Abstract
RANKL is a stromal cell-derived tumor necrosis factor (TNF)-like factor that plays a primary role in osteoclast formation and function. Recent studies suggest that 1,25(OH)2D3 induces Rankl expression via vitamin D receptor (VDR) interaction at several enhancers located up to 76 kb upstream of the gene’s transcriptional start site (TSS). In the current studies, we explored these interactions further using ChIP-chip and RNA analysis. We confirm VDR and RXR binding to the five enhancers described previously and identify two additional sites, one located within the Rankl coding region. We also show that RNA polymerase II is recruited to these enhancers, most likely through transcription factors TBP, TFIIB and TAFII250. Interestingly, the recruitment of these factors leads to the production of RNA transcripts, although their role at present is unknown. We also discovered that histone H4 acetylation (H4ac) marks many upstream Rankl enhancers under basal conditions and that H4ac is increased upon 1,25(OH)2D3 treatment. Surprisingly, the hormone also induces C/EBPβ binding across the Rankl locus. C/EBPβ binding correlates directly with increased H4ac activity following 1,25(OH)2D3 treatment. Finally, elevated H4ac is restricted to an extended region located between two potential insulator sites occupied by CTCF and Rad21. These data suggest a mechanism whereby 1,25(OH)2D3 functions via the VDR and C/EBPβ to upregulate Rankl expression.
Keywords: Transactivation, RankL, ChIP analysis, VDR, RNA pol II, chromatin, histone acetylation
INTRODUCTION
Skeletal remodeling in adults occurs via the opposing actions of bone-forming osteoblasts and bone-resorbing osteoclasts [Harris and Heaney, 1969]. Osteoclasts represent terminally differentiated, multinucleated cells of the monocyte-macrophage lineage [Teitelbaum, 2000]. Osteoclastogenesis is orchestrated by a number of growth factors, steroidal components and cytokines [Suda et al., 1999]. Many of these regulatory factors are secreted by adjacent cells that include stromal cells and cells of the osteoblast lineage. While multiple components are critical for bone remodeling, the aberrant production of many of them can be pathological and lead to focal and/or systemic bone disease [Mundy, 2000].
Although a number of factors participate in osteoclastogenesis, the molecule that is considered to be both necessary and sufficient for this process in vivo and in vitro is receptor activator of NF-κB ligand (RANKL). RANKL is a TNF-like factor that is expressed on the surface of stromal cells and osteoblasts as well as numerous other cell types [Lacey et al., 1998]. RANKL interacts with receptor activator of NF-κB (RANK) and is required for not only osteoclast differentiation, but also for the cell’s full bone- resorbing activity and for its survival [Jimi et al., 1999a]. Evidence for the essentiality of RANKL and its receptor in osteoclast formation is most strongly supported by the skeletal phenotypes of both Rankl- and Rank-null mice. Neither strain is capable of producing osteoclasts in vivo [Hsu et al., 1999; Kong et al., 1999] and both lead to an osteoclast-dependent failure of tooth eruption and severe systemic osteopetrosis. The expression of Rankl is modulated by a myriad of factors, many of which are essential for physiologic bone turnover. These include the two hormones integral to calcium homeostasis, 1,25- dihydroxyvitamin D3 (1,25(OH)2D3) [Takahashi et al., 1988b] and parathyroid hormone (PTH) [Groyer et al., 1987; Liu et al., 1998]. In addition, expression of Rankl is also induced by the inflammatory cytokines IL-1β [Jimi et al., 1999b], TNFα, IL-6 [Romas et al., 1996; Tamura et al., 1993], and the prostaglandin PGE2 both in vitro and in vivo [Wani et al., 1999]. Thus, enhanced expression of RANKL results in osteopathy’s ranging from osteolytic disease caused by metastatic cancer to systemic osteoporosis.
Kitazawa et al. were the first to explore the regulation of the Rankl gene at the molecular level [Kitazawa and Kitazawa, 2002; Kitazawa et al., 2003]. These studies, using Rankl proximal promoter constructs, revealed a modest response to both vitamin D and glucocorticoids following transfection into mouse ST2 stromal cells. These studies led to the identification of several regulatory elements for the vitamin D receptor (VDR) and the glucocorticoid receptor (GR) [Kitazawa and Kitazawa, 2002; Kitazawa et al., 2003]. Neither we nor O’Brien and colleagues, using larger promoter-reporter constructs containing up to 7 kb of 5′-flanking DNA relative to the Rankl gene, were able to confirm these modest inductions using either 1,25(OH)2D3 or dexamethasone [Fan et al., 2004; Kabe et al., 2005; O’Brien et al., 2002]. Further exploration of the Rankl locus using ChIP-chip analysis, however, revealed that 1,25(OH)2D3 was capable of inducing strong VDR and RXR binding to five enhancers designated D1– D5 and located −16 (D1), −22 (D2), −60 (D3), −69 (D4) and −76 (D5) kb upstream of the gene’s transcriptional start site (TSS) [Kim et al., 2007b]. The D1, D2 and D5 regions each displayed inducible reporter activity in transfection assays whereas the TSS was relatively inactive [Kim et al., 2007b]. Further studies, by us and O’Brien and colleagues demonstrated that these regions, particularly an extended region around D5 termed D5b also mediated the actions of cAMP/forskolin/PTH and the gp130 cytokine OSM as well [Fu et al., 2006; Kim et al., 2007b]. Importantly, deletion of this extended D5 region reduced the response to both 1,25(OH)2D3 and PTH in the contexts of both a large BAC clone containing the entire mouse RANKL gene locus in cell culture [Fu et al., 2006] and of the mouse genome in vivo [Galli et al., 2008]. In the latter case, cells derived from D5-null mice displayed not only reduced response to the two hormones, but also a detectable bone phenotype. In addition to the modest response to 1,25(OH)2D3 and the glucocorticoids identified by Kitazawa and colleagues [Kitazawa and Kitazawa, 2002; Kitazawa et al., 2003], subsequent studies of the proximal Rankl promoter suggested that this region was also capable of mediating response to prolactin, CART and heat shock proteins [Elefteriou et al., 2005; Roccisana et al., 2004; Srivastava et al., 2003]. The ability of these regulators to modulate Rankl expression from the five distal enhancers has not been examined.
Deciphering transcriptional mechanisms is central to the development of therapies designed to inhibit Rankl mRNA production in a cell type-specific manner. In the current studies, we further explored the capacity of 1,25(OH)2D3 to induce Rankl, focusing upon both the properties of VDR/RXR binding to previously identified enhancers and the mechanisms through which they regulate transcription [Kim et al., 2006a]. Thus, these studies provide important new insight into the overall structure of the mouse Rankl gene and its regulation by 1,25(OH)2D3.
MATERIALS AND METHODS
Reagents
General biochemicals were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma Chemical Co. (St. Louis, MO). 1,25(OH)2D3 was obtained from Solvay (da Weesp, The Netherlands). Oligonucleotide primers were obtained from IDT (Coralville, IA). Anti-VDR (sc-1008), -TBP (TFIID) (sc-273), -TFIIB (sc-225X), -TAFII250 (sc-17134), and -C/EBPβ (sc-150) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-tetra-acetyl (K5, K8, K12, and K16) H4 antibody (06-866) and anti-CTCF antibodies (07–729) were obtained from Millipore Corporation (Billerica, MA). Anti-RNA polymerase II antibody (8WG16, MMS-126R) and antibody to RNA polymerase II phosphorylated at Ser-5 in the C-terminal domain (CTD) (H14, MMS-134R) were obtained from Covance (Princeton, NJ). Anti-Rad21 (AB-992) was obtained from Abcam (Cambridge, MA). AffiniPure Rabbit Anti-Mouse IgM, μ chain-specific secondary antibody was obtained from Jackson ImmunoResearch Laboratories, Inc (West Grove, PA). Rabbit anti-Goat IgG (G4018) was obtained from Sigma (St. Louis, MO). Lipofectamine Plus was obtained from Invitrogen Corporation (Carlsbad, CA).
Cell Culture
Mouse ST2 osteoblastic cells were cultured in MEMα (GIBCO, 12000-022) supplemented with 10% heat-inactivated FBS (HyClone, Logan, Utah) and 100 U/ml Pen/ 10U Strep (HyClone). RAW264.7 cells were grown in DMEM (12800-066) with 1.5 g/L NaHCO3, 10% defined FBS (Hyclone) and 1X HEPES (15630-080). All ligands were added in ethanol (0.1% maximum final concentration).
RNA Isolation/mRNA Analysis
Total RNA was isolated using TriReagent obtained from MRC (Cincinnati, OH) as previously described with the addition of a secondary precipitation step with LiCl. Isolated RNA was reverse transcribed using the SuperScript III RNase H reverse transcriptase kit from Invitrogen (Carlsbad, CA) and subjected to PCR or RT-PCR analysis. Primers used include mouse β-actin (forward, 5′-TGTTTGAGACCTTCAACACCC-3′, reverse, 5′-CGTTGCCAATAGTGATGACCT-3′). Primers for mRNA spanned exon-exon boundaries whenever possible. Real-time PCR (qPCR) was performed using Eppendorf Mastercycler Realplex with Power SYBR GREEN (ABI). Relative expression levels were determined from a standard curve of serial dilutions of cDNA from 1,25(OH)2D3 -treated ST2 cells.
Fractionation of Nuclear and Cytoplasmic RNA
ST2 cells were treated with vehicle or 1,25(OH)2D3 for 6 hr. Cells were trypsinized and washed with cold PBS. The cell pellet was resuspended in Buffer 1 (10 mM Tris (pH 7.5), 140 mM NaCl, 5 mM KCL, 1% NP-40, 1 mM dithiothreitol, and 2 U/ μl RNase Out (Invitrogen)). Cells were lysed on ice for 10 min and nuclei were collected by centrifugation at 270 x g for 5 min. Nuclear pellets were washed once with Buffer 1 to remove cytoplasmic carryover. The supernatant from the initial centrifugation (cytoplasmic fraction) was cleared at 14,000 × g for 5 min at 4°C to remove residual contamination. RNA from the nuclear and cytoplasmic fractions was then isolated using TriReagent using a sample:TriReagent ratio of at least 1:10. The abundance of transcripts was evaluated using standard PCR. All primer sequences are available upon request.
