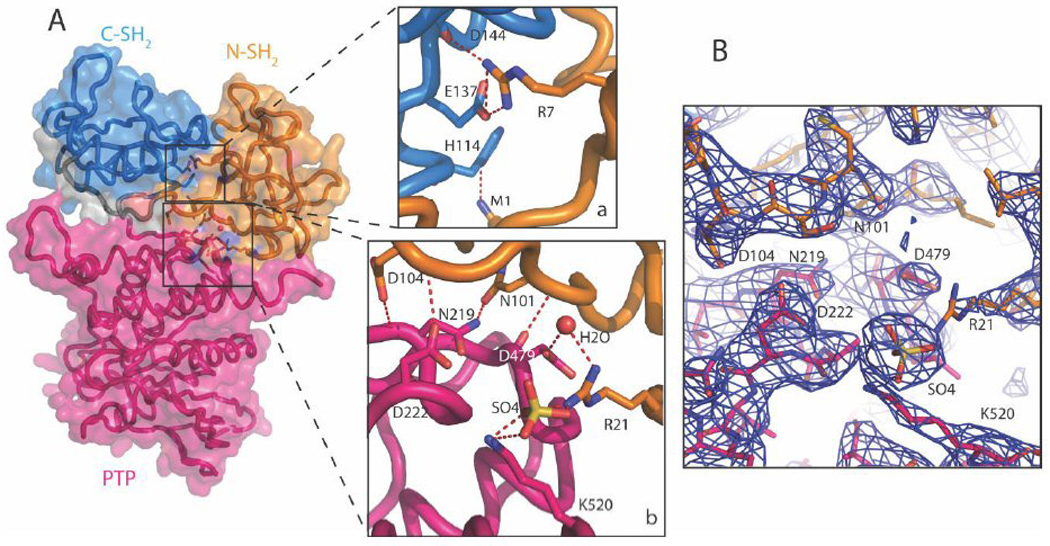
The open form of the SHP-1 is stabilized by the inter-domain interactions. A(a). The interaction between the N-SH2 and C-SH2 domains. Residues M1, R7 from the N-SH2 domain interact with H114, E137 and D144 from the C-SH2 domain. A(b). The interaction between the N-SH2 and the PTP domains. A sulfate ion and a water molecule are involved in this interaction. The side chains of N101 and D104 form hydrogen bonds with the side chain and the main chain of N219, respectively; the main chain of D104 also forms a hydrogen bond with the side chain of D222. In addition, the side chain of R21 interacts with the side chain of D479 through a water molecule, and interacts with the side chain of K520 through a sulfate ion. The backbone of D479 also forms a hydrogen bond with that of N101. B. Electron density map of the interface between the N-SH2 and the PTP domains. The 2Fo-Fc map is contoured at 1σ level.
A set of new interactions between the N-SH2 domain and the catalytic domain was also observed in the current structure (Fig. 3A (b), B). Several residues from the N-SH2 domain, located on the solvent side in the tail-truncated SHP-1 structure (opposite side to the Nβ4–Nβ5 hairpin loop that interacts with the catalytic domain and keeps the enzyme in the inactive conformation), show extensive interactions with a set of residues, on the surface of PTP domain. The side chains of N101 and D104 form hydrogen bonds with the side chain and the main chain of N219, respectively; The main chain of D104 also forms a hydrogen bond with the side chain of D222 and the side chain of R21 interacts with the side chain of D479 through a water molecule, and interacts with the side chain of K520 through a sulfate ion. The backbone of D479 also forms a hydrogen bond with that of N101 (Fig.3A (b)). This set of hydrogen bonds may be crucial in maintaining the SHP-1 structure in the open conformation.
