Abstract
In this work we provide an up-to-date short review of computational magnetic resonance imaging (MRI) and software tools that are widely used to process and analyze diffusion-weighted MRI data. A review of different methods used to acquire, model and analyze diffusion-weighted imaging data (DWI) is first provided with focus on diffusion tensor imaging (DTI). The major preprocessing, processing and post-processing procedures applied to DTI data are discussed. A list of freely available software packages to analyze diffusion MRI data is also provided.
Keywords: diffusion tensor imaging, diffusion spectrum imaging, diffusion kurtosis, quantification, tractography, segmentation, connectivity, brain mapping, DTI, DSI, DKI, Q-ball, HYDI, atlas, AFNI, ICBM, TBSS, VBA, SPM, FreeSurfer, FSL, DTI software
1. Introduction
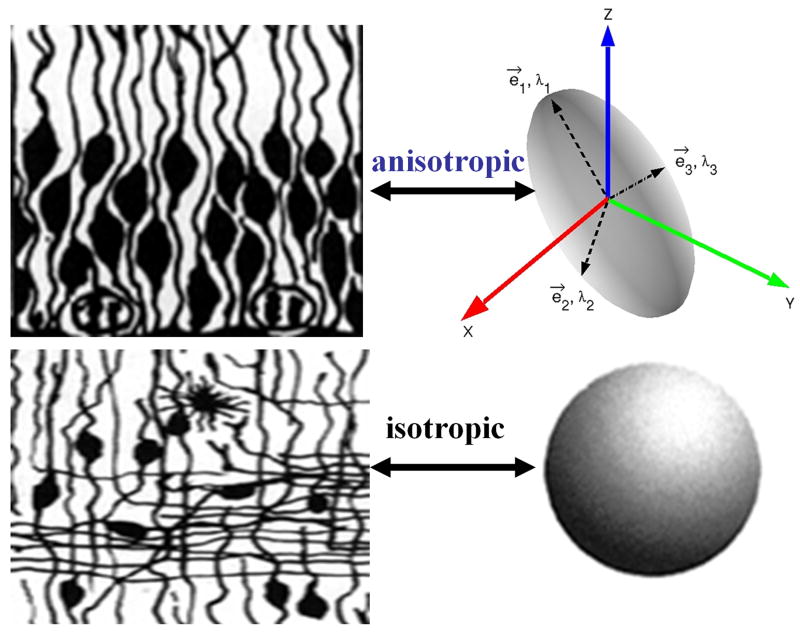
Whereas conventional magnetic resonance imaging (cMRI) provides methods to map the anatomy or tissue volume, diffusion-weighted imaging (DWI) of random translational water molecules offers quantitative anisotropy and orientation information that has been utilized to map the integrity or architecture of the soft tissue in the central nervous system [1-6]. Contributors to diffusion tensor anisotropy include cellular membranes, axons, myelin sheaths, and other factors [7]. Water molecular diffusion in cerebral white matter is less restricted along the axon than perpendicular to the compact bundles and hence it is termed anisotropic (see Figure 1). Gray matter is less anisotropic, while diffusion in barrier free tissue (e.g. edema, cerebrospinal fluid) is isotropic [8-10].
1.
Illustration of diffusion anisotropy using the ellipsoid representation of the single tensor model.
2. Mathematical Background
In general, DWI data are acquired on a prescribed volume (e.g. brain) by repeating the acquisition while altering the magnitude or orientation of the diffusion-sensitizing gradients. Hence, the DWI data acquired are generally multidimensional and can always be pooled as 4D data (e.g. in space x, y, z and diffusion encoding). Diffusion-weighted data are occasionally repeated in time and magnitude-averaged to enhance the signal-to-noise ratio (SNR). This data averaging can be done by the scanner software. Depending on the acquisition protocol, the DWI data may undergo model-based single or multiple diffusion tensor imaging (DTI) or model-free analysis to obtain scalar and vector metrics that can be used to map the tissue connectivity [11-13]. Currently, there are three diffusion MRI books [11-13] and several reviews on diffusion MRI [3-6, 14-19]. The sections below provide a short overview of the basics of diffusion MRI applied to whole brain human brain mapping in health and disease.
2.1 Model-based Diffusion Tensor Imaging
Mathematically, the k-th signal Sk obtained from a volume element upon applying a diffusion-weighting or b-factor bk along the unit vector gk can be modeled using the Gaussian mixture model (GMM) [5] as the superposition of different slowly exchanging positive-definite and symmetric tensors Dn each with a population fraction fn:
| (1) |
Where Dn refers to a second rank and positive definite tensor with three unique diagonal and three unique off-diagonal elements that can be represented by a 3×3 symmetric matrix [1-3, 11-13]:
| (2) |
In Equation 1, S0 refers to the reference signal obtained without diffusion-sensitization (e.g. b=0). The sum of all population fractions is unity:
| (3) |
In Equation 1, the apparent diffusion coefficient (ADC) can be defined as:
| (4) |
The system of equations described in Equation 1 can be expressed as a matrix equation after defining yk=Sk/S0
| (5) |
This system of equations obtained from all measurements can be solved using constrained or regularized non-linear least-squares fit methods for the unknown diffusivities for the fractions [20-27]; this analysis is the basis for single or multiple diffusion tensor imaging (DTI). For a system with a single unknown tensor (N=1), Equation 1 can be simplified by taking the logarithm to obtain a linear system of equations
| (6a) |
This equation can also be written as:
| (6b) |
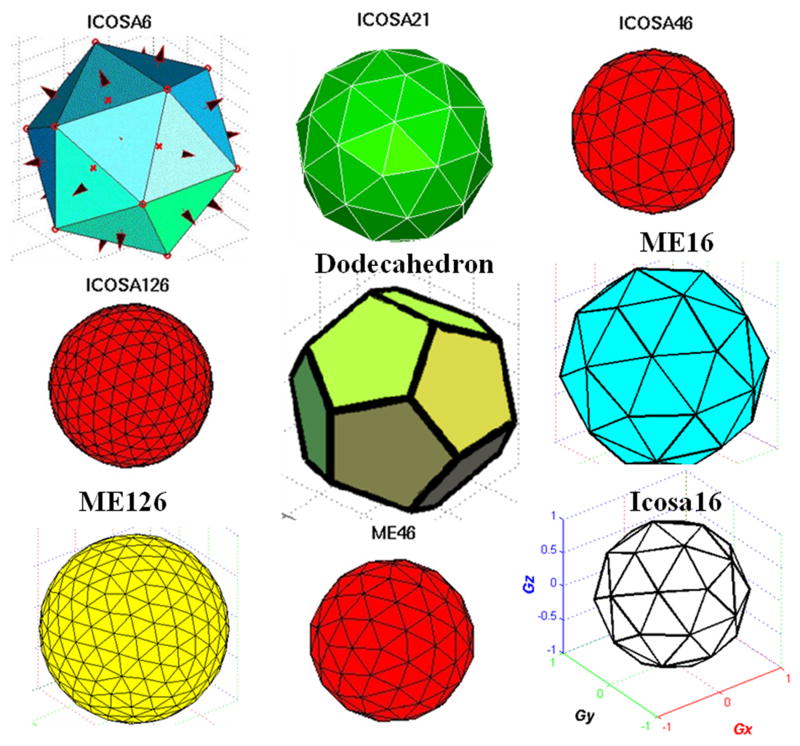
A generalization of this equation assumes the presence of high order tensors (HOT) and forms the basis of generalized DTI [28-30]. To solve this linear system of equations for the 6 diffusion tensor elements, a minimum of six independent diffusion-weighted measurements are needed in addition to the reference map (S0). In general, more than seven measurements are acquired with different diffusion b-factors and non-collinear orientations. Examples of non-collinear or uniformly distributed diffusion encoding sets are provided in Figure 2.
2.
Illustration of several uniformly distributed minimum energy and icoshderal diffusion encoding sets.
The over-determined system described by Equation 6 can be solved by least-squares and singular value decomposition (SVD) methods [28-30].
