Abstract
The vitamin D receptor (VDR) gene has been associated with cancer risk, but only a few polymorphisms have been studied in relation to melanoma risk and the results have been inconsistent. We examined 38 VDR gene SNPs in a large international multi-center population-based case-control study of melanoma.
Buccal DNAs were obtained from 1207 people with incident multiple primary melanoma and 2469 with incident single primary melanoma. SNPs with known or suspected impact on VDR activity, htSNPs with ≥10% MAF in Caucasians, and SNPs reported as significant in other association studies were examined. Logistic regression was used to calculate the relative risks conferred by the individual SNP.
Eight of 38 SNPs in the promoter, coding, and 3’ gene regions were individually significantly associated with multiple primary melanoma after adjusting for covariates. The estimated increase in risk for individuals who were homozygous for the minor allele ranged from 25% to 33% for 6 polymorphisms: rs10875712 (OR 1.28; 95%CI, 1.01–1.62), rs4760674 (OR 1.33; 95% CI, 1.06–1.67), rs7139166 (OR 1.26; 95%CI, 1.02–1.56), rs4516035 (OR 1.25; 95%CI, 1.01–1.55), rs11168287 (OR 1.27; 95%CI, 1.03–1.57), rs1544410 (OR 1.30; 95%CI, 1.04–1.63); for 2 polymorphisms, homozygous carriers had a decreased risk: rs7305032 (OR 0.81; 95%CI 0.65–1.02), rs7965281 (OR, 0.78; 95%CI, 0.62–0.99). We recognize the potential false positive findings due to multiple comparisons; however the 8 significant SNPs in this study outnumbered the 2 significant tests expected to occur by chance. The vitamin D receptor may play a role in melanomagenesis.
Keywords: VDR, SNP, melanoma, polymorphism, vitamin D
INTRODUCTION
Vitamin D is hypothesized to lower the risk for or mortality from several cancers, including melanoma. In vitro and in vivo assays show that vitamin D modulates cell proliferation, differentiation and programmed cell death 1–5, supporting its anti-carcinogenic potential. A number of observational studies in humans have also suggested that vitamin D, or putative surrogates for vitamin D status such as season, geographic latitude and evidence of chronic sun exposure, is associated with better outcomes in melanoma patients, despite the fact that UVB exposure, which activates the precursor of vitamin D present in skin, is a known risk factor for melanomagenesis 6–9. This adds to the plausibility of the hypothesis that vitamin D affects melanoma risk.
The active form of Vitamin D, 1,25-dihydroxyvitamin D, exerts its biological function by binding to the vitamin D receptor (VDR), a nuclear steroid hormone receptor expressed in many tissues and organs including skin 10. This receptor is encoded by the vitamin D receptor gene (ID 7421, OMIM 601769) which spans approximately 100 Kb on chromosome 12q13.11, with a large non-coding region spanning exons 1F to 1C, and with exons 2 to 9 encoding a 424 aminoacid protein 11. The VDR gene contains numerous variants, some of which are hypothesized to influence the expression and/or function of the VDR protein product, and some of which have been associated with complex traits and diseases such as osteoporosis, diabetes, small stature, and cancer 12, 13.
Despite the large number of SNPs, to date only a few common polymorphisms in the VDR gene have been studied in relation to melanoma risk: FokI, BsmI, TaqI, ApaI, A-1012G, and Cdx2, and the results for some of the SNPs have not been consistent across different studies 14–23. For example, the FokI (rs2228570) minor allele T (f, M4) results in production of a VDR protein that is less effective as a transcriptional activator 24–26 and has been linked to an increased risk for melanoma in several studies 14,16,21,23, and in pooled analyses 23,27,28, but has been observed to have no effect according to other reports 19,20,22,23. Some of the inconsistencies may be due to differences in the study design, study populations, or in the exposures or adjustments for covariates during the analyses. It is also possible, especially when the size of the VDR gene region is considered, that the causal genetic variant or key group of variants remains to be identified.
Nejentsev et al. (2004)12 identified the minimal set of tag SNPs and then evaluated the informativeness of the most commonly studied VDR SNPs in relation to disease association. The authors found that FokI was not in LD with any other SNP, while ApaI, TaqI, and BsmI predicted only 38% of the SNPs in block B with R2>0.8. Therefore, in studies that genotyped only FokI, TaqI, BsmI or ApaI, information on a large fraction of common SNPs was not captured. The aim of this study was to conduct a comprehensive analysis of common genetic variants including less studied SNPs across the VDR gene and to determine their effects on melanoma risk.
MATERIAL AND METHODS
Study population
Subjects were recruited between 1998 and 2003 through an international multi-center population-based case control study of multiple primary melanoma, the Genes, Environment and Melanoma Study (GEM). The GEM population consists of incident cases of multiple primary melanoma identified in eight population- based registries and one hospital center in Australia, Canada, Italy and USA. Patients with incident single primary melanoma were used as controls. This design is, in effect, a case-control study of melanoma conducted in the high risk population of individuals who have already experienced a melanoma diagnosis. Further details of the study design and its rationale have been published 29,30.
The study was approved by the institutional review boards at all centers. All study participants signed informed consent, provided a buccal sample for extraction of germline DNA, and completed a detailed interview, providing information on sun exposure history, sun sensitivity, and phenotypic characteristics.
Selection of VDR SNPs
DNA samples from 1207 individuals with multiple primary melanoma and 2469 with single primary melanoma were available for genotyping. A total of 42 SNPs located on average at 3.6Kb from each other were selected. These included htSNPs with MAF>10% in Caucasians 12,13, SNPs with known or suspected impact on the transcription, stability, and/or activity of the VDR, and SNPs reported as significant in other association studies. Specifically, rs11568820 located on the promoter has been found to affect transcription of VDR by interfering with the binding site of the transcription factor Cdx2 11. SNPs rs7139166 (1A-1521) and rs4516035 (1A-1012), when the haplotype 1521G/-1012A is present, were shown to affect promoter activity 31. The polymorphism rs2228570 (FokI), responsible for an alternative earlier start and a less active protein was included. The commonly studied polymorphisms rs731236 (TaqI) and rs1544410 (BsmI) were also included although their functional impact is less apparent.
Laboratory methods
The isolation of DNA was previously described 32. We used the Sequenom MassARRAY iPLEX genotyping Platform (Sequenom Inc., San Diego, CA) 33 to test 36 SNPs and pyrosequencing and melting temperature analysis to test 2 SNPs. For iPLEX, experiments were designed with the RealSNP Assay Database and the MassArray software v.3.4. Five to 10 ng of genomic DNA were amplified using specific primers, reagents, and cycling conditions detailed in the Supplementary Material (Table S1). Products were spotted onto 384 SpectroCHIP bioarrays (Sequenom). Cluster plots were evaluated with the TyperAnalyzer application (MassARRAY v.3.4). Assays were considered optimal according to degree of clustering, absence of signal in the blanks, and when sequencing of representative samples within the clusters confirmed the genotypes. Four SNPs were rejected during the assay design or during the wet testing due to non-specific signals (rs2238139, rs739837) or very low signals and call rates (rs2408876, rs3847987). SNPs rs2228570 (267bp) and rs1544410 (209bp) were genotyped using single SNP genotyping methods: melting temperature analysis34 coupled to the LightTyper (Roche Applied Science, Indianapolis, USA) and pyrosequencing 35 with the PSQ™ MA instrument (Biotage AB, Uppsala, Sweden). Specific primers and probes, reaction and cycling conditions are available in Table S1. Quality control (QC) procedures included use of barcodes on samples and plates, inclusion of internal laboratory controls, randomly selected repeats (5% for MassArray and 10% for pyrosequencing and for melting temperature analyses), pre-PCR dedicated materials and working space, and at least two independent readers for review and interpretation of cluster plots, pyrograms, melting curves, and results. A plate containing discordant results for at least one pair of duplicate samples or internal controls was newly assayed. Samples with weak signals and outliers were repeated. QC for data entry included an additional review of 20% of genotype calls. There was 100% agreement in the genotyping calls between two independent laboratory members. Direct sequencing was performed on an independently amplified PCR fragment to confirm the genotype of laboratory control samples and when a new SNP was identified by different sequence pattern in the pyrogram, or different melting profile using standard methods 36.
Statistical analysis
We analyzed a total of 38 VDR SNPs in relation to risk of melanoma using conventional methods for case-control studies. The “variant” allele was defined to be the less frequently occurring allele. For each SNP, logistic regression was used to calculate adjusted odds ratios (OR) with 95% confidence intervals (95% CI). Heterozygous and homozygous carriers of the variant allele were separately compared to homozygous carriers of the most prevalent or “reference” allele within the same SNP-specific logistic regression model. An ordinal variable for the three possible genotypes was also created to evaluate linear trend. A multiplicative age-sex interaction term was included in all models to control for the fact that the age incidence curves for melanoma differ for men and women. In addition to age, sex, and age-sex interaction, study center was included as a covariate in the multivariate analysis to account for potential differences among populations. Analyses further adjusting for known melanoma risk factors, such as mole count, hair and eye color, and other “phenotypic” characteristics, were also conducted, but the results are not materially different and are not presented. Hardy-Weinberg equilibrium (HWE) tests were performed to identify possible gross systematic genotyping problems; since our control group consists of melanoma cases, absence of HWE could be expected in SNPs that confer risk. We evaluated the issue of multiple comparisons using techniques described in Abramovich and Benjamini (2006)37. We also employed quantile-quantile (Q-Q) plots 38 for comparing the distribution of p-values obtained in the logistic regression against those expected if the data occurred randomly and not specifically due to associations between SNPs and melanoma risk. All analyses were performed using SAS statistical software version 9.2 (SAS Institute, Cary, NC).