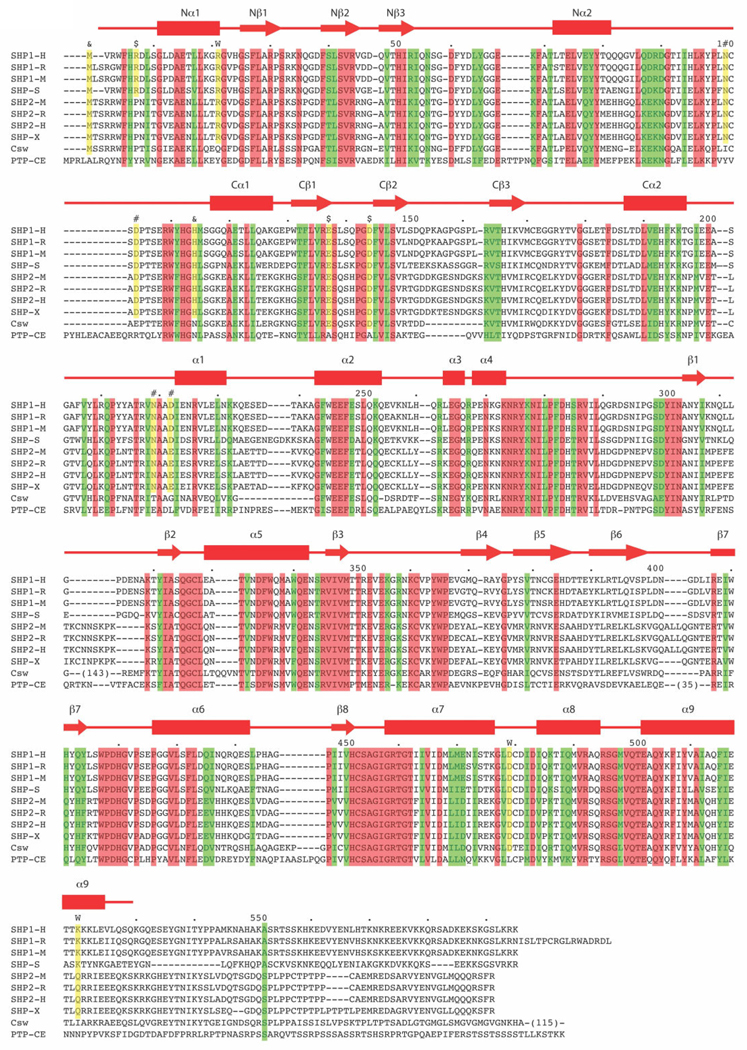
To examine whether or not the amino acids involved in the stabilization of the open conformation are conserved in other SHP proteins, we have compared the sequences of SHP-1 and SHP-2 from human, rat and mouse, of SHP protein from salmon, Xenopus, of Corkscrew (Csw) from Drosophila, and of PTP from C. elegans using ClustalW (Thompson et al., 1994). As shown in Figure 4, Residues N101, D104, N219 and N222 (marked as #) involved in direct interactions between the N-SH2 and the catalytic domains are conserved in all SHP proteins except those in Drosophila and C. elegans. The same pattern is observed for R21, D479, K520 (marked W) involved in the water molecule–mediated N-SH2 and catalytic domain interactions. Residues involved in the N-SH2 and C-SH2 domain interactions, M1, H114 (marked &) and E137, D144 (marked $), are conserved in all SHP proteins except those in Drosophila and C. elegans. R7 is conserved in SHP-1 proteins from human, rat, mice and in SHP protein from salmon.
Figure 4.
Multiple sequence alignment of SHP-1 homologs. Protein sequences of SHP-1 from human (SHP1-H), rat (SHP1-R) and mice (SHP1-M), SHP2 from human (SHP2-H), rat (SHP2-R) and mice (SHP2-M), SHP from salmon (SHP-S), Xenopus (SHP-X), Corkscrew (Csw) from Drosophila and PTP from C. elegans (PTP) are aligned with ClustalW program (35). Residues identical in all proteins are in red columns and those with conserved substitutions are in green columns. Conserved residues involved in the interactions between two SH2 domains and between N-SH2 and catalytic domains are in yellow columns. Resides involved in interactions between two SH2 domains are labeled with & and $; those involved in direct interactions between N-SH2 and catalytic domains are labeled with #, and those involved in the water molecule – mediated interactions are labeled with w. The secondary structural elements of SHP-1 from current stucture are represented by helices as cylinders, strands as arrows, and others as lines.