In the case where a single-b-factor is selected based on some known range of ADC, the orientation of the encoding vectors have to be uniformly distributed in three dimensional space [31-43]. The optimization of diffusion encoding schemes for white matter fibers with selected orientations such as skeletal muscle or spinal cord has been discussed by Peng and Arfanakis [44]. In the case where non-zero b-factors are acquired along with at least 15 encoding measurements for each b-factor, diffusion kurtosis imaging (DKI) methods may be used [45-46]. Additional data-driven methods such as principal component analysis (PCA) or independent component analysis (ICA) may use the moments of the measured data [47-50].
In the general case where N diffusion tensors with rank two are sought, 6*N variables need to be determined in addition to N-1 unknown population fractions subject to Equation 3 or a total of 6N+(N-1) unknowns.
Analysis of diffusion-weighted data acquired at finite SNR, angular and spatial resolutions according to the multi-tensor model may lead generally to unstable results as the exact number (N), population fractions (f), diffusion tensor orientation, and magnitude are unknown. The two-tensor case has received some attention as it appeals to determining the extent of fast and slow diffusion compartments in a voxel [51-53] or the interesting case of within or intravoxel crossing fibers [54-57]. The fast and slow diffusion tensor decoupling requires diffusion measurements with different b-factors (e.g. b=1000 and 4000 s mm-2), while the case of multiple crossing-fibers was modeled with uniformly distributed orientations at clinically attainable b-factors (e.g b ∼ 1000 s mm-2). In general, the two-tensor modeling problem can be solved if sufficient data are acquired at acceptable SNR using non-linear fitting approaches or can be regularized to reduce the number of unknowns by assuming cylindrical symmetry of the unknown fibers [20-27].
2.2 Model-Free Approaches
Data-driven or model-free approaches may require longer acquisition times and the diffusion-weighted data have to be acquired according to prescribed paradigms [13]. Data-driven approaches with high angular resolution diffusion (HARDI) measurements with two b-factors may use spherical harmonic decomposition (SHD) [58-63] which is based on the expansion of the measured apparent diffusion coefficient data in terms of a complete orthonormal set (e.g. spherical Legendre polynomials Ylm). Mathematically, this can be expressed as:
| (7) |
It has been shown that single fibers correspond to l=0, 2 and two fibers may be represented by l=0, 2, 4, while acquisition artifacts may be represented by odd number cases (e.g. l=1, l=3). Other model-free methods such as Q-ball imaging [64-67], diffusion spectrum imaging (DSI) [68-74] and hybrid diffusion imaging (HYDI) [75] may use mathematical operations such as Funk-Radon transform (FRT) and fast Fourier transforms (FFT) to characterize the magnitude and orientation distribution function (ODF) as detailed elsewhere [13].
2.3 The Single Diffusion Tensor Model
Currently, there are several methods developed to process or model the signal obtained according to specific acquisition paradigms. Despite its simplicity, the single tensor model remains the most popular model adopted in clinical research where healthy and patient groups are compared. The single tensor model requires a minimum of seven measurements and hence it is time-efficient. Due to SNR considerations, repeated acquisition of DWI data or acquisitions reduce noise variance. Data acquisition at higher magnetic fields [76-78] increases the intrinsic signal and offers a more time-efficient approach to acquire thinner slices [79-80] that can be used to reduce partial volume averaging effects [79-80]. It has been argued in published DTI literature that at constant imaging time acquiring data with more encoding directions is better than repeated averaging of the minimally needed amount of data [43].
2.4 Diffusion Tensor Diagonalization and Scalar Metrics
The single tensor or 3×3 matrix can be diagonalized (e.g. D = EΛEt) to obtain three orthonormal eigenvectors (E) and corresponding eigenvalues (λ1, λ2 and λ3). Several scalar functions or invariants can be defined from the diffusion eigenvalues such as mean or average diffusivity (Dav), relative anisotropy (RA), fractional anisotropy (FA), linear anisotropy (CL), planar anisotropy (CP) and spherical anisotropy (CS) [14, 81]:
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
Note that RA and FA are related analytically [50]:
| (14) |
In the special case of cylindrical symmetry [81] or λ1 > λ2 = λ3
| (15) |
The principal eigenvector (e1) is usually used in fiber tracking algorithms [12-13, 82-91] along with anisotropy measures.
2.5 Example of a DTI Acquisition Protocol
In general, DWI data are acquired covering the entire human brain at ∼ 1-3 mm axial sections with no gap and can be accomplished in 7-15 minutes. Table 1 describes a typical DTI acquisition paradigm implemented at a 3 T clinical scanner equipped with parallel imaging powerful gradient and data sampling technology. A total of 50 volumes with 8 reference (magnitude-averaged S0) and 42 diffusion-encoded volumes at b=1000 s mm-2 are acquired on a unit diffusion-encoding shell. Since 43 measurements are acquired, modeling the contributions from multiple tensors (e.g. N=2) should be feasible, in principle. Alternatively, the time spent to acquire the 42 encoding orientations could have been partitioned into 6 uniformly-distributed or icosahedral orientations each with 7 b-factors (0, 143, 286, 429, 571, 714, 857, 1000 s mm-2) keeping the number of S0 images, echo time, maximum b-factor and total scan time constant.
Table 1.
A tabulation of the acquisition parameters in a typical isotropic diffusion tensor imaging protocol.
| Data Acquisition Parameter | Value |
|---|---|
| Magnetic Field | 3T |
| Gradient Systems | 4G/cm per channel |
| Gradient Slew Rate | 200 mT/m/ms |
| Coil | Head/Parallel imaging |
| Acquistion Mode | 2D-axial: Fast spin-echo-EPI |
| Whole Brain | Inferior-to-superior |
| Field-of-view | 256mm × 256mm |
| Voxel size | 2mm × 2mm × 2mm |
| Acquistion Matrix | 128×64 |
| Acceleration | R=2 |
| Zero filling | k-space |
| Image Matrix | 256×256×70slices |
| Diffusion b-Factors | 1000 s mm-2 |
| Reference S0 | 8 |
| Signal-To-Noise Ratio | ∼ 50 |
| Encoding Directions | 42 (icosahedral: alternating Icosa21b) |
| Echo Time (TE) | 65 ms |
| Replication Time (TR) | 12,000 ms |
| Scan Time | 7 minutes |
Acquiring DWI data using different or arbitrary diffusion encoding orientations and multiple b-factors can be rather complicated and involves trade-offs between spatial and angular resolutions. For example, acquiring data with high isotropic spatial resolution at 1mm × 1mm × 1mm and using multiple b-factors and encoding directions may be best used to model the contributions from multiple fibers in a voxel, but this may not be time-efficient and SNR will be much smaller than a whole brain 2mm × 2mm × 2mm protocol that can be acquired under 7 minutes. Acquiring data with high b-factor sensitization and high angular diffusion allows multiple tensor compartment construction (e.g. fast and slow diffusion) and modeling of crossing fibers [51-57], with a trade-off of reduced SNR and increased scan time for whole brain protocols. As illustrated in Figure 3, noisy DWI measurements lead to an overestimation of anisotropy in isotropic systems [41, 92] whereas noise reduces the estimated anisotropy in highly anisotropic systems [93]. The estimated apparent or effective tensor anisotropy in regions of crossing fibers is reduced [94].
3.
An illustration of the effect of noise and crossing fibers on the estimated diffusion anisotropy.