Meta-analyses
We conducted a comprehensive, systematic search of the medical literature to identify relevant studies on cutaneous melanoma in which genotyping results were available for each of the SNPs identified as statistically significant in our study. The literature search was limited to studies in humans, included all years up to November 2010, and was performed using the following databases: PubMed, ISI Web of Science, ISI Web of Knowledge, Cochrane, and Embase. Search terms included ‘Vitamin D Receptor’ and separately, ‘VDR’, in combination with ‘melanoma’, ‘allele’, ‘gene’, ‘polymorphism’, and ‘risk’. We searched for publications with frequencies or estimates of relative risk for cutaneous melanoma, with 95% CI. Review articles were considered as sources of additional relevant citations. We extracted information on the characteristics of the study population, study design, year of publication, and source of controls (Table S2). The homozygous occurrence of the most common allele for each polymorphism was considered the reference for our analyses. Hardy-Weinberg tests were performed among controls. For one study in which genotype frequencies were not reported 20, we contacted the authors to obtain this information. Among the articles retrieved, two papers 17,21 presented overlapping data on rs731236 and we therefore included results of the report with the largest number of subjects 21. The variation across different studies attributable to heterogeneity rather than chance was assessed using the I-squared measure 39. We considered that heterogeneity was present when the overall I2 p-value was <0.05. Calculation of summary odds ratios, 95% confidence intervals and study heterogeneity p-values for meta-analysis was done in STATA (version 11.0; STATA Corp, College Station, TX) using the metan function. Publication bias was evaluated using the adjusted rank correlation test within the metabias function of STATA 40.
Bioinformatics
The SNPs analyzed in this study do not affect the amino acid sequence. Therefore, we evaluated their potential functional relevance on the VDR transcription or activity by searching for overlaps between the SNP genomic location and transcription factor binding sites, splice sites, sequences complementary to miRNA seed regions, and by considering conservation across species. We obtained information from the UCSC’s genome browser tables (accessed October 2009), and each SNP, when information was available, was further annotated with conservation information, and miRNA and transcription factor binding site overlaps. Information on conservation was obtained from the UCSC’s runs of the PHAST package on primates, vertebrates and mammals. The miRNA annotations were obtained from Target Scan, and information on transcription factor binding from the binding site conservation across the human/mouse/rat alignment. Further information can be obtained from: http://genome.ucsc.edu/cgi-bin/hgTables 41. We also used an alternative method for detecting potential overlaps between VDR SNPs and miRNAs. Specifically, we looked for miRNA seed regions by searching for exact matches between the 2–7 nucleotide regions of known mature human miRNAs and the reverse complement of these substrings in the 3’UTR of the VDR gene. Selbach et al. (2008) 42 has shown that this 6-mer miRNA seed is essential for its function. Consequently, a SNP located in the miRNA target site which is complementary to the miRNA seed region is likely to deregulate the expression of the target gene.
RESULTS
Descriptive characteristics of the study participants are provided in Table 1. The distribution of melanoma risk factors such as age, gender, and number of moles on the back differed between GEM cases and GEM controls. Further details on the associations between participants’ characteristics and melanoma risk in this study have been published elsewhere 43.
Table 1.
Selected characteristics of study participants
| MPM (GEM cases) N (%) |
SPM (GEM controls) N (%) |
||
|---|---|---|---|
| Age | |||
| <50 | 169 (14.0) | 917 (37.1) | |
| 50+ | 1038 (86.0) | 1552 (62.9) | |
| Gender | |||
| Male | 796 (65.9) | 1277 (51.7) | |
| Female | 411 (34.1) | 1192 (48.3) | |
| Breslow thickness (mm) | |||
| 0 (in situ) | 431 (35.7) | 0 (0.0) | |
| <1 | 543 (45.0) | 1666 (67.5) | |
| 1–2 | 108 (8.9) | 447 (18.1) | |
| 2–4 | 49 (4.1) | 194 (7.9) | |
| >4 | 74 (6.1) | 157 (6.4) | |
| Site | |||
| head & neck | 295 (24.4) | 381 (15.5) | |
| trunk | 493 (40.9) | 1091 (44.4) | |
| arms | 207 (17.2) | 460 (18.7) | |
| legs | 210 (17.4) | 520 (21.2) | |
| other | 2 (0.2) | 4 (0.2) | |
| Histology | |||
| superficial spreading | 352 (29.5) | 1347 (54.6) | |
| nodular melanoma | 62 (5.2) | 220 (8.9) | |
| lentigo maligna | 89 (7.5) | 174 (7.1) | |
| in situ | 428 (35.9) | 0 (0.0) | |
| other | 263 (22.0) | 726 (29.4) | |
| Number of Moles | |||
| 0–4 | 390 (33.2) | 850 (35.2) | |
| 5–10 | 256 (21.8) | 545 (22.5) | |
| 11–25 | 279 (23.8) | 530 (21.9) | |
| > 25 | 249 (21.2) | 492 (20.4) | |
| Total | 1207 | 2469 | |
The individual call rates for the SNPs evaluated in this study, their distribution in the population according to HapMap_CEU and other databases, and the genotype frequencies obtained in the GEM cases and controls are shown in Table 2. After excluding four SNPs with poor assay performance, the average call rate was 0.98 for the remaining 38 SNPs. For SNPs rs11568820 and rs7305032 the failure rates were 0.12 and 0.07, respectively. However the proportion of failures did not differ significantly between cases and controls (p=0.33 and p=0.34). The proportion of failures was also similar between cases and controls for SNPs rs2228570 (p=0.85) and rs1544410 (p=0.33). All other SNPs displayed failure rates lower than 0.03. Genotype frequencies were similar to those reported in dbSNP in Caucasians or Europeans (HapMap-CEU or AFD_EUR_PANEL), with few exceptions (rs11168275, rs12370156, and rs7305032). For rs11168275 we and others 44 have found that G, and not A, is the minor allele in Caucasians. All three SNPs are in HWE (Table S1). The following SNPs were in LD with each other (D’ > 0.96): rs7139166 and rs4516035 (D’ = 0.972; R2 = 0.942); rs2544027 and rs2544028 (D’ = 0.968, R2 = 0.708); rs2544027 and rs2544038 (D’ = 0.974, R2 = 0.815); rs2544028 and rs2544038 (D’ = 0.974, R2 = 0.815); rs1544410 and rs731236 (D’=0.957, R2=0.912).
Table 2.
Genotype frequency of VDR variants in individuals with multiple primary (GEM cases) and single primary (GEM controls) melanoma and adjusted odds ratios by genotype.
| Relative Position |
RefSeq | Location1 | Genotype | Call Rate (%) |
Frequency in dbSNP (%)2 |
Cases N (%) |
Controls N (%) |
OR (95% CI)3 | p-value3 |
|---|---|---|---|---|---|---|---|---|---|
| Promoter Region | |||||||||
| 1 | rs2071358 | 1F−29607 | CC | 98.4 | 56.7 | 842 (70.7) | 1656 (68.2) | 1 | 0.1 |
| CA | 36.7 | 314 (26.4) | 682 (28.1) | 0.91 (0.77–1.08) | |||||
| AA | 6.7 | 35 (2.9) | 89 (3.7) | 0.73 (0.47–1.11) | |||||
| 2 | rs10875712 | 1F−26412 | GG | 98.1 | 41.7 | 455 (38.5) | 1005 (41.5) | 1 | 0.02 |
| 41.7 | 554 (46.8) | 1088 (44.9) | 1.16 (0.99–1.37) | ||||||
| CC | 16.7 | 174 (14.7) | 331 (13.7) | 1.28 (1.01–1.62) | |||||
| 3 | rs6823 | 1F−25571 | CC | 98.4 | 27.1 | 369 (31.1) | 765 (31.5) | 1 | 0.33 |
| CG | 52.5 | 565 (47.6) | 1165 (47.9) | 1.04 (0.88–1.24) | |||||
| GG | 20.3 | 253 (21.3) | 500 (20.6) | 1.11 (0.90–1.37) | |||||
| 4 | rs4760674 | 1F−20173 | CC | 98.9 | 40 | 424 (35.4) | 965 (39.6) | 1 | 0.002 |