Comparisons of the current SHP-1 structure with the structures of the tail-truncated SHP-1 and SHP-2
To understand regulatory mechanism of SHP-1 and SHP-2, we compared the current structure with those of tail-truncated SHP-1 and SHP-2 (Fig. 5). Relative to the tail-truncated SHPs’ structures, the N-SH2 domain undergoes a huge relocation, moving from the catalytic side of the PTP domain to the distal side resulting in the exposure of the catalytic active site (Fig. 5A). The N-SH2 domain's movement brings it toward the other side of the C-SH2 domain as compared with the tail-truncated structures. Coupled with the displacement of N-SH2 domain, the C-SH2 domain is rotated anticlockwise by about 110° (Fig. 5B).
Comparisons of various N-SH2 domains of SHPs reveal that the N-SH2 domain in current structure is in the “Active” state
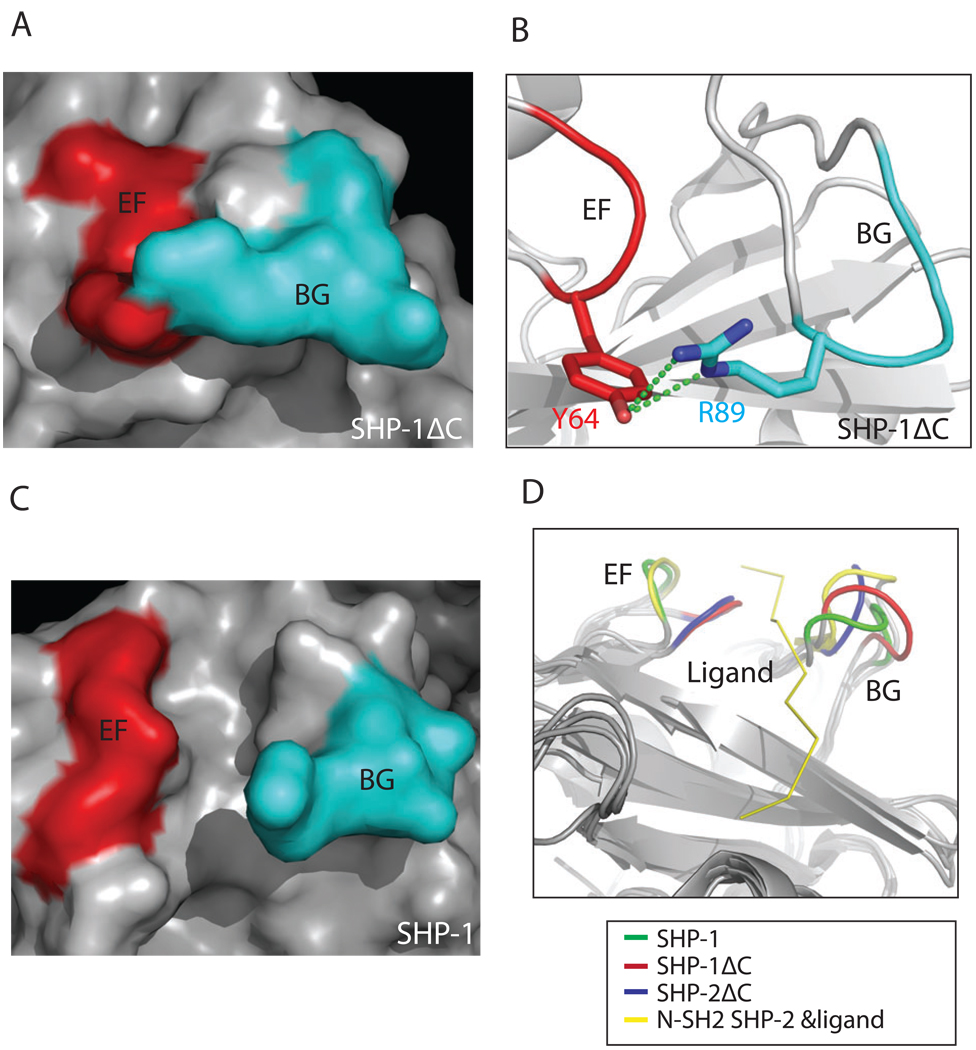
Previous comparison has revealed that the N-SH2 domains in the tail-truncated SHP-1 and SHP-2 structures are closed in the resting inhibited state (“I” state) through the interaction of the EF loop with the BG loop; upon an activation signal the EF loop separates from the BG loop and changes to the active state (“A” state), so that the N-SH2 domain is ready for ligand binding and the subsequent activation of SHPs (Hof et al., 1998). The tight association between the EF loop and the BG loop has been proposed as a mechanism to restrict the ligand binding to the N-SH2 domain in the SH2 domain-containing enzymes (Zvelebil et al., 1995; Barford and Neel, 1998; Porter et al., 2007; Kaneko et al., 2010). To characterize the state of the N-SH2 domain in the open conformation, we compared the N-SH2 domain in the current structure with those from the tail-truncated SHP-1 and SHP-2 and from the complex structure of the N-SH2 domain of SHP-2 with the phosphotyrosine peptides (Lee et al., 1994) (Fig. 6). Unlike the “I” state shown in the truncated SHP-1 structure in which the EF and BG loops interact through the hydrogen bond between the Y64 from the EF loop and the R89 from the BG loop (Fig 6A, B, D), two loops are apart from each other in the current structure (Fig. 6C, D), suggesting that the N-SH2 domain in current SHP-1 structure adopts an “A” state. The separation of two loops in the current structure further facilitates sulfate ion binding in the ligand binding pocket, as shown in figure 2.