RNA Analysis on Tiling Arrays
The abundance of the RNA transcripts generated at or surrounding the Rankl gene locus was evaluated using the ChIP-chip tiling arrays. Total RNA was isolated as described above. 9 μg of DNaseI-treated RNA was used to generate dscDNA using the Invitrogen Superscript Double-Stranded cDNA Synthesis Kit according to the protocol supplied by NimbleGen Arrays User’s Guide: Gene Expression Analysis v3.0. The protocol was modified to include a DNaseI treatment and the use of a combination of random hexamers and oligo-(dT) as primers for first strand synthesis. Sonicated genomic DNA was labeled with Cy3 and the dscDNA samples were labeled with Cy5 as described in the NimbleGen labeling protocol. 6 μg each of Cy3-labeled genomic DNA and Cy5-labeled dscDNA were co-hybridized to the custom microarray.
siRNA Studies
All small interfering RNA (siRNA) duplexes were obtained from Dharmacon RNA Technologies (Lafayette, CO). ST2 cells were seeded into six-well plates at a concentration of 1.5 × 105 cells/well and transfected approximately 24 h later using Lipofectamine Plus in serum and antibiotic-free medium. 20 nM mVDR siRNA (D-058923-01), non-targeting siRNA pool (D-001206-13) or a Rankl siRNA (M-050280-000) were used for transfection. After transfection, the cells were cultured in medium supplemented with 10% FBS for 48 h prior to treatment with 10−7 M 1,25(OH)2D3 for 6 h. RNA isolation and real-time PCR analysis were carried out as described above.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation was performed as described previously [Kim et al., 2005a; Kim et al., 2007b; Kim et al., 2006b]. Briefly, ST2 cells were treated for 6 hrs with vehicle or the indicated experimental conditions. The cells were washed twice with PBS and then cross-linked for 15 minutes with 1.5% formaldehyde/PBS. Following isolation, the cell extracts were sonicated to prepare chromatin fragments (average DNA size of 500–600 bp DNA as assessed by agarose gel electrophoresis) using a Fisher Model 100 Sonic Dismembranator at a power setting of 2. Pre-cleared samples were then subjected to immunoprecipitation using either a control IgG antibody or the indicated experimental antibody. The precipitated DNA was washed, the cross-links were reversed and the DNA fragments purified using Qiagen QIAquick PCR Purification Kits (Valencia, CA). The isolated DNA (or DNA acquired prior to precipitation (input) was subjected to quantitative RT-PCR (qRT-PCR) using primers designed to amplify fragments of the murine Rankl locus and/or amplified for ChIP-chip microarray analysis. All primer sequences used for amplification of ChIP DNA gene segments are available upon request.
ChIP-chip Microarray Analysis
Output DNA from the ChIP assay was amplified using a ligation-mediated PCR protocol optimized by Ren [Ren et al., 2000] and Farnham [Oberley and Farnham, 2003; Ren et al., 2000] and then assayed for integrity and signal generation (data not shown). These samples were then labeled according to the manufacturer’s ChIP-chip labeling recommendation (NimbleGen Systems, Madison, WI). Briefly, experimental or reference DNA samples (1.5 μg) were labeled with 1 O.D. of 5′-Cy5 or 5′-Cy3 random nonamer wobble primers (TriLink BioTechnologies, San Diego, CA), respectively. 6 μg of each labeled sample were combined with 2x Hybridization Buffer and Hyb Component A (NimbleGen Systems), denatured at 95°C for 5 minutes and then hybridized to custom NimbleGen DNA microarrays overnight at 42°C using X1 mixer lids (NimbleGen Systems) and a MAUI hybridization system (BioMicro, Salt Lake City, UT). Our custom arrays were designed to tile across the mouse Rankl transcription unit from the gene’s downstream to its upstream neighbors, a span of almost 500 kb. The 385k feature array contained many additional genes that are not considered here. All oligonucleotide sequences for the array were generated using NimbleGen’s design standard, Tm-balanced oligomers (50- to 75-mers) with an approximate spacing of 70 bp. Low complexity and repeat regions were excluded from tiling. Following hybridization, arrays were washed using a Hybridization Kit (NimbleGen Systems) and scanned at 5 μm using the GenePix 4000B Scanner (Axon/Molecular Devices, Sunnyvale, CA). Data and peaks were extracted and analyzed using the NimbleScan (version 2.4) software. The log2 ratios, where log2(n) = ln(n)/ln(2) = log(n)/log(2) of a control versus experimental test were calculated for each point. Peaks were called and statistically evaluated using NimbleScan algorithms (version 2.4).
Real-time PCR Analysis
ChIP assays were analyzed using the Eppendorf Mastercycler Realplex with Power SYBR GREEN (ABI). 2 μl of eluted ChIP samples were analyzed in a 25 μl reaction with primers at a final concentration of 100 nm. All dissociation (melt) curves yielded appropriate single peaks.
Plasmids
The pCH110-β-galactosidase (pCH110-βgal) reporter plasmid and the pcDNA-hVDR vector were previously described [Yamamoto et al., 2003]. The parent thymidine kinase (TK) and luciferase (luc) vector pTK-luc was utilized in subsequent cloning efforts. The cloning of mRLD1 (−16.4 to −15.2), mRLD2 (−23.1 to −21.5), mRLD3 (−60.4 to −59.3), mRLD4 (−69.0 to −68.1), and mRLD5 (76045 to −74973) into the pTK-luc vector were previously described [Kim et al., 2007b]. These segments were also cloned into the promoter-less pGL3 vector to generate pGL3-mD1, pGL3-mD2, pGL3-mD3, pGL3-mD4, pGL3-D5a, pGL3-D5b and pGL3-D5a/b. mD5a, mD5b and mD5a/b were also introduced into pGL3 in the reverse orientation to generate pGL3-mD5aR, pGL3-mD5bR, and pGL3-mD5a/bR.
Transient Transfection Assays
ST2 cells were seeded into 24-well plates at appropriate densities and cultured in α-MEM containing 10% FBS. Cells were transfected 24 hr later with Lipofectamine Plus (Invitrogen) in serum and antibiotic-free medium. Individual wells were transfected with 250 ng of a luciferase reporter vector, 50 ng of pCH110-βgal reporter plasmid and 50 ng of pcDNA-hVDR (the latter was routinely transfected with all luciferase reporters unless otherwise indicated). After transfection, the cells were cultured for 48 h in medium supplemented with 10% FBS and then for an additional 24 h with or without 10−7 M 1,25(OH)2D3. Cells were harvested and the lysates assayed for luciferase and β-galactosidase activities as previously described [Yamamoto et al., 2003]. Luciferase activity was normalized to β-galactosidase activity in all cases.
RESULTS
ChIP/chip Analysis Reveals Multiple VDR/RXR Binding Sites in the Mouse Rankl Gene Locus
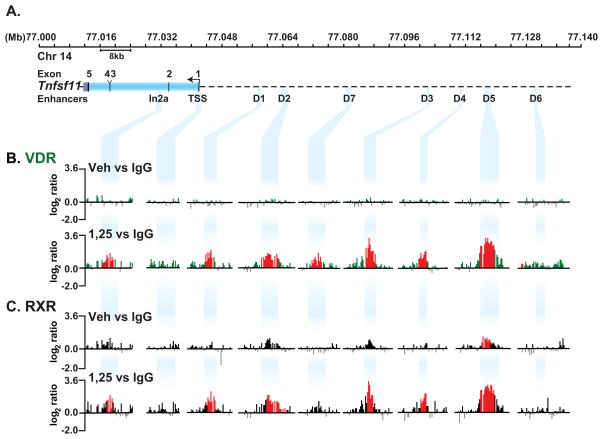
Previous studies using ChIP-chip analysis revealed the presence of five highly conserved regions in the Rankl gene locus that bound the VDR and mediated the actions of 1,25(OH)2D3 [Kim et al., 2007b]. These regions, termed mRLD1-mRLD5, together with three additional regions (mRLD6, mRLD7 and mRLIn2a) are depicted in a schematic of the mouse Rankl gene locus located on chromosome 14 (Fig. 1A). Applying improvements in tiled-array design made since we first explored this gene by ChIP-chip analysis, we re-examined VDR and RXR binding at this locus in the absence and presence of 1,25(OH)2D3. ST2 cells were treated with vehicle or 1,25(OH)2D3 for 6 hr and subjected to ChIP-chip analysis. Data tracks from these analyses represent the log2 ratios of fluorescence obtained from vehicle- treated samples precipitated with antibody to VDR or control IgG (VDRveh vs IgG) (basal VDR) and from 1,25(OH)2D3-treated samples precipitated with the same antibodies (VDR1,25 vs IgG) (inducible VDR) (Fig. 1B). We focused upon regulatory regions identified within the Rankl transcription unit and upstream of the TSS. The complete data tracks are documented in supplemental Fig. S1. Peaks identified using NimbleScan 2.4 at an FDR of <0.05 are shown in red (see Materials and Methods). Data obtained on chromosome 14 from nucleotides 77,226,400 to77,741,622 had no significant VDR binding in untreated and 1,25(OH)2D3-treated cells (data not shown). VDR was undetectable at the Rankl gene locus in the absence of 1,25(OH)2D3, but was strongly induced at multiple sites in its presence (Fig. 1B). These data confirm our previous studies which demonstrated VDR localization to mRLD1-mRLD5 in response to 1,25(OH)2D3, but also reveal several additional sites, one designated mRLD7 at −40 kb and one in Rankl intron2 at +10 kb designated mRLIn2a. VDR motifs are predicted in each of the peak regions detected by ChiP-chip analysis as annotated in supplemental Table 1. RXR occupancy at the Rankl locus in untreated and 1,25(OH)2D3-treated cells was conducted in the same manner described for VDR. Statistically significant peaks of RXR were detected at the mRLD5 region. Pronounced, yet statistically insignificant enrichments were also detected at the mRLD2 and mRLD3 regions in untreated cells. RXR binding was profoundly upregulated at each of the VDR-occupied enhancers (mRLD1-mRLD5, and perhaps at mRLD7) in the presence of ligand (Fig. 1C). Neither VDR nor RXR could be detected at the TSS regardless of treatment. We conclude that 1,25(OH)2D3 is required for VDR binding at enhancers in the Rankl locus. RXR binding, on the other hand, is enhanced together with the VDR at most sites in response to 1,25(OH)2D3, but is pre-bound at mRLD5 and perhaps at mRLD2 and mRLD3. The latter is not surprising, since RXR is a strong DNA binding protein that forms both homo- and heterodimers with various nuclear receptors (NR) that are activated by intracellular ligands [Mangelsdorf and Evans, 1995].