3.0 Overview of Computational Procedures applied to Diffusion MRI Data
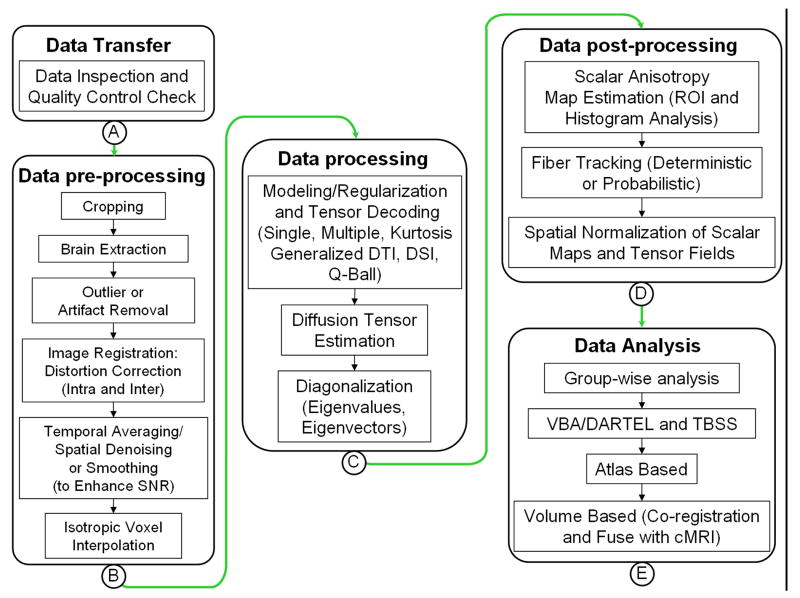
Freely available MRI computational [13] and DTI software packages [95-103] summarized below require the user to upload raw data along with the data acquisition parameters that include spatial and temporal image and diffusion acquisition parameters. The preprocessing and analysis conducted by these packages depend on the software primary design and models adopted to decode the diffusion-weighted data. Figure 4 summarizes some possible steps that can be applied on the DWI data.
4.
A schematic representation of several preprocessing, processing and post-processing procedures that can be performed on the raw DWI data or processed diffusion tensor maps.
3.1 Diffusion-weighted Imaging Preprocessing
The major preprocessing steps applied on a typical DWI data are summarized in Figure 4. The acquired DWI data may undergo several steps that depend on the clinical scanner, acquisition parameters and image quality. Most clinical MRI vendors provide the data in 2D-image DICOM (digital imaging and communications in medicine) format or other special data formats which may need to be converted to the format readable by the software package of interest.
For anatomical MRI and DWI data format preparation, conversion, inspection, tissue segmentation, and visualization, Table 2 provides a list of useful web sites with freely downloadable software packages. MRIcro is a popular free and easy to use package that has a user-friendly graphical user interface (GUI) on all operating systems to convert data files from 2D (DICOM http://medical.nema.org/) to 3D (e.g. Analyze http://mayoresearch.mayo.edu/mayo/research/robb_lab/analyze.cfm) or 4D neuroimaging informatics technology initiative (NIFTI http://nifti.nimh.nih.gov/). Other packages such as AFNI, FreeSurfer, FSL, SPM offer command-line conversion capabilities that enable users to prepare DWI and batch process [104] large data sets.
Table 2.
A list of MRI image analysis tools that can be used for data conversion, preparion and tissue segmentation.
In general, the acquired DWI volume data need to be co-aligned with the reference b0 volume and distortion correction need to be applied carefully [105,106]. Several sophistical image registration packages (e.g. AIR, ART, FNIRT) summarized in Table 3, discussed and compared elsewhere [107-108] can handle data coalignment, and spatial normalization, distortion correction [109-111]. Post registration DWI data may be subjected to simple geometric operations to reduce data storage such as cropping. In addition, brain extraction may follow to remove skull and non-brain tissues [112]. Diffusion-weighted MRI data can subsequently undergo spatial denoising (e.g. median or anisotropic edge-preserving filtering) to enhance SNR or interpolated to attain isotropic voxel dimensions as needed [113,114].
Table 3.
A list of software packages used for image registration and distortion correction.
| Volume Registration Package | Purpose | Web Location |
|---|---|---|
| AFNI | Analysis of Functional NeuroImages | http://afni.nimh.nih.gov/afni/doc/misc/ |
| AIR | Volume registartion Spatial Normalisation |
http://bishopw.loni.ucla.edu/ http://www.loni.ucla.edu/Software/AIR |
| ANTs | http://www.picsl.upenn.edu/ANTS/ | |
| ART | Registration Toolbox | http://www.nitrc.org/proiects/art/ |
|
FSL FLIRT/FNIRT |
Registration |
http://www.fmrib.ox.ac.uk/fsl/flirt/ http://www.fmrib.ox.ac.uk/fsl/fnirt/ |
| HAMMER | http://www.nitrc.org/projects/hammer/ | |
| IRTK | http://www.doc.ic.ac.uk/∼dr/software/ | |
| ITK | http://www.itk.org/ | |
|
SPM Dartel |
spatial normalization registration |
http://www.fil.ion.ucl.ac.uk/spm/ http://www.fil.ion.ucl.ac.uk/spm/ext/#toolboxes |
3.2 Diffusion-weighted Imaging Processing
Depending on the data acquisition, the prepared and encoded DWI data may subsequently get decoded to estimate the diffusion tensor per voxel using the acquisition encoding table (e.g. encoding orientation and b-factors). The diffusion tensor D may be diagonalized further to obtain the eigenvalues and eigenvectors. The eigenvalues may be used to obtain the mean diffusivity and anisotropy maps as defined above. Figure 5 illustrates the steps applied on some DWI data acquired using the Icosa21 encoding scheme on a single section.
5.
A pictorial illustration of the processing steps applied to a representative DWI data set using the icosa21 encoding scheme.
3.3 Diffusion Post-Processing
Most DTI quantitative packages produce diffusion tensor, FA, eigenvalue and principal eigenvector volumes from the DWI data acquired on each subject. Table 4 provides a list of free diffusion MRI software packages that have been reported in the literature. Additional MRI and diffusion MRI packages are listed on the neuroimaging tools and resources home page (http://www.nitrc.org/). Clinical MRI scanner vendor proprietary packages and commercial packages with DTI capabilities such as BrainVoyager (http://www.brainvoyager.com/index.html) and Analyze direct (http://www.analyzedirect.com/products/mridti.asp) are not discussed below.
Table 4.
A list of freely downloadable diffusion MRI software packages.
The first level of DTI quantification uses region-of-interest (ROI) to measure diffusivity and anisotropy (see Figure 6). A second level may use the estimated diffusion tensors, FA or diffusivities for group-wise analyses such as voxel-based analysis (VBA) [115-121], tract-based statistics (TBSS) [13, 122-125] or tensor-based that require multiple subject data alignment and spatial normalization or warping to a template [126-128]. A third level of analysis may use the principal eigenvector combined with anisotropy and multiple ROIs to perform deterministic fiber tracking [12-13, 82-91, 129-130]. A fourth level of analysis may involve using the anisotropy and diffusivity maps to segment the data into white and gray matter which can subsequently be used to perform volume or atlas-based analyses using the international consortium for brain mapping (ICBM http://www.loni.ucla.edu/ICBM/Downloads/Downloads_Atlases.shtml) atlases [131-138]. The regional results of the latter step can be used as masks for probabilistic fiber tracking [139-145]. An extension of the atlas-based approach utilizes the cMRI or T1-weighted data which can be used to generate brain volumes that can be warped unto the DTI data or other data sets such as relaxation, functional MRI, PET and MEG [146- 150].
6.
An illustration of the ROI and fiber tracking in three dimensional space.