Figure 6.
Structural comparison of N-SH2 domain in various conformations. A. The surface of N-SH2 domain in the tail-truncated SHP-1 structure. The EF (red) and BG (cyan) loops are in “I” state through interactions shown in figure 6B. B. Residue Y64 from the EF loop interacts with residue R89 from the BG loop in the tail-truncated SHP-1 structure. C. The surface of N-SH2 domain in the current structure. The EF (red) and BG (cyan) loops are in “A” state. D. Comparison of the EF and BG loops shows that the two loops are dissociated in the N-SH2 domains of SHP-1 (green) and ligand-bound SHP-2 N-SH2 (yellow), while they interact in the tail-truncated SHP-1 (red) and SHP-2 (blue).
DISCUSSION
To study the regulatory mechanism of SHP-1, we have crystallized the full length SHP-1 and solved its structure. Although the tail was disordered, the structural features indicate that the current SHP-1 structure is in an open conformation. Firstly, the N-SH2 domain, functioning as an intramolecular switch, is shifted away from the active site allowing exposure of the active site to facilitate the substrate binding (Fig. 2A, C, and Fig. 5A). In addition, a sulfate ion, an analog of phosphate, is identified in the binding site of each domain (Fig. 2). Therefore, the current work provides the first crystal structure of the SH2 domain-containing PTPs in an open form.
The substantial movement of the N-SH2 domain results in a conformational change of all the domains within the protein. Consequently new interactions between the two SH2 domains, and between the N-SH2 domain and the PTP domain are formed (Fig. 3). These interactions could be critical for stabilizing the SHP-1 structure in the open conformation. Sequence alignment of SHP-1 and SHP-2 shows that most of the residues on the new interfaces are conserved (Fig. 4), indicating the importance of these residues in the activation of SHPs. The conserved sequences suggest that SHP-2 and other SHP proteins have similar open conformation as the current SHP-1 structure.
