Abstract
At present several entirely different explanatory approaches compete to illuminate the mechanisms by which animal body plans have evolved. Their respective relevance is briefly considered here in the light of modern knowledge of genomes and the regulatory processes by which development is controlled. Just as development is a system property of the regulatory genome, so causal explanation of evolutionary change in developmental process must be considered at a system level. Here I enumerate some mechanistic consequences that follow from the conclusion that evolution of the body plan has occurred by alteration of the structure of developmental gene regulatory networks. The hierarchy and multiple additional design features of these networks act to produce Boolean regulatory state specification functions at upstream phases of development of the body plan. These are created by the logic outputs of network subcircuits, and in modern animals these outputs are impervious to continuous adaptive variation unlike genes operating more peripherally in the network.
Keywords: GRN evolution, GRN selection
Introduction
Never in the modern history of evolutionary bioscience have such essentially different ideas about how to understand evolution of the animal body plan been simultaneously current. Of the many different aspects of evolution, we are here to be concerned with how the developmental mechanisms generating the body plan architectures recognized in Linnaen systematics at the level of Phylum and Class evolve, and how these mechanisms have been maintained, often since the Cambrian or Ordovician. Ideas about the nature of the underlying evolutionary mechanisms, and what to do to study them, generally associate with one of several paradigmatic views. Two of these views, though mutually incompatible, share the conviction that evolution of the body plan can be illuminated by study of adaptive evolution of detailed properties of modern organisms that are generated by far downstream developmental processes such as terminal differentiation. The first is the classic neo-Darwinian concept that evolution of animal morphology occurs by means of small continuous changes in primary protein sequence which in general require homozygosity to effect phenotype. The second paradigm holds that evolution at all levels can be illuminated by detailed analysis of cis-regulatory changes in genes that are direct targets of sequence level selection, in that they control variation of immediate adaptive significance. Both approaches often focus on changes at single gene loci, and both are framed within the concepts of population genetics. An entirely different way of thinking is that the evolution of animal body plans is a system level property of the developmental gene regulatory networks (dGRNs) which control ontogeny of the body plan. It follows that gross morphological novelty required dramatic alterations in dGRN architecture, always involving multiple regulatory genes, and typically affecting the deployment of whole network subcircuits. Because dGRNs are deeply hierarchical, and it is the upper levels of these GRNs that control major morphological features in development, a question dealt with below in this essay arises: how can we think about selection in respect to dGRN organization? The answers lie in the architecture of dGRNs and the developmental logic they generate at the system level, far from micro-evolutionary mechanism. While adaptive evolutionary variation occurs constantly in modern animals at the periphery of dGRNs, the stability over geological epochs of the developmental properties that define the major attributes of their body plans requires special explanations rooted deep in the structure/function relations of dGRNs.
Views of body plan evolution
Of the first of these approaches (e.g., Hoekstra and Coyne, 2007), I shall have nothing to say, as mechanistic developmental biology has shown that its fundamental concepts are largely irrelevant to the process by which the body plan is formed in ontogeny. In addition it gives rise to lethal errors in respect to evolutionary process. Neo-Darwinian evolution is uniformitarian in that it assumes that all process works the same way, so that evolution of enzymes or flower colors can be used as current proxies for study of evolution of the body plan. It erroneously assumes that change in protein coding sequence is the basic cause of change in developmental program; and it erroneously assumes that evolutionary change in body plan morphology occurs by a continuous process. All of these assumptions are basically counterfactual. This cannot be surprising, since the Neo-Darwinian Synthesis from which these ideas stem was a pre-molecular biology concoction focused on population genetics and adaptation natural history, neither of which have any direct mechanistic import for the genomic regulatory systems that drive embryonic development of the body plan.
