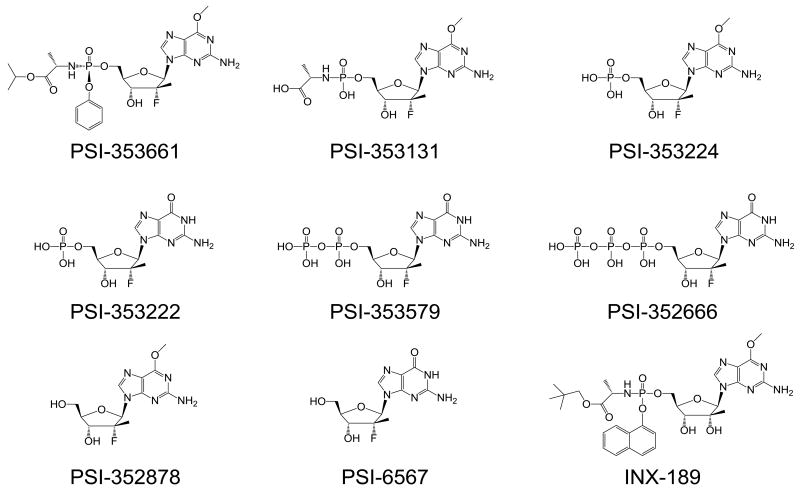
Abstract
PSI-353661, a phosphoramidate prodrug of 2′-deoxy-2′-fluoro-2′-C-methylguanosine-5′-monophosphate, is a highly active inhibitor of genotype 1a, 1b, and 2a HCV RNA replication in the replicon assay and of genotype 1a and 2a infectious virus replication. PSI-353661 is active against replicons harboring the NS5B S282T or S96T/N142T amino acid alterations that confer decreased susceptibility to nucleoside/tide analogs as well as mutations that confer resistance to non-nucleoside inhibitors of NS5B. Replicon clearance studies show that PSI-353661 was able to clear cells of HCV replicon RNA and prevent a rebound in replicon RNA. PSI-353661 showed no toxicity toward bone marrow stem cells or mitochondrial toxicity. The metabolism to the active 5′-triphosphate involves hydrolysis of the carboxyl ester by cathepsin A (Cat A) and carboxylesterase 1 (CES1) followed by a putative nucleophilic attack on the phosphorus by the carboxyl group resulting in the elimination of phenol and the alaninyl phosphate metabolite, PSI-353131. Histidine triad nucleotide-binding protein 1 (Hint 1) then removes the amino acid moiety, which is followed by hydrolysis of the methoxyl group at the O6-position of the guanine base by adenosine deaminase-like protein 1 (ADAL1) to give 2′-deoxy-2′-fluoro-2′-C-methylguanosine-5′-monophosphate. The monophosphate is phosphorylated to the diphosphate by guanylate kinase. Nucleoside diphosphate kinase is the primary enzyme involved in phosphorylation of the diphosphate to the active triphosphate, PSI-352666. PSI-352666 is equally active against wild-type NS5B and NS5B containing the S282T amino acid alteration.
Keywords: Hepatitis C virus, antiviral, nucleotide analog, prodrug, phosphoramidate, 2′ -deoxy-2′ -fluoro-2′ -C-methylguanosine-5′ -monophosphate
1. Introduction
Selective inhibitors of HCV replication that target the NS3 protease and the NS5B RNA-dependent RNA polymerase (RdRp) in particular have been pursued as potential new therapies for chronic HCV infection. Nucleoside and nucleotide inhibitors of HCV are a desirable approach to treating HCV infection because of their high barrier to resistance and pan-genotype activity (Herlihy et al., 2008; McCown et al., 2008). The active triphosphates of nucleoside analogue inhibitors of HCV replication act as non-obligate chain terminators of the polymerization process. Several nucleoside polymerase inhibitors have been in development. 2′-α-OH-2′-β-C-methylcytidine, the active component of valopicitabine, was the first polymerase inhibitor to enter clinical trials, but further development of this compound was terminated due to limited efficacy and side effects (Afdahl et al., 2004). R1626, the prodrug of 4′-azidocytidine (R1479), and RG7128, the diisobutyrate prodrug of 2′-deoxy-2′-α-fluoro-2′-β-C-methylcytidine (PSI-6130), were shown to have efficacy against HCV in clinical studies. However, the development of R1626 was stopped because of hematological side effects (Nelson et al., 2008). RG7128 is the most advanced anti-HCV nucleoside and is currently in Phase IIb clinical studies. We recently reported in vitro results for PSI-7851, a phosphoramidate prodrug of 2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine 5′-monophosphate (Lam et al., 2010). PSI-7851 was synthesized as a way of circumventing the lack of activity associated with 2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine resulting from the inability of the compound to be phosphorylated to the corresponding 5′-monophosphate (Murakami et al., 2008). PSI-7851 consists of a mixture of two diastereoisomers, PSI-7976 and PSI-7977. Both PSI-7851 and the purified isomers demonstrated potent, specific and broad genotypic anti-HCV activity in replicon assays and infectious virus assays with PSI-7977 being the more active of the two isomers (Lam et al., 2010; Sofia et al., 2010). PSI-7977 is currently being evaluated in Phase II clinical trials.
A number of 2′-deoxy-2′-α-fluoro-2′-β-C-methylpurine analogs, including 2′-deoxy-2′-α-fluoro-2′-β-C -methylguanosine (PSI-6567), have weak activity in the replicon assay. This lack of activity was most likely due to the failure of cellular enzymes to efficiently metabolize the compound to the corresponding 5′-triphosphate, a result of the inefficient phosphorylation of the nucleoside to the 5′-monophosphate. Therefore, phosphoramidate prodrug methodology was employed as an approach to bypass the non-productive first phosphorylation step and to intracellularly deliver 2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine 5′-monophosphate. Our subsequent work led to the identification of the phosphoramidate prodrug PSI-352879, (S)-2-{[(2R,3R,4R,5R)-5-(2-amino-6-methoxy-purin-9-yl)-4-fluoro-3-hydroxy-4-methyl-tetrahydro-furan-2-ylmethoxy]-phenoxy-phosphorylamino}-propionic acid isopropyl ester), which has potent activity in the HCV replicon assay (Chang et al., 2010).