FIGURE 1. ChIP-chip analysis reveals localization of VDR and RXR to the Rankl gene locus in response to 1,25(OH)2D3.
ST2 cells were treated with vehicle or 1,25(OH)2D3 for 6 hours and then subjected to ChIP analysis using antibodies to VDR, RXR or IgG as described in Materials and Methods. A, The upper track depicts the Rankl gene locus. Nucleotide positions (Mb) are shown on chromosome 14 (February 2006 assembly) while the Rankl gene and the positions of the distal enhancer regions are indicated below the chromosomal position and designated by descending blue bands. B, Interaction of VDR with the Rankl gene locus in response to vehicle or 1,25(OH)2D3. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either VDR or IgG (VDRveh vs IgG) (basal, upper track) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either VDR or IgG (VDR1,25 vs IgG) (total inducible VDR, lower track). Only relevant data segments are shown. C, Interaction of RXR with the Rankl gene locus in response to 1,25(OH)2D3 as described in B. All peaks highlighted in red are statistically significant (FDR<0.05).
1,25(OH)2D3 Induces the Recruitment of RNA Polymerase II (RNA pol II) to Sites of VDR Occupancy at the Rankl Locus
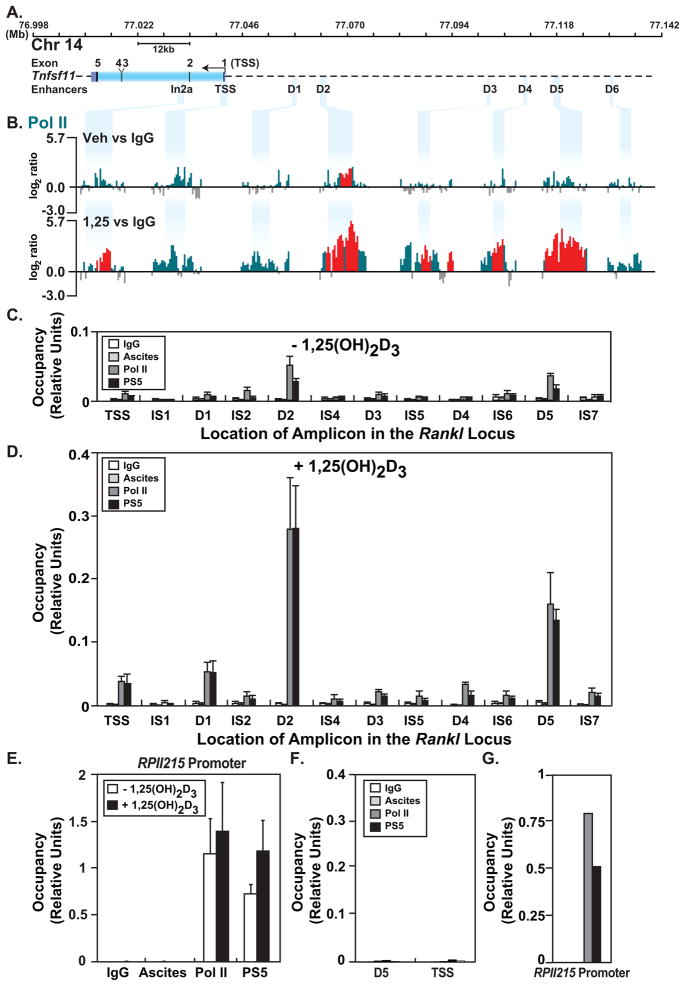
Genome-wide ChIP-chip and ChIP-seq analyses have shown that RNA pol II is often associated not only with transcriptional start sites but with distal enhancers and intergenic sites as well [Feng et al., 2008; Gilchrist et al., 2009; Kim et al., 2005b]. Indeed, our studies using ChIP-chip analysis revealed that RNA pol II accumulated at several Rankl enhancers in the presence of 1,25(OH)2D3 [Kim et al., 2007b]. To explore further sites of RNA pol II binding across the entire Rankl gene locus, we conducted ChIP-chip analyses from genomic samples derived from ST2 cells treated for 6 hr with either vehicle or 1,25(OH)2D3. We assayed RNA pol II occupancy using the 8WG16 RNA pol II antibody which preferentially recognizes hypo-phosphorylated RNA pol II, although this antibody does recognize RNA pol II that is partially phosphorylated at the CTD [Jones et al., 2004]. We focused the data at the regulatory sites identified within the Rankl transcription unit and upstream of the TSS (Fig. 2A). Only minor peaks of RNA pol II were detected outside of the regulatory regions shown (supplemental Fig. S2). In the absence of 1,25(OH)2D3, significant levels of RNA pol II were detected at the mRLD2 and a statistically insignificant enrichment was observed at the TSS (Fig. 2B, upper track). 1,25(OH)2D3 induced a striking increase of RNA pol II at the mRLD2 and mRLD5 enhancers and a de novo increase at additional sites including mRLIn2a, mRLD3 and mRLD4 (Fig. 2B, lower track). Modest increases were also observed at mRLD1 and the TSS. The transition of RNA pol II from a stalled to an elongation-competent polymerase involves phosphorylation of Serine 5 in the carboxyl terminal domain (CTD) [Lin et al., 2002; Marshall et al., 1996]. To determine whether RNA pol II at the Rankl enhancers was phosphorylated at this site, we conducted a direct ChIP analysis of these genomic regions using the H14 antibody specific for phosphoserine 5 in the CTD of RNA pol II (PS5). Like hypo-phosphorylated RNA pol II, the PS5 RNA pol II was also detected predominantly at mRLD2 and mRLD5 in untreated ST2 cells (Fig. 2C). PS5 RNA pol II was also strikingly upregulated at mRLD1, mRLD2 and mRLD5 in 1,25(OH)2D3-treated ST2 cells, mirroring the pattern of RNA pol II occupancy (Fig. 2D). Surprisingly, RNA pol II occupancy at the TSS was lower than at the enhancers. We also examined the RNA pol II promoter (RPII215) as a control in ST2 cells. While both hypo-phosphorylated and PS5 RNA pol II were detected at this promoter, neither were upregulated with 1,25(OH)2D3 treatment (Fig. 2E). While neither form of RNA pol II was detected at the Rankl locus (TSS or mRLD5) in an irrelevant cell line (RAW264.7), both forms of RNA pol II were observed at the RPII215 promoter (Fig. 2F and G). These studies reveal that RNA pol II is pre-bound to the mRLD2 and the mRLD5 Rankl distal enhancers yet strikingly elevated at almost all of the Rankl enhancer regions following treatment with 1,25(OH)2D3.
FIGURE 2. ChIP-chip analysis reveals recruitment of S-5 phosphorylated RNA pol II at the Rankl gene locus in response to 1,25(OH)2D3.
ST2 cells were treated as in Fig. 1 and then subjected to ChIP analysis using antibodies to hypo-phosphorylated RNA pol II (pol II), pol II phosphorylated at serine-5 (PS5) or IgG. A, Schematic of the Rankl locus as in Fig. 1A. B, Recruitment of RNA pol II to the Rankl gene locus in response to 1,25(OH)2D3. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either RNA pol II or IgG (RNA pol IIveh vs IgG) (basal) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either RNA pol II or IgG (RNA pol II1,25 vs IgG) (total inducible). Only relevant data segments are shown. All peaks highlighted in red are statistically significant (FDR<0.05). C, ST2 cells were treated with vehicle or D, 10−7 M 1,25(OH)2D3 for 6 hr and then subjected to direct ChIP assay using antibodies to hypo-phosphorylated RNA pol II (pol II) or serine-5 phosphorylated (PS5) RNA pol II. Class-specific IgGs were used as controls (RNA pol II, IgG; PS5, ascites). Precipitated DNA was analyzed by q-PCR and the products quantitated using a standard curve consisting of dilutions of the corresponding input DNA. E, qPCR analysis of RNA pol II and PS5 recruitment to the RPII215 promoter in untreated and 1,25(OH)2D3-treated ST2 cells. F and G, qPCR analysis of RNA pol II and PS5 recruitment to the mRLD5 and TSS regions of the Rankl locus or to the RPII215 promoter in control RAW264.7 macrophage cells, respectively. All results represent the average of three independent experiments ± SD.