3.4 Illustration of Quantitative Diffusion MRI Software Output
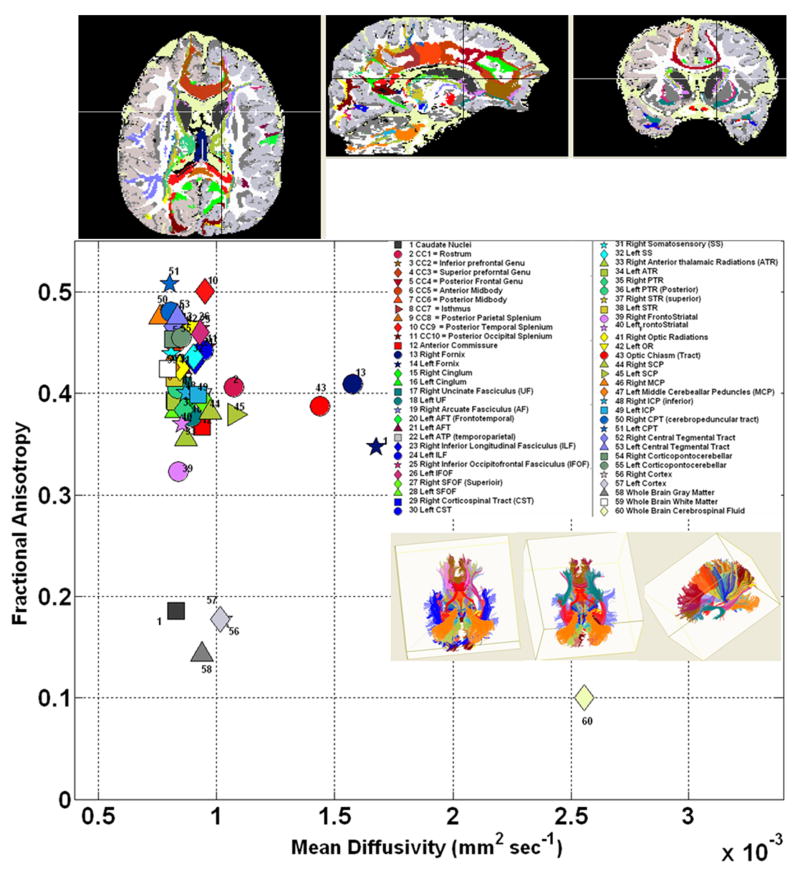
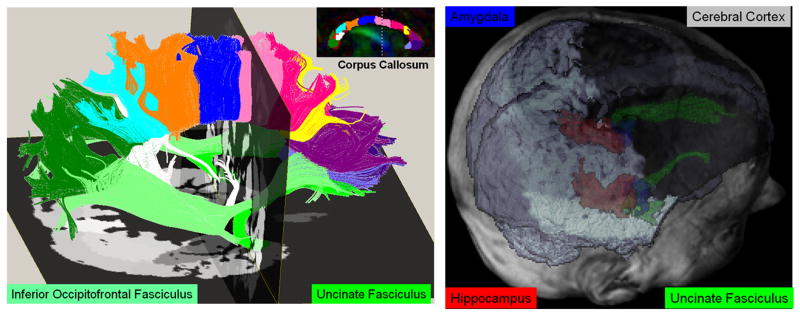
We illustrate DTI quantification and ability to cluster different tissue types. In Figure 7, we have used deterministic fiber tracking using multiple ROIs implemented in DTIstudio [95] to track several association, projection and commissural pathways. These pathways were fused with the anatomical cMRI-parcellated results using FreeSurfer. The cMRI and DTI maps were coaligned in SPM and viewed in MRIcro after saving all results as ANALYZE 3D volume files. Figure 8 illustrates the ability to use DTI data to segment the cortical and deep gray matter (e.g. hippocampus, amygdale) and corresponding connections (e.g. uncinate fasciculus) using a DTI atlas-based approach [138].
7.
An illustration of quantitative DTI data obtained using white matter fiber tracking and cortical and regional (e.g. caudate) gray matter segmentation. Several tissue types are represented in the mean diffusivity vs. FA space.
8.
An illustration of DTI atlas-based gray matter segmentation and fusion with fiber tracking and anatomical MRI data after registration.
4. Conclusions
In this work, we have provided an up-to-date review of computational methods applied to diffusion MRI data with focus on the single diffusion tensor model. We have also listed a host of software packages that have been reported in the diffusion MRI literature. To-date there has been no general consensus on the optimal diffusion tensor acquisition protocol (e.g. ∼ 7 minute single tensor vs. ∼ 30 minute DSI). Attention must be paid to data acquisition details (e.g. SNR and spatial resolution) while minimizing image distortion and motion artifacts and keeping the scan time tolerable for imaging children and patients. Quality assurance measures to ensure consistency and accuracy of the DTI data need to be implemented before comparing healthy control and patient data collected for cross-sectional and serial analyses. There are trade-offs, strengths and weaknesses associated with the choice of diffusion data analysis strategy (e.g. deterministic vs. probabilistic tracking, ROI, atlas-based, VBA vs. TBSS).
Acknowledgments
This work is funded by the NIH-NINDS Grant R01 NS052505-04 and the Dunn Fund.
Footnotes
Conflict of Interest Statement: None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Basser PJ. New histological and physiological stains derived from diffusion-tensor MR images. Ann N Y Acad Sci. 1997;820:123–138. doi: 10.1111/j.1749-6632.1997.tb46192.x. Review. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, Le Bihan D. Estimation of the effective self diffusion tensor from NMR spin echo. J Magn Reson B. 1994;123:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 3.Le Bihan D, Mangin JF, Poupon C, Clark C, Pappata S, Molko N, Chabriat H. Diffusion Tensor Imaging: concepts and applications. J Magn Reson Imaging. 2001;13:534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 4.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nature Rev. Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 5.Basser PJ, Jones DK. Diffusion-tensor MRI: theory, experimental design and data analysis - a technical review. NMR Biomed. 2002;15:456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- 6.Mori S, Zhang J. Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron. 2006;51:527–539. doi: 10.1016/j.neuron.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. Review. [DOI] [PubMed] [Google Scholar]
- 8.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Di Chiro G. Diffusion tensor MR imaging of the human brain. Radiology. 1996;201:637–48. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 9.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161:401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 10.Fieremans E, De Deene Y, Delputte S, Ozdemir MS, D'Asseler Y, Vlassenbroeck J, Deblaere K, Achten E, Lemahieu I. Simulation and experimental verification of the diffusion in an anisotropic fiber phantom. J Magn Reson. 2008;190:189–199. doi: 10.1016/j.jmr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat Rev Neurosci. 2003;4(6):469–80. doi: 10.1038/nrn1119. Review. [DOI] [PubMed] [Google Scholar]
- 12.Mori S. Introduction to Diffusion Tensor Imaging. Elsiever; Amsterdam: 2007. [Google Scholar]
- 13.Johansen-Berg H, Behrens TEJ. Diffusion MRI: From quantitative measurements to in vivo neuroanatomy. Academic Press; London: 2009. [Google Scholar]
- 14.Westin CF, Maier SE, Magmata H, Nabavi A, Jolesz FA, Kikinis R. Processing and visualization for diffusion tensor MRI. Med Image Anal. 2002;6:93–108. doi: 10.1016/s1361-8415(02)00053-1. Review. [DOI] [PubMed] [Google Scholar]
- 15.Hagmann P, Jonasson L, Maeder P, Thiran JP, Wedeen VJ, Meuli R. Understanding diffusion MR imaging techniques: from scalar diffusion-weighted imaging to diffusion tensor imaging and beyond. Radiographics. 2006;26:S205–23. doi: 10.1148/rg.26si065510. [DOI] [PubMed] [Google Scholar]
- 16.Hasan KM. Fundamentals of Diffusion Tensor Imaging of the Entire Human Brain: Review of Basic Theory, Data Acquisition, Processing and Potential Applications at 1.5 T and 3.0 T. In: Chen FJ, editor. Progress in Brain Mapping Research. Chapter 1. Nova Science Publishers; Hauppauge NY: 2006. pp. 1–80. [Google Scholar]
- 17.Minati L, Weglarz WP. Physical foundations, models, and methods of diffusion magnetic resonance imaging of the brain: a review. Concept Magn Reson – A. 2007;30A:278–307. [Google Scholar]
- 18.Lenglet C, Campbell JS, Descoteaux M, Haro G, Savadjiev P, Wassermann D, Anwander A, Deriche R, Pike GB, Sapiro G, Siddiqi K, Thompson PM. Mathematical methods for diffusion MRI processing. Neuroimage. 2009;45:S111–S122. doi: 10.1016/j.neuroimage.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander AL, Lee JE, Lazar M, Field AS. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4:316–29. doi: 10.1016/j.nurt.2007.05.011. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreher BW, Schneider JF, Mader I, Martin E, Hening J, llyasov KA. Multitensor approach for analysis and tracking of complex fiber configurations. Magn Reson Med. 2005;54:1216–25. doi: 10.1002/mrm.20670. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Bammer R, Moseley ME. Limitations of apparent diffusion coefficient-based models in characterizing non-gaussian diffusion. Magn Reson Med. 2005;54:419–428. doi: 10.1002/mrm.20579. [DOI] [PubMed] [Google Scholar]
- 22.Peled S, Friman O, Jolesz F, Westin CF. Geometrically constrained two-tensor model for crossing tracts in DWI. Magn Reson Imaging. 2006;24:1263–1270. doi: 10.1016/j.mri.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada K, Sakai K, Hoogenraad FG, Holthuizen R, Akazawa K, Ito H, Oouchi H, Matsushima S, Kubota T, Sasajima H, Mineura K, Nishimura T. Multitensor tractography enables better depiction of motor pathways: initial clinical experience using diffusion-weighted MR imaging with standard b-value. AJNR Am J Neuroradiol. 2007;28:1668–1673. doi: 10.3174/ajnr.A0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez-Manzanares A, Rivera M, Vemuri BC, Carney P, Mareci T. Diffusion basis functions decomposition for estimating white matter intravoxel fiber geometry. IEEE Trans Med Imaging. 2007;26:1091–1102. doi: 10.1109/TMI.2007.900461. [DOI] [PubMed] [Google Scholar]