PSI-352879 is a mixture of two diastereoisomers, of which PSI-353661 ((S)-2-{(S)- [(1R,4R,5R)-5-(2-Amino-6-methoxy-purin-9-yl)-4-(R)-fluoro-3-hydroxy-4-methyl-tetrahydro-furan-2-ylmethoxy]-phenoxy-phosphorylamino}-propionic acid isopropyl ester)) is the more active isomer in the replicon assay (Chang et al., 2010). Herein we describe the in vitro activity and metabolic pathway of the single isomer PSI-353661, a novel phosphoramidate prodrug of β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyl-6-methoxyguanosine-5′-monophosphate that has broad genotypic activity. Furthermore PSI-353661 remains active against HCV replicons containing the S282T mutation, a mutation that confers resistance to 2′-β-C-methyl-nucleoside/nucleotide analogs including PSI-6130, PSI-7851 and PSI-7977 (Lam et al., 2010; Sofia et al., 2010; Stuyver et al., 2006).
2. Materials and Methods
2.1. Compounds
The following compounds were synthesized at Pharmasset and their purity determined by proton NMR, MS and HPLC analysis: PSI-353661 ((S)-2-{(S)- [(1R,4R,5R)-5-(2-amino-6-methoxy-purin-9-yl)-4-(R)-fluoro-3-hydroxy-4-methyl-tetrahydro-furan-2-ylmethoxy]-phenoxy-phosphorylamino}-propionic acid isopropyl ester (98.9% pure), PSI-6567 (2′-F-2′-C-methylguanosine) (>99% pure), PSI-353131 ((S)-2-{[(2R,3R,4R,5R)-5-(2-amino-6-methoxy-purin-9-yl)-4-fluoro-3-hydroxy-4-methyl-tetrahydro-furan-2-ylmethoxy]-hydroxy-phosphorylamino}-propionic acid) (96.3% pure), PSI-353224 (2-amino-6-methoxypurine 2′-deoxy-2′-α-fluoro-2-β-C-methylribose-5′-monophosphate) (99.8% pure), PSI-353222 (2′-deoxy-2-α-fluoro-2-β-C-methylguanosine-5′-monophosphate) (99.3% pure), PSI-353579 (2′-deoxy-2′-α-fluoro-2′-β-C –methylguanosine-5′-diphosphate) (99.7% pure), PSI-352878 ((2R,3R,4R,5R)-5-(2-amino-6-methoxy-purin-9-yl)-4-fluoro-2-hydroxymethyl-4-methyl-tetrahydro-furan-3-ol) (>97% pure), R1479 (4′-azidocytidine) (>95% pure), INX-08189 (a phophoramidate prodrug of 2′-α-OH-2′-β-C–methylguanosine) (>99% pure), and the non-nucleoside NS5B inhibitors that include HCV-796 (5-cyclopropyl-2-(4-fluoro-phenyl)-6-[(2-hydroxy-ethyl)-methanesulfonyl-amino]-benzofuran-3-carboxylic acid methylamide) (>96% pure), a thiophene compound (3-((1r,4r)-N-isopropyl-4-methylcyclohexanecarboxamido)-5-phenylthiophene-2-carboxylic acid) (>99% pure), and a benzothiadiazine compound (N-{3-[4-Hydroxy-1-(3-methyl-butyl)-2-oxo-1,2-dihydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,4-dihydro-1λ6-benzo[1,2,4]thiadiazin-7-yl}-N-methyl-methanesulfonamide) (>99% pure). A 2-phenol indole compound, 5a-amino-12-cyclohexyl-N-(N,N-dimethylsulfamoyl)-3-methoxy-4b,5,5a,6-tetrahydrobenzo [3,4] cyclopropa [5,6] azepino [1,2-a] indole-9-carboxamide, was synthesized by WuXi Apptec (Shanghai, China). PSI-352666 (2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-triphosphate) was synthesized by NuBlocks (Vista, CA). [3H]-labeled PSI-353661 was synthesized by Moravek (Brea, CA) and [α-32P]-UTP was purchased from Perkin Elmer (Waltham, MA). Gemcitabine (2′-deoxy-2′-difluorocytidine) was purchased from Hetero Drugs Ltd (Hyderabad, India). Zalcitabine (2′,3′-dideoxycytidine, ddC) was purchased from RI Chemicals (Orange, CA). 3′-dCTP was purchased from Trilink Biotechnology (San Diego, CA). Telaprevir was synthesized by ACME Biosciences (Palo Alto, CA) and bis(4-nitrophenyl)phosphate (BNPP) was obtained from Sigma-Aldrich Corp.(St. Louis, MO).
2.2. Cells and Viruses
The Clone A HCV genotype 1b (Con1 strain: GenBank accession no. AJ238799.1) replicon cells (Apath LLC, Brooklyn, NY) were maintained in culture medium containing Dulbecco's Modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 4 mM L-glutamine, 0.1 mM non-essential amino acids (NEAA), 100 units/mL penicillin and 100 μg/mL streptomycin and 1 mg/mL G418 (Invitrogen, Carlsbad, CA). The genotype 1b derived ET replicon (Con1 strain) with adaptative mutations E1202G, T1280I and K1846T (Lohmann et al., 2003) and the Lunet cell line (Koutsoudakis et al., 2007) were kindly provided by Dr. R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). Plasmid DNA containing the genotype 1a replicon (H77 strain: NCBI reference NC_004102.1) with adaptive mutations P1496L and S2204I (Blight et al., 2003), and the genotype 2a J6/JFH-1 replicon were licensed from Apath. The J6/JFH-1 replicon encodes a partial core (first 19 amino acids) and 3′-nontranslated region (NTR) from the J6 strain (GenBank accession no. AF177036) and 5′-NTR and NS3 to NS5B region from the JFH-1 strain (GenBank accession no. AB047639). Plasmids containing ET, H77 and J6/JFH-1 replicon were linearized with ScaI, HpaI or XbaI, respectively. In vitro transcription and electroporation of the replicon RNA into Lunet cells was performed as described previously (Lam et al., 2010). Cells electroporated with the H77 replicon were selected in medium containing 0.75 mg/ml G418. Cells electroporated with the ET or J6/JFH-1 replicon were selected with medium containing 0.25 mg/ml G418. Huh7 and HepG2 cells (ATCC, Manassas, VA) were maintained in DMEM supplemented with 10% FBS, 4 mM L-glutamine, 0.1 mM NEAA, 100 units/mL penicillin and 100 μg/mL streptomycin. CEM and BxPC3 cells (ATCC) were maintained in RPMI-1640 (Invitrogen) supplemented with 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin.