TBP, TFIIB and TAFII250 are Recruited to mRLD2, mRLD5 and the mRLIn2a Regions in the Rankl Locus
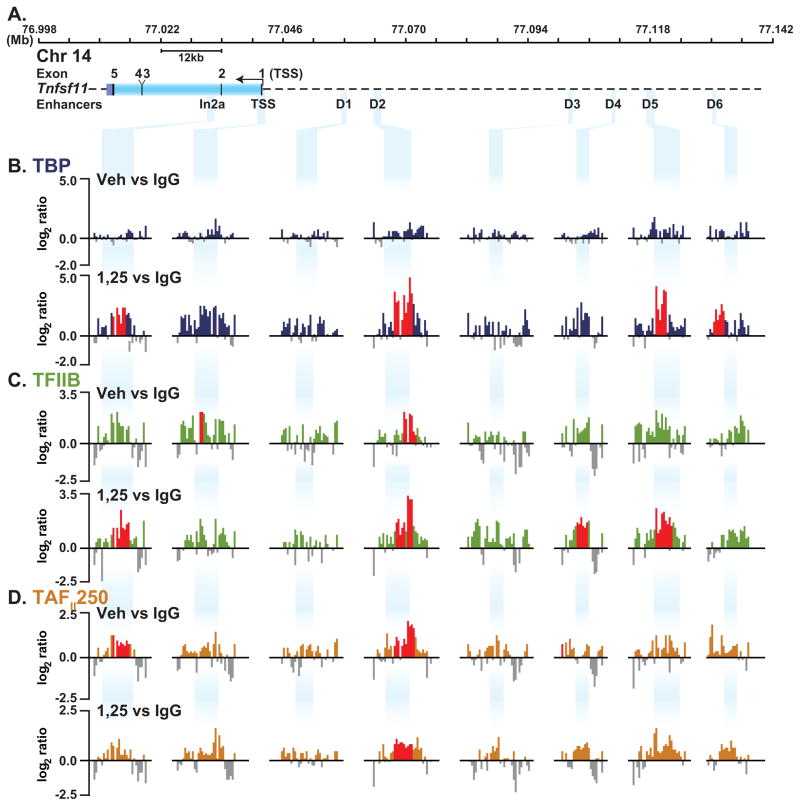
Pre-initiation complex (PIC) assembly at promoters is a sequential process initiated by TATA binding protein (TBP) binding directly to the DNA. TAFII250 represents a subunit of TFIID and has been implicated in RNA pol II promoter selection [Verrijzer et al., 1995]. TAFII250 acts as a scaffold for the assembly of the basal transcriptional machinery and bridges activators to the PIC [Wassarman and Sauer, 2001]. TAFII250 retains kinase [Dikstein et al., 1996], histone acetyltransferase [Mizzen et al., 1996] and ubiquitin-activating and -conjugating activities [Pham and Sauer, 2000]. TFIIB provides a subsequent bridge between TBP and RNA pol II. To explore the underlying mechanism responsible for both the presence and inducibility of RNA pol II at the Rankl locus and in particular at the upstream enhancers, we treated ST2 cells with either vehicle or 1,25(OH)2D3 and conducted a ChIP-chip analysis using antibodies to TBP, TFIIB or TAFII250. In untreated ST2 cells, no significant TBP is detected at the regulatory regions (Fig. 3B, upper track). TBP is recruited to In2a, D2, D5, and D6 at statistically significant levels, and is elevated at the TSS upon 1,25(OH)2D3 treatment (Fig. 3B, lower track). Significant but low levels of TFIIB were detected at the TSS and D2 in untreated cells and TFIIB was enriched at In2a, D3, D4 and D5 with 1,25(OH)2D3 treatment (Fig. 3C). TAFII250 was detected at significant levels at In2a and D2 in untreated cells and was relatively unchanged with 1,25(OH)2D3 treatment (Fig. 3D). Figure 3 documents the results we obtained for each of these transcription factors at discrete sites across the locus. The complete analysis is seen in supplemental Fig. S2. Interestingly, significant levels of TFIIB (Fig. 3C) and TAFII250 (Fig. 3D) but not TBP (Fig. 3B) binding were observed in the absence of ligand, only in association with mRLD2, a finding similar to that observed for RNA pol II. Levels of these transcription factors were increased in response to 1,25(OH)2D3. Thus, while levels of TBP, TFIIB and perhaps TAFII250 were upregulated at mRLD2 and in the case of TFIIB induced at mRLD4 and mRLD5, these transcription factors were not present in significant amounts at the other regions. Surprisingly, only statistically insignificant levels of TBP, TFIIB and TAFII250 were present at the TSS, a finding similar to that for RNA pol II. This may be a reflection of the sensitivity of the ChIP-chip assay, as others have noted a similar lack of PIC components at active promoters with ChIP-chip assays [Kim et al., 2005b]. Our studies suggest a relationship between RNA pol II and TBP, TFIIB and TAFII250 at the Rankl locus. Although it is unclear why TBP, TFIIB, TAFII250 and RNA pol II are only weakly detected at the TSS, it suggests that the upstream enhancers in the Rankl locus may play a key role in the recruitment of the basic transcriptional apparatus.
FIGURE 3. Detection of components of the pre-initiation complex at mRLD2 and mRLD5 in response to 1,25(OH)2D3 at the Rankl gene locus.
ST2 cells were treated as in Fig. 1 and then subjected to ChIP analysis using antibodies to TBP, TFIIB, TAFII250. A, Schematic of the Rankl locus as in Fig. 1 A. B, C, and D, Recruitment of TBP, TFIIB, TAFII250 under treatment conditions as described in Fig 1B. Only relevant data segments are shown. All peaks highlighted in red are statistically significant (FDR<0.05).
The mRLD5 Region of the Mouse Rankl Gene Exhibits 1,25(OH)2D3-dependent Enhancer Activity in the Context of a Heterologous Promoter
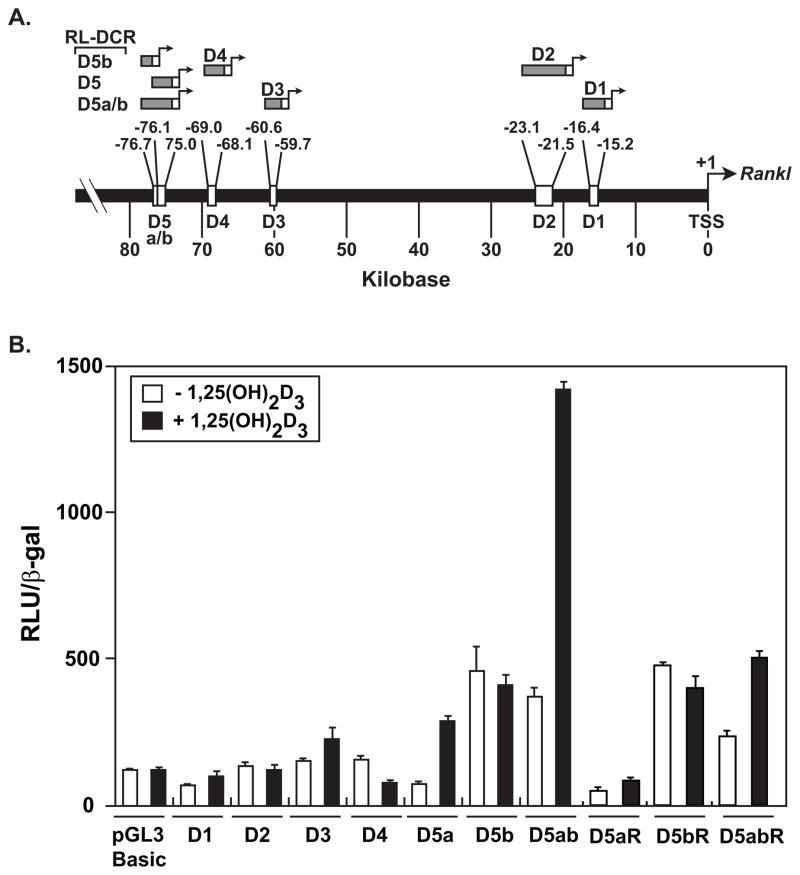
Recent studies have suggested that the level of transcription across genomes is much greater than previously measured. Indeed, as much as 90% of the human genome, by some estimates, is believed to be transcribed [Birney et al., 2007]. The presence of RNA pol II together with transcription factors such as TBP, TFIIB, TAFII250 at many of the Rankl enhancers prompted us to examine whether these regions possessed independent “promoter” activity and whether they were also capable of transcription. To assess the former, we transfected promoter-less constructs containing mRLD1-mRLD4, mRLD5a, mRLD5b, and mRLD5a/b into ST2 cells and examined both their basal and inducible activities in response to 1,25(OH)2D3 (Fig. 4A and B). We also examined the activity of mRLD5a, mRLD5b, and mRLD5a/b cloned in the reverse orientation (R) (Fig. 4B). mRLD1-mRLD3 and mRLD4 display limited basal activity and are not induced by 1,25(OH)2D3 (Fig. 4B). mRLD5b displayed higher basal activity but was similarly resistant to 1,25(OH)2D3 (Fig. 4B). mRLD1 and mRLD2, but not mRLD5b, have been shown to contain vitamin D response elements (VDRE) and to mediate the activity of 1,25(OH)2D3 when cloned into pTK-luc reporter constructs, although induction levels were modest [Nerenz et al., 2008]. Both mRLD5a and mRLD5a/b were induced by 1,25(OH)2D3 (Fig. 4B). These same segments were also inducible in the reverse orientation (Fig. 4B). Like mRLD5b, reversed mRLD5b was similarly unresponsive to 1,25(OH)2D3 (Fig. 4B). We conclude that the enhancer segments of the Rankl locus may be capable of producing RNA transcripts. The absolute activities of the forward constructs were similar, however, to constructs in which the D segments were cloned upstream of the mouse Rankl minimal promoter.
FIGURE 4. Assessment of promoter-independent Rankl enhancer activity in ST2 cells.
A, Location of distal enhancers in the Rankl locus (Feb 2006 Assembly) and the segments examined for promoter-independent transcriptional activity. B, Transcriptional activity of the Rankl enhancers. ST2 cells were transiently transfected with 250 ng pGL3 control vector, pGL3-mRLD1 - pGL3-mRLD5, pGL3-mRLD5a, pGL3-mRLD5b, pGL3-mRLD5a/b, pGL3-mRLD5aR, pGL3-mRLD5bR, or pGL3-mRLD5a/bR together with 50 ng pcDNA-hVDR and pCH110-βgal using lipofectamine. Cells were treated with either vehicle or 1,25(OH)2D3, (10−7 M) and then evaluated 24 hr later for luciferase and β-gal activity as described in Materials and Methods. Each point represents the normalized RLU average ± SEM of a triplicate set of transfections. These data are representative of three or more replicate experiments.
Expression of Rankl RNA Transcripts can be Detected on Tiled Microarrays
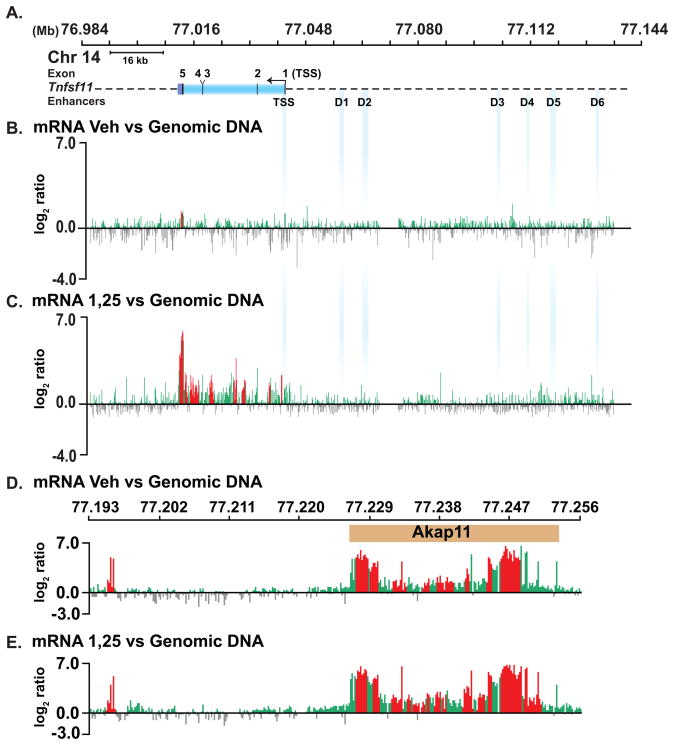
Based upon the above observations, we explored the possibility that RNA might be produced at these regions in the Rankl locus. As a first approach, we isolated total RNA from vehicle- or 1,25(OH)2D3-treated ST2 cells, generated dscDNA from these samples, labeled this DNA with Cy5 and co-hybridized the samples with Cy3-labeled sonicated genomic DNA for normalization on the same tiled microarrays used for ChIP-chip analysis. Little RNA expression was detected across the Rankl locus (Fig. 5A) when derived from vehicle-treated ST2 cells (Fig. 5B); however, RNA was detected at housekeeping genes on the custom chip (data not shown). A striking upregulation of transcripts was detected, however, when RNA was isolated from cells treated for 6 hr with 1,25(OH)2D3 (Fig. 5C). This increase corresponded to the upregulation of Rankl observed using RT-PCR analysis (data not shown). Upregulation was not observed at Akap11, the gene located approximately 200 kb immediately upstream of Rankl (Fig. 5D and 5E). No increase in intergenic transcription was observed at any of the upstream regulatory regions, despite the fact that intergenic transcription at other sites on the custom microarray was apparent (data not shown). Analysis of RNA derived exclusively from either cytoplasmic or nuclear preparations led to similar results. We conclude from these studies that transcription from the upstream enhancers, if present, is likely to be of low abundance and perhaps sensitive to degradation.