It is still unclear what causes the current SHP-1 structure to be in the open form. The lipid-mimicking detergent, deoxy Big CHAP used to optimize the crystal growth, may act as the activator, as SHP-1 can be activated by lipids (Zhao et al., 1993; Frank et al., 1999). Indeed, detergent deoxy Big CHAP can activate SHP-1 by 2–3 fold compared to the basal condition when using p-NPP as substrate (data not shown). Although the activation of SHP-1 by detergent is relatively low, it could be enough to render the enzyme active. As a negative regulator, SHP-1 is more likely than SHP-2 to have some activity under basal conditions (Jones et al., 2004; Poole and Jones, 2005). Although it is disordered in the current structure, the C-terminal tail of SHP-1 may contribute to the observed conformational rearrangement, by interacting with either the detergent, or the SH2 domains, or both. In addition, incorporation of the sulfate ions into the binding sites of all three domains indicates that the sulfate could be important in stabilizing the structure in the open form. Further studies are needed to understand the regulatory mechanism of the SHP-1 as in the open conformation.
The N-SH2 ligand binding site is in “A” state in the current structure (Fig. 6C, D), and plays a more active role than the C-SH2 domain in SHPs activation, as demonstrated from biochemical and structural studies (Pregel et al., 1995; Pei et al., 1996; Barford and Neel, 1998; Hof et al., 1998; O'Reilly et al., 1998; Yang et al., 2003). However, it was not clear what role the C-SH2 domain plays in the activation regulation of SHPs. In the current structure, a sulfate ion was observed in the C-SH2 domain and the rotational movement of C-SH2 domain appears to be associated with the long-distance relocation of the N-SH2 domain. These data suggest that the C-SH2 domain is also critical in the activation of SHP-1.
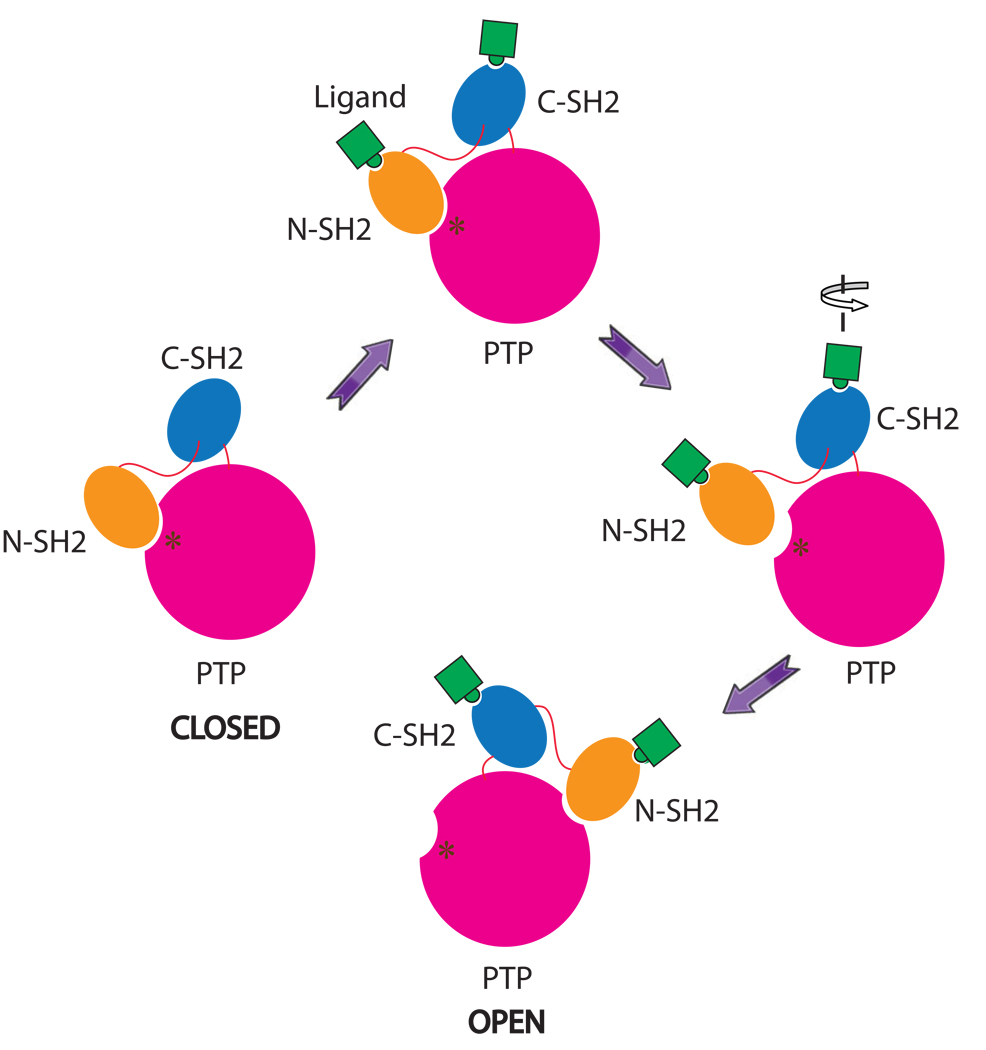