2.3. HCV RNA replication and viral inhibition assays
HCV replicon assays using Clone A or ET-Lunet cells were performed as described previously (Stuyver et al., 2006). H77-Lunet and J6/JFH-1-Lunet cells were tested in a similar manner. Briefly, replicon-containing cells were seeded at a density of 1500 or 3000 cells per well in a 96-well plate and were incubated with serially diluted test compounds prepared in culture medium without G418 so that the final DMSO concentration was 0.5%. Plates were incubated at 37°C in a 5% CO2 atmosphere for 4 days. Inhibition of HCV RNA replication was determined by real time PCR (RT-PCR) (Stuyver et al., 2003) or by measuring the levels of luminescence expressed via the firefly luciferase (ET replicon) encoded within the replicon using the Bright-Glo luciferase reagent (Promega, Madison, WI). For the RT-PCR assay, total RNA was extracted using the RNeasy-96 kit as recommended by the Manufacturer (Qiagen, Valencia, CA), reversed transcribed into cDNA, and amplified using a primer and probe mix for HCV 5′-NTR RNA and human ribosomal RNA (rRNA) in a multiplex RT-PCR reaction as described previously (Stuyver et al., 2003). Primers and probes were designed for the HCV IRES region: sense primer 5′-AGC CAT GGC GTT AGT ATG AGT GT, antisense primer 5′-TTC CGC AGA CCA CTA TGG, and probes 5′-FAM-CCT CCA GGA CCC CCC CTC CC-TAM (GT 1b), 5′-FAM-CCT CCA GGC CCC CCC CTC CC-TAM (GT 2a).
Anti-HCV assays against the H77sV2 and JFH-1 infectious viruses were performed as previously described (Yi et al., 2006). Briefly, increasing concentrations of HCV inhibitors were added to En5-3 cells infected with the H77Sv2 or JFH-1 virus. Fresh compound-containing medium was replaced every day for three days, after which cells were fixed and incubated with a primary HCV-core mouse monoclonal IgG1 antigen (Affinity BioReagents, Golden, CO), which was reacted with a FITC-labeled secondary antibody to mouse IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD). Clusters of infected cells were considered to constitute a single infectious focus. Foci counts were analyzed with a Zeiss LSM 510 laser scanning confocal microscope and EC50 and EC90 values were calculated using Graphpad Prism software (San Diego, CA).
2.4. Cytotoxicity assays
Cytotoxicity and mitochondrial toxicity assays with PSI-353661 were performed as described previously (Stuyver et al., 2002). For the cytotoxicity assay, Huh7 (2 × 103 cells/well), HepG2 (2 × 103 cells/well), BxPC3 (2 × 103 cells/well), or CEM (5 × 103 cells/well) cells were incubated with either PSI-353661, INX-08189 or Gemcitabine, for 8 days at 37°C. At the end of the growth period, MTS dye from the CellTiter 96 Aqueous One Solution Cell Proliferation assay kit (Promega) was added to each well and the absorbance at 490 nm measured using a Victor3 plate reader (Perkin Elmer, Boston, MA).
To assess the effect of PSI-353661 and INX-08189 on mitochondrial DNA synthesis, both compounds and ddC were serially diluted from 100 μM in assay medium containing DMSO and added to HepG2 or CEM cells seeded at 1 × 104 cells/well in a 24-well plate. Cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 14 days, after which cells were harvested and total cellular DNA was extracted to perform a multiplex quantitative RT-PCR assay measuring the levels of the mitochondrial cytochrome C oxidase subunit II (COXII) gene and ribosomal DNA. The ΔCt of mitochondrial COXII DNA (mtDNA) and ΔCt of ribosomal DNA (rDNA) for each sample were determined as described previously (Stuyver et al., 2002). The fold difference in mitochondrial DNA normalized for ribosomal DNA relative to the no-drug control was calculated.
Lactic acid quantification was performed using the EnzyChromTM L-Lactate Assay Kit (Bioassay System, Hayward, CA). Following a seven day incubation at 37° C in the presence of various concentrations of PSI-353661, INX-08189, or ddC (0, 0.1 1, 10 and 50 μM), the level of lactic acid in each sample of cell culture medium was determined as described in the Manufacturer's instructions. The total amount of lactic acid produced and the level of rDNA for each sample was determined as a percent of the untreated control. The fold change in lactic acid production was calculated by dividing the percent of lactic acid by the percent of rDNA.
The human bone marrow stem cell toxicity assay was performed by Reachbio (Seattle, WA). Clonogenic progenitor cells of the erythroid (CFU-E and BFU-E) and myeloid (CFU-GM) lineages were set up in a methylcellulose-based medium. PSI-353661 was added to give final concentrations ranging from 0.5 to 50 μM. 5-Fluorouracil was used as a positive control at concentrations of 1, 0.1, and 0.01 μg/mL. The cultures were set up in triplicate at 2 × 104 cells per culture. Following 14 days of incubation the number of colonies was assessed.