The second paradigm holds that general evolutionary process will be revealed by studies of continuous variation in cis-regulatory modules affecting expression of adaptively meaningful genes. Its experimental application has indeed been enormously revealing in respect to the sequence level cis-regulatory mechanisms by which much natural variation arises. For example, very clear examples of functional evolutionary changes in cis-regulatory modules have come from recent studies on regulation of pigmentation genes between and within various Drosophila species. Among these are adaptively significant variations in regulation of the yellow gene, which accounts for a variety of spatial pigmentation patterns in higher Dipteran wings and body surfaces including sexually dimorphic markings (Gompel et al., 2005; Prud’homme et al., 2006; 2007; Jeong et al., 2006); and of the ebony gene, which controls the degree of melanization in differently pigmented populations living in diverse Ugandan environments (Rebeiz et al., 2009). Sequence level changes in cis-regulatory modules controlling expression of these genes are demonstrated to be the cause of these variations, and in general they operate by altering the response of the cis-regulatory module to the pleisiomorphic spatial landscape of regulatory states. Evolutionary change in a cis-regulatory module controlling downstream gene expression is of course far less pleiotropically dangerous to the whole system than if either the coding region of the gene had been mutated, or if the upstream regulatory landscape had been altered (Prud’homme et al., 2007). Another, essentially similar, recent demonstration of cis-regulatory evolutionary change in an adult trait concerned some detailed pattern differences in trichome distribution that distinguish Drosophila species (McGregor et al., 2007). Trichome assembly is controlled by genes requiring expression of the regulatory gene shavenbaby (Chalnut-Delalande et al., 2006), and several cis-regulatory modules determine the exact spatial expression of this gene in response to the upstream regulatory landscape. Evolutionarily arising differences in the DNA sequences of these modules collectively determine differences in spatial expression of this gene, and thereby generate the species specific differences in trichome pattern. Prud’homme et al. (2007; op. cit.) explicitly proposed that experimental analysis of functional cis-regulatory differences affecting adaptive traits among related species will illuminate larger evolutionary changes in development of the body plan, just as such analysis illuminates selective changes in intra- or inter- specific color patterns or surface morphology (another uniformitarian view). The arguments are that essentially all evolutionary changes in morphology are at root cis-regulatory, which is indeed basically true; and that intra-modular mechanisms of cis-regulatory evolution will operate on similar principles wherever it occurs, also true. But these assumptions do not suffice to support the uniformitarian conclusion about body plan evolution: when the properties of the gene regulatory networks that actually generate body plans and body parts are taken into account, it can be seen that many entirely new and different mechanistic factors come into play. The result is that just as the paleontological record of evolutionary change in animal morphology is the opposite of uniformitarian (see the paper of D. Erwin in this collection), so, for very good reasons that are embedded in their structure/function relations, are the mechanisms of dGRN evolution.
Suppose that we begin with the following syllogism, which to a systems developmental biologist seems inescapable: Since dGRNs control ontogeny of the body plan, and since evolution of the body plan requires genomic alteration of the developmental program, then relevant explanations must be couched in terms of those genomic alterations that change the structure and function of dGRNs. This rather obvious argument gives rise to additional specific consequences, which taken together provide a new set of principles that apply to the mechanisms of body plan evolution (Britten and Davidson, 1971; Davidson and Erwin, 2006; Peter and Davidson, 2011). They are new in that none are specifically predicted by classical evolutionary theory. In the interests of conciseness these principles are summarized in Table 1, and briefly discussed in the following.
Table 1.
Evolution of Animal Body Plans as Change and Conservation of Developmental Gene Regulatory Network (dGRN) Structure: Mechanistic Consequences
| Premise | Consequence |
|---|---|
| Since dGRN structure depends on cis-regulatory linkages at nodes: | 1. Change in dGRN structure occurs by co-optive redeployment of cis-regulatory modules controlling regulatory gene expression |
| Since co-optive cis-regulatory redeployments are gain of function changes: | 2. Co-optive redeployment of regulatory gene expression will generally be haplosufficient and act dominantly |
| Since dGRNs are deeply hierarchical: | 3. Effects of given cis-regulatory mutations (including co-options) depend specifically on their location in dGRN |
| Since dGRNs are deeply hierarchical: | 4. Subcircuits operating at upper levels (early in developmental process) preclude certain downstream linkages, and mediate others, i.e., canalize dGRN structure (and developmental process). |
| Since dGRNs are deeply hierarchical: | 5. Conserved upper level subcircuits should produce patterns of canalization that reflect phylogenetic distribution of the developmental processes that generate clade specific body parts (prediction of Kernels) |
| Since flexibility at given dGRN nodes depends on their upstream and downstream linkages: | 6. dGRN structure should contain information for prediction of evolutionary hotspots vs. evolutionarily conserved structural features |
| Since dGRNs are modular, i.e., given functions are executed by given subcircuits: | 7. Evolution of new developmental outcomes must often involve co-optive gain of function changes that cause redeployment of whole dGRN subcircuits |
| Since redeployment of dGRN subcircuits is a mechanism of evolution of developmental novelty: | 8. Evolutionary change must occur in dGRN linkages controlling subcircuit deployment, i.e., in signal presentation and reception, regulatory switches, and inter-subcircuit inputs. |
Some principles that emerge from the precept that evolution of the animal body plan occurs by alteration of genomic developmental GRNs
Many of the arguments referred to in Table 1 have been presented earlier, as indicated. At the outset, the main point of difference between this and all other approaches to understanding evolution of the body plan is that this is a system approach to developmental evolution, in which answers derive from the topologies of regulatory gene interaction circuitry. No observations on single genes can ever illuminate the overall mechanisms of development of the body plan or of body parts except at the minute and always partial, if not wholly illusory, level of the worm’s eye view. The same must be true as well of major evolutionary change in the body plan or in body parts.