Based on the comparison of current SHP-1 structure with that of the tail-truncated SHP-1, we propose a model for the SHP-1’s activation (Fig. 7). In this model, the N-SH2 domain associates with the PTP domain in the resting state. Ligand binding to the SH2 domains will cause conformational change of the N-SH2 domain, resulting in the dissociation of the N-SH2 domain from the PTP domain. Due to the flexibility of the C-SH2 domain, the release of the N-SH2 domain triggers the rotation of the C-SH2 domain, which facilitates the movement of the N-SH2 domain to the other side of the C-SH2 domain and the opening-up of the active site. This open conformation is further stabilized by the new inter-domain interactions identified from current structure (Fig. 7).
Figure 7.
Cartoon representation of a proposed model for the activation of SHP-1. The N-SH2 domains are in orange, the C-SH2 domains are in marine, the PTP domains are in hot pink, and the linkers between them are in red. The dark red asterisks show the position of the active site resides. The ligand is in green. The N-SH2 domain blocks the active site of the PTP domain in the closed conformation. Ligand binding to SH2 domains will cause conformational changes, resulting in the release of the N-SH2 domain from the PTP domain. This will trigger the rotation of the C-SH2 domain, the repositioning of the N-SH2 domain and consequent exposure of the active site of the catalytic domain.
In conclusion, we have determined a crystal structure of SHP-1 in the open conformation. This first image of the open conformation of a cytosolic PTP revealed a long-distance movement of the N-SH2 domain, with the help of the C-SH2 domain, from blocking to unblocking the active site of the PTP domain. The movements of SH2 domains are sufficient to completely expose the active site of the PTP domain for substrate binding. Meanwhile, the ligand-binding sites of all three domains are occupied by sulfate ions. This open conformation structure provides important information for understanding the SHPs’ regulatory mechanism.
ACKNOWLEDGMENTS
We gratefully acknowledge funding by the National Institutes of Health (grants AL45858 to G.W. Z. and HL079441 to J. Z. Z.). We thank Guanyuan Fu and Vicky Yang for their help on different stages of the project. HDB was partially supported by the Louisiana Governor's Biotechnology Initiative.
Grant sponsor: National Institutes of Health; Grant numbers: AL45858 and HL079441 The coordinates and structure factors have been deposited in the Protein Data Bank (http://www.pdb.org/) with the accession code of 3PS5.