| CA | 41.7 | 583 (48.7) | 1110 (45.6) | 1.32 (1.12–1.56) | |||||
| AA | 18.3 | 191 (15.9) | 361 (14.8) | 1.33 (1.06–1.67) | |||||
| 5 | rs1015390 | 1F−7196 | CC | 98.9 | 75 | 876 (73.3) | 1781 (73.0) | 1 | 0.52 |
| CT | 25 | 291 (24.4) | 601 (24.6) | 0.94 (0.79–1.12) | |||||
| TT | 28 (2.3) | 57 (2.3) | 0.96 (0.58–1.58) | ||||||
| 6 | rs4237856 | 1F−1198 | AA | 97.8 | 65 | 683 (57.9) | 1363 (56.4) | 1 | 0.22 |
| CA | 26.7 | 427 (36.2) | 888 (36.7) | 0.94 (0.80–1.10) | |||||
| CC | 8.3 | 69 (5.9) | 166 (6.9) | 0.83 (0.60–1.14) | |||||
| 7 | rs4073729 | 1F−217 | CC | 98.7 | 70 | 862 (72.4) | 1780 (73.0) | 1 | 0.89 |
| TC | 30 | 300 (25.2) | 604 (24.8) | 0.99 (0.83–1.17) | |||||
| TT | 28 (2.4) | 55 (2.3) | 0.99 (0.60–1.64) | ||||||
| 8 | rs11168314 | 1F+6225 | GG | 98.3 | 68.3 | 771 (65.0) | 1574 (64.8) | 1 | 0.74 |
| GA | 31.7 | 357 (30.1) | 742 (30.5) | 0.95 (0.80–1.11) | |||||
| AA | 58 (4.9) | 113 (4.7) | 1.04 (0.73–1.49) | ||||||
| 9 | rs10459217 | 1F+20593 | TT | 98.4 | 66.7 | 752 (63.7) | 1545 (63.4) | 1 | 0.68 |
| TC | 31.7 | 372 (31.5) | 783 (32.1) | 0.93 (0.79–1.10) | |||||
| CC | 1.7 | 56 (4.7) | 110 (4.5) | 1.06 (0.74–1.52) | |||||
| 10 | rs11568820 | 1E−1739 | GG | 88.4 | 68.3 | 742 (68.4) | 1512 (69.9) | 1 | 0.79 |
| Cdx2 | AG | 30 | 295 (27.2) | 559 (25.8) | 0.97 (0.81–1.16) | ||||
| AA | 1.7 | 48 (4.4) | 93 (4.3) | 1.18 (0.79–1.75) | |||||
| 11 | rs7139166 | 1A−1521 | CC | 97 | 30 | 381 (32.7) | 814 (33.9) | 1 | 0.04 |
| GC | 50 | 534 (45.8) | 1137 (47.4) | 1.06 (0.89–1.26) | |||||
| GG | 20 | 251 (21.5) | 450 (18.7) | 1.26 (1.02–1.56) | |||||
| 12 | rs4516035 | 1A−1012 | TT(AA) | 98.5 | 30 | 382 (32.2) | 816 (33.5) | 1 | 0.05 |
| EcoRV | CT(GA) | 50 | 551 (46.5) | 1165 (47.8) | 1.07 (0.90–1.26) | ||||
| CC(GG) | 20 | 252 (21.3) | 454 (18.6) | 1.25 (1.01–1.55) | |||||
| 13 | rs7299460 | 1D+2175 | CC | 98.1 | 45.8 | 593 (50.1) | 1189 (49.1) | 1 | 0.99 |
| (1B−2528) | CT | 49.2 | 466 (39.4) | 1000 (41.3) | 0.96 (0.82–1.13) | ||||
| TT | 5.1 | 125 (10.6) | 234 (9.7) | 1.04 (0.81–1.35) | |||||
| 14 | rs11168287 | −12516 | GG | 98.1 | 16.7 | 292 (24.6) | 639 (26.4) | 1 | 0.03 |
| AG | 50 | 577 (48.7) | 1187 (49.0) | 1.16 (0.97–1.40) | |||||
| AA | 33.3 | 316 (26.7) | 594 (24.5) | 1.27 (1.03–1.57) | |||||
| 15 | rs11168284 | −10151 | AA | 98.1 | n/a | 501 (42.4) | 979 (40.4) | 1 | 0.37 |
| GA | 534 (45.2) | 1146 (47.3) | 0.93 (0.79–1.10) | ||||||
| GG | 146 (12.4) | 299 (12.3) | 0.92 (0.72–1.17) | ||||||
| 16 | rs10875694 | −8762 | TT | 99.6 | n/a | 836 (69.5) | 1703 (69.3) | 1 | 0.82 |
| TA | 331 (27.5) | 671 (27.3) | 1.03 (0.87–1.22) | ||||||
| AA | 36 (3.0) | 84 (3.4) | 0.83 (0.54–1.28) | ||||||
| 17 | rs4760648 | −7767 | CC | 98.5 | 25 | 405 (34.1) | 726 (29.9) | 1 | 0.06 |
| CT | 56.7 | 569 (47.9) | 1262 (51.9) | 0.83 (0.70–0.98) | |||||
| TT | 18.3 | 215 (18.1) | 444 (18.3) | 0.84 (0.67–1.04) | |||||
| 18 | rs2238135 | −5292 | GG | 99 | 60.6 | 711 (59.3) | 1388 (56.8) | 1 | 0.18 |
| (1C−1663) | GC | 31 | 414 (34.6) | 895 (36.6) | 0.93 (0.79–1.09) | ||||
| CC | 8.5 | 73 (6.1) | 160 (6.5) | 0.84 (0.61–1.15) | |||||
| 19 | rs1989969 | −5112 | CC | 99.3 | 43.7 | 417 (34.8) | 923 (37.6) | 1 | 0.08 |
| (1C−1453) | CT | 46.5 | 574 (48.0) | 1151 (46.9) | 1.16 (0.98–1.37) | ||||
| TT | 9.9 | 206 (17.2) | 378 (15.4) | 1.18 (0.94–1.47) | |||||
| Coding Region | |||||||||
| 20 | rs2228570 | Start-3 | CC (FF) | 95 | 35 | 421 (36.7) | 873 (37.2) | 1 | 0.71 |
| FokI | (Ex2) | CT (Ff) | 41.7 | 548 (47.8) | 1117 (47.6) | 1.01 (0.85–1.19) | |||
| TT (ff) | 23.3 | 177 (15.4) | 355 (15.1) | 1.05 (0.83–1.33) | |||||
| 21 | rs11168275 | Intron 2+476 | AA | 99.5 | 3.3^ | 687 (57.1) | 1401 (57.1) | 1 | 0.87 |
| GA | 33.3^ | 445 (37.0) | 899(36.6) | 1.01 (0.86–1.18) | |||||
| GG | 63.3^ | 71 (5.9) | 155 (6.3) | 0.94 (0.68–1.29) | |||||
| 22 | rs7974708 | Intron 2+2586 | TT | 98.9 | 480 (40.1) | 1047 (42.9) | 1 | 0.11 | |
| CT | n/a | 558 (46.6) | 1127 (46.2) | 1.07 (0.91–1.25) | |||||
| CC | 159 (13.3) | 265 (10.9) | 1.22 (0.96–1.56) | ||||||
| 23 | rs3782905 | Intron 2+6584 | CC | 99 | 53.3 | 523 (43.7) | 1128 (46.2) | 1 | 0.08 |
| CG | 36.7 | 529 (44.2) | 1083 (44.3) | 1.08 (0.92–1.26) | |||||
| GG | 10 | 145 (12.1) | 233 (9.5) | 1.25 (0.97–1.60) | |||||
| 24 | rs2189480 | Intron 2+8923 | CC | 98.1 | 40 | 514 (43.4) | 986 (40.7) | 1 | 0.08 |
| CA | 41.7 | 518 (43.8) | 1100 (45.4) | 0.89 (0.76–1.05) | |||||
| AA | 18.3 | 152 (12.8) | 337 (13.9) | 0.84 (0.66–1.06) | |||||
| 25 | rs886441 | Intron 2+4004 | TT | 98.5 | 66.7 | 793 (66.8) | 1574 (64.7) | 1 | 0.64 |
| CT | 28.3 | 344 (29.0) | 754 (31.0) | 0.95 (0.81–1.12) | |||||
| CC | 5 | 50 (4.2) | 106 (4.4) | 0.98 (0.68–1.43) | |||||
| 26 | rs2239181 | Intron 3+2881 | TT | 98.8 | 80 | 972 (81.3) | 1938 (79.6) | 1 | 0.47 |
| GT | 18.3 | 214 (17.9) | 474 (19.5) | 0.95 (0.78–1.15) | |||||
| GG | 1.7 | 9 (0.8) | 24 (1.0) | 0.79 (0.34–1.82) | |||||
| 27 | rs2107301 | Intron 3+3260 | CC | 98.9 | 48.3 | 649 (54.2) | 1253 (51.4) | 1 | 0.32 |
| CT | 46.7 | 461 (38.5) | 995 (40.8) | 0.94 (0.80–1.10) | |||||
| TT | 5 | 87 (7.3) | 190 (7.8) | 0.89 (0.66–1.19) | |||||
| 28 | rs2239182 | Intron 3+3419 | GG | 97 | 25 | 354 (30.2) | 644 (26.9) | 1 | 0.08 |
| AG | 46.7 | 555 (47.4) | 1170 (48.9) | 0.89 (0.74–1.06) | |||||
| AA | 28.3 | 263 (22.4) | 579 (24.2) | 0.83 (0.67–1.03) | |||||
| 29 | rs12370156 | Intron 3+4697 | TT | 98.7 | 25 | 337 (28.2) | 641 (26.4) | 1 | 0.33 |
| CT | 37.5 | 576 (48.2) | 1179 (48.5) | 0.90 (0.76–1.08) | |||||
| CC | 37.5 | 283 (23.7) | 612 (25.2) | 0.90 (0.73–1.11) | |||||
| 30 | rs2238140 | Intron 3+6166 | TT | 98.9 | 339 (28.4) | 637 (26.1) | 1 | 0.23 | |
| CT | n/a | 573 (47.9) | 1193 (48.9) | 0.88 (0.74–1.05) | |||||
| CC | 283 (23.7) | 612 (25.1) | 0.88 (0.72–1.09) | ||||||
| 31 | rs7305032 | Intron 5+1052 | AA | 93.3 | 41.7 | 351 (30.7) | 612 (26.7) | 1 | 0.05 |
| AG | 36.7 | 577 (50.5) | 1197 (52.3) | 0.83 (0.70–0.99) | |||||
| GG | 21.7 | 214 (18.7) | 479 (20.9) | 0.81 (0.65–1.02) | |||||
| 32 | rs1544410 | Intron 8 | GG (bb) | 95.8 | 35 | 390 (34.2) | 881 (37.0) | 1 | 0.03 |
| BsmI | (Ex8+284) | GA (bB) | 41.7 | 534 (46.8) | 1136 (47.7) | 1.05 (0.89–1.25) | |||
| AA (BB) | 23.3 | 216 (18.9) | 363 (15.3) | 1.30 (1.04–1.63) | |||||
| 33 | rs731236 | Exon 9−32 | TT (TT) | 97.9 | 34.5 | 407 (34.5) | 897 (37.1) | 1 | 0.06 |
| TaqI | TC (Tt) | 43.1 | 544 (46.1) | 1140 (47.1) | 1.01 (0.86–1.20) | ||||
| CC (tt) | 22.4 | 229 (19.4) | 381 (15.8) | 1.27 (1.02–1.58) | |||||
| 3' UTR | |||||||||
| 34 | rs11574139 | Ex9+3234 | AA | 99 | 95.4 | 1107 (92.7) | 2269 (92.8) | 1 | 0.76 |
| (U−2978) | TA | 4.6 | 85 (7.1) | 168 (6.9) | 1.12 (0.83–1.50) | ||||
| TT | 2 (0.2) | 8 (0.3) | 0.46 (0.09–2.34) | ||||||
| 35 | rs7965281 | UTR+3713 | GG | 93.8 | 36.7 | 292 (25.8) | 545 (23.5) | 1 | 0.04 |
| AG | 40 | 623 (55.1) | 1271 (54.9) | 0.87 (0.72–1.04) | |||||
| AA | 23.3 | 215 (19.0) | 501 (21.6) | 0.78 (0.62–0.99) | |||||
| 36 | rs2544027 | UTR+18799 | CC | 99 | 318 (26.6) | 691 (28.3) | 1 | 0.16 | |
| CT | n/a | 562 (47.0) | 1154 (47.2) | 1.15 (0.96–1.37) | |||||
| TT | 315 (26.4) | 601 (24.6) | 1.16 (0.94–1.42) | ||||||
| 37 | rs2544028 | UTR+18898 | TT | 98.5 | 40 | 401 (33.7) | 872 (35.9) | 1 | 0.08 |
| TA | 45 | 550 (46.2) | 1129 (46.4) | 1.10 (0.93–1.30) | |||||
| AA | 15 | 239 (20.1) | 430 (17.7) | 1.21 (0.98–1.50) | |||||
| 38 | rs2544038 | UTR+20095 | TT | 98.1 | 365 (31.0) | 780 (32.1) | 1 | 0.26 | |
| CT | n/a | 544 (46.2) | 1145 (47.1) | 1.08 (0.91–1.28) | |||||
| CC | 269 (22.8) | 505 (20.8) | 1.12 (0.91–1.39) | ||||||
Positions according to Nejentsev et al., 2004 and Fang et al., 2005.