- 25.Sotiropoulos SN, Bai L, Morgan PS, Auer DP, Constantinescu CS, Tench CR. A regularized two-tensor model fit to low angular resolution diffusion images using basis directions. J Magn Reson Imaging. 2008;28:199–209. doi: 10.1002/jmri.21380. [DOI] [PubMed] [Google Scholar]
- 26.Kabasawa H, Masutani Y, Abe O, Aoki S, Ohtomo K. Quantitative diffusion tensor analysis using multiple tensor ellipsoids model and tensor field interpolation at fiber crossing. Acad Radiol. 2008;15:84–92. doi: 10.1016/j.acra.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Landman BA, Wan H, Bogovic JA, Bazin PL, Prince JL. Resolution of Crossing Fibers with Constrained Compressed Sensing using Traditional Diffusion Tensor MRI. Proc Soc Photo Opt Instrum Eng. 2010;7623:76231H. doi: 10.1117/12.844171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Özarslan E, Mareci TH. Generalized diffusion tensor imaging and analytical relationships between diffusion tensor imaging and high angular resolution diffusion imaging. Magn Reson Med. 2003;50:955–965. doi: 10.1002/mrm.10596. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Bammer R, Acar B, Moseley ME. Characterizing non-Gaussian diffusion by using generalized diffusion tensors. Magn Reson Med. 2004;51:924–937. doi: 10.1002/mrm.20071. [DOI] [PubMed] [Google Scholar]
- 30.Akkerman EM. The direct tensor solution and higher-order acquisition schemes for generalized diffusion tensor imaging. J Magn Reson. 2010;206:9–19. doi: 10.1016/j.jmr.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Xing D, Papadakis NG, Huang CL, Lee VM, Carpenter TA, Hall LD. Optimised diffusion-weighting for measurement of apparent diffusion coefficient (ADC) in human brain. Magn Reson Imaging. 1997;15:771–784. doi: 10.1016/s0730-725x(97)00037-4. [DOI] [PubMed] [Google Scholar]
- 32.Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–525. [PubMed] [Google Scholar]
- 33.Papadakis NG, Xing D, Huang CL, Hall LD, Carpenter TA. A comparative study of acquisition schemes for diffusion tensor imaging using MRI. J Magn Reson. 1999;137:67–82. doi: 10.1006/jmre.1998.1673. [DOI] [PubMed] [Google Scholar]
- 34.Skare S, Hedehus M, Moseley ME, Li TQ. Condition number as a measure of noise performance of diffusion tensor data acquisition schemes with MRI. J Magn Reson. 2000;147:340–352. doi: 10.1006/jmre.2000.2209. [DOI] [PubMed] [Google Scholar]
- 35.Hasan KM, Parker DL, Alexander AL. Comparison of gradient encoding schemes for diffusion-tensor MRI. J Magn Reson Imaging. 2001;13:769–780. doi: 10.1002/jmri.1107. [DOI] [PubMed] [Google Scholar]
- 36.Batchelor PG, Atkinson D, Hill DLG, Calamante F, Connelly A. Anisotropic noise propagation in diffusion tensor MRI sampling schemes. Magn Reson Med. 2003;49:1143–1151. doi: 10.1002/mrm.10491. [DOI] [PubMed] [Google Scholar]
- 37.Alexander DC, Barker GJ. Optimal imaging parameters for fiber-orientation estimation in diffusion MRI. NeuroImage. 2005;27:357–367. doi: 10.1016/j.neuroimage.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Mang SC, Gembris D, Grodd W, Klose U. Comparison of gradient encoding directions for higher order tensor diffusion data. Magn Reson Med. 2009;61:335–343. doi: 10.1002/mrm.21797. [DOI] [PubMed] [Google Scholar]
- 39.Farrell JA, Landman BA, Jones CK, Smith SA, Prince JL, van Zijl PC, Mori S. Effects of signal-to-noise ratio on the accuracy and reproducibility of diffusion tensor imaging-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5 T. J Magn Reson Imaging. 2007;26:756–767. doi: 10.1002/jmri.21053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landman BA, Farrell JA, Jones CK, Smith SA, Prince JL, Mori S. Effects of diffusion weighting schemes on the reproducibility of dti-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5 T. NeuroImage. 2007;36:1123–1138. doi: 10.1016/j.neuroimage.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 41.Hasan KM. A framework for quality control and parameter optimization in diffusion tensor imaging: theoretical analysis and validation. Magn Reson Imaging. 2007;25:1196–1202. doi: 10.1016/j.mri.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan L, Leow AD, Jahanshad N, Chiang MC, Barysheva M, Lee AD, Toga AW, McMahon KL, de Zubicaray GI, Wright MJ, Thompson PM. How does angular resolution affect diffusion imaging measures? Neuroimage. 2010;49:1357–1371. doi: 10.1016/j.neuroimage.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni H, Kavcic V, Zhu T, Ekholm S, Zhong J. Effects of number of diffusion gradient directions on derived diffusion tensor imaging indices in human brain. AJNR Am J Neuroradiol. 2006;27:1776–1781. [PMC free article] [PubMed] [Google Scholar]
- 44.Peng H, Arfanakis K. Diffusion tensor encoding schemes optimized for white matter fibers with selected orientations. Magn Reson Imaging. 2007;25:147–153. doi: 10.1016/j.mri.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Jensen JH, Helpern JA, Ramani A, Lu H, Kaczynski K. Diffusional kurtosis imaging: the quantification of non-Gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53:1432–1440. doi: 10.1002/mrm.20508. [DOI] [PubMed] [Google Scholar]
- 46.Jensen JH, Helpern JA. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR in biomed. 2010 doi: 10.1002/nbm.1518. (2010) 10.1002/nbm.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadakis NG, Zheng Y, Wilkinson ID. Analysis of diffusion tensor magnetic resonance imaging data using principal component analysis. Phys Med Biol. 2003;48:343–350. doi: 10.1088/0031-9155/48/24/n01. [DOI] [PubMed] [Google Scholar]
- 48.Arfanakis K, Cordes D, Haughton VM, Carew JD, Meyerand ME. Independent component analysis applied to diffusion tensor MRI. Magn Reson Med. 2002;47:354–363. doi: 10.1002/mrm.10046. [DOI] [PubMed] [Google Scholar]
- 49.Akkerman EM. Efficient measurement and calculation of MR diffusion anisotropy images using the Platonic variance method. Magn Reson Med. 2003;49:599–604. doi: 10.1002/mrm.10365. [DOI] [PubMed] [Google Scholar]
- 50.Hasan KM, Narayana PA. Computation of the fractional anisotropy and mean diffusivity maps without tensor decoding and diagonalization: Theoretical analysis and validation. Magn Reson Med. 2003;50:589–598. doi: 10.1002/mrm.10552. [DOI] [PubMed] [Google Scholar]
- 51.Maier SE, Vajapeyam S, Mamata H, Westin CF, Jolesz FA, Mulkern RV. Biexponential diffusion tensor analysis of human brain diffusion data. Magn Reson Med. 2004;51:321–330. doi: 10.1002/mrm.10685. [DOI] [PubMed] [Google Scholar]
- 52.Clark CA, Hedehus M, Moseley ME. In vivo mapping of the fast and slow diffusion tensors in human brain. Magn Reson Med. 2002;47:623–628. doi: 10.1002/mrm.10118. [DOI] [PubMed] [Google Scholar]
- 53.Inglis BA, Bossart EL, Buckley DL, Wirth ED, 3rd, Mareci TH. Visualization of neural tissue water compartments using biexponential diffusion tensor MRI. Magn Reson Med. 2001;45:580–587. doi: 10.1002/mrm.1079. [DOI] [PubMed] [Google Scholar]
- 54.Wiegell MR, Larsson HB, Wedeen VJ. Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology. 2000;217:897–903. doi: 10.1148/radiology.217.3.r00nv43897. [DOI] [PubMed] [Google Scholar]