FIGURE 5. Evaluation of RNA transcription across the Rankl locus using tiled DNA microarrays.
A, Schematic of the mRankl locus. B-E, dscDNA was generated from RNA isolated from ST2 cells treated for 6 hrs with either vehicle or 1,25(OH)2D3 10−7 M. dscDNA and total genomic DNA were labeled with Cy5 and Cy3, respectively, and co-hybridized to custom microarrays identical to those used for ChIP-chip analysis. B and C, analysis across the Rankl gene locus. D and E, analysis across the neighboring Akap11 gene locus.
Expression of Transcripts from the Enhancer Regions of the Rankl Locus
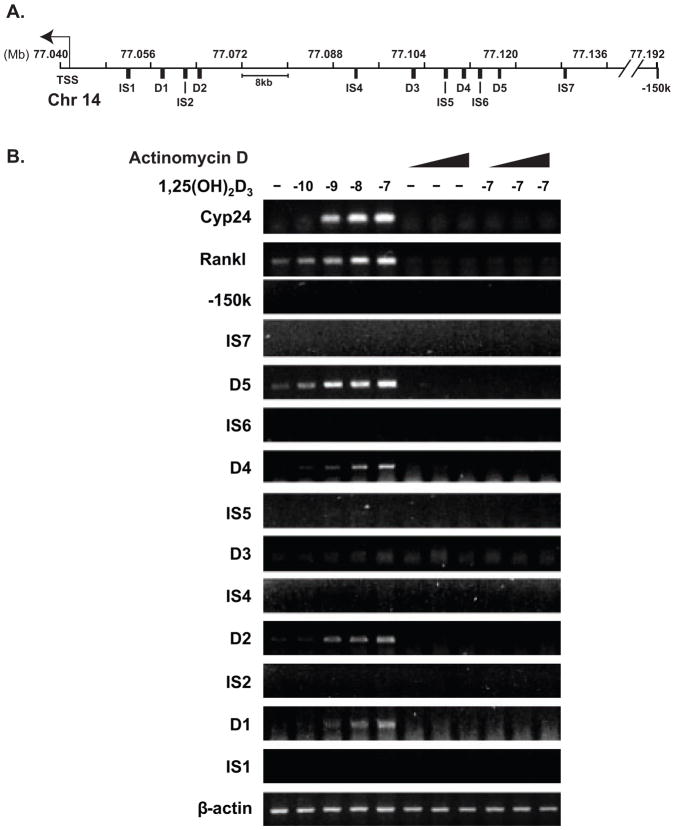
As a result of the above analysis, we searched for the presence of intergenic transcripts using traditional and more sensitive RT-PCR. cDNA was synthesized from DNaseI-treated RNA isolated from vehicle or 1,25(OH)2D3-treated ST2 cells using a combination of oligo-(dT) and random hexamers as primers. The cDNA was evaluated using primer sets located within and between the regulatory regions (Fig. 6A). Dose-dependent 1,25(OH)2D3-inducible RNA transcripts were observed using primers to mRLD1, mRLD2, mRLD4 and mRLD5, but not to any of the intervening sequences (IS) (Fig. 6B). Importantly, RNA was detected using primers to both Rankl and Cyp24a1, the latter a strongly induced target for 1,25(OH)2D3. Importantly, the induction of these RNAs were sensitive to increasing concentrations of actinomycin D (ActD) (0.5, 1, and 5 μg/ml) indicating that the transcripts accumulate as a result of transcriptional activation and not stabilization (Fig. 6B). Moreover, RNase treatment of the samples prior to cDNA synthesis prevented detection (data not shown). Further analysis using a series of PCR primers suggested that the transcript produced at mRLD5 was at least 1.8 kb in size (data not shown). Since open reading frames were not apparent in the mRLD1-mRLD5 regions, we conclude that these transcripts likely represent noncoding RNAs (ncRNAs).
FIGURE 6. RNA transcripts are generated from the intergenic regulatory regions of the Rankl locus.
ST2 cells were treated for 6 hrs with vehicle, increasing concentrations of 1,25(OH)2D3 (10−10 to 10−7 M) or increasing concentrations of ActD (0.5, 1, or 5 μg/ml) in the absence or presence of 10−7 M 1,25(OH)2D3. RT-PCR analysis was used to detect transcripts generated from Rankl and Cyp24a1 or from Rankl enhancers and intervening regions. A, Schematic of the regions across the Rankl locus examined by PCR. B, Detection of RNA transcripts. Cyp24a1 mRNA was examined as a positive control. These results are representative of three or more similar experiments.
The Intergenic Transcript Originating from the mRLD5 Region is Not Spliced to the Rankl mRNA Transcript
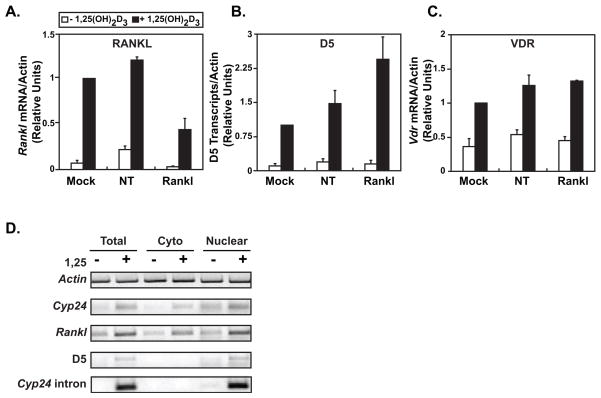
Given the above properties, it is possible that the upstream enhancers we have identified represent distal promoters for the Rankl gene. Thus, RNA transcribed from these regions might comprise unprocessed components of a primary transcript from the Rankl gene and contain exons that are linked directly to the mature Rankl mRNA. While no evidence exists in either the literature or current databases to support this hypothesis and transcription between the distal enhancers has not been detected, we explored this possibility by examining the impact of Rankl mRNA knock down on the levels of the transcript measured upstream. We focused our attention on mRLD5, since this region manifests the most robust transcriptional activity despite the low abundance of RNA transcripts. We hypothesized that if an upstream exon and the authentic Rankl mRNA were linked, degradation of the authentic transcript would result in a decrease in upstream exon abundance. The reverse experiment, although perhaps more attractive, was not feasible due to a lack of sufficient information regarding the mRLD5 transcript itself. Accordingly, we either mock transfected, or transfected ST2 cells with a non-specific control siRNA or Rankl siRNA. After 48 hrs, we treated the cells for 6 hrs with either vehicle or 1,25(OH)2D3 and then examined total RNA for levels of Rankl, mRLD5 and control Vdr transcripts using qRT-PCR. RANKL siRNA treatment effectively reduced both basal and 1,25(OH)2D3-inducible levels of Rankl mRNA (Fig. 7A). Neither mRLD5 nor VDR transcript levels were influenced by this maneuver (Fig. 7B and C). Indeed, if anything, mRLD5 levels were increased. This analysis suggests that neither the mRLD5 transcript nor an exon derived from this transcript is spliced to the Rankl mRNA. Similarly, attempts to measure a mRLD5/Rankl junction by PCR using primers to the mRLD5 region and within the Rankl mRNA did not detect a mRLD5/Rankl transcript (data not shown). Thus, we conclude that the mRLD5 transcript is not spliced to mRankl mRNA and that mRLD5 does not represent an upstream promoter. Interestingly, while no transcripts have been detected at mRLD5, an expressed sequence tag (est) contained within the mRLD1 (accession numbers BG794861 and BG796385) and mRLD2 (accession number CR516436) regions have been identified. This also supports our contention that the mRLD regions do not represent distal Rankl promoters.
FIGURE 7. The mRLD5 transcript is not linked to the expression of Rankl mRNA and is localized to the nucleus.
Knockdown of Rankl mRNA does not affect the level of mRLD5 transcripts. ST2 cells were transfected with 20 nM nontargeting (NT) siRNA or Rankl siRNA. After 48 h, the cells were treated with 10−7 M 1,25(OH)2D3. Total RNA was isolated and the relative concentrations of transcripts for A, mRankl, B, mRLD5, and C, VDR (a control gene) were assayed by qRT-PCR. The results represent the average of three independent experiments ± SEM. D, mRLD5 transcripts are predominantly localized to the nucleus. ST2 cells were treated with either vehicle or 10−7 M 1,25(OH)2D3 for 6 hrs and then used directly to isolate total RNA or to obtain cytoplasmic or nuclear enriched fractions from which RNA was isolated. The relative abundance of transcripts for β-actin, Cyp24a1, a Cyp24a1 intron, Rankl and the mRLD5 enhancer was then assessed using primers documented in Materials and Methods. These results are representative of at least three similar experiments.