The purpose of Table 1 is to indicate the specific consequences for considerations of evolutionary process that derive from dGRN structure/function relationships (cf. Davidson, 2010 for review of this subject). The first principle in Table 1 is that the mechanism underlying structural change in dGRNs is re-deployment of cis-regulatory modules, due to sequence changes that result in co-option of regulatory gene expression to a new spatial and/or temporal domain of the developing animal. This tells us where to look in the regulatory system for differences in developmental patterning. Co-option can occur by various mechanisms at the genomic sequence level. An important point is that while these mechanisms include gradual, continuous, and reversible SNP mutations, they also (and perhaps more importantly) encompass irreversible and discontinuous mutational events such as transposon-mediated sequence insertion and other mechanisms of sequence change that cannot be accommodated in neo-Darwinian algorithms (for current review, Peter and Davidson, 2011).
Principle 2 follows from the point that such co-optive changes in general belong to the cis-regulatory gain of function class. As initially pointed out by Ruvkun et al. (1991), laboratory experiments show that where the genes affected are regulatory genes operating in embryonic development, these are almost always haplosufficient mutations; one copy expressed in a new location does the job (otherwise, of course, none of the regulatory genes isolated by haploid recessive screens would have been found!). The fundamental importance of haplosufficiency is that in evolution an individual bearing such a mutation will become a clonal founder of a novel population expressing a new developmental regulatory state (Davidson and Erwin, 2010), unless it is developmentally deleterious. To make a long story short, it follows that change in dGRN structure does not require the population genetics functions that result in homozygosity; that such co-optive dGRN change is likely to happen, and that it could happen at a relatively high rate were there not stabilizing circuitry in dGRNs that precludes alternative outcomes and locks down regulatory states once they are established (Davidson and Erwin, 2009; 2010; Peter and Davidson, 2011). In addition, as discussed below, dGRNs are insensitive to quantitative regulatory state changes.
A distinguishing feature of dGRNs is their deep hierarchy, which essentially stems from the long sequence of successive spatial regulatory states required to be installed in building first the axial embryonic/larval body plan, and then constructing individual body parts (Peter and Davidson, 2011; Davidson, 2010). Principles 3–5 derive from the hierarchical characteristic of dGRNs. Principle 3 is to the effect that the significance or functionality of any given cis-regulatory mutation affecting expression of a regulatory gene will depend entirely on where in the dGRN is located the affected cis-regulatory node (Erwin and Davidson, 2009). The effects of given cis-regulatory DNA sequence changes on GRN function cannot be inferred simply from results obtained in the “flat” regulatory landscape where the phenomenon studied is the effects of SNPs or small indels on either protein coding sequence, or on cis-regulatory function in the control of expression of peripheral effector genes.
Implicit in the hierarchical structure of GRNs is the mechanism of evolutionary canalization, as indicated in Table 1 at Principle 4. The subcircuits at each level provide feeds to the next level in the same or, via signaling, in other specified spatial domains. But each subcircuit produces a finite set of inputs for the next level, and only recipient nodes that contain target site combinations can respond to those particular inputs. Thus the universe of possible responses is vastly constrained by dGRN hierarchy at each level transition, inevitably resulting in what was classically termed “canalization” of the developmental process (Waddington, 1957; Gibson and Wagner, 2000). A few years ago remarkably conserved subcircuits, termed network “kernels” that operate high in the dGRN hierarchy were discovered (Davidson and Erwin, 2006). These produce regulatory states in the fields of cells that will later in development give rise to specific body parts (e.g., a pan-bilaterian heart progenitor field kernel; Davidson, 2006). A testable theory to explain the hierarchical shape of Linnean bilaterian phylogeny (Superphylum, Phylum, Class, etc), or what Erwin (this collection) terms the “clumpiness” of the phylogenetic distribution of animal morphologies, is based on kernels (Davidson and Erwin, 2006; 2009). The conservation of developmental process within each animal clade generates the phylogenetic distribution of the morphologies these processes generate. The prediction follows that the underlying cause is the phylogenetic distribution of dGRN kernels conserved within all members of a Superphylum or Phylum or Class; that is, these shared kernels would account for the shared morphogenetic characters of each clade. The argument is commutative. This theory requires that the kernels similarly canalize downstream developmental process in each member of each given clade. But since on first principles hierarchical dGRNs must produce canalization (Principle 4), then in order to account for the phylogenetic distribution of shared morphological characters, the existence of kernels could have been predicted, as stated in Principle 5 of Table 1.