2.5. Cross resistance studies
HCV Clone A replicon cells containing the S282T amino acid alteration in NS5B were established previously by selecting the cells in the presence of increasing concentrations of 2′-C-methyladenosine (Stuyver et al., 2006). The ET-S96T/N142T replicon was generated by site directed mutagenesis as previously described (Lam et al., 2010). Site directed mutagenesis was also used to generate the ET-C316Y, ET-M414T, ET-M423T, and ET-P495L mutations using the following primers and their complements: C316Y: 5′-CTG CAC GAT GCT CGT ATA CGG AGA CGA CCT TGT CG; M414T: 5′-CTA GGC AAC ATC ATC ACG TAT GCG CCC ACC, M423T: 5′-CCT TGT GGG CAA GGA CGA TCC TGA TGA CTC ATT TC, P495L: 5′-GGA AAC TTG GGG TAC TGC CCT TGC GAG TC (Integrated DNA Technologies, Coralville, IA). Cell lines containing each of these replicon mutants were generated by electroporation of the replicon RNA into Lunet cells and selecting for stable cells using 0.25 mg/ml G418. The anti-HCV activity of PSI-353661 against the Clone A wild type replicon and the replicon harboring the S282T amino acid alteration was evaluated using RT-PCR. The activity against replicons containing the S96T/N142T, C3316Y, M414T, M423T, or P495L amino acid alteration was evaluated using genotype 1b ET-replicon cells and a firefly luciferase-based assay.
2.6. HCV replicon clearance and rebound studies
ET-Lunet replicon cells were seeded at 1 × 105 cells per well in a 6-well plate in culture medium without G418. PSI-353661 was added to cells at 0, 2, 5, 10 and 20-fold over its EC50 value (4 nM) so that the final DMSO concentration was 0.5% in each well. When the cells reached approximately 90% confluency, they were passaged at ratios of 1:4 or 1:5 every 3 to 4 days for 2 weeks, and replenished with fresh medium containing the appropriate amount of PSI-353661. At each passage an aliquot of cells were harvested for RNA analysis. After 2 weeks, cells were passaged into culture medium containing 0.25 mg/mL G418 without inhibitor and cultured for an additional 2 weeks without passaging. At the end of the experiment, total RNA was extracted from all cell samples using the RNeasy-96 kit (Qiagen). Levels of HCV RNA and ribosomal RNA (rRNA) were determined using RT-PCR as described above. To determine relative log HCV RNA reduction, HCV RNA was normalized to rRNA by calculating ΔCt (HCV Ct – rRNA Ct). ΔΔCt was calculated by subtracting the average DMSO controls from each ΔCt value. The level of HCV RNA was calculated as log(2-ΔΔCt). Results were expressed as log HCV RNA change in the compound-treated cells compared to the HCV RNA level in DMSO treated control cells. The effect of compound treatment was also examined by colony formation. Cells were washed with PBS, fixed using 7% formaldehyde, and stained with 1.25% crystal violet at the end of the four weeks experiment.
2.7. PSI-353661 metabolism studies in primary human hepatocytes
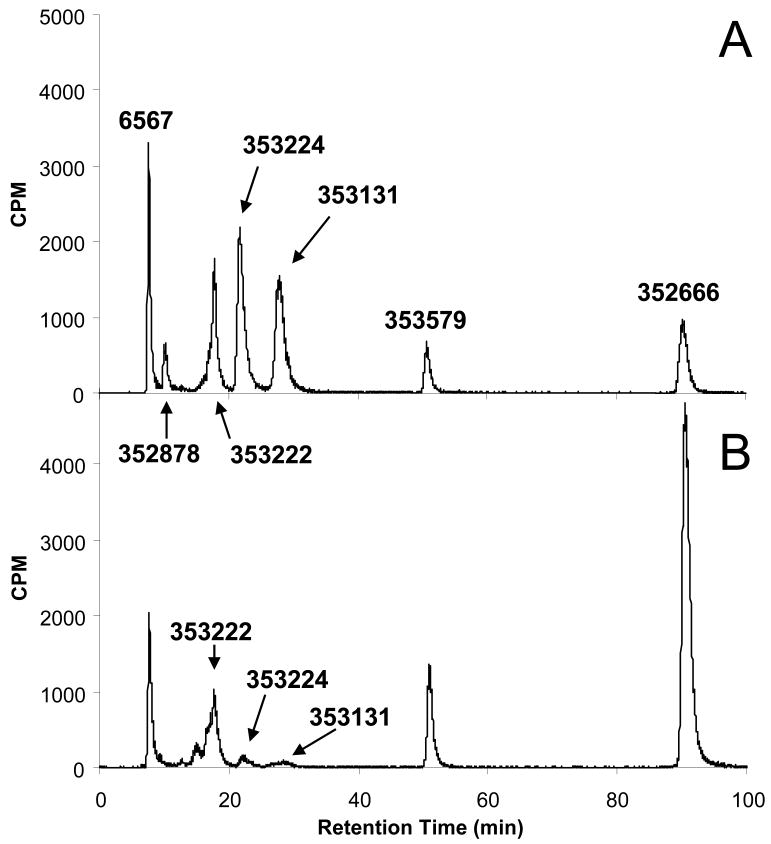
Primary human hepatocytes (CellzDirect, Inc, Durham, NC) were seeded in cell plating medium (CellzDirect, Inc) into 6-well plates at about 1 × 106 cells/well. After overnight incubation to allow the cells to attach, cells were incubated with [3H]-PSI-353661 in cell maintenance medium (CellzDirect, Inc) for up to 24 hours at 37°C in a 5% CO2 atmosphere. At selected times, the medium was removed and the cell layer was washed with cold PBS. After trypsinization, cells were counted and centrifuged at 1,200 rpm for 5 min. The cell pellets were suspended in 1mL of cold 60% methanol and incubated overnight at -20°C. The samples were centrifuged at 14,000 rpm for 5 min and the supernatants were collected and dried using a SpeedVac Concentrator (Thermo Electron Corporation), and stored at -20°C until they were analyzed by high performance liquid chromatography (HPLC). Residues were suspended in 100 μL of water, and 50 μL aliquots were injected onto the HPLC column. PSI-353661 and metabolites were separated by ion exchange HPLC with a Whatman 10 μm SAX column (Whatman, Maidstone, England) using a Series 200 HPLC system (PerkinElmer, Waltham, MA). The mobile phase consisted of buffer A (0.02 M KH2PO4, pH 3.5) and buffer B (1 M KH2PO4, pH 3.5). Elution was performed using a linear gradient of buffer B from 0 to 100% for 100 min. Radioactivity was analyzed using a 610TR Radiometric Flow Scintillation Analyzer (Perkin Elmer). PSI-353661 and its respective metabolites were identified based on the retention time of synthesized standards (Figure 1). The intracellular concentration (pmol/106 cells) of the metabolites was converted to μM based on a 3 μL cell volume per 106 cells for normal human liver parenchymal cells (Duarte et al., 1989).