To further explore the nature of the transcript produced at the mRLD5 region, we isolated RNA from the crude nuclear and cytoplasmic fractions and contrasted the abundance of RNA transcripts for both Rankl and mRLD5. Transcripts for β-actin, Cyp24a1 (exon to exon) and a Cyp24a1 intron were used as controls. While Rankl mRNA is distributed in cytoplasmic and nuclear fractions, as might be expected of typical mRNAs, transcripts from mRLD5 appear largely nuclear (Fig. 7D). The quality of the fractionation procedure is highlighted by the almost exclusive localization of the Cyp24a1 intron in the nucleus. These results suggest that the mRLD5 transcript is not exported from the nucleus. While we hypothesize that these transcripts may play a role in Rankl gene expression, their low abundance has prevented further analysis of their function.
1,25(OH)2D3 Induces Histone H4 Acetylation (H4ac) at the Regulatory Regions of the Rankl Locus
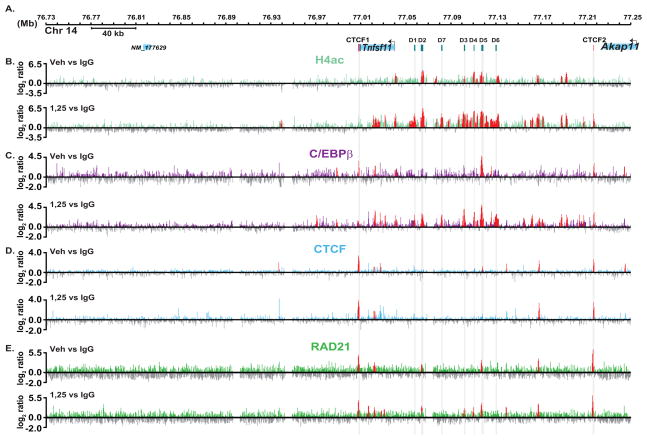
Acetylation of multiple sites on histone tails is generally, although not always, associated with transcriptional activation [Brownell and Allis, 1996]. We previously used ChIP coupled with standard PCR to evaluate H4ac at limited sites within the Rankl locus in a time-dependent manner following 1,25(OH)2D3 treatment [Kim et al., 2007b]. These assays were limited, however, as they did not provide perspective across the entire gene locus. To explore this phenomenon further, we treated ST2 cells with either vehicle or 1,25(OH)2D3 and performed ChIP-chip analysis using an antibody to tetra-acetylated H4. Data tracks from these analyses representing the log2 ratios of fluorescence obtained from 1) vehicle-treated samples precipitated with antibody to H4ac or control IgG (H4acveh vs IgG) (basal H4ac), 2) 1,25(OH)2D3-treated samples precipitated with antibody to H4ac or control IgG (H4ac1,25 vs IgG) (total induced H4ac) and 3) 1,25(OH)2D3-treated or vehicle treated samples precipitated with antibody to H4ac (H4ac1,25 vs H4acveh) (net inducible H4ac) (Fig 8A and B). Although Rankl expression is undetectable in ST2 cells in the absence of 1,25(OH)2D3, many sites across the gene were significantly acetylated (Fig. 8B, upper track). Interestingly, many of these sites highlight the regions to which the VDR and its partner RXR bind upon treatment including mRLD2 and mRLD5. Basal H4ac levels are observed at the TSS and at specific sites within the next 150 kb of the upstream intergenic sequence. On the other hand, basal H4ac was not detected downstream for over +330 kb, demonstrating that basal acetylation is restricted to the Rankl locus. Importantly, 1,25(OH)2D3 treatment induces a striking upregulation in H4ac at all of these sites and de novo at mRLD1, mRLD7, and perhaps others (Fig. 8B, upper and lower tracks). Substantial levels of H4ac exist at many of the Rankl distal enhancers prior to induction by 1,25(OH)2D3, suggesting that these regions may be epigenetically marked and thus retain conformations accessible to regulatory proteins such as the VDR. Clearly, however, H4ac upregulation upon 1,25(OH)2D3 treatment indicates a role for the VDR/RXR heterodimer in this event across the locus.
FIGURE 8. ChIP-chip analysis reveals a large domain of basal and 1,25(OH)2D3-inducible H4ac and C/EBPβ across the mRankl locus and whose limits are defined by occupied CTCF and Rad21 sites.
ST2 cells were treated as in Figure 1 and then subjected to ChIP analysis using antibodies tetra-acetylated H4, C/EBPβ, CTCF, Rad21 or IgG. A, Schematic of the Rankl locus as in Fig. 1A. B, Detection of H4ac across the Rankl locus in response to vehicle or 1,25(OH)2D3. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either H4ac or IgG (H4acveh vs IgG) (basal) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either H4ac or IgG (H4ac1,25 vs IgG) (total inducible H4ac). C, Detection of binding sites for C/EBPβ. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either C/EBPβ or IgG (C/EBPβveh vs IgG) (basal) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either C/EBPβ or IgG (C/EBPβ,25 vs IgG) (total inducible C/EBPβ). D, Detection of binding sites for CTCF. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either CTCF or IgG (CTCF veh vs IgG) (basal) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either CTCF or IgG (CTCF1,25 vs IgG) (total inducible CTCF). The positions of the most striking CTCF sites are highlighted in grey columns. E, Detection of binding sites for Rad21. Data tracks represent the log2 ratios of fluorescence obtained from 1) a vehicle-treated sample precipitated with antibodies to either Rad21 or IgG (Rad21 veh vs IgG) (basal) and 2) an 1,25(OH)2D3-treated sample precipitated with antibodies to either Rad21 or IgG (Rad211,25 vs IgG) (total inducible Rad21). In all tracks, peaks highlighted in red are statistically significant (FDR<0.05).
C/EBPβ Occupies mRLD5 and is Recruited to Additional Sites in Response to 1,25(OH)2D3 Independently of VDR Occupancy
1,25(OH)2D3 treatment of osteoblastic and kidney cells increases C/EBPβ mRNA and protein levels and knockdown of C/EBPβ blunts 1,25(OH)2D3-mediated transcriptional activation of Cyp24a1 [Dhawan et al., 2005]. Our own RT-PCR, microarray and Western blot analyses suggest that 1,25(OH)2D3 induces expression of C/EBPβ as well (data not shown). In addition, C/EBPβ-null mice display delayed boned development which show defects in chondrocyte maturation and osteoblast differentiation [Tominaga et al., 2008]. Based upon these observations, we explored the possibility that in addition to its direct activation of the VDR, 1,25(OH)2D3 might also induce C/EBPβ binding to the Rankl locus. ST2 cells were treated with either vehicle or 1,25(OH)2D3 and lysates subjected to ChIP-chip analysis using an antibody to C/EBPβ. Figure 8C documents the data tracks from these analyses under the following conditions: 1) vehicle-treated samples precipitated with antibody to C/EBPβ or control IgG (C/EBPβveh vs IgG) (basal C/EBPβ) and 2) 1,25(OH)2D3-treated samples precipitated with antibody to C/EBPβ or control IgG (C/EBPβ1,25 vs IgG) (total induced C/EBPβ). The presence of C/EBPβ was detected in untreated ST2 cells at the Rankl TSS and prominently at the mRLD5 region (Fig. 8C, upper track). 1,25(OH)2D3 treatment also induced a striking increase in C/EBPβ binding at multiple sites throughout this locus (Fig. 8C, lower track). These sites included not only those bound to the VDR, but many additional sites as well. Since C/EBPβ and VDR binding sites overlap but are not identical, it seems likely that C/EBPβ may play an independent role at the Rankl gene locus. Importantly, C/EBPβ binding outside these regions was almost completely absent (data not shown), indicating a high specificity for C/EBPβ at the Rankl locus. Interestingly, this pattern of C/EBPβ occupancy closely reflected the pattern of H4ac observed following treatment with 1,25(OH)2D3, indicating that the activation of Rankl gene expression may be mediated not only by the VDR, but by C/EBPβ as well. In addition, C/EBPβ binding motifs are predicted at each of the regulatory regions (S. Fig. 3 and S. Table 1). Thus, VDR and C/EBPβ may act cooperatively in response to 1,25(OH)2D3 to regulate the expression of the Rankl gene.
CTCF Occupied Sites Define the Boundaries of the Rankl Chromatin Domain
Elevated levels of H4ac highlight a region that begins at the 3′ end of the Rankl gene and extends upstream over 210 kb, ending approximately 20 kb downstream of the neighboring gene Akap11 (Fig. 8A and B). As indicated earlier, the ability of 1,25(OH)2D3 to induce an increase in H4ac and to promote C/EBPβ binding is restricted to this region. To determine whether this extended region defined the complete Rankl gene locus, we searched for sites occupied by CTCF across this region. ST2 cells were treated with either vehicle or 1,25(OH)2D3 and then subjected to ChIP-chip analysis using an antibody to CTCF. CTCF data tracks are shown from these analyses under the following conditions: 1) vehicle- treated samples precipitated with antibody to CTCF or control IgG (CTCF veh vs IgG) (basal CTCF) and 2) 1,25(OH)2D3-treated samples precipitated with antibody to CTCF or control IgG (CTCF1,25 vs IgG) ( total induced CTCF) (Fig. 8D). As can be seen, prominent CTCF binding activity was observed immediately downstream of the 3′ UTR (CTCF1) and at the upstream boundary of inducible H4ac activity (CTCF2) (Fig 8D). 1,25(OH)2D3 had no effect on the level or the location of CTCF binding. The CTCF2 also aligns with the most distal of the induced C/EBPβ binding sites. Both CTCF sites represent regions of high H4ac and contain sequences predicted to bind CTCF by Genomatix (S. Fig. 3 and S. Table 1). Several CTCF sites were also observed within these boundaries, perhaps highlighting the multiple activities defined earlier for CTCF [Filippova, 2008]. Recently, Rad21, A component of the cohesin complex, has been shown to co-occupy CTCF sites [Wendt and Peters, 2009; Wendt et al., 2008]. We therefore conducted Rad21 ChIP-chip in the same manner at CTCF as described above. Similar to results of others, the “major” CTCF peaks were co-occupied by Rad21, whereas “minor” Rad21 peaks were present at D2, D3, and D4 independently of CTCF (Fig. 8E). Each site was confirmed through ChIP-chip analysis in other mouse cell lines and at highly conserved regions in the human genome as well (data not shown). While we have not proven that these CTCF sites function directly as insulators, the limited activity that occurs outside these sites supports the hypothesis that this region of over 200 kb defines the Rankl locus.