On purely internal considerations, some aspects of dGRN structure appear much more impervious to change than others. For example, a frequently encountered type of subcircuit in upstream regions of dGRNs consists of two or three genes locked together by feedback inputs (Davidson, 2010). These feedback structures act to stabilize regulatory states, and there is a high penalty to change, in that interference with the dynamic expression of any one of the genes causes the collapse of expression of all, and the total loss from the system of their contributions to the regulatory state. On the other hand, peripheral far downstream subcircuits such as differentiation gene batteries can change freely without affecting major patterning functions or causing network collapse (Davidson and Erwin, 2006; Erwin and Davidson, 2009]). Generalizing, if we knew enough about the structure and functions of the constituent subcircuits, and their contextual upstream and downstream linkages, the architecture of the dGRN should predict its evolutionarily flexible and its evolutionarily less flexible linkages (Peter and Davidson, 2011), leading to Principle 6 in Table 1. Other features often thought of as properties of single genes, such as pleiotropy or epistasis, are likewise due to the positions genes occupy in network topology. Principle 7 states the self-evident: since no one gene produces body parts or executes a whole element of the developmental process, while on the other hand such functions are executed by dGRN subcircuits, the most powerful form of evolutionary change in dGRN structure should be those co-optive alterations that result in re-deployment of whole subcircuits. A very good example is the evident redeployment of an adult skeletogenic GRN to an embryological address in sea urchin evolution, at least a large part of the mechanism by which the “modern” sea urchins acquired skeletogenic function in their embryonic micromere lineages (Gao et al., 2008). Putting Principle 6 together with Principle 7 we see that an important place in dGRN structure to look for evolutionary change is in linkages that control subcircuit deployment: as Principle 8 indicates, such linkages include those that determine where signal ligands will be expressed; those that link one subcircuit to another; and those that serve as switches on the outside of morphogenetic subcircuits, so to speak, allowing or prohibiting their expression. As reviewed by Peter and Davidson (2011), much evidence indicates that hox gene functions often fall into this latter class. An ancillary point is that these kinds of linkage usually lack the feedback relations that act to stabilize developmental state (and evolutionary status); rather they are often wired as one way connections, and are likely to be intrinsically less resistant to change without catastrophe.
dGRN hierarchy and selection
In dGRNs the effector genes that constitute terminal differentiation and morphogenetic gene batteries, and their immediate controllers, lie at the network periphery (Davidson 2006; 2010). Their functions are terminal from the genetic control point of view, in that they lie at the ends of upstream cascades of regulatory steps, and they lack direct transcriptional feedbacks directed upstream. The same is true of many quantitative developmental traits which affect post-embryonic developmental process. The cis-regulatory modules for which functionally adaptive evolutionary sequence variation has been demonstrated, such as in the paradigmatic studies cited above on the yellow, the ebony, and the shavenbaby genes of Drosophila, all lie at such peripheral positions in the respective dGRNs. Here we can readily perceive continuous Darwinian processes of sequence change, and selective adaptive variation in cis-regulatory output, as shown explicitly in the cited studies, among many other less well worked out examples. Throughout the dGRN, at every level of hierarchy, the processes of sequence change in cis-regulatory modules must be the same. Yet the outputs of the upper level pattern formation circuits of dGRNs which specify the overall body plan, and the clade specific organization of individual body parts, do not display continuous variation in the types of forms they generate. Thus the disposition and morphologies of the major components of the body plan are invariant at the levels which define unequivocally the Phylum, Class, Order, to which an animal belongs; and thus, the development of an embryo is extremely canonical even though, as in sea urchins, the exact size of the egg, the temperature, or the amounts of many regulatory gene transcripts (Materna et al., 2010) may vary considerably. Or consider the particular example used by Prud’homme et al. (2007) to argue for the uniformity of evolutionary process at all levels of dGRN hierarchy, viz. the repression of wing patterning functions in the haltere imaginal disc by Ubx in Diptera (Weatherbee et al., 1998; Galant et al., 2002). In fact we do not see variation in the amount of “wingness” vs. “haltereness” displayed in development of this imaginal disc; in bees, which have four wings, Ubx has different cis-regulatory targets than in flies (Weatherbee et al., 1999), and there is either the one morphological output, four wings, or the other, two wings and two halteres, across this region of insect phylogeny. Whatever continuous variation occurs at individual cis-regulatory sequences, the dGRN circuit output preserves its Boolean morphogenetic character.