Figure 1. Chemical structures of PSI-353661 and its metabolites.
2.8. Hydrolysis of the carboxyl ester of PSI-353661
Hydrolysis of the carboxyl ester of PSI-353661 was evaluated using two enzymes, human cathepsin A (Cat A) (R&D Systems, Minneapolis, MN) and human carboxylesterase 1 (CES-1) (R&D Systems). For the CatA assay the enzyme was activated following the Manufacturer's instructions. Briefly, CatA (10 μg/mL) was first activated by incubating with 1 μg/mL cathepsin L (R&D Systems) in 25 mM MES pH 6.0, ,5 mM DTT for 30 minutes at 37 °C. Cat L was then inactivated by adding 10 μM of the protease inhibitor E64. The hydrolysis of PSI-353661 by activated CatA was performed in reactions (100 μL) containing 100 μM PSI-353661 and 0.1 μg of CatA in 25 mM MES pH 6.0, 0.1 M NaCl, 1 mM DTT, and 0.1 % NP40. Hydrolysis of PSI-353661 by CES-1 was performed in reactions (100 μL) containing 100 μM PSI-353661 and 0.5 μg of CES-1 in 50 mM Tris/HCl pH 7.5. After incubation at 37 °C for 30, 60, 90, 120 and 150 minutes, the reaction mixture was applied to an Ultracell 10 filter (Millipore, Billerica, MA) to remove the protein. The flow-through from the filter was collected and analyzed by reverse phase HPLC using a Gemini 5 μm C18 column (Phenomenex, Torrance, CA). The mobile phase consisted of buffer A (water + 0.1% formic acid) and buffer B (acetonitrile + 0.1% formic acid). Elution was performed using a linear gradient of buffer B from 0 to 50% for 40 min. Amount of product formed was calculated based on the ratio of the peak areas of substrate and product, PSI-353131.
2.9. Hint1 studies
Hint1 was cloned, expressed and purified as described previously (Murakami et al., 2010). Hint1 reactions were performed at 37 °C in a reaction volume of 50 μL containing 50 mM HEPES pH 7.2, 1 mM MgCl2, 0.5 μM Hint1, and varied concentrations of PSI-353131 (0.2 mM to 8.0 mM). After incubating the Hint1 reaction for different times (1, 2, 3, 4, and 5 hours), the L-alanine dehydrogenase (Ala-DH) reaction was initiated by mixing with 150 μL of 67 mM sodium carbonate buffer pH 10.0 containing 1.33 mM β-NAD and 0.067 U Ala-DH. After mixing, the pH of the solution was 9.5 - 10. It was confirmed that at this pH Hint1 activity was significantly reduced (< 10 % remaining). The initial rate of β-NADH product formation was measured. Amount of L-alanine in the mixture was calculated based on the rate of the reaction and the steady-state kinetic parameters for L-alanine (Km and kcat),. Hint1 reaction rates were determined using varied concentrations of PSI-353131 based on the plots of the time dependent L-alanine formation. Steady-state kinetic parameters for PSI-353131 with Hint1 were determined by fitting the data to the Michaelis-Menten equation.
2.10. Human adenosine deaminase like protein1 studies
The cDNA fragment encoding ADAL1 was amplified from Huh7 cells using primers described by Schinkmanová et al. (Schinkmanova et al., 2008): 5′-ATG ATA GAG GCA GAA GAG CAA CAG CCT TGC-3′ and 5′-TTA AAT ATG TAA CAC TCT GGG CTT CAG GTG-3′. The amplified PCR product was confirmed by sequencing, cloned into the pET28a+ vector (Novagen, La Jolla, CA) and transformed into BL21-gold (DE3) cells (Stratagene, La Jolla, CA) for expression. ADAL1 was purified using a 1 mL Ni-affinity column (GE Healthcare, Piscataway, NJ) and eluted using a linear gradient (20 mM to 400 mM) of imidazole in buffer A, followed by a MonoQ 5/50 GL column (1 mL) (GE Healthcare) using a linear gradient of 0 to 1 M NaCl in 20 mM Tris/HCl pH 8.0, 10 % glycerol, and 1 mM DTT. The fractions containing ADAL1 were identified by activity assay and SDS-PAGE analysis, which showed >95 % purity. Protein concentration was determined based on the extinction coefficient (30,160 M-1cm-1) and the molecular weight of 40,263 Da using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE).
The ADAL1 assay was conducted in a 1 mL reaction mixture containing 50 mM potassium phosphate buffer pH 6.7, 2 mM DTT, 100 μg/mL BSA, and varying concentrations of the test compound. The reactions were started by addition of the appropriate amount of ADAL1 and incubated at 37°C in a Lambda 35 UV-vis spectrophotometer (Perkin Elmer). The reaction was followed by measuring the UV absorbance change at 258 nm for PSI-353224 (Δε = 6,110 M-1cm-1) and at 262 nm for O6-methyl-GMP (Δε = 4,438 M-1cm-1) and O6-methyl-dGMP (Δε = 4,791 M-1cm-1).