DISCUSSION
The central importance of RANKL to the formation of osteoclasts and to the bone remodeling process in the adult skeleton is highlighted by the absence of such bone resorbing cells and a striking osteopetrotic phenotype in the Rankl −/− mouse [Kong et al., 1999]. While numerous systemic agents regulate the expression of Rankl, perhaps the most important are PTH [Kondo et al., 2002; Liu et al., 1998] and 1,25(OH)2D3 [Takahashi et al., 1988a]. Our previous studies identified a complex mechanism whereby the expression of Rankl is modulated by both of these hormones and involves multiple enhancers located −16 to −76 kb upstream of the gene’s TSS [Kim et al., 2007b; Kim et al., 2006b]. In this report, we explore this mechanism of activation further using ChIP-chip analysis, an approach that allowed us to scan the entire Rankl gene locus for not only 1,25(OH)2D3-induced VDR binding activity, but for changes in histone modification and for the presence of other factors resulting from this binding activity. Our results both confirm and significantly extend the observations we previously reported.
The present studies confirm previously discovered Rankl enhancers for the VDR and RXR at mRLD1-mRLD5 and identify new sites of action at mRLD7 and mRLIn2a. mRLIn2a is the first regulatory region to be found within the Rankl transcription unit. These sites display characteristics similar to those identified for mRLD1-mRLD5 including an ability to recruit RNA pol II and to mediate an increase in H4ac in response to 1,25(OH)2D3. A new site identified as mRLD6 as well as mRLD7 and mRLIn2a also bind 1,25(OH)2D3-induced C/EBPβ (see Discussion below).
Our studies have revealed several significant insights regarding the general actions of the vitamin D hormone. First, it has been suggested that the VDR/RXR heterodimer is normally bound to regulatory elements at gene targets in the absence of 1,25(OH)2D3 where it functions to suppress gene expression via the recruitment of co-repressors [Perissi et al., 1999]. Thus, the role of the hormone is not to promote DNA binding, but rather to induce a conformational change in the receptor that causes the dismissal of bound co-repressors and the recruitment of co-activators, similar to that suggested for RAR, TR, and PPAR [Perissi et al., 1999]. Our results, which emerged from an examination of VDR and RXR binding at the Rankl locus in the absence and presence of 1,25(OH)2D3, do not support this hypothesis. Thus, we find that the VDR is highly dependent upon 1,25(OH)2D3 for DNA binding activity. RXR, on the other hand, although strongly induced to bind to Rankl enhancers by 1,25(OH)2D3, is present at several of the enhancers in the absence of the hormone. We suggest that pre-bound RXR may mark sites for potential VDR binding. Alternatively, it is possible that RXR is bound at sites in the Rankl locus as a heterodimer with another nuclear receptor partner. Despite our results at the Rankl locus, however, it is premature to conclude that identical results will emerge for all VDR target genes. Certainly, each gene is unique both with respect to the VDREs to which the VDR binds and to its surrounding chromatin environment. It is also likely that VDR concentrations may impact ligand-independent binding as well. Indeed, we and others have detected the VDR at other genes in the absence of ligand, albeit at lower levels [Ewing et al., 2007; Kitagawa et al., 2003]. Genome-wide studies will be necessary to determine to what extent this principle holds at a complete cellular collection of VDR target genes. Our preliminary studies at this level suggest that the degree to which unliganded VDR and RXR binds to multiple gene targets is highly context dependent.
Second, we were surprised to find that in addition to the VDR, 1,25(OH)2D3 also induced significant C/EBPβ binding at the Rankl enhancers. This binding activity was extensive, and was observed at all the VDR occupied regions and at additional sites. All of these sites contained potential C/EBPβ response elements, although none have been explored for functionality. C/EBPβ binding activity is also found at other 1,25(OH)2D3 target genes, including those for Cyp24a1 and the Vdr itself [Dhawan et al., 2005]. Thus, Christakos and colleagues, for example, demonstrated that cotransfection of C/EBPβ together with the Cyp24a1 and VDR gene promoters enhance overall reporter activity in bone cells [Dhawan et al., 2005]. Our observations are unique, as they show that the actions of 1,25(OH)2D3 can directly increase endogenous C/EBPβ binding at a target gene. Studies of Cyp24a1 and other genes using the ChIP-chip approach indicate that Rankl is not the only gene target activated by 1,25(OH)2D3 in this manner (data not shown). The involvement of C/EBPβ in 1,25(OH)2D3-induced gene expression adds an unexpected dimension to this hormone’s actions. Thus, it is clear that 1,25(OH)2D3 can enhance transcription at certain target genes by inducing not only the VDR, but C/EBPβ as well. Interestingly, these data also raise the possibility that C/EBPβ alone may mediate 1,25(OH)2D3 activityat genes that are responsive exclusively to C/EBPβ and not directly to the VDR. Finally, this transcription factor is also a direct target of protein kinase-mediated signal transduction pathways known to directly phosphorylate C/EBPβ at multiple sites [Trautwein et al., 1993]. Thus, activation of C/EBPβ could potentiate 1,25(OH)2D3 response under some circumstances, but dampen the response in others. The mechanism through which 1,25(OH)2D3 induces C/EBPβ binding at the Rankl gene (and to other genes) is unknown. While 1,25(OH)2D3 promotes an upregulation of C/EBPβ mRNA in bone cells [Dhawan et al., 2005], it is unclear whether this upregulation is sufficient to enhance C/EBPβ DNA binding at target genes or to activate the protein. 1,25(OH)2D3 is also known to activate membrane-associated kinases [Zanello and Norman, 2006]. Thus, C/EBPβ binding at the Rankl locus may be induced by 1,25(OH)2D3-dependent phosphorylation of C/EBPβ and then potentiated by the ability of the hormone to promote a secondary upregulation of the gene as well. Regardless, increased H4ac levels at the sites to which C/EBPβ binds in the absence of the VDR suggests that this factor is transcriptionally active.
Our initial studies indicated that RNA pol II is recruited to the Rankl locus, although the site-specific nature of direct ChIP assays provided limited detail [Kim et al., 2007b]. Our ChIP-chip studies provide much greater insight, however. RNA pol II is present at mRLD2 and mRLD5 but not the TSS. Following treatment, elongation-competent RNA pol II is increased at these enhancers and is also induced at the other sites. Surprisingly, limited occupancy was discovered at the TSS. Perhaps RNA pol II recruited to the TSS becomes rapidly engaged, thus preventing detection. RNA pol II occupancy at the Rankl enhancers is accompanied by the recruitment of TBP, TFIIB and TAFII250. The recruitment of these factors together with RNA pol II at mRLIn2a suggests that this region does not represent an RNA pol II transcription pause site, in contrast to our earlier suggestion, but rather an intronic enhancer element. Our findings also suggest that while Rankl enhancers are promoter-like, they are not simply alternative Rankl promoters. This hypothesis is supported by the overall transcriptional activities manifested by these regions in transient transfection studies and by the detection of noncoding nuclear RNA transcripts that appear to be synthesized from these regions. The functional role of this class of RNAs remains to be identified, although several RNAs, including HOTAIRE, are known to manifest regulatory control at adjacent genes [Guttman et al., 2009; Rinn et al., 2007]. Due to the low abundance of the RNAs identified here, however, we have not yet been able to ascribe a function.
ChIP-chip analysis has enabled us to examine both the basal and inducible acetylation state of H4 at the Rankl locus. Previous studies using site- directed ChIP analysis suggested differences in H4ac across the locus, although we were unable to place these differences in a high resolution context [Kim et al., 2007a]. The results obtained here indicate that specific regions of the Rankl locus are defined by increased levels of H4ac under conditions in which the gene is not expressed. Interestingly, this basal H4ac activity is localized to many of the enhancers for the Rankl gene. Importantly, 1,25(OH)2D3 treatment causes a significant upregulation of H4ac across the locus. This upregulation appears to be initiated at all sites to which the VDR binds, but also at sites to which C/EBPβ binds as well. The broad nature of the induced H4ac across the locus and its definition by the CTCF sites defines the overall locus of the Rankl gene. These observations suggest the possibility that this region represents an important chromatin domain whose overall architecture may be altered through extensive transcription factor binding. Future studies using chromosome conformation capture (3C) techniques will be necessary to define directly the interrelationships that might exist and whether they are altered upon treatment with 1,25(OH)2D3. Despite this, the presence of both basal and inducible acetylation at H4 supports our suggestion that these distal regions play a central role in Rankl activation.
In summary, we have shown that 1,25(OH)2D3 induces the Rankl gene via simultaneous induction of two transcription factors at multiple enhancers, the bulk of which are located significant distances upstream of the Rankl TSS. Transcription factor binding induces the recruitment of not only RNA pol II, but basal transcriptional machinery as well. The presence of these factors is likely responsible for the production of small amounts of noncoding RNA that emanate for these sites. We also show that regions that bind VDR and C/EBPβ also undergo an increase in H4ac. Finally, virtually all activity induced by hormone is confined to a region between two occupied CTCF/Rad21 sites, suggesting that these sites represent the boundaries of the Rankl locus.
Supplementary Material
Acknowledgments
We thank members of the Pike laboratory for insightful discussion. The early technical contributions of Drs. Sungtae Kim and Miwa Yamazaki are gratefully acknowledged.