2 Genotype frequencies were obtained from HapMap-CEU, AFD_EUR_PANEL, or VDR-LD-Caucasian. Note (^), this study and the recent report44 found that G is the minor allele in Caucasians.
Adjusted for age, sex, age-sex interaction, and center. Significant associations (p-values for trend ≤ 0.05) are shown in bold font.
The genotype distribution among cases and controls, and the relative risks based on individual VDR genotypes are presented in Table 2. Nominally significant results (at the 5% significance level) are in bold type. Eight SNPs displayed statistically significant associations (p-value for trend p≤0.05) after adjusting for age, sex, age-sex interaction, and study center. The estimated increase in risk for individuals who were homozygous for the variant allele ranged from 25% to 33% for 6 polymorphisms: rs10875712, rs4760674, rs7139166, rs4516035, rs11168287, rs1544410; for 2 polymorphisms, homozygous variant carriers had a decreased risk: rs7305032, rs7965281. Results of all significant and marginally significant SNPs are displayed in Figure 1, in which the 8 significant SNPs appear in bold typeface. Figure 2 depicts the Q-Q plot, which should track with the 45° line in the absence of association across the locus. The shape of the curve exhibits a strong departure from what we would expect in the absence of association. Five promoter SNPs, two coding SNPs, and one 3’UTR SNP showed significant associations. With respect to previously described haplotype blocks, five of the significant SNPs lie within ‘block C’ and two of these overlap with ‘block 2’ 13; while three SNPs lie within ‘block B’ 12.
Figure 1.
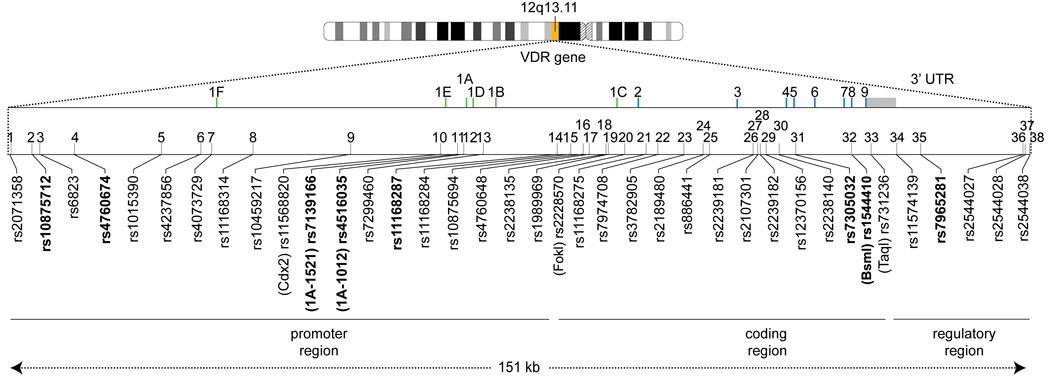
Position of the SNPs studied GEM in relation to the VDR structure. SNPs with significant associations in multivariate analyses after adjusting for age, sex, age-sex interaction, center are shown in bold font. The vertical bars represent promoters 1F – 1C green) and exons (blue). The numbers 1 to 36 refer to the SNP’s relative position from 5’ to 3’ (Table 2). The polymorphism TaqI is located near the stop codon on exon 9, however is strongly linked to the 3’ regulatory region. The grey box represents the 3’ UTR.
Figure 2.
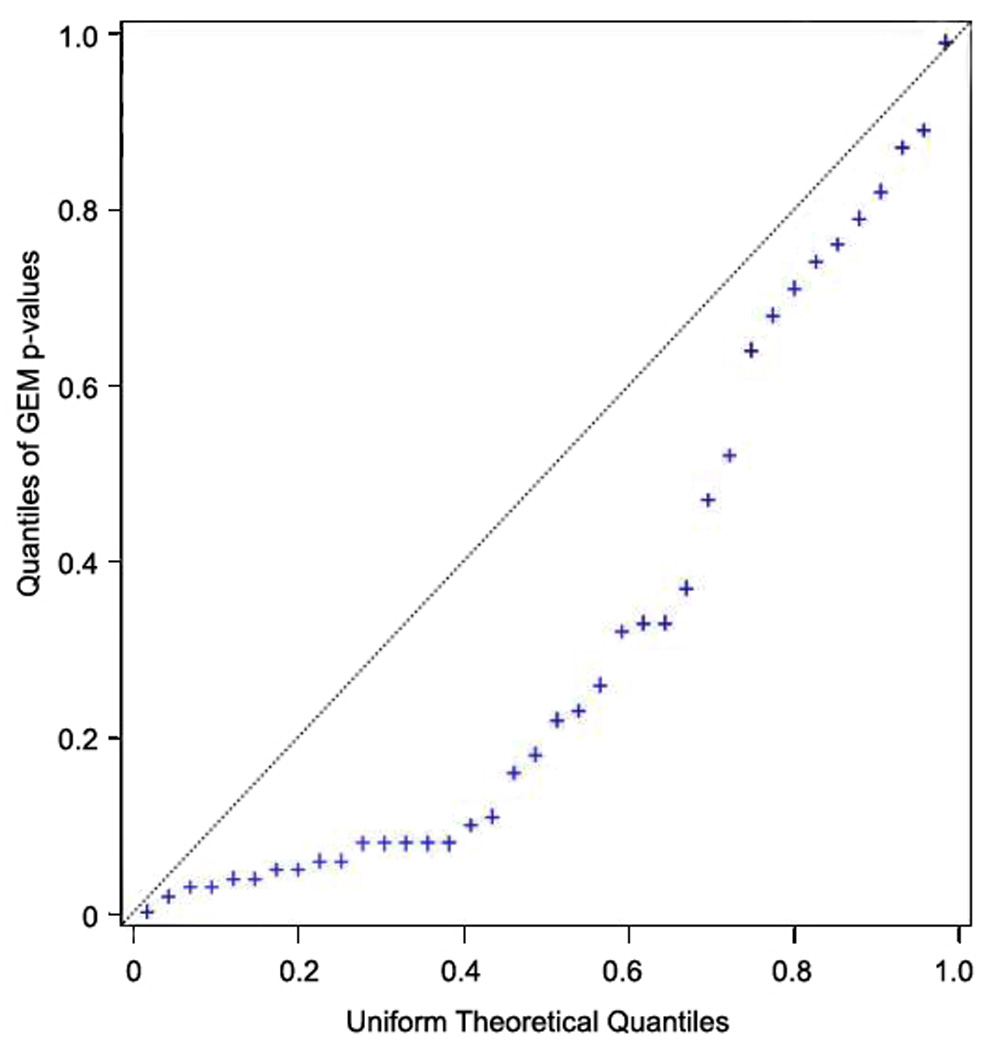
Uniform Q-Q Plot for the VDR SNPs risk trend p-values. Quantiles of the obtained p-values were plotted against the theoretical uniform quantiles.
The VDR SNPs tested in this study included promoter, intronic, and 3’UTR variants and therefore do not directly affect the protein sequence. We evaluated their potential functional significance by searching for overlaps between genomic localization of SNPs and putative transcription factor binding sites, and miRNA seed regions. We also evaluated the conservation across species. We found that rs2228570 localizes to a nucleotide conserved among primates, vertebrates, and mammals (conservation scores 392, 418, and 379 respectively); SNPs rs731236 and rs2544028 localize to nucleotides conserved among vertebrates (conservation scores 272, 480) and mammals (conservation scores 275, 443); and SNPs rs4073729 and rs731236 localize to nucleotides conserved among primates (conservation scores of 293 and 407). The genomic locations of two SNPs overlap with transcription factor binding sites: rs4237856 (V$HNF4_01 HNF-4alpha 1 and 2, transcription z-score 1.98; and V$PPARG_01 PPARgamma, transcription z-score 2.58); and rs1544410 (V$IK3_01 Ikaros 3, transcription z-score 2.77; and V$IK1_01 Ikaros 1, transcription z-score 2.14), although the z-scores for HNF-4alpha 1 and 2 and for Ikaros 1 fall below the threshold of 2.33. SNP rs11568820 is known to interfere with the binding site of the transcription factor Cdx2 11. We did not find any overlaps with seed regions of known miRNAs.