2.11. Phosphorylation of 2′-deoxy-2′-α-2′ -fluoro-2′-β -C-methylguanosine 5′-monophosphate (PSI-353222) by human guanylate kinase
Human guanylate kinase (hGUK1) was cloned from hGUK1 full-length cDNA obtained from Open Biosystems (Huntsville, AL) using a forward primer with a Nde I restriction site: 5′-CAG CCA TAT GTC GGG CCC CAG GCC TGT GGT GC-3′, and a reverse primer with a Hind III site: 5′- CCG CAA GCT TCA GGC GCC GGT CCT TTG AGC-3′. The PCR product was digested and cloned into the pET28a+ vector (Novagen), expressed and purified by affinity chromatography as described above for ADAL1. The purified protein showed >95 % purity based on SDS-PAGE analysis. hGUK1 concentration was measured using a spectrophotometer and calculated using an extinction coefficient of 7,450 M-1cm-1 and the molecular weight (21.73 kDa). The purified protein was dialyzed against Buffer B (20 mM Tris/HCl pH 8.0, 150 mM NaCl, 10 % glycerol, and 3 mM DTT) using a D-Tube Dialyzer Midi, MWCO 6–8 kDa (Novagen) overnight at 4 °C.
Phosphorylation of PSI-353222 by hGUK1 was studied spectrophotometrically as described previously (Murakami et al., 2007). The reaction was coupled with pyruvate kinase (PK) (Sigma-Aldrich) and lactate dehydrogenase (LDH) (Sigma-Aldrich). Oxidation of NADH (Sigma-Aldrich) was followed at 340 nm using a Lambda 35 UV/VIS Spectrometer (Perkin Elmer). All assays were performed at 37°C in a 1 mL reaction containing 64 mM Tris-HCl pH 7.5, 3.8 mM EDTA, 180 mM KCl, 12.8 mM MgCl2, 24 mM (NH4)2SO4, 1 mM ATP, 0.5 mM phosphoenolpyruvate, 0.1 mM NADH, 5 IU/ml PK, 13.8 IU/ml LDH, nucleoside substrate, and purified hGUK1. The final enzyme concentration was 610 nM in reactions containing PSI-353222 and PSI-353221 as substrate and was 6.1nM for GMP reactions. The reaction rate at different concentrations of the nucleotide substrate was determined and steady-state parameters were using GraphFit program version 5 (Erithacus Software, Horley, Surrey, UK).
2.12. Phosphorylation of 2′-deoxy-2′-α-2′ -fluoro-2′-β -C-methylguanosine-5′-diphosphate (PSI-353579)
Human nucleoside diphosphate kinase (NDPK) was cloned, expressed, and purified as described previously (Murakami et al., 2007). The phosphorylation of GDP and PSI-353579 by NDPK was studied spectrophotometrically using a coupled reaction as described above for hGUK1. The reaction was performed at 25 °C and the final concentration of NDPK in the reaction was 34.6 nM.
The phosphorylation of PSI-353579 by pyruvate kinase (PK) was studied spectrophotometrically. The reaction was followed at 340 nm using a Lambda 35 UV/VIS Spectrometer (Perkin Elmer). All assays were performed at 25 °C in a 1 mL reaction mixture containing 50 mM imidazole, pH 7.5, 100 mM KCl, 60 mM MgSO4, 0.2 mM NADH, 1.5 mM phosphoenol pyruvate (PEP) and 5 U/ml of LDH, PSI-353579 and rabbit muscle PK. The final PK concentration in the reaction was 21 nM.
3-phosphoglycerate kinase (3-PGK) from bakers yeast was purchased from Sigma-Aldrich. The 3-PGK activity was measured using coupled reactions with a buffer containing 50 mM Tris-acetate (pH 7.5), 5 mM MgCl2, 1 mM NaF, 1mM DTT, 10 mM sodium phosphate, 4 mM β-NAD, and 4 mM D,L-glyceraldehyde-3-phosphate as described previously (Krishnan et al., 2002). Prior to starting the reaction, phosphate donor (1.3-biphosphoglycerate) for the reaction was generated by preincubation with 5 units/ml of GAPDH (Sigma-Aldrich) for 20 min at 25 °C. The 3-PGK reaction was initiated by adding PSI-353579 (50, 200, and 500 μM) and 0.5 units/mL of 3-PGK and was incubated at 37 °C. After 0, 2, 5, 10, 20, and 40 min, a 100 μL aliquot was taken and the reaction was stopped on dry ice. The samples were thawed and filtered through 3K Amicon Ultra-0.5 mL centrifugal filters (Millipore) at 4 °C. Phosphorylation of PSI-353579 was analyzed by HPLC (Perkin Elmer).
The reaction rate at different concentrations of the nucleotide substrate was determined and the steady-state parameters were determined with GraphFit. Since the NDPK reaction was coupled with the PK reaction, in order to determine NDPK-mediated phosphorylation rates, the rate for the PK reaction was determined at each concentration of substrate and subtracted from the rate of the NDPK-coupled reaction.