- 55.Alexander DC, Barker GJ, Arridge SR. Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magn Reson Med. 2002;48:331–340. doi: 10.1002/mrm.10209. [DOI] [PubMed] [Google Scholar]
- 56.Staempfli P, Jaermann T, Crelier GR, Kollias S, Valavanis A, Boesiger P. Resolving fiber crossing using advanced fast marching tractography based on diffusion tensor imaging. Neuroimage. 2006;30:110–120. doi: 10.1016/j.neuroimage.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 57.Alexander DC. Multiple-fiber reconstruction algorithms for diffusion MRI. Ann N Y Acad Sci. 2005;1064:113–133. doi: 10.1196/annals.1340.018. Review. [DOI] [PubMed] [Google Scholar]
- 58.Frank LR. Anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2001;45:935–939. doi: 10.1002/mrm.1125. [DOI] [PubMed] [Google Scholar]
- 59.Frank LR. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magn Reson Med. 2002;47:1083–1099. doi: 10.1002/mrm.10156. [DOI] [PubMed] [Google Scholar]
- 60.Alexander DC, Barker GJ, Arridge SR. Detection and modelling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magnetic Resonance in Medicine. 2002;48:331–340. doi: 10.1002/mrm.10209. [DOI] [PubMed] [Google Scholar]
- 61.Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002;48:577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 62.Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using sphericalde convolution. Neuroimage. 2004;23:1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 63.Jian B, Vemuri BC. A unified computational framework for deconvolution to reconstruct multiple fibers from diffusion weighted MRI. IEEE Trans Med Imaging. 2007;26:1464–1471. doi: 10.1109/TMI.2007.907552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minat L, Aquino D, Rampoldi S, Papa S, Grisoli M, Bruzzone MG, Maccagnano E. Biexponential and diffusional kurtosis imaging, and generalised diffusion-tensor imaging (GDTI) with rank-4 tensors: a study in a group of healthy subjects. MAGMA. 2007;20:241–253. doi: 10.1007/s10334-007-0091-1. [DOI] [PubMed] [Google Scholar]
- 65.Tuch DS. Q-ball imaging. Magn Reson Med. 2004;52:1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- 66.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40:885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 67.Barnett A. Theory of Q-ball imaging redux: Implications for fiber tracking. Magn Reson Med. 2009;62:910–923. doi: 10.1002/mrm.22073. [DOI] [PubMed] [Google Scholar]
- 68.Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Agmann PH, Arceuil HD, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 69.Wedeen VJ, Hagmann P, Tseng WY, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005;54:1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 70.Kuo LW, Chen JH, Wedeen VJ, Tseng WY. Optimization of diffusion spectrum imaging and q-ball imaging on clinical MRI system. Neuroimage. 2008;41:7–18. doi: 10.1016/j.neuroimage.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 71.Schmahmann JD, Pandya DN, Wang R, Dai G, D'Arceuil HE, de Crespigny AJ, Wedeen VJ. Association fibre pathways of the brain: parallel observations from diffusion spectrum imaging and autoradiography. Brain. 2007;130:630–653. doi: 10.1093/brain/awl359. [DOI] [PubMed] [Google Scholar]
- 72.Granziera C, Schmahmann JD, Hadjikhani N, Meyer H, Meuli R, Wedeen V, Krueger G. Diffusion spectrum imaging shows the structural basis of functional cerebellar circuits in the human cerebellum in vivo. PLoS One. 2009;4:e5101. doi: 10.1371/journal.pone.0005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi E, Dai G, Wang R, Ohki K, Rosen GD, Galaburda AM, Grant PE, Wedeen VJ. Development of cerebral fiber pathways in cats revealed by diffusion spectrum imaging. Neuroimage. 2010;49:1231–1240. doi: 10.1016/j.neuroimage.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nezamzadeh M, Wedeen VJ, Wang R, Zhang Y, Zhan W, Young K, Meyerhoff DJ, Weiner MW, Schuff N. In-vivo investigation of the human cingulum bundle using the optimization of MR diffusion spectrum imaging. Eur J Radiol. 2010;75:e29–36. doi: 10.1016/j.ejrad.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexander AL, Wu YC, Venkat PC. Hybrid diffusion imaging (HYDI) Conf Proc IEEE Eng Med Biol Soc. 2006;1:2245–2248. doi: 10.1109/IEMBS.2006.259453. [DOI] [PubMed] [Google Scholar]
- 76.Jaermann T, Crelier G, Pruessmann KP, Golay X, Netsch T, van Muiswinkel AM, Mori S, van Zijl PC, Valavanis A, Kollias S, Boesiger P. SENSE-DTI at 3 T. Magn Reson Med. 2004;51:230–236. doi: 10.1002/mrm.10707. [DOI] [PubMed] [Google Scholar]
- 77.Roebroeck A, Galuske R, Formisano E, Chiry O, Bratzke H, Ronen I, Kim DS, Goebel R. High-resolution diffusion tensor imaging and tractography of the human optic chiasm at 9.4 T. Neuroimage. 2008;39:157–68. doi: 10.1016/j.neuroimage.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 78.Mukerjee P, Hess CP, Xu D, Han ET, Kelley DA, Vigneron DB. Development and initial evaluation of 7-T q-ball imaging of the human brain. Magn Reson Imaging. 2008;26:171–180. doi: 10.1016/j.mri.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jaermann T, De Zanche N, Staempfli P, Pruessmann KP, Valavanis A, Boesiger PP, Kollias SS. Preliminary experience with visualization of intracortical fibers by focused high-resolution diffusion tensor imaging. AJNR Am J Neuroradiol. 2008;29:146–150. doi: 10.3174/ajnr.A0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hasan KM, Kamali A, Kramer LA. Mapping the human brain white matter tracts relative to cortical and deep gray matter using diffusion tensor imaging at high spatial resolution. Magn Reson Imaging. 2009;27:631–636. doi: 10.1016/j.mri.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 81.Hasan KM, Basser PJ, Parker DL, Alexander AL. Analytical computation of the eigenvalues and eigenvectors in DT-MRI. J Magn Reson. 2001;152:41–47. doi: 10.1006/jmre.2001.2400. [DOI] [PubMed] [Google Scholar]
- 82.Mori S, Wakana S, Nagae-Poetscher LM, van Zijl PCM. MRI Atlas of Human Whitte Matter. Elsevier; 2005. [Google Scholar]
- 83.Conturo TE, Lori NF, Cull TS, Akbudak E, Snyder AZ, Shimony JS, McKinstry RC, Burton H, Raichle ME. Tracking neuronal fiber pathways in the living human brain. Proc Natl Acad Sci U S A. 1999;18:10422–10427. doi: 10.1073/pnas.96.18.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mori S, van Zijl PC. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. Review. [DOI] [PubMed] [Google Scholar]
- 85.Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- 86.Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- 87.Aralasmak A, Ulmer JL, Kocak M, Salvan CV, Hillis AE, Yousem DM. Association, commissural, and projection pathways and their functional deficit reported in literature. J Comput Assist Tomogr. 2006;30:695–715. doi: 10.1097/01.rct.0000226397.43235.8b. Review. [DOI] [PubMed] [Google Scholar]