At the time of the preparation of this manuscript, thirteen original reports addressed VDR SNPs and the risk of melanoma, and eleven articles containing a total of 13 series reported results for three of our significant or borderline significant SNPs: rs4516035, rs1544410, and rs731236. Of these, two reports overlapped with a subsequent publication from the same investigators and were not considered for the current meta-analyses 17,21. The main study characteristics and the distribution of genotypes among cases and controls are listed in the supplemental material (Table S2; Table S3). The individual unadjusted ORs and summary odds ratio for heterozygotes and for carriers of the homozygote variant compared to the homozygote common allele are presented in Table 3 and in Figure 3.
Table 3.
Individual odds ratios and summary odds ratios for heterozygote and homozygote variants versus common alleles for SNPs rs4516035 and rs731236
| rs4516035 | Study (Series) | OR (95% CI) TC(AG) vs. TT(AA) |
% weight | OR (95% CI) CC(GG) vs. TT(AA) |
% weight |
|---|---|---|---|---|---|
| Gapska, 2009 (CC1) | 0.86 (0.68–1.09) | 29.94 | 0.95 (0.71–1.28) | 29.1 | |
| Randerson-Moor, 2009 (CC1) | 1.10 (0.85–1.43) | 21.19 | 0.96 (0.69–1.34) | 22.29 | |
| Randerson-Moor, 2009 (CC2) | 1.32 (0.95–1.82) | 12.76 | 1.37 (0.92–2.06) | 12.38 | |
| Barroso, 2008 | 1.08 (0.73–1.60) | 9.38 | 1.51 (0.90–2.53) | 7.38 | |
| Povey, 2007 (CC1) | 1.01 (0.76–1.34) | 18.3 | 0.79 (0.55–1.14) | 20.61 | |
| Santonocito, 2007 | 1.39 (0.76–2.54) | 3.54 | 1.42 (0.60–3.37) | 2.7 | |
| Halsall, 2004 | 0.73 (0.40–1.34) | 4.89 | 0.33 (0.14–0.78) | 5.54 | |
| Summary | 1.03 (0.91–1.16) | 100 | 0.99 (0.85–1.16) | 100 | |
| rs1544410 | Study (Series) |
OR (95% CI) GA (‘bB’) vs. GG (‘bb’) |
% weight |
OR (95% CI) AA (‘BB’) vs. GG (‘bb’) |
% weight |
| Gapska, 2009 (CC1) | 0.91 (0.73–1.13) | 25.83 | 0.92 (0.67–1.27) | 21.6 | |
| Randerson-Moor, 2009 (CC1) | 0.93 (0.72–1.20) | 18.01 | 1.00 (0.71–1.41) | 17.96 | |
| Randerson-Moor, 2009 (CC2) | 0.81 (0.59–1.11) | 13.03 | 0.69 (0.45–1.05) | 14.44 | |
| Li, 2008 | 0.74 (0.60–0.92) | 28.45 | 0.78 (0.59–1.04) | 29.77 | |
| Santonocito, 2007 | 0.74 (0.40–1.40) | 3.34 | 0.29 (0.12–0.71) | 5.12 | |
| Han, 2007 | 0.87 (0.62–1.20) | 11.33 | 0.82 (0.51–1.31) | 11.11 | |
| Summary | 0.84 (0.75–0.94) | 100 | 0.82 (0.70–0.95) | 100 | |
| rs731236 | Study (Series) |
OR (95% CI) TC('Tt') vs. CC('tt') |
% weight |
OR (95% CI) TT ('TT') vs. CC ('tt') |
% weight |
| Gapska, 2009 (CC1) | 1.03 (0.83–1.28) | 25.78 | 1.10 (0.79–1.53) | 20.28 | |
| Randerson-Moor, 2009 (CC1) | 0.97 (0.75–1.26) | 18.83 | 1.07 (0.76–1.51) | 18.86 | |
| Randerson-Moor, 2009 (CC2) | 0.96 (0.70–1.31) | 12.76 | 0.73 (0.48–1.13) | 14.74 | |
| Barroso, 2008 | 1.26 (0.85–1.87) | 6.91 | 1.09 (0.64–1.84) | 7.98 | |
| Li, 2008 | 0.69 (0.55–0.85) | 31.71 | 0.65 (0.49–0.87) | 34.21 | |
| Hutchinson, 2000 | 1.29 (0.77–2.15) | 4.01 | 1.28 (0.62–2.65) | 3.95 | |
| Summary | 0.93 (0.83–1.04) | 100 | 0.89 (0.76–1.04) | 100 | |
CC1 or CC2, case-control series; OR, unadjusted odds ratio; CI, confidence intervals. Weights were calculated using Mantel-Haenszel methods and represent the individual contribution of each study to the overall OR
Figure 3.
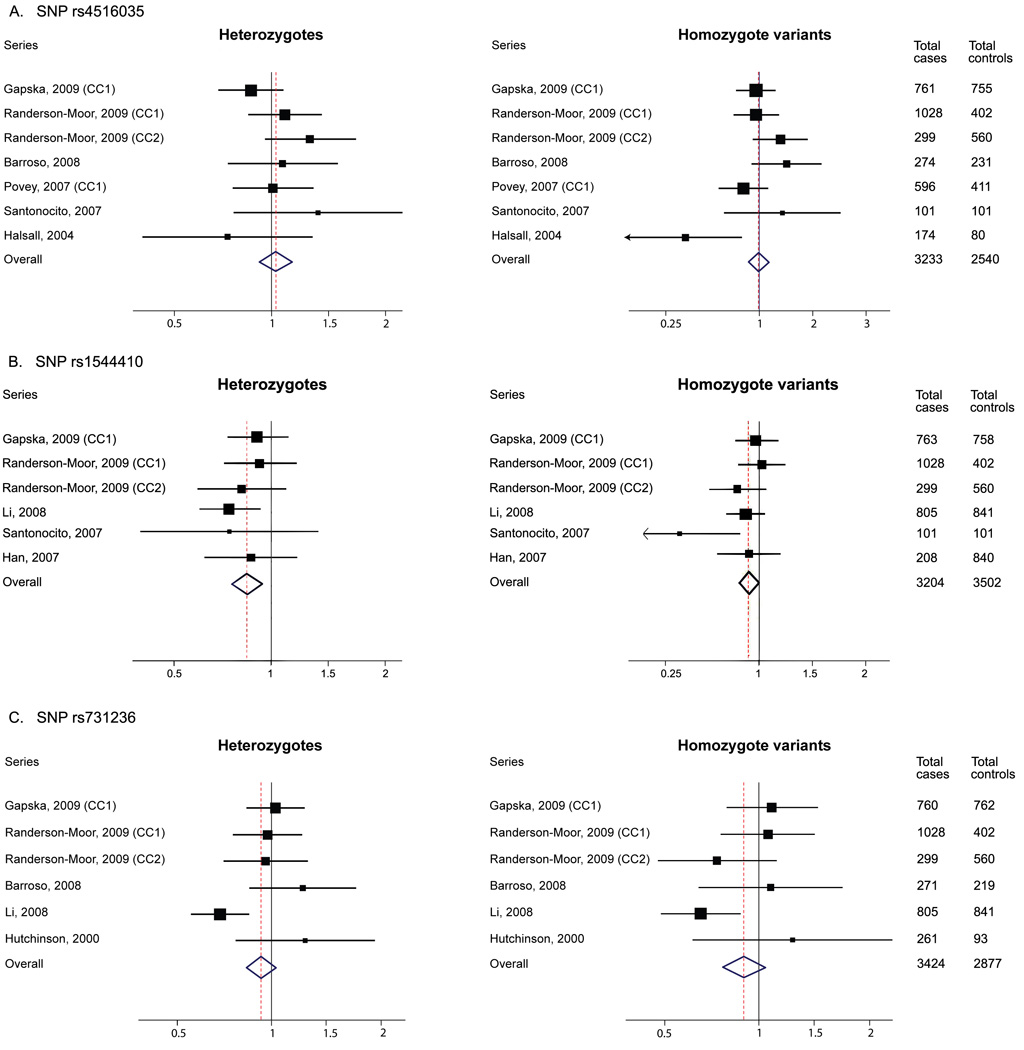
Meta-analysis of VDR SNPs reported in other investigations of melanoma in relation to risk. (A) SNP rs4516035 (variant allele G), (B) SNP rs1544410 (variant allele A or ‘B’); and (C) SNP rs731236 (variant allele C or ‘t’). Unadjusted ORs and 95% CI are shown.
For studies that addressed rs4516035 (1A-1012), the adjustment for covariates varied substantially. Specifically, four studies 18,19,22,23 matched cases and controls by age and gender; three studies 14,15,21 included age and sex in multivariate analyses; while one study 20 adjusted for phenotypic features. The variant allele G produces a null association with melanoma. There was evidence of heterogeneity for the observed risks of the homozygous variant (p=0.04) but not for heterozygote (p=0.31), and no evidence of publication bias (p=0.88 and p=0.65, respectively). For rs1544410 (BsmI) the allele A (‘B’) showed a statistically negative association with melanoma (Figure 3-B, Table 3). There was no evidence of heterogeneity for the observed risks of the heterozygous variant (p=0.78) or for the homozygote variant (p=0.18), and no evidence of publication bias (p=0.45 and p=0.45, respectively). The rs731236 (TaqI) allele C (‘t’) showed a statistically non-significant negative association with melanoma (Figure 3-C, Table 3). There was evidence of heterogeneity for the observed risks of the heterozygous variant (p=0.03) but not for the heterozygote variant (p=0.10), and no evidence of publication bias (p=0.19 and p=0.85, respectively). We did not observe departure from HWE in any of the series (Tables S3).