ABBREVIATIONS
- M
mouse
- h
human
- VDR
vitamin D receptor
- Pol II
RNA polymerase II
- PS5-Pol II
RNA polymerase II phosphorylated at Serine 5 of the carboxyl terminal domain
- C/EBPβ
CCAAT enhancer binding protein beta
- ChIP-chip
chromatin immunoprecipitation-DNA microarray
- CTCF
CCCTC-binding factor
- DMSO
dimethyl sulfoxide
- Cyp24a1
25-hydroxy-vitamin D3-24 hydroxylase gene
- RANKL
receptor activator of NF-κB ligand
- TBP
TATA binding protein
- TFIIB
transcription factor IIB
- TFIID
transcription factor IID
- TNF
tumor necrosis factor
- TSS
transcriptional start site
- TAFII250
TATA binding protein associated factor II250
- PTH
parathyroid hormone
- 1,25(OH)2D3
1,25-dihydroxyvitamin D3
- VDR
vitamin D receptor
- RNA pol II
RNA polymerase II
- RT-PCR
reverse transcription-polymerase chain reaction
References
- Birney E, Stamatoyannopoulos J, Dutta A, Guigo R, Gingeras T, Margulies E, Weng Z, Snyder M, Dermitzakis E, Thurman R, Kuehn M, Taylor C, Neph S, Koch C, Asthana S, Malhotra A, Adzhubei I, Greenbaum J, Andrews R, Flicek P, Boyle P, Cao H, Carter N, Clelland G, Davis S, Day N, Dhami P, Dillon S, Dorschner M, Fiegler H, Giresi P, Goldy J, Hawrylycz M, Haydock A, Humbert R, James K, Johnson B, Johnson E, Frum T, Rosenzweig E, Karnani N, Lee K, Lefebvre G, Navas P, Neri F, Parker S, Sabo P, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins F, Dekker J, Lieb J, Tullius T, Crawford G, Sunyaev S, Noble W, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker I, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch H, Sekinger E, Lagarde J, Abril J, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen J, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson M, Thomas D, Weirauch M, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J, Allis C. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–84. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Peng X, Sutton AL, MacDonald PN, Croniger CM, Trautwein C, Centrella M, McCarthy TL, Christakos S. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol Cell Biol. 2005;25:472–87. doi: 10.1128/MCB.25.1.472-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell. 1996;84:781–90. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Ahn JD, Takeda S, Starbuck M, Yang X, Liu X, Kondo H, Richards WG, Bannon TW, Noda M, Clement K, Vaisse C, Karsenty G. Leptin regulation of bone resorption by the sympathetic nervous system and CART. Nature. 2005;434:514–20. doi: 10.1038/nature03398. [DOI] [PubMed] [Google Scholar]
- Ewing A, Attner M, Chakravarti D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol Endocrinol. 2007;21:1791–806. doi: 10.1210/me.2007-0095. [DOI] [PubMed] [Google Scholar]
- Fan X, Roy EM, Murphy TC, Nanes MS, Kim S, Pike JW, Rubin J. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J Cell Biochem. 2004;93:807–18. doi: 10.1002/jcb.20217. [DOI] [PubMed] [Google Scholar]
- Feng W, Liu Y, Wu J, Nephew KP, Huang TH, Li L. A Poisson mixture model to identify changes in RNA polymerase II binding quantity using high-throughput sequencing technology. BMC Genomics. 2008;9(Suppl 2):S23. doi: 10.1186/1471-2164-9-S2-S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–60. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26:6453–68. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C, Zella LA, Fretz JA, Fu Q, Pike JW, Weinstein RS, Manolagas SC, O’Brien CA. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–53. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fargo DC, Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groyer A, Schweizer-Groyer G, Cadepond F, Mariller M, Baulieu EE. Antiglucocorticosteroid effects suggest why steroid hormone is required for receptors to bind DNA in vivo but not in vitro. Nature. 1987;328:624–6. doi: 10.1038/328624a0. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin M, Feldser D, Huarte M, Zuk O, Carey B, Cassady J, Cabili M, Jaenisch R, Mikkelsen T, Jacks T, Hacohen N, Bernstein B, Kellis M, Regev A, Rinn J, Lander E. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WH, Heaney RP. Skeletal renewal and metabolic bone disease. N Engl J Med. 1969;280:303–11. doi: 10.1056/NEJM196902062800605. concl. [DOI] [PubMed] [Google Scholar]
- Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96:3540–5. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimi E, Akiyama S, Tsurukai T, Okahashi N, Kobayashi K, Udagawa N, Nishihara T, Takahashi N, Suda T. Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J Immunol. 1999a;163:434–42. [PubMed] [Google Scholar]
- Jimi E, Nakamura I, Duong L, Ikebe T, Takahashi N, Rodan G, Suda T. Interleukin 1 induces multinucleation and bone-resorbing activity of osteoclasts in the absence of osteoblasts/stromal cells. Exp Cell Res. 1999b;247:84–93. doi: 10.1006/excr.1998.4320. [DOI] [PubMed] [Google Scholar]
- Jones J, Phatnani H, Haystead T, MacDonald J, Alam S, Greenleaf A. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–64. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–22. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005a;20:305–17. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde N, Pike J. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol. 2007a;21:197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Shevde NK, Pike JW. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol. 2007b;21:197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella L, Shevde N, Pike J. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006a;26:6469–86. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF- kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006b;26:6469–86. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–17. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Kitazawa R, Kitazawa S. Vitamin D(3) augments osteoclastogenesis via vitamin D-responsive element of mouse RANKL gene promoter. Biochem Biophys Res Commun. 2002;290:650–5. doi: 10.1006/bbrc.2001.6251. [DOI] [PubMed] [Google Scholar]
- Kitazawa S, Kajimoto K, Kondo T, Kitazawa R. Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J Cell Biochem. 2003;89:771–7. doi: 10.1002/jcb.10567. [DOI] [PubMed] [Google Scholar]
- Kondo H, Guo J, Bringhurst F. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J Bone Miner Res. 2002;17:1667–79. doi: 10.1359/jbmr.2002.17.9.1667. [DOI] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–23. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- Lin PS, Marshall NF, Dahmus ME. CTD phosphatase: role in RNA polymerase II cycling and the regulation of transcript elongation. Prog Nucleic Acid Res Mol Biol. 2002;72:333–65. doi: 10.1016/s0079-6603(02)72074-6. [DOI] [PubMed] [Google Scholar]
- Liu BY, Guo J, Lanske B, Divieti P, Kronenberg HM, Bringhurst FR. Conditionally immortalized murine bone marrow stromal cells mediate parathyroid hormone-dependent osteoclastogenesis in vitro. Endocrinology. 1998;139:1952–64. doi: 10.1210/endo.139.4.5895. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–83. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Mizzen C, Yang X, Kokubo T, Brownell J, Bannister A, Owen-Hughes T, Workman J, Wang L, Berger S, Kouzarides T, Nakatani Y, Allis C. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–70. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Mundy GR. Pathogenesis of osteoporosis and challenges for drug delivery. Adv Drug Deliv Rev. 2000;42:165–73. doi: 10.1016/s0169-409x(00)00060-0. [DOI] [PubMed] [Google Scholar]
- Nerenz RD, Martowicz ML, Pike JW. An Enhancer 20 kb Upstream of the Human Receptor Activator of NF-{kappa}B Ligand Gene Mediates Dominant Activation by 1,25-Dihydroxyvitamin D3. Mol Endocrinol. 2008 doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien CA, Kern B, Gubrij I, Karsenty G, Manolagas SC. Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone. 2002;30:453–62. doi: 10.1016/s8756-3282(01)00692-5. [DOI] [PubMed] [Google Scholar]
- Oberley MJ, Farnham PJ. Probing chromatin immunoprecipitates with CpG-island microarrays to identify genomic sites occupied by DNA-binding proteins. Methods Enzymol. 2003;371:577–96. doi: 10.1016/S0076-6879(03)71043-X. [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–60. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rinn J, Kertesz M, Wang J, Squazzo S, Xu X, Brugmann S, Goodnough L, Helms J, Farnham P, Segal E, Chang H. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccisana J, Kawanabe N, Kajiya H, Koide M, Roodman G, Reddy S. Functional role for heat shock factors in the transcriptional regulation of human RANK ligand gene expression in stromal/osteoblast cells. J Biol Chem. 2004;279:10500–7. doi: 10.1074/jbc.M303727200. [DOI] [PubMed] [Google Scholar]
- Romas E, Udagawa N, Zhou H, Tamura T, Saito M, Taga T, Hilton D, Suda T, Ng K, Martin T. The role of gp130-mediated signals in osteoclast development: regulation of interleukin 11 production by osteoblasts and distribution of its receptor in bone marrow cultures. J Exp Med. 1996;183:2581–91. doi: 10.1084/jem.183.6.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Matsuda M, Hou Z, Bailey JP, Kitazawa R, Herbst MP, Horseman ND. Receptor activator of NF-kappaB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J Biol Chem. 2003;278:46171–8. doi: 10.1074/jbc.M308545200. [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Udagawa N, Jimi E, Gillespie MT, Martin TJ. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr Rev. 1999;20:345–57. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Akatsu T, Sasaki T, Nicholson G, Moseley J, Martin T, Suda T. Induction of calcitonin receptors by 1 alpha, 25-dihydroxyvitamin D3 in osteoclast-like multinucleated cells formed from mouse bone marrow cells. Endocrinology. 1988a;123:1504–10. doi: 10.1210/endo-123-3-1504. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Akatsu T, Sasaki T, Nicholson GC, Moseley JM, Martin TJ, Suda T. Induction of calcitonin receptors by 1 alpha, 25-dihydroxyvitamin D3 in osteoclast-like multinucleated cells formed from mouse bone marrow cells. Endocrinology. 1988b;123:1504–10. doi: 10.1210/endo-123-3-1504. [DOI] [PubMed] [Google Scholar]
- Tamura T, Udagawa N, Takahashi N, Miyaura C, Tanaka S, Yamada Y, Koishihara Y, Ohsugi Y, Kumaki K, Taga T. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc Natl Acad Sci U S A. 1993;90:11924–8. doi: 10.1073/pnas.90.24.11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–8. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- Tominaga H, Maeda S, Hayashi M, Takeda S, Akira S, Komiya S, Nakamura T, Akiyama H, Imamura T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol Biol Cell. 2008;19:5373–86. doi: 10.1091/mbc.E08-03-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein C, Caelles C, van der Geer P, Hunter T, Karin M, Chojkier M. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature. 1993;364:544–7. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- Verrijzer C, Chen J, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–25. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- Wani M, Fuller K, Kim N, Choi Y, Chambers T. Prostaglandin E2 cooperates with TRANCE in osteoclast induction from hemopoietic precursors: synergistic activation of differentiation, cell spreading, and fusion. Endocrinology. 1999;140:1927–35. doi: 10.1210/endo.140.4.6647. [DOI] [PubMed] [Google Scholar]
- Wassarman D, Sauer F. TAF(II)250: a transcription toolbox. J Cell Sci. 2001;114:2895–902. doi: 10.1242/jcs.114.16.2895. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–14. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Shevde NK, Warrier A, Plum LA, DeLuca HF, Pike JW. 2-Methylene-19-nor-(20S)- 1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J Biol Chem. 2003;278:31756–65. doi: 10.1074/jbc.M304737200. [DOI] [PubMed] [Google Scholar]
- Zanello L, Norman A. 1alpha,25(OH)2 Vitamin D3 actions on ion channels in osteoblasts. Steroids. 2006;71:291–7. doi: 10.1016/j.steroids.2005.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.