- 88.Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl PC, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hasan KM, Kamali A, Abid H, Kramer LA, Fletcher JM, Ewing-Cobbs L. Quantification of the spatiotemporal micro structural organization of the human brain association, projection and commissural pathways across the lifespan using diffusion tensor tractography. Brain Struct Funct. 2010;214:361–373. doi: 10.1007/s00429-009-0238-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hasan KM, Iftikhar A, Kamali A, Kramer LA, Ashtari M, Cirino PT, Papanicolaou AC, Fletcher JM, Ewing-Cobbs L. Development and aging of the healthy human brain uncinate fasciculus across the lifespan using diffusion tensor tractography. Brain Res. 2009;1276:67–76. doi: 10.1016/j.brainres.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hasan KM, Kamali A, Iftikhar A, Kramer LA, Papanicolaou AC, Fletcher JM, Ewing-Cobbs L. Diffusion tensor tractography quantification of the human corpus callosum fiber pathways across the lifespan. Brain Res. 2009;1249:91–100. doi: 10.1016/j.brainres.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson AW. Theoretical analysis of the effects of noise on diffusion tensor imaging. Magn Reson Med. 2001;46:1174–1188. doi: 10.1002/mrm.1315. [DOI] [PubMed] [Google Scholar]
- 93.Mamata H, Jolesz FA, Maier SE. Characterization of central nervous system structures by magnetic resonance diffusion anisotropy. Neurochem Int. 2004;45:553–560. doi: 10.1016/j.neuint.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 94.Oouchi H, Yamada K, Sakai K, Kizu O, Kubota T, Ito H, Nishimura T. Diffusion anisotropy measurement of brain white matter is affected by voxel size: underestimation occurs in areas with crossing fibers. AJNR Am J Neuroradiol. 2007;28:1102–116. doi: 10.3174/ajnr.A0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang H, van Zijl PC, Kim J, Pearlson GD, Mori S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81:106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 96.Cook PA, Bai Y, Nedjati-Gilani S, Seunarine KK, Hall MG, Parker GJ, Alexander DC. Camino: Open-Source Diffusion-MRI Reconstruction and Processing. 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine; Seattle, WA, USA. May (2006); p. 2759. [Google Scholar]
- 97.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–19. doi: 10.1016/j.neuroimage.2004.07.051. Review. [DOI] [PubMed] [Google Scholar]
- 98.Ashburner L. Computational anatomy with the SPM software. Magn Reson Imaging. 2009;27:1163–1174. doi: 10.1016/j.mri.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 99.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuro anatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 100.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 101.Lucas BC, Bogovic JA, Carass A, Bazin PL, Prince JL, Pham DL, Landman BA. The Java Image Science Toolkit (JIST) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics. 2010;8:5–17. doi: 10.1007/s12021-009-9061-2. Erratum in: Neuroinformatics 8 (2010) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cárdenes R, Muñoz-Moreno E, Tristan-Vega A, Martin-Fernandez M. Saturn: a software application of tensor utilities for research in neuroimaging. Comput Methods Programs Biomed. 2010;97:264–279. doi: 10.1016/j.cmpb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 103.Cointepas Y, Poupon C, Maroy R, Rivière D, LeBihan D, Mangin JF. A freely available Anatomist/BrainVISA package for analysis of diffusion MR data. NeuroImage; Proceedings of the Ninth HBM Scientific Meeting; New York. 2003. p. 810. http://brainvisainfo. [Google Scholar]
- 104.Karmonik C, York M, Grossman R, Kakkar E, Patel K, Haykal H, King D. An image analysis pipeline for the semi-automated analysis of clinical fMRI images based on freely available software. Comput Biol Med. 2010;40(3):279–87. doi: 10.1016/j.compbiomed.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 105.Chang LC, Jones DK, Pierpaoli C. RESTORE: Robust estimation of tensors by outlier rejection. Magnetic Resonance in Medicine. 2005;53:1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- 106.Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med. 2009;61:1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- 107.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 109.Andersson JLR, Skare S. A model-based method for retrospective correction of geometric distortions in diffusion-weighted EPI. NeuroImage. 2002;16:177–199. doi: 10.1006/nimg.2001.1039. [DOI] [PubMed] [Google Scholar]
- 110.Ardekani S, Sinha U. Geometric distortion correction of high-resolution 3 T diffusion tensor brain images. Magn Reson Med. 2005;54:1163–1171. doi: 10.1002/mrm.20651. [DOI] [PubMed] [Google Scholar]
- 111.Netsch T, van Muiswinkel A. Quantitative evaluation of image-based distortion correction in diffusion tensor imaging. IEEE Trans Med Imaging. 2004;23:789–98. doi: 10.1109/TMI.2004.827479. [DOI] [PubMed] [Google Scholar]
- 112.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ding Z, Gore JC, Anderson AW. Reduction of noise in diffusion tensor images using anisotropic smoothing. Magn Reson Med. 2005;53:485–490. doi: 10.1002/mrm.20339. [DOI] [PubMed] [Google Scholar]
- 114.Hahn KR, Prigarin S, Rodenacker K, Hasan KM. Denoising for Diffusion Tensor Imaging with low Signal to Noise Ratios: Method and Monte Carlo Validation. International Journal for Biostatistics and Biomathematics. 2010;1:63–81. [Google Scholar]
- 115.Jones DK, Griffin LD, Alexander DC, Catani M, Horsfield MA, Howard R, Williams SC. Spatial normalization and averaging of diffusion tensor MRI data sets. Neuroimage. 2002;17:592–617. [PubMed] [Google Scholar]
- 116.Jones DK, Symms MR, Cercignani M, Howard RJ. The effect of filter size on VBM analyses of DT-MRI data. NeuroImage. 2005;26:546–554. doi: 10.1016/j.neuroimage.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 117.Salat DH, Tuch DS, Greve DN, van der Kouwe AJ, Hevelone ND, Zaleta AK, Rosen BR, Fischl B, Corkin S, Rosas HD, Dale AM. Age-related alterations in white matter microstructure measured by diffusion tensor imaging. Neurobiol Aging. 2005;26:1215–1227. doi: 10.1016/j.neurobiolaging.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 118.Snook L, Plewes C, Beaulieu C. Voxel based versus region of interest analysis in diffusion tensor imaging of neurodevelopment. NeuroImage. 2007;34:243–252. doi: 10.1016/j.neuroimage.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 119.Ashtari M, Cervellione K, Cottone J, Ardekani BA, Sevy S, Kumra S. Diffusion abnormalities in adolescents and young adults with a history of heavy cannabis use. J Psychiatr Res. 2009;43:189–204. doi: 10.1016/j.jpsychires.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vangberg TR, Skranes J, Dale AM, Martinussen M, Brubakk AM, Haraldseth O. Changes in white matter diffusion anisotropy in adolescents born prematurely. Neuroimage. 2006;32:1538–1548. doi: 10.1016/j.neuroimage.2006.04.230. [DOI] [PubMed] [Google Scholar]
- 121.Keller TA, Just MA. Altering cortical connectivity: remediation-induced changes in the white matter of poor readers. Neuron. 2009;64:624–631. doi: 10.1016/j.neuron.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 123.Smith SM, Johansen-Berg H, Jenkinson M, Rueckert D, Nichols TE, Miller KL, Robson MD, Jones DK, Klein JC, Bartsch AJ, Behrens TE. Acquisition and voxelwise analysis of multi-subject diffusion data with tract-based spatial statistics. Nat Protoc. 2007;2:499–503. doi: 10.1038/nprot.2007.45. [DOI] [PubMed] [Google Scholar]