Therefore the action of selection differs across dGRN structure. Selection does not operate to produce continuous adaptive change except at the dGRN periphery. The lack of continuous variation in morphogenetic traits defining Class and Phylum level clades is obvious in the striking evolutionary stasis revealed by the fossil record (Davidson and Erwin, 2006; 2009; Erwin, 2011). In other words, while cis-regulatory sequence variation may have continuing adaptive significance at the dGRN periphery, at upper levels of the dGRN hierarchy it does not have the same significance because the system level output is very impervious to change, except for catastrophic loss of the body part or loss of viability altogether. As long realized and much discussed in a non-mechanistic way in advance of actual knowledge of dGRN structure and function (for review see Gibson and Wagner, 2000), this imperviousness has something to do with whatever processes generate canalization and/or “buffering” of the genetic control system. We can now begin to understand canalization mechanistically in terms of dGRN hierarchy and subcircuit structure, as above, but in so far as “buffering” is taken to mean protection against “environmental fluctuations” as in many evolutionary mathematical models, it is irrelevant to animal embryonic processes, since in the main these depend not at all upon environmental inputs.
Then what structural features of dGRN design do account for the imperviousness of upper level system output to continuous cis-regulatory variation and to continuous selective functional change? Or, a very closely related question, what accounts for the evolutionary stasis over geologic time of body plan phylogeny in Bilateria (Davidson and Erwin, 2009)? A dramatic illustration of such stasis is reproduced in Fig. 1 (Bottjer et al., 2006): here we see the real time distribution of fossil variants of echinoderm body plans. The early Cambrian was a period of (relatively) rapid evolutionary exploration of diverse developmental pathways as the programs directing the formation of crown group echinoderm characters were stepwise added into the stem group dGRNs. But following the period of morphological change the definitive properties of the five surviving echinoderm Classes have remained stable essentially since the Cambrian and Ordovician (cf. Erwin, 2011). The answer to the questions posed at the beginning of this paragraph is that there are multiple intrinsic design features of modern dGRN structure that all contribute at the system level to imperviousness to continuous variation and to evolutionary morphogenetic stasis. A short discussion of such features follows, and in the final section of this paper are some further considerations of the meaning of the most interesting of these dGRN design properties.
Fig. 1.
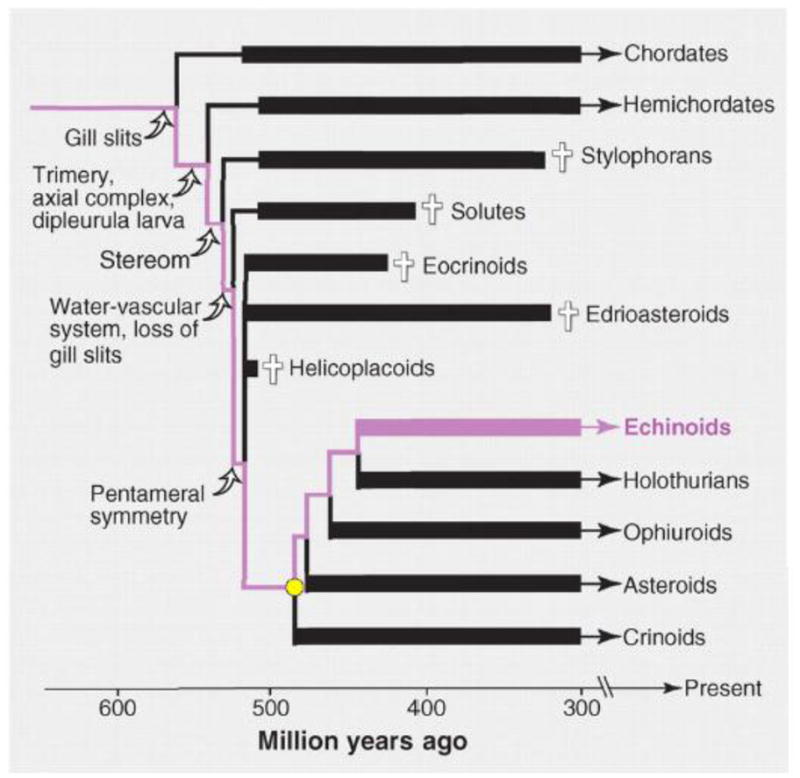
Evolutionary history of the major echinoderm groups. Cambrian echinoderms are recognized by the possession of stereom, but the phylogenetically most basal groups (such as stylophorans) lack the water vascular system, are highly asymmetrical, and possess gill slits. Pentameral symmetry is seen in two major Early Cambrian lineages, the edrioasteroids and eocrinoids. All stem-group echinoderm lineages became extinct by the Carboniferous (indicated with crosses). Crown-group echinoderms, indicated by the yellow circle, consist of the five major extant lineages in addition to numerous extinct lineages not shown. Most class-level crown groups first appear in the latest Paleozoic–early Mesozoic, including echinoids. The lineage leading to echinoids is indicated in purple. Known stratigraphic ranges are shown with thick lines, and inferred range extensions are shown with thin lines. Reproduced from Bottjer et al., 2006. Copyright (2006) AAAS.