2.13. NS5B polymerase assay
IC50 values were determined for PSI-352666 or for 2′-C-methylguanosine 5′-triphosphate using recombinant HCV NS5B from wild-type or S282T genotype 1b NS5B (Con1 strain). Reactions were performed in a 20 μL mixture containing varying concentrations of the test compound, 5 μM of the four natural ribonucleotides, [α-32P]UTP, 20 ng/μL of genotype 1b (-) IRES RNA template, 1 unit/μL of SUPERase•In (Ambion, Austin, TX), 40 ng/μL of NS5B, 1 mM MgCl2, 0.75 mM MnCl2, and 2 mM DTT in 50 mM HEPES buffer (pH 7.5). The reaction was quenched by adding 80 μL of stop solution (12.5 mM EDTA, 2.25 M NaCl, and 225 mM sodium citrate) after incubating at 27 °C for 30 minutes. The radioactive RNA products were separated from unreacted substrates by passing the quenched reaction mixture through a Hybond N+ membrane (GE Healthcare) using a dot-blot apparatus. The RNA products were retained on the membrane and the free nucleotides were removed by washing the membrane four times with a solution containing 0.6 M NaCl and 60 mM sodium citrate. After rinsing the membrane with water followed by ethanol, the membrane was exposed to a phosphorscreen and the products were visualized and quantified using a phosphorimager. The IC50 values were calculated using GraphFit. All the reactions were performed in triplicate and the results were reported as IC50 ± standard deviation (S.D.). PSI-352666 was also tested for activity against recombinant NS5B from genotypes 2a, 3a, and 4a. The genotype 1b (Con1 strain) enzyme and 3a and 4a enzymes (cloned from genotype 3a or 4a HCV human serum samples obtained from SeraCare Life Sciences, Milford, MA) were prepared internally at Pharmasset as previously described (Lam et al., 2010). Genotype 2a enzyme (JFH-1 strain) was kindly provided by Dr. David Frick and Dr. Julie Heck at New York Medical College (Valhalla, NY). The experiments were essentially the same as described above except that the reactions contained 1 μM of the four natural ribonucleotides and the reaction times were 20 minutes for genotype 1b, 2 hours for genotype 2a, and 3 hours for genotypes 3a and 4a NS5B.
3. Results
3.1. Inhibition of HCV replicon RNA and infectious viral replication
PSI-6567 (2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine) (Figure 1) was shown previously to be a poor inhibitor of HCV replicon RNA replication (Clark et al., 2006). The reduced activity of PSI-6567 was subsequently demonstrated to be the result of the inability of cellular kinases to efficiently phosphorylate the compound to the 5′-monophosphate (Murakami, E., unpublished data). Therefore the phosphoramidate prodrug, PSI-353661 (Figure 1), of the 2′-α-F-2′-β-C-methylguanosine-5′-monophosphate was synthesized as an approach to bypassing this non-productive phosphorylation step. We compared the anti-HCV activity of PSI-353661 to that of PSI-6567 and of the recently described pro-drug of 2′-α-OH-2′-β-C-methylguanosine 5′-monophosphate (INX-08189, Figure 1) (McGuigan et al., 2010) (Table 1). When incubated for 4 days (96 h) with genotype 1b replicon cells, the EC50 and EC90 values for PSI-353661 were 0.0030 ± 0.0014 μM and 0.0085 ± 0.0007 μM, respectively, whereas the EC50 and EC90 values for PSI-6567, the parent nucleoside, were 22.0 ± 7.7 μM and 69.2 ± 19.3 μM, respectively. The EC50 and EC90 values for INX-08189 were similar to those of PSI-353661. PSI-353661 also demonstrated potent activity against a genotype 1a H77 subgenomic replicon and a genotype 2a J6/JFH-1 subgenomic replicon (Table 1). In addition, the compound showed potent anti-HCV activity in the HCV 1a H77Sv2 and HCV 2a JFH-1 infectious virus assays (Table 2).
Table 1. Activity of PSI-353661, PSI-6567 and INX-08189 against genotype 1a, 1b, and 2a replicons.
| Replicon | PSI-353661 | PSI-6567 | INX-08189 | |||
|---|---|---|---|---|---|---|
| EC50, nM | EC90, nM | EC50, μM | EC90, μM | EC50, nM | EC90, nM | |
| GT1a_H77 | 4.5 ± 0.7 | 10.5 ± 0.7 | 19.0 ± 8.6 | 53.4 ± 12.8 | 1.4 ± 0.6 | 4.0 ± 0.7 |
| GT1b_Con1 | 3.0 ± 1.4 | 8.5 ± 0.7 | 22.0 ± 7.7 | 69.2 ± 19.3 | 2.6 ± 0.5 | 4.6 ± 3.2 |
| GT2a_JFH-1 | 6.5 ± 2.1 | 14.0 ± 1.4 | 38.3 ± 18.6 | 60.0 ± 14.8 | 7.3 ± 2.9 | 12.8 ± 3.5 |
Cells were treated with PSI-353661, PSI-6567, or INX-08189 for four days prior to determining HCV inhibition. Quantitative real time PCR or a luciferase-based reporter assay was used to quantify levels of HCV inhibition. Values are reported as the mean ± S.D. from at least three independent experiments performed in duplicate.
Table 2.
Activity of PSI-353661 against genotype 1a and 2a infectious virus.
| Infectious virus | EC50 (nM) | EC90 (nM) |
|---|---|---|
| GT1a_H77 | 3.5 | 8.6 |
| GT2a_JFH-1 | 1.6 | 11.2 |
Values are reported as the average of duplicate experiments. Infectious foci were determined using a primary HCV-core mouse monoclonal antibody reacted with a FITC-labeled secondary antibody.
3.2. In vitro safety profile of PSI-353661
PSI-353661 and INX-08189 were evaluated for cytotoxicity in an 8-day assay using four different human derived cell lines: Huh7 (human hepatoma), HepG2 (human hepatoma), CEM (human T lymphocyte), and BxPC3 (human pancreatic) cells. Gemcitabine was included as a positive control for cytotoxicity (Table 3). The results showed no measurable cytotoxicity for PSI-353661 toward HepG2, BxPC-3, or CEM cells at 100 μM, the highest concentration tested (CC50 >100 μM). The CC50 value determined for PSI-353661 with Huh 7 cells was 80.0 ± 6.0 μM. In contrast, INX-08189 was found to be significantly more cytotoxic than PSI-353661 with CC50 values of 0.32 ± 0.03, 2.23 ± 0.10, 2.81 ± 1.52, and 15.0 ± 2.0 μM in Huh7, HepG2, BxPC3, and CEM cells, respectively.