- 124.Focke NK, Yogarajah M, Bonelli SB, Bartlett PA, Symms MR, Duncan JS. Voxel-based diffusion tensor imaging in patients with mesial temporal lobe epilepsy and hippocampal sclerosis. Neuroimage. 2008;40:728–737. doi: 10.1016/j.neuroimage.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 125.Abe O, Takao H, Gonoi W, Sasaki H, Murakami M, Kabasawa H, Kawaguchi H, Goto M, Yamada H, Yamasue H, Kasai K, Aoki S, Ohtomo K. Voxel-based analysis of the diffusion tensor. Neuroradiology. 2010;52:699–710. doi: 10.1007/s00234-010-0716-3. [DOI] [PubMed] [Google Scholar]
- 126.Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial Transformations of Diffusion Tensor Magnetic Resonance Images IEEE Trans. Medical Imaging. 2001;20:1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- 127.Xu D, Mori S, Shen D, van Zijl PC, Davatzikos C. Spatial normalization of diffusion tensor fields. Magnetic Resonance in Medicine. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- 128.Peng H, Orlichenko A, Dawe RJ, Agam G, Zhang S, Arfanakis K. Development of a human brain diffusion tensor template. Neuroimage. 2009;46:967–980. doi: 10.1016/j.neuroimage.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kamali A, Kramer LA, Hasan KM. Feasibility of prefronto-caudate pathway tractography using high resolution diffusion tensor tractography data at 3T. Journal of Neuroscience Methods. 2010;191:249–254. doi: 10.1016/j.jneumeth.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamamoto T, Yamada K, Nishimura T, Kinoshita S. Tractography to depict three layers of visual field trajectories to the calcarine gyri. Am J Ophthalmol. 2005;140:781–785. doi: 10.1016/j.ajo.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 131.Hua K, Zhang J, Wakana S, Jiang H, Li X, Reich DS, Calabresi PA, Pekar JJ, van Zijl PC, Mori S. Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification. Neuroimage. 2008;39:336–347. doi: 10.1016/j.neuroimage.2007.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike GB, Evans A, Toga AW, Woods R, Mazziotta JC, Miller MI, van Zijl PC, Mori S. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage. 2010;52:1289–1301. doi: 10.1016/j.neuroimage.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Faria AV, Zhang J, Oishi K, Li X, Jiang H, Akhter K, Hermoye L, Lee SK, Hoon A, Stashinko E, Miller MI, van Zijl PC, Mori S. Atlas-based analysis of neurodevelopment from infancy to adulthood using diffusion tensor imaging and applications for automated abnormality detection. Neuroimage. 2010;52:415–428. doi: 10.1016/j.neuroimage.2010.04.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lawes IN, Barrick TR, Murugam V, Spierings N, Evans DR, Song M, Clark CA. Atlas-based segmentation of white matter tracts of the human brain using diffusion tensor tractography and comparison with classical dissection. Neuroimage. 2008;39:62–79. doi: 10.1016/j.neuroimage.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 135.Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW, Pike GB, Neto PR, Evans A, Zhang J, Huang H, Miller MI, van Zijl P, Mazziotta J. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40:570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hagler DJ, Jr, Ahmadi ME, Kuperman J, Holland D, McDonald CR, Halgren E, Dale AM. Automated white-matter tractography using probabilistic diffusion tensor atlas: application temporal lobe epilepsy. Hum Brain Mapp. 2009;30:1535–1547. doi: 10.1002/hbm.20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Verhoeven JS, Sage CA, Leemans A, Van Hecke W, Callaert D, Peeters R, De Cock P, Lagae PL, Sunaert S. Construction of a stereotaxic DTI atlas with full diffusion tensor information for studying white matter maturation from childhood to adolescence using tractography-based segmentations. Hum Brain Mapp. 2010;31:470–486. doi: 10.1002/hbm.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hasan KM, Frye RE. Diffusion Tensor based Regional Gray Matter Tissue Segmentation Using the International Consortium for Brain Mapping Atlases. Human Brain Mapping. 2010 doi: 10.1002/hbm.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Parker GJM, Haroon HA, Wheeler-Kingshott CAM. A Framework for a Streamline-Based Probabilistic Index of Connectivity (PICo) using a Structural Interpretation of MRI Diffusion Measurements. Journal of Magnetic Resonance Imaging. 2003;18:242–254. doi: 10.1002/jmri.10350. [DOI] [PubMed] [Google Scholar]
- 140.Parker GJM, Alexander DC. Probabilistic anatomical connectivity derived from the microscopic persistent angular structure of cerebral tissue. Philos Trans R Soc B Biol Sci. 2005;360:893–902. doi: 10.1098/rstb.2005.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Park HJ, Kim JJ, Lee SK, Seok JH, Chun J, Kim DI, Lee JD. Corpus callosal connection mapping using cortical gray matter parcellation and DT-MRI. Hum Brain Mapp. 2008;29:503–516. doi: 10.1002/hbm.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pannek K, Mathias JL, Bigler ED, Brown G, Taylor JD, Rose S. An automated strategy for the delineation and parcellation of commissural pathways suitable for clinical populations utilising high angular resolution diffusion imaging tractography. Neuroimage. 2010;50:1044–1053. doi: 10.1016/j.neuroimage.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 143.Behrens TE, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott CA, Boulby PA, Barker GJ, Sillery EL, Sheehan K, Ciccarelli O, Thompson AJ, Brady JM, Matthews PM. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- 144.Zhang D, Snyder AZ, Shimony JS, Fox MD, Raichle ME. Noninvasive functional and structural connectivity mapping of the human thalamocortical system. Cereb Cortex. 2010;20:1187–1194. doi: 10.1093/cercor/bhp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Helmich RC, Derikx LC, Bakker M, Scheeringa R, Bloem BR, Toni I. Spatial remapping of cortico-striatal connectivity in Parkinson's disease. Cereb Cortex. 2010;20:1175–1186. doi: 10.1093/cercor/bhp178. [DOI] [PubMed] [Google Scholar]
- 146.Studholme C. Dense feature deformation morphometry: Incorporating DTI data into conventional MRI morphometry. Med Image Anal. 2008;12:742–751. doi: 10.1016/j.media.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Staempfli P, Reischauer C, Jaermann T, Valavanis A, Kollias S, Boesiger P. Combining fMRI and DTI: a framework for exploring the limits of fMRI-guided DTI fiber tracking and for verifying DTI-based fiber tractography results. Neuroimage. 2008;39:119–126. doi: 10.1016/j.neuroimage.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 148.Inoue K, Ito H, Uchida S, Taki Y, Kinomura S, Tsuji I, Sato S, Horie K, Kawashima R, Ito M, Fukuda H. Decrease in glucose metabolism in frontal cortex associated with deterioration of microstructure of corpus callosum measured by diffusion tensor imaging in healthy elderly. Hum Brain Mapp. 2008;29:375–384. doi: 10.1002/hbm.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boss A, Kolb A, Hofmann M, Bisdas S, Nägele T, Ernemann U, Stegger L, Rossi C, Schlemmer HP, Fannenberg CP, Reimold M, Claussen CD, Pichler BJ, Klose U. Diffusion tensor imaging in a human PET/MR hybrid system. Invest Radiol. 2010;45:270–274. doi: 10.1097/RLI.0b013e3181dc3671. [DOI] [PubMed] [Google Scholar]
- 150.Hasan KM, Walimuni IS, Kramer LA, Frye RE. Human Brain Atlas-based Volumetry and Relaxometry: Application to Healthy Development and Natural Aging. Magn Reson Med. 2010 doi: 10.1002/mrm.22515. [DOI] [PubMed] [Google Scholar]