DISCUSSION
In this large population based case-control study comprising 3676 people with incident single and multiple primary melanoma we evaluated 38 VDR gene single nucleotide polymorphisms for their impact in the development of skin melanoma and found that in addition to the BsmI (rs1544410), seven SNPs (rs10875712, rs4760674, rs7139166, rs4516035, rs11168287, rs7305032, rs7965281) were significantly associated with risk. Because we conducted 38 individual statistical tests we recognize the increased risk of false positive findings due to these “multiple comparisons”; we would expect about 2 of the tests to be significant simply due to chance. The fact that we observed 8 significant SNPs, reflected by the skewness in the Q-Q plot (Figure 2), suggests that the evidence broadly favors the hypothesis that the gene harbors at least some causal variants.
Promoter SNPs
Most of the VDR SNPs identified in the last several years are of unknown functional significance; however in vitro studies suggest that at least some SNPs affect the VDR mRNA and/or protein levels 13,24,25,26. Such is the case for two of the SNPs possibly associated with multiple primary melanoma in our study: rs7139166 and rs4516035, located in the promoter region in positions 1A-1521 and 1A-1012. The genotype frequencies in our study are similar to those reported in dbSNP in Caucasians and to those reported by others for rs4516035 15,18,22,23,45. These two neighboring SNPs are in strong LD (D’ = 0.968), therefore it is not surprising that the genotype frequencies and the ORs conferred by each of the minor alleles are very similar in our study (Table 2). A previous report suggested a link between these SNPs and stature 31 although results from a larger study did not support this finding 46. We found that the G allele of rs4516035 appears to confer risk. Recently, Randerson-Moor et al. (2009) 23 in a population-based study of 1028 melanoma cases and 402 controls found no association for rs4516035, while in a second case-control study of 299 melanoma cases and 560 unaffected females, the authors reported an increased risk associated with the same allele (G), though the results fall short of statistical significance. We further compared our results to those obtained in our meta-analysis of a total of six published reports on this SNP and melanoma and found no evidence overall that this SNP is associated with melanoma risk, although the results are inconsistent, with significant heterogeneity in the observed relative risks across studies.
Our findings are consistent with the in vitro data for rs4516035 13,31. Specifically, d’Alesio et al. (2005)31 observed that one base-change in any of the two variant sites located in positions 1A-1521 or 1A-1012 led to a dramatic change in protein-DNA complex formation, and that the promoter activity of the VDR was nearly doubled in cells carrying the haplotype rs7139166-G/rs4516035-A when compared to those with the rs7139166-C/rs4516035-G haplotype.
It has been recently hypothesized that the A allele in position 1A-1012 (rs4516035) indirectly promotes a GATA-3 driven T-cell switch of naïve T cells to T-helper 2 cells and that this allele may play different roles in susceptibility and in metastasis possibly as a function of the transcription factors secreted by different cellular backgrounds 13,45,47. Our results do not support an adverse role of the allele ‘A’ in position 1A-1012 in relation to melanoma risk, and in support of our findings, the qualitative analyses of the meta-analyses conducted on published melanoma case-control series suggests that ‘G’ may represent the risk allele.
We found four significant VDR promoter SNPs (rs10875712, rs4760674, rs7139166, rs11168287) that have not been previously reported in relation to melanoma risk or progression.
Coding region and 3’UTR SNPs
Three adjacent RFLP SNPs for BsmI (rs1544410), ApaI (rs7975232), and TaqI (rs731236) respectively, at the 3’ end of the gene, have been the most frequently studied in cancer and in melanoma. The functional characterization of these SNPs has produced inconsistent results across studies, and if these SNPs are non-functional, the effect observed in some investigations may be due to truly relevant SNPs in strong LD located elsewhere in the gene. Here we report that in this gene region, in addition to the minor allele of the BsmI polymorphism (rs1544410) which increases risk, the minor allele of rs7965281, a SNP which has never been studied in relation to melanoma prior to this work, seems to confer some reduction in risk. We also report that two additional SNPs in this location may modify risk: rs7305032, which was not previously reported in relation to melanoma, and rs731236 (TaqI), although the latter, which is in high LD with BsmI and maps to a nucleotide conserved across species, exhibited only borderline significance (p-value 0.06, Table 2). Some investigations targeted to determine the expression differences for polymorphisms in the 3’ end of the VDR gene have shown higher levels of mRNA expression for the BsmI-ApaI-TaqI haplotype ‘BAt’ (rs1544410-A/rs7975232-A/rs731236-C) than for the haplotype ‘baT’(rs1544410-G/rs7975232-C/rs731236-T), however, the opposite results have been found not only for the VDR mRNA expression but also for the mRNA stability and transactivation in human fibroblasts, and in leukemia and prostate cancer cell lines 11. Our findings for rs1544410 suggest that the A (‘B’) might be the adverse allele in relation to melanoma risk which differs from what other studies have found with one exception 16,19,21–23. Specifically, three studies were null, one study reported reduction of risk in heterozygous bB individuals but not in homozygotes BB, and one small hospital-based study conducted in 101 cases and 101 controls found a protective effect for the B allele; it is of note that the reported MAF among controls was higher than the one reported in dbSNP and in other studies including ours. A more recent population-based study of 763 cases and 763 controls conducted in a Polish population found that haplotypes containing the B allele increased risk for melanoma 22. Results from our meta-analysis conducted on five previously published studies, like in previous meta-analyses 23,27–28, suggest a protective role for the B allele; however, considering the results from our large study after adjusting for covariates, we argue that the results from the meta-analyses should be considered with caution and that BsmI should be further characterized in large population-ascertained cohorts and in functional assays preferentially using melanoma and/or skin models. Our findings for rs731236 suggest that the C (‘t’) allele might be the adverse allele in relation to melanoma risk. Recent reports have found similar genotype frequencies to those in the present work, however investigators found either no significant associations with melanoma for rs731236 14,20,22,23, or, in one study, a significant association for the contralateral allele T (‘T’) 21. Our meta-analysis conducted on five publications does not provide sufficient evidence for an effect of this SNP and melanoma, however it is important to note that our meta-analyses were unadjusted for covariates, and that the results may differ in a multivariate logistic regression.
In relation to bioinformatics predictions, there are two opposing theories concerning conservation of sequences across species in regards to functional relevance. The first supports the idea of a plausible biological function, and as an extension of this idea, one would expect to uncover melanoma risk SNPs in this region. A second opposing hypothesis proposes that variants in a conserved region are subjected to negative selection, and those that persist will more likely be non functional. Interestingly, two of the investigated SNPs (rs731236 and rs2544028) are located on the boundary of the conserved region and although the associations do not reach statistical significance, the variants increase the risk by 21–27% (p-values 0.06 and 0.08, respectively). SNP rs11568820 has been extensively characterized in in vitro studies and overlaps with the binding site of Cdx2. One would expect to find a significant role for this polymorphism in melanoma risk; however it is possible that its effect is cell-specific. In fact, Cdx2 is differentially expressed in normal stomach and intestine and seems progressively decreased in gastric intestinal metaplasia, dysplasia and cancer 17,48,49. SNP rs4237856 overlaps with the binding site of PPAR-gamma. The PPARs are known to heterodimerize with the retinoid X receptor and with the vitamin D bound to its receptor. PPARs may be of importance in cell growth regulation in melanocytes; however expression of PPAR-gamma is much weaker in melanocytes, at least based on observations in MeWo cells 50; therefore, one could speculate that even if this SNP can facilitate or hinder the binding of PPAR-gamma, the contribution of this binding in melanoma risk may not be as significant as with other PPARs.
A major limitation so far of association studies using VDR SNPs in relation to complex-diseases such as cancer is the limited number of variants studied. The VDR gene is very polymorphic; however information on the existing gene variants was initially available from the rather insensitive search of polymorphic patterns within a limited region using restriction enzymes. Additional polymorphisms have been identified during the search for mutation in tumors 24. In more recent years, the studies of Nejentsev et al. (2004)12 and Fang et al. (2005)13 have contributed to the field by systematically scanning and characterizing variations across the entire VDR gene, and by describing patterns of linkage disequilibrium between alleles in different ethnic groups. The main strengths of our study are the comprehensive evaluation of VDR and the large number of study subjects, as well as the availability of data on known melanoma risk confounders. The overall effect on melanoma risk found here was small. Furthermore the results overall do not reach significance if evaluated either by the Bonferroni correction or by restricting the false discovery rate to 5% 37. However eight of 38 (~21%) investigated SNPs revealed statistically significant associations at the 5% significance level (versus 1.9 expected), and 16 of 38 are significant at the 10% level (versus 3.8 expected), proportions considerably higher than would be expected if there is in fact no association between VDR genotype and melanoma risk. Thus we believe that the results, collectively, are strongly suggestive that VDR is a risk locus for melanoma.
Our case-control study design was novel since the controls have melanoma and the cases are required to have experienced melanoma twice. The premise is that risk factors that affect melanoma must affect the occurrence of a second melanoma in a population of individuals with melanoma, and this is borne out empirically in earlier results from this and other studies, although there is evidence that the magnitude of the observed relative risks may be attenuated29. The rationale for this design is that it delivers much greater statistical power for the evaluation of rare, strong risk factors than a conventional case-control design with healthy controls 30. In such a design the double primary cases are typically older than the single primary controls, and in the case of melanoma the gender distribution is altered because of the strong interaction between age and gender on melanoma incidence, and so we adjusted for these factors in our analyses. A further complication is that many second primaries are detected as in situ lesions due to the careful clinical surveillance that melanoma patients experience, and we elected to include these individuals as cases on the assumption that they would ultimately have been diagnosed with an invasive lesion.