REFERENCES
- Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263(5152):1397–1404. [PubMed] [Google Scholar]
- Barford D, Neel BG. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6(3):249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- Bonaparte KL, Hudson CA, Wu C, Massa PT. Inverse regulation of inducible nitric oxide synthase (iNOS) and arginase I by the protein tyrosine phosphatase SHP-1 in CNS glia. Glia. 2006;53(8):827–835. doi: 10.1002/glia.20344. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Croker BA, Lawson BR, Rutschmann S, Berger M, Eidenschenk C, Blasius AL, Moresco EM, Sovath S, Cengia L, Shultz LD, Theofilopoulos AN, Pettersson S, Beutler BA. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc Natl Acad Sci U S A. 2008;105(39):15028–15033. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck MJ, Pluskey S, Trüb T, Harrison SC, Shoelson SE. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature. 1996;379(6562):277–280. doi: 10.1038/379277a0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer EH, Charbonneau H, Tonks NK. Protein tyrosine phosphatases: a diverse family of intracellular and transmembrane enzymes. Science. 1991;253(5018):401–406. doi: 10.1126/science.1650499. [DOI] [PubMed] [Google Scholar]
- Frank C, Keilhack H, Opitz F, Zschörnig O, Böhmer FD. Binding of phosphatidic acid to the protein-tyrosine phosphatase SHP-1 as a basis for activity modulation. Biochemistry. 1999;38(37):11993–12002. doi: 10.1021/bi982586w. [DOI] [PubMed] [Google Scholar]
- Green MC, Shultz LD. Motheaten, an immunodeficient mutant of the mouse. I. Genetics and pathology. J Hered. 1975;66(5):250–258. doi: 10.1093/oxfordjournals.jhered.a108625. [DOI] [PubMed] [Google Scholar]
- Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92(4):441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- Jones ML, Craik JD, Gibbins JM, Poole AW. Regulation of SHP-1 tyrosine phosphatase in human platelets by serine phosphorylation at its C terminus. J Biol Chem. 2004;279(39):40475–40483. doi: 10.1074/jbc.M402970200. [DOI] [PubMed] [Google Scholar]
- Kamata T, Yamashita M, Kimura M, Murata K, Inami M, Shimizu C, Sugaya K, Wang CR, Taniguchi M, Nakayama T. Src homology 2 domain-containing tyrosine phosphatase SHP-1 controls the development of allergic airway inflammation. J Clin Invest. 2003;111(1):109–119. doi: 10.1172/JCI15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Huang H, Zhao B, Li L, Liu H, Voss CK, Wu C, Schiller MR, Li SS. Loops Govern SH2 Domain Specificity by Controlling Access to Binding Pockets. Sci Signal. 2010;3(120):ra34. doi: 10.1126/scisignal.2000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo E, Park DK, Tzvetanova ID, Leiton CV, Cho BS, Colognato H. Tyrosine phosphatases Shp1 and Shp2 have unique and opposing roles in oligodendrocyte development. J Neurochem. 2010;113(1):200–212. doi: 10.1111/j.1471-4159.2010.06596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J. Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure. 1994;2(5):423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- Liang X, Meng W, Niu T, Zhao Z, Zhou GW. Expression, purification, and crystallization of the catalytic domain of protein tyrosine phosphatase SHP-1. J Struct Biol. 1997;120(2):201–203. doi: 10.1006/jsbi.1997.3927. [DOI] [PubMed] [Google Scholar]
- Lorenz U. SHP-1 and SHP-2 in T cells: two phosphatases functioning at many levels. Immunol Rev. 2009;228(1):342–359. doi: 10.1111/j.1600-065X.2008.00760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U, Ravichandran K, Pei D, Walsh CT, Burakoff SJ, Neel BG. Lck-dependent tyrosyl phosphorylation of the phosphotyrosine phosphatase SH-PTP1 in murine T cells. Mol Cell Biol. 1994;14(3):1824–1834. doi: 10.1128/mcb.14.3.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero MB, Venema VJ, Ju H, Eaton DC, Venema RC. Regulation of angiotensin II-induced JAK2 tyrosine phosphorylation: roles of SHP-1 and SHP-2. Am J Physiol. 1998;275(5 Pt 1):C1216–C1223. doi: 10.1152/ajpcell.1998.275.5.C1216. [DOI] [PubMed] [Google Scholar]