To consider this question we must first remind ourselves what is the main function of upper level dGRNs for body plan formation. This has been discussed in detail in developmental (Peter and Davidson, 2009; Davidson, 2010) and evolutionary (Peter and Davidson, 2011) contexts; a very brief summary is that the fundamental role of upper level dGRNs is to set up in embryonic space a progressive series of regulatory states, which functionally define first the regions of the body with respect to its axes; then the location of the progenitor fields of the body parts; then the subparts of each body part. At each stage the output is a mosaic of sharply bounded regional regulatory states. This constitutes a Boolean checkerboard of diverse dGRN subcircuit expressions. Our problem thus resolves into understanding the system properties that “booleanize” dGRN subcircuit output, thus converting quantitatively and qualitatively varying sets of inputs into the same spatial regulatory state checkerboards for each member of the species at each stage. There are at least six different aspects to the solution to this problem.
i. Transcriptional dynamics of developmental gene regulatory cascades
In embryonic development the transcriptional processes mediated by dGRNs are intrinsically insensitive to varying cis-regulatory input levels. First, from the basic physical chemistry of target site occupancy, we know that modest changes in transcription factor concentration have little effect on target site occupancy; and second, as shown by Bolouri and Davidson (2003) in a dynamic analysis, in a typical embryonic gene cascade target genes are activated long before input factors approach steady state. This means that these “forward drive” systems operate over a great range of input concentrations, in contrast to typical physiological or biochemical macromolecular pathways in which quantitative output is usually mediated by exact control of steady state input levels.
ii. dGRN subcircuits controlling spatial regulatory state in development which execute Boolean logic transactions
Such subcircuits include the “X, 1-X” processors of Peter and Davidson (2009[Febb Lett]); these set up given regulatory states in a domain “X” and completely prohibit the expression of the given regulatory state everywhere else. For example, Tcf/β-catenin-mediated Wnt signaling operates to permit expression of target genes in cells receiving the signal but in all other cells, the dominant repressor Groucho replaces the Tcf cofactor β-catenin and transcriptionally represses the same target genes (for multiple examples see Peter and Davidson, 2010). Other subcircuits set sharp boundaries of expression by a variety of design devices; others mutually exclude regulatory states; etc. As Peter and Davidson (2009) showed, Boolean truth tables can be used to represent the function of each such subcircuit.
iii. Transcriptional repression, utilized in most spatial control dGRN subcircuits
While some mechanisms of repression merely result in decreasing rate of output, others dominantly silence gene expression in a given cell. There are many and diverse biochemical mechanisms of transcriptional repression but a prominent feature of dominant developmental repression is that it is a multistep, non-equilibrium, one-way process which, following the initial appearance of the sequence-specific transcriptional repressor, alters the configuration of the transcription complex so it can no longer function even after the transcriptional repressor has disappeared. Thus inclusion of repression in subcircuit topology increases all-or-nothing behavior.
iv. Specific feedback state lockdowns
Noticed when dGRN circuitry first began to be revealed experimentally (Davidson et al., 2002), it is an almost invariant observation that after a transient specification function first installs a spatial regulatory state, a feedback circuit is soon set up such that genes of the regulatory state are locked into a dynamic positive mutual embrace and the state is now stabilized (for review, Davidson, 2006). This general design feature clearly contributes to imperviousness to input variation since once these “stabilization motors” are activated they enable the system to forget upstream events so long as they worked at all, and the feedback circuitry has the capacity to strongly amplify the dGRN output. New levels of expression are established irrespective of the initial inputs. As development proceeds, such “reloading” and “restabilizing” devices are brought into play in each region of the organism, often at each stage.
v. Evolutionary inflexibility due to highly conserved canalizing dGRN kernels
As discussed above these subcircuits operate at upper levels of dGRN hierarchy so as to affect characters of the body plan that are definitive for upper level taxa, i.e., they control the early stages of just the types of developmental process of which the invariance per taxon constitutes our problem. Since they preclude developmental alternatives, they may act to “booleanize” the evolutionary selective process: either body part specification works the way it is supposed to or the animal fails to generate the body part and does not exist.