In summary, we found that eight VDR SNPs, located in the promoter, coding and 3’ gene regions, were associated with melanoma and confer a modest but statistically significant increased (rs10875712, rs4760674, rs7139166, rs4516035, rs11168287, rs1544410) or decreased (rs7305032, rs7965281) risk of becoming a multiple primary melanoma. To our knowledge, this constitutes the largest study of VDR polymorphisms in melanoma to date. Our results provide some evidence in support of the hypothesis that the vitamin D pathway may play an important role in melanoma genesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Laetitia Borsu for assistance with the Sequenom assays; Javier Cotignola for help provided during the initial stages of the iPLEX assay design and genotyping; Brian Clas and Concetta Perretta for their help during the individual SNP genotyping; Aaron Gabow and the MSKCC Bioinformatics Core for providing assistance with the computational characterization of SNPs; Joseph Kanik for assistance with graphics; and all the individuals who agreed to participate in the study.
The study was conducted by the GEM Study Group: Marianne Berwick (PI, University of New Mexico), Memorial Sloan-Kettering Cancer Center, New York, NY, USA: Colin Begg (Co-PI), Irene Orlow (Co-Investigator), Urvi Mujumdar (Project Coordinator), Klaus Busam (Dermatopathologist), Pampa Roy (Laboratory Technician). Study Centers: The University of Sydney and The Cancer Council New South Wales, Sydney (Australia): Bruce Armstrong (PI), Anne Kricker (co-PI), Melisa Litchfield (Study Coordinator). Menzies Research Institute, University of Tasmania, Hobart (Australia): Terence Dwyer (PI, currently at the Murdoch Childrens Research Institute, Melbourne, Victoria), Paul Tucker (Dermatopathologist), Alison Venn (co-Investigator), Nicola Stephens (Study Coordinator). British Columbia Cancer Agency, Vancouver (Canada): Richard Gallagher (PI), Teresa Switzer (Coordinator). Cancer Care Ontario, Toronto (Canada): Loraine Marrett (PI), Elizabeth Theis (Co-Investigator), Lynn From (Dermatopathologist), Noori Chowdhury (Coordinator), Louise Vanasse (Coordinator). Centro per la Prevenzione Oncologia Torino, Piemonte (Italy): Stefano Rosso (PI), Roberto Zanetti (co-PI), Carlotta Sacerdote (Coordinator). University of California, Irvine (USA): Hoda Anton-Culver (PI), Nancy Leighton (Coordinator). University of Michigan, Ann Arbor (USA): Stephen Gruber (PI), Joanne Jeter (Coordinator). New Jersey Department of Health and Senior Services, Trenton (USA): Judith Klotz (PI), Homer Wilcox (Co-PI), Helen Weiss (Coordinator). University of North Carolina, Chapel Hill (USA): Robert Millikan (PI), Nancy Thomas (Co-Investigator), Dianne Mattingly (Coordinator), Jon Player (Laboratory Technician). University of Pennsylvania, Philadelphia, PA (USA): Timothy Rebbeck (PI), Peter Kanetsky (Co-Investigator), Amy Walker (Laboratory Manager), Saarene Panossian (Laboratory Technician). Consultants: Julia Lee Taylor and Sasha Madronich, National Centre for Atmospheric Research, Boulder, Colorado (USA).
This work is supported by the National Cancer Institute, award CA112524. The MSKCC Sequenom facility is supported by the Anbinder Fund.
Abbreviations used
- SNP
single nucleotide polymorphism
- VDR
vitamin D receptor
- GEM
Genes, environment and melanoma study
- rs
reference sequence
- UTR
untranslated
- MAF
minor allele frequency
- LD
linkage disequilibrium
- htSNP
haplotype tagging SNP
Footnotes
Novelty and impact: We analyzed 38 common vitamin D receptor (VDR) gene polymorphisms in relation to melanoma risk in a large population-based case control study comprised of 3676 individuals with incident primary melanoma and found modest but statistically significant associations between 8 SNPs and the risk for developing subsequent new primary melanomas, including 6 SNPs investigated for the first time in relation to melanoma. These results support the hypothesis that the vitamin D pathway plays an important role in the genesis of melanomas. To our knowledge, this is the largest and most comprehensive study of VDR polymorphisms in melanoma to date.
REFERENCES
- 1.Colston K, Colston MJ, Feldman D. 1,25-dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology. 1981;108:1083–1086. doi: 10.1210/endo-108-3-1083. [DOI] [PubMed] [Google Scholar]
- 2.Danielsson C, Fehsel K, Polly P, Carlberg C. Differential apoptotic response of human melanoma cells to 1 alpha,25-dihydroxyvitamin D3 and its analogues. Cell Death Differ. 1998;5:946–952. doi: 10.1038/sj.cdd.4400437. [DOI] [PubMed] [Google Scholar]
- 3.Evans SR, Houghton AM, Schumaker L, Brenner RV, Buras RR, Davoodi F, Nauta RJ, Shabahang M. Vitamin D receptor and growth inhibition by 1,25-dihydroxyvitamin D3 in human malignant melanoma cell lines. J Surg Res. 1996;61:127–133. doi: 10.1006/jsre.1996.0092. [DOI] [PubMed] [Google Scholar]
- 4.Ranson M, Posen S, Mason RS. Human melanocytes as a target tissue for hormones: in vitro studies with 1 alpha-25, dihydroxyvitamin D3, alpha-melanocyte stimulating hormone, and beta-estradiol. J Invest Dermatol. 1988;91:593–598. doi: 10.1111/1523-1747.ep12477126. [DOI] [PubMed] [Google Scholar]
- 5.Yudoh K, Matsuno H, Kimura T. 1alpha,25-dihydroxyvitamin D3 inhibits in vitro invasiveness through the extracellular matrix and in vivo pulmonary metastasis of B16 mouse melanoma. J Lab Clin Med. 1999;133:120–128. doi: 10.1016/s0022-2143(99)90004-5. [DOI] [PubMed] [Google Scholar]
- 6.Berwick M, Armstrong BK, Ben-Porat L, Fine J, Kricker A, Eberle C, Barnhill R. Sun exposure and mortality from melanoma. J Natl Cancer Inst. 2005;97:195–199. doi: 10.1093/jnci/dji019. [DOI] [PubMed] [Google Scholar]
- 7.Moan J, Porojnicu AC, Dahlback A, Setlow RB. Addressing the health benefits and risks, involving vitamin D or skin cancer, of increased sun exposure. Proc Natl Acad Sci U S A. 2008;105:668–673. doi: 10.1073/pnas.0710615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosso S, Sera F, Segnan N, Zanetti R. Sun exposure prior to diagnosis is associated with improved survival in melanoma patients: results from a long-term follow-up study of Italian patients. Eur J Cancer. 2008;44:1275–1281. doi: 10.1016/j.ejca.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Newton-Bishop JA, Beswick S, Randerson-Moor J, Chang YM, Affleck P, Elliott F, Chan M, Leake S, Karpavicius B, Haynes S, Kukalizch K, Whitaker L, et al. Serum 25-hydroxyvitamin D3 levels are associated with breslow thickness at presentation and survival from melanoma. J Clin Oncol. 2009;27:5439–5444. doi: 10.1200/JCO.2009.22.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpf WE, Sar M, Reid FA, Tanaka Y, DeLuca HF. Target cells for 1,25-dihydroxyvitamin D3 in intestinal tract, stomach, kidney, skin, pituitary, and parathyroid. Science. 1979;206:1188–1190. doi: 10.1126/science.505004. [DOI] [PubMed] [Google Scholar]
- 11.Uitterlinden AG, Fang Y, Van Meurs JB, Pols HA, Van Leeuwen JP. Genetics and biology of vitamin D receptor polymorphisms. Gene. 2004;338:143–156. doi: 10.1016/j.gene.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Nejentsev S, Godfrey L, Snook H, Rance H, Nutland S, Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Tuomilehto-Wolf E, et al. Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet. 2004;13:1633–1639. doi: 10.1093/hmg/ddh169. [DOI] [PubMed] [Google Scholar]
- 13.Fang Y, van Meurs JB, d'Alesio A, Jhamai M, Zhao H, Rivadeneira F, Hofman A, van Leeuwen JP, Jehan F, Pols HA, Uitterlinden AG. Promoter and 3'-untranslated-region haplotypes in the vitamin d receptor gene predispose to osteoporotic fracture: the Rotterdam study. Am J Hum Genet. 2005;77:807–823. doi: 10.1086/497438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson PE, Osborne JE, Lear JT, Smith AG, Bowers PW, Morris PN, Jones PW, York C, Strange RC, Fryer AA. Vitamin D receptor polymorphisms are associated with altered prognosis in patients with malignant melanoma. Clin Cancer Res. 2000;6:498–504. [PubMed] [Google Scholar]
- 15.Halsall JA, Osborne JE, Potter L, Pringle JH, Hutchinson PE. A novel polymorphism in the 1A promoter region of the vitamin D receptor is associated with altered susceptibilty and prognosis in malignant melanoma. Br J Cancer. 2004;91:765–770. doi: 10.1038/sj.bjc.6602006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, Colditz GA, Hunter DJ. Polymorphisms in the MTHFR and VDR genes and skin cancer risk. Carcinogenesis. 2007;28:390–397. doi: 10.1093/carcin/bgl156. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Liu Z, Zhang Z, Strom SS, Gershenwald JE, Prieto VG, Lee JE, Ross MI, Mansfield PF, Cormier JN, Duvic M, Grimm EA, et al. Genetic variants of the vitamin D receptor gene alter risk of cutaneous melanoma. J Invest Dermatol. 2007;127:276–280. doi: 10.1038/sj.jid.5700544. [DOI] [PubMed] [Google Scholar]