- Marsh HN, Dubreuil CI, Quevedo C, Lee A, Majdan M, Walsh GS, Hausdorff S, Said FA, Zoueva O, Kozlowski M, Siminovitch K, Neel BG, Miller FD, Kaplan DR. SHP-1 negatively regulates neuronal survival by functioning as a TrkA phosphatase. J Cell Biol. 2003;163(5):999–1010. doi: 10.1083/jcb.200309036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Tsuji S, Inui M, Sakamoto Y, Endo S, Ito Y, Fujimura S, Koga T, Nakamura A, Takayanagi H, Itoi E, Takai T. Inhibitory immunoglobulin-like receptors LILRB and PIR-B negatively regulate osteoclast development. J Immunol. 2008;181(7):4742–4751. doi: 10.4049/jimmunol.181.7.4742. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- O'Reilly AM, Neel BG. Structural determinants of SHP-2 function and specificity in Xenopus mesoderm induction. Mol Cell Biol. 1998;18(1):161–177. doi: 10.1128/mcb.18.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pei D, Lorenz U, Klingmüller U, Neel BG, Walsh CT. Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: a new function for Src homology 2 domains. Biochemistry. 1994;33(51):15483–15493. doi: 10.1021/bi00255a030. [DOI] [PubMed] [Google Scholar]
- Pei D, Wang J, Walsh CT. Differential functions of the two Src homology 2 domains in protein tyrosine phosphatase SH-PTP1. Proc Natl Acad Sci U S A. 1996;93(3):1141–1145. doi: 10.1073/pnas.93.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AW, Jones ML. A SHPing tale: perspectives on the regulation of SHP-1 and SHP-2 tyrosine phosphatases by the C-terminal tail. Cell Signal. 2005;17(11):1323–1332. doi: 10.1016/j.cellsig.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Porter CJ, Matthews J, Mackay JP, Pursglove SE, Schmidberger JW, Leedman PJ, Pero SC, Krag DN, Wilce MC, Wilce JA. Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation. BMC Struct Biol. 2007;7:58. doi: 10.1186/1472-6807-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregel MJ, Shen SH, Storer AC. Regulation of protein tyrosine phosphatase 1C: opposing effects of the two src homology 2 domains. Protein Eng. 1995;8(12):1309–1316. doi: 10.1093/protein/8.12.1309. [DOI] [PubMed] [Google Scholar]
- Salmond RJ, Alexander DR. SHP2 forecast for the immune system: fog gradually clearing. Trends Immunol. 2006;27(3):154–160. doi: 10.1016/j.it.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Shi ZQ, Yu DH, Park M, Marshall M, Feng GS. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol. 2000;20(5):1526–1536. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Schweitzer PA, Rajan TV, Yi T, Ihle JN, Matthews RJ, Thomas ML, Beier DR. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell. 1993;73(7):1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature. 1994;370(6490):571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HW, Siminovitch KA, de Souza L, Tsui FW. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993;4(2):124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003;306:1–12. doi: 10.1016/s0378-1119(03)00400-1. [DOI] [PubMed] [Google Scholar]
- Yang J, Liang X, Niu T, Meng W, Zhao Z, Zhou GW. Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1. J Biol Chem. 1998;273(43):28199–28199. doi: 10.1074/jbc.273.43.28199. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu L, He D, Song X, Liang X, Zhao ZJ, Zhou GW. Crystal structure of human protein-tyrosine phosphatase SHP-1. J Biol Chem. 2003;278(8):6516–6520. doi: 10.1074/jbc.M210430200. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Shen SH, Fischer EH. Stimulation by phospholipids of a protein-tyrosine-phosphatase containing two src homology 2 domains. Proc Natl Acad Sci U S A. 1993;90(9):4251–4255. doi: 10.1073/pnas.90.9.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu Rev Pharmacol Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- Zvelebil MJ, Panayotou G, Linacre J, Waterfield MD. Prediction and analysis of SH2 domain-phosphopeptide interactions. Protein Eng. 1995;8(6):527–533. doi: 10.1093/protein/8.6.527. [DOI] [PubMed] [Google Scholar]