vi. Multiplicity of dGRN subcircuits ensuring given developmental outcomes
The characteristic tempo of evolutionary change illustrated in Fig. 1, in which a period of intense morphogenetic novelty is succeeded by long epochs of body plan stasis, suggests that early in clade history dGRNs were in some way different from crown group dGRNs (Davidson and Erwin, 2009; Erwin, 2011). This is of course another prima facie contradiction of the uniformitarian assumption that current observations on adaptive evolutionary change in specific peripheral cis-regulatory systems can illuminate early animal evolution. One way of thinking about this is to imagine that the evolutionary stability of crown group dGRN structure is due to the addition of more and more circuitry to control developmental pathways and exclude alternatives. These changes would have affected control of those embryonic stages at which the body plan is being specified by regional installation of regulatory states (Davidson and Erwin, 2009; Peter and Davidson, 2011). The implication is that stem group dGRNs, for example those of the early Cambrian echinoderms of Fig. 1, were structured differently from modern crown group dGRNs in respect to the multiplicity of the subcircuits brought to bear on each phase of the developmental process.
The significance of crown group dGRN design
At first glance subcircuit deployment in dGRNs can appear “overwired” or even redundant. Typically a regulatory state is installed in a given domain by a signal, or a gate of one sort or another; and the same state is not just activated exclusively in the right place but also specifically repressed everywhere else; domains are set up by alternative regional activation and their boundaries are then enforced by cross boundary repressive signaling and/or specific repressive exclusion of possible alternative regulatory states; dynamic feedback loops stabilize and enforce regulatory states; and not uncommonly many of the above devices are all deployed together in the same dGRN (for examples, Oliveri et al., 2008; Peter and Davidson, 2009, 2011; Smith and Davidson, 2009; Davidson, 2010). Though multiple such devices lead to the given overall developmental outcome, on principle they cannot be redundant, and in fact they never are when tested experimentally. That is, interference with expression of any of the key genes of these subcircuits always causes an immediate loss of function phenotype, such as ectopic expression if a spatial repression function is interrupted in cis (by mutation of repressor target sites) or trans (by application of a morpholino). For instance in the sea urchin embryo the regulatory genes of the initial endoderm specific dGRN are all activated by means of a Wnt signaling gate mediated by β-catenin/Tcf, because their cis-regulatory modules include essential Tcf target sites (Peter and Davidson, 2010; 2011). The requisite Wnt signal and its biochemical response in recipient cells, nuclearized β-catenin, are present only in the appropriate vegetal cell lineages of the embryo, and this might be thought quite sufficient to ensure expression of the endoderm genes only in those cells. However, in all other cells, as noted above, in the absence of nuclearized β-catenin the same endoderm specific genes are actively repressed outside the prospective endoderm by the alternative Tcf co-factor Groucho. Logically this could be regarded as a redundant spatial control, but it is clearly not, since if the Tcf sites of the cis-regulatory modules governing expression of endoderm genes are mutated, wild ectopic expression results (e.g., Ben-Tabou de-Leon et al., 2010; Smith and Davidson., 2008). This result is instructive: we see that the wiring enables these genes to utilize powerful ubiquitous activators in addition to their spatial control gates, though eventually control is handed off to the spatially confined cross-regulatory endoderm specific dGRN (Peter and Davidson, 2009; Ben-Tabou de-Leon et al., 2010). As a second example, in the skeletogenic micromere lineage the gcm gene is inactive, while gcm is directly turned on as a result of Notch signaling in the adjacent mesoderm cells in response to Delta expression in the skeletogenic cells (Ransick and Davidson, 2006). On top of this, an additional element of circuitry ensures independently that gcm is not expressed in the skeletogenic cells, a negative consequence of skeletogenic alx1 expression (Oliveri et al., 2008). But nor is this a redundant spatial control: if alx1 expression is prevented, gcm is indeed transcribed in skeletogenic cells, and so we learn that Delta signals among the micromeres would trigger gcm expression if not prevented from doing so. Examples could easily be multiplied, but without doing so their import can be generally summarized. Each apparently redundant spatial control mechanism turns out to have a special function, often not evident a priori. The overall control principle is that the embryonic process is finely divided into precise little “jobs” to be done, and each is assigned to a specific subcircuit or wiring feature in the upper level dGRN. No subcircuit functions are redundant with another, and that is why there is always an observable consequence if a dGRN subcircuit is interrupted. Since these consequences are always catastrophically bad, flexibility is minimal, and since the subcircuits are all interconnected, the whole network partakes of the quality that there is only one way for things to work. And indeed the embryos of each species develop in only one way.