- 18.Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekky M, Rees J, Doherty V, Kavanagh G, Anderson N, Campbell H, MacKie RM, Melton DW. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis. 2007;28:1087–1093. doi: 10.1093/carcin/bgl257. [DOI] [PubMed] [Google Scholar]
- 19.Santonocito C, Capizzi R, Concolino P, Lavieri MM, Paradisi A, Gentileschi S, Torti E, Rutella S, Rocchetti S, Di Carlo A, Di Stasio E, Ameglio F, et al. Association between cutaneous melanoma, Breslow thickness and vitamin D receptor BsmI polymorphism. Br J Dermatol. 2007;156:277–282. doi: 10.1111/j.1365-2133.2006.07620.x. [DOI] [PubMed] [Google Scholar]
- 20.Barroso E, Fernandez LP, Milne RL, Pita G, Sendagorta E, Floristan U, Feito M, Aviles JA, Martin-Gonzalez M, Arias JI, Zamora P, Blanco M, et al. Genetic analysis of the vitamin D receptor gene in two epithelial cancers: melanoma and breast cancer case-control studies. BMC Cancer. 2008;8:385. doi: 10.1186/1471-2407-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Liu Z, Wang LE, Gershenwald JE, Lee JE, Prieto VG, Duvic M, Grimm EA, Wei Q. Haplotype and genotypes of the VDR gene and cutaneous melanoma risk in non-Hispanic whites in Texas: a case-control study. Int J Cancer. 2008;122:2077–2084. doi: 10.1002/ijc.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gapska P, Scott RJ, Serrano-Fernandez P, Mirecka A, Rassoud I, Gorski B, Cybulski C, Huzarski T, Byrski T, Nagay L, Maleszka R, Sulikowski M, et al. Vitamin D receptor variants and the malignant melanoma risk: a population-based study. Cancer Epidemiol. 2009;33:103–107. doi: 10.1016/j.canep.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Randerson-Moor JA, Taylor JC, Elliott F, Chang YM, Beswick S, Kukalizch K, Affleck P, Leake S, Haynes S, Karpavicius B, Marsden J, Gerry E, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur J Cancer. 2009;45:3271–3281. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai H, Miyamoto KI, Yoshida M, Yamamoto H, Taketani Y, Morita K, Kubota M, Yoshida S, Ikeda M, Watabe F, Kanemasa Y, Takeda E. The polymorphism in the caudal-related homeodomain protein Cdx-2 binding element in the human vitamin D receptor gene. J Bone Miner Res. 2001;16:1256–1264. doi: 10.1359/jbmr.2001.16.7.1256. [DOI] [PubMed] [Google Scholar]
- 25.Jurutka PW, Remus LS, Whitfield GK, Thompson PD, Hsieh JC, Zitzer H, Tavakkoli P, Galligan MA, Dang HT, Haussler CA, Haussler MR. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol. 2000;14:401–420. doi: 10.1210/mend.14.3.0435. [DOI] [PubMed] [Google Scholar]
- 26.Colin EM, Weel AE, Uitterlinden AG, Buurman CJ, Birkenhager JC, Pols HA, Van Leeuwen JP. Consequences of vitamin D receptor gene polymorphisms for growth inhibition of cultured human peripheral blood mononuclear cells by 1, 25-dihydroxyvitamin D3. Clin Endocrinol (Oxf) 2000;52:211–216. doi: 10.1046/j.1365-2265.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- 27.Mocellin S, Nitti D. Vitamin D receptor polymorphisms and the risk of cutaneous melanoma: a systematic review and meta-analysis. Cancer. 2008;113:2398–2407. doi: 10.1002/cncr.23867. [DOI] [PubMed] [Google Scholar]
- 28.Gandini S, Raimondi S, Gnagnarella P, Dore JF, Maisonneuve P, Testori A. Vitamin D and skin cancer: a meta-analysis. Eur J Cancer. 2009;45:634–641. doi: 10.1016/j.ejca.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Begg CB, Hummer AJ, Mujumdar U, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Culver HA, Zanetti R, Gallagher RP, Dwyer T, et al. A design for cancer case-control studies using only incident cases: experience with the GEM study of melanoma. Int J Epidemiol. 2006;35:756–764. doi: 10.1093/ije/dyl044. [DOI] [PubMed] [Google Scholar]
- 30.Begg CB, Berwick M. A note on the estimation of relative risks of rare genetic susceptibility markers. Cancer Epidemiol Biomarkers Prev. 1997;6:99–103. [PubMed] [Google Scholar]
- 31.d'Alesio A, Garabedian M, Sabatier JP, Guaydier-Souquieres G, Marcelli C, Lemacon A, Walrant-Debray O, Jehan F. Two single-nucleotide polymorphisms in the human vitamin D receptor promoter change protein-DNA complex formation and are associated with height and vitamin D status in adolescent girls. Hum Mol Genet. 2005;14:3539–3548. doi: 10.1093/hmg/ddi382. [DOI] [PubMed] [Google Scholar]
- 32.Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Anton-Culver H, Zanetti R, Gallagher RP, Dwyer T, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst. 2005;97:1507–1515. doi: 10.1093/jnci/dji312. [DOI] [PubMed] [Google Scholar]
- 33.Nakai K, Habano W, Fujita T, Schnackenberg J, Kawazoe K, Suwabe A, Itoh C. Highly multiplexed genotyping of coronary artery disease-associated SNPs using MALDI-TOF mass spectrometry. Hum Mutat. 2002;20:133–138. doi: 10.1002/humu.10099. [DOI] [PubMed] [Google Scholar]
- 34.Bennett CD, Campbell MN, Cook CJ, Eyre DJ, Nay LM, Nielsen DR, Rasmussen RP, Bernard PS. The LightTyper: high-throughput genotyping using fluorescent melting curve analysis. Biotechniques. 34(6):1288–1292. doi: 10.2144/03346pf01. 1294-2003. [DOI] [PubMed] [Google Scholar]
- 35.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281(5375):363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 36.Cotignola J, Reva B, Mitra N, Ishill N, Chuai S, Patel A, Shah S, Vanderbeek G, Coit D, Busam K, Halpern A, Houghton A, Sander C, Berwick M, Orlow I. Matrix Metalloproteinase-9 (MMP-9) polymorphisms in patients with cutaneous malignant melanoma. BMC Med Genet. 2007 Mar 8;8:10. doi: 10.1186/1471-2350-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abramovich F, Benjamini Y. False Discovery Rate. In: Balakrishnan N, editor. Encyclopedia of Statistical Sciences. Vol. 4. New Jersey: Wiley & Sons, Inc.; 2006. pp. 2240–2243. [Google Scholar]
- 38.Wilk MB, Gnanadesikan R. Probability plotting methods for the analysis of data. Biometrika. 1968 Mar;55(1):1–17. [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 41.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 43.Berwick M, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Anton-Culver H, Zanetti R, Gallagher RP, Dwyer T, et al. The prevalence of CDKN2A germ-line mutations and relative risk for cutaneous malignant melanoma: an international population-based study. Cancer Epidemiol Biomarkers Prev. 2006;15:1520–1525. doi: 10.1158/1055-9965.EPI-06-0270. [DOI] [PubMed] [Google Scholar]
- 44.Martin RJ, McKnight AJ, Patterson CC, Sadlier DM, Maxwell AP. A rare haplotype of the vitamin D receptor gene is protective against diabetic nephropathy. Nephrol Dial Transplant. 2010;25:497–503. doi: 10.1093/ndt/gfp515. [DOI] [PubMed] [Google Scholar]
- 45.Halsall JA, Osborne JE, Epstein MP, Pringle JH, Hutchinson PE. The unfavorable effect of the A allele of the vitamin D receptor promoter polymorphism A-1012G has different mechanisms related to susceptibility and outcome of malignant melanoma. Dermatoendocrinol. 2009;1:54–57. doi: 10.4161/derm.1.1.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macgregor S, Hottenga JJ, Lind PA, Suchiman HE, Willemsen G, Slagboom PE, Montgomery GW, Martin NG, Visscher PM, Boomsma DI. Vitamin D receptor gene polymorphisms have negligible effect on human height. Twin Res Hum Genet. 2008;11:488–494. doi: 10.1375/twin.11.5.488. [DOI] [PubMed] [Google Scholar]
- 47.Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21:479–483. doi: 10.1016/s0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto H, Miyamoto K, Li B, Taketani Y, Kitano M, Inoue Y, Morita K, Pike JW, Takeda E. The caudal-related homeodomain protein Cdx-2 regulates vitamin D receptor gene expression in the small intestine. J Bone Miner Res. 1999;14:240–247. doi: 10.1359/jbmr.1999.14.2.240. [DOI] [PubMed] [Google Scholar]
- 49.Rozek LS, Lipkin SM, Fearon ER, Hanash S, Giordano TJ, Greenson JK, Kuick R, Misek DE, Taylor JM, Douglas JA, Rennert G, Gruber SB. CDX2 polymorphisms, RNA expression, and risk of colorectal cancer. Cancer Res. 2005;65:5488–5492. doi: 10.1158/0008-5472.CAN-04-3645. [DOI] [PubMed] [Google Scholar]
- 50.Sertznig P, Dunlop T, Seifert M, Tilgen W, Reichrath J. Cross-talk between vitamin D receptor (VDR)- and peroxisome proliferator-activated receptor (PPAR)-signaling in melanoma cells. Anticancer Res. 2009;29:3647–3658. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