Thus we can think of a crown group dGRN as an evolutionarily terminal, finely divided, extremely elegant control system that allows continuing alteration, variation, and evolutionary experimentation only after the body plan per se has formed, i.e., in structural terms, at the dGRN periphery, and in developmental terms, late in the process. It is no surprise, from this point of view, that cell type re-specification by insertion of alternative differentiation drivers is change only at the dGRN periphery, quite a different matter from altering body plan. In terms of their general hierarchical depth, the dGRNs of all living (non-degenerate) bilaterians are probably approximately similar (Peter and Davidson, 2011), though the number of subcircuits required at each given developmental stage or dGRN level to complete the body plan is likely much greater for some forms than others.
Deconstructing the evolutionary process by which stem group body plans were stepwise formulated will require us to traverse the conceptual pathway to dGRN elegance, beginning where no modern dGRN provides a model. The basic control features of the initial dGRNs of the Precambrian and early Cambrian must have differed in fundamental respects from those now being unraveled in our laboratories. The earliest ones were likely hierarchically shallow rather than deep, so that in the beginning adaptive selection could operate on a larger portion of their linkages. Furthermore, we can deduce that the outputs of their subcircuits must have been polyfunctional rather than finely divided and functionally dedicated, as in modern crown group dGRNs. A general result of these arguments is that considerations of evolutionary change in dGRN structure may at last provide a unified conceptual framework for understanding the stages of crown group evolution, and in the same breath the sequential history of change that has produced the different hierarchical levels of animal dGRNs.
But some things never change, and a principle that must have obtained from early in metazoan evolution is that developmental jobs are controlled through the logic outputs of genetic subcircuits. Thus how evolution of the animal body plan has occurred is a question that in the end can only be addressed in the terms of transcriptional regulatory systems biology.
Acknowledgments
I am extremely grateful to Dr. Isabelle S. Peter and to Dr. Doug Erwin for their critical and helpful reviews of this manuscript. This work was supported by NSF Grant IOS-0641398 and NIH Grant HD-37105.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ben-Tabou de-Leon S, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc Natl Acad Sci USA. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: Initial rates, not steady state, determine network kinetics. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314:956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Quart Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S. Savenbably couples patterning to epidermal cell shape control. PLoS Biol. 2006;4:1549–1561. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Gene Regulatory Networks in Development and Evolution. Academic Press/Elsevier; San Diego: 2006. The Regulatory Genome. [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010 doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. An integrated view of Precambrian eumetazoan evolution. Cold Spring Harbor Symp Quant Biol. 2010;74:65–80. doi: 10.1101/sqb.2009.74.042. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee P, Revilla R, Rust AG, Pan ZJ, Schilstra MJ, Clarke PJC, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Erwin DH. Evolutionary uniformitarianism. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.01.020. in press. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nature Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB. Hox expression of a target gene: Extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129:3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci USA. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G, Wagner G. Canalization in evolutionary genetics: a stabilizing theory? Bioessays. 2000;22:372–380. doi: 10.1002/(SICI)1521-1878(200004)22:4<372::AID-BIES7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rokas A, Carroll SB. Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell. 2006;125:1387–1399. doi: 10.1016/j.cell.2006.04.043. [DOI] [PubMed] [Google Scholar]
- Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expression Patterns. 2010;10:177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. Nobel Symposium 146. FEBS Lett. 2009;583:3948–3958. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Evolution of developmental gene regulatory networks. Cell. 2011 In press. [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Repeated morphological evolution through cis- regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. cis-Regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Davidson EH. A new method, using cis-regulatory control, for blocking embryonic gene expression. Dev Biol. 2008;318:360–3658. doi: 10.1016/j.ydbio.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Regulative recovery in the sea urchin embryo, and the stabilizing role of fail-safe gene network wiring. Proc Nat, Acad Sci USA. 2009;106:18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. Allen & Unwin; London: 1957. [Google Scholar]
- Weatherbee SD, Halder G, Kim J, Hudson A, Carroll SB. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998;9:3038–3050. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Nijhout HF, Grunert LW, Halder G, Galant R, Selegue J, Carroll S. Ultrabithorax function in butterfly wings and the evolution of insect wing patterns. Curr Biol. 1999;9:109–115. doi: 10.1016/s0960-9822(99)80064-5. [DOI] [PubMed] [Google Scholar]