Table 3. Cytotoxicity of PSI-353661 and INX-08189.
| Compounds | CC50 (μM) | |||
|---|---|---|---|---|
| Huh7 | HepG2 | BxPC3 | CEM | |
| PSI-353661 | 80.0 ± 6.0 | >100 | >100 | >100 |
| INX-08189 | 0.32 ± 0.03 | 2.23 ± 0.1 | 2.81 ± 1.52 | 15.0 ± 2.0 |
| Gemcitabine | <1 | <1 | <1 | <1 |
Cells were incubated with PSI-353661 or INX-08189 (both up to 100 μM) for eight days prior to determining the effect of these compounds in cell viability using a MTS assay. Gemcitabine (1 μM) was included as positive control. Values are reported as the mean ± S.D. from at least three independent experiments performed in triplicate.
Mitochondrial toxicity has been associated with long-term use of some nucleoside analogs (Fleischer and Lok, 2009; Lewis et al., 2003; Moyle, 2000; Tanji et al., 2001). As a measure of mitochondrial toxicity we assessed the effect of PSI-353661 and INX-08189 on mitochondrial DNA content and lactic acid production (Table 4). Exposing CEM and HepG2 cells to PSI-353661 at concentrations up to 100 μM, the highest concentration tested, for 14 days did not affect mitochondrial DNA content. Because of the cytotoxicity associated with INX-08189 (as measured by reduction in rDNA), it was difficult to determine if the compound affected mitochondrial DNA synthesis. Dideoxycytidine (ddC), the positive control for mitochondrial toxicity, reduced mitochondrial DNA levels in CEM and HepG2 cells by more than one log10 at the lowest concentration tested (0.5 μM).
Table 4. Mitochondrial toxicity of PSI-353661 and INX-08189 in CEM and HepG2 cells.
| CC90 (μM)a | Lactic acidb (Fold change from untreated cells) | |||||
|---|---|---|---|---|---|---|
| HepG2 cells | mtDNA | rDNA | 0.1 μM | 1.0 μM | 10 μM | 50 μM |
| PSI-353661 | >100 | >100 | 1.2 ± 0.35 | 1.2 ± 0.29 | 1.1 ± 0.06 | 1.3 ± 0.31 |
| INX-08189 | 0.97 ± 0.62 | 0.75 ± 0.27 | 1.1 ± 0.21 | 3.1 ± 0.61 | Toxic | Toxic. |
| ddC | <0.5 | 12.9 ± 9.1 | 1.6 ± 0.68 | 1.4 ± 0.12 | 3.9 ± 0.93 | Toxic. |
| CEM cells | ||||||
| PSI-353661 | >100 | >100 | 1.4 ± 0.36 | 1.4 ± 0.10 | 1.4 ± 0.31 | 1.3 ± 0.40 |
| INX-08189 | 16.9 ± 1.7 | 16.3 ± 4.1 | 1.4 ± 0.35 | 1.4 ± 0.69 | 1.5 ± 0.50 | Toxic. |
| ddC | <0.5 | 22.9 ± 11.9 | 0.9 ± 0.3 | 2.2 ± 0.81 | 4.2 ±1.0 | Toxic |
Cells were incubated with PSI-353661 (up to 100 μM) or INX-08189 (up to 50 μM) for fourteen days prior to determination of mitochondria COXII DNA (mtDNA) and ribosomal DNA (rDNA) using real time PCR. ddC (up to 100 μM) was included as positive control.
Lactic acid levels were measure after 7-days using a colorimetric assay. The effect on lactic acid values are expressed as the ratio of percent change in lactic acid divided by the percent change in rDNA in order to account for the toxicity of the compounds. The fold change is expressed as the mean ± S.D. of the ratio of treated to untreated from at least three independent experiments performed in triplicate.
To test the effect of PSI-353661 on mitochondrial function, the level of lactic acid in the cell culture medium was quantified. When CEM and HepG2 cells were incubated with PSI-353661 for seven days no increase in lactic acid was seen at any of the concentrations tested (Table 4). A 3.1 ± 0.61 fold increase in lactic acid was seen when HepG2 cells were incubated with 1.0 μM INX-08189 (Table 4). No increase in lactic acid was observed when CEM cells were incubated with up to 10 μM INX-08189. An increase in lactic acid was seen when CEM and HepG2 cells were incubated with ddC at 1 and 10 μM (CEM cells) and 10 μM (HepG2 cells) (Table 4). Because of the cytotoxicity associated with INX-08189 and ddC (as measured by reduction in rDNA), it was not possible to accurately quantify the level of lactic acid at the higher concentrations.
The effect of PSI-353661 on the proliferation of human erythroid and myeloid progenitor cells was evaluated using a 14-day colony formation assay. An IC50 of >50 μM was calculated for the erythroid progenitor cells (CFU-E and BFU-E) and for myeloid progenitor cells (CFU-GM) the IC50 was calculated to be 45 ± 2 μM, suggesting that PSI-353661 was not significantly toxic toward bone marrow progenitor cells. At 1 μg/mL (4 μM) the positive control, 5-fluorouracil, inhibited CFU-E progenitor cell colony formation by >80 % and completely inhibited BFU-E and CFU-GM colony formation.
3.3. Cross resistance studies
PSI-353661 was assessed for activity against replicons harboring the NS5B S282T or S96T/N142T amino acid alterations that confer decreased susceptibility to nucleoside/tide analogs. The S96T/N142T amino acid alterations confer resistance to 4′-azidocytidine (R1479) (Le Pogam et al., 2006a), while the S282T amino acid alteration confers resistance to various 2′-β-C-methyl-modified nucleoside analogs as well as the pyrimidine analogs PSI-6130, PSI-7851, and PSI-7977, which have a 2′-α-fluoro-2′-β-C-methyl substitution on the ribose (Lam et al., 2010; Sofia et al., 2010; Stuyver et al., 2006). As shown in Table 5, neither the S96T/N142T amino acid alterations nor the S282T amino acid alteration conferred cross resistance to PSI-353661. The EC90 value for the wild-type replicon and the S96T/N142T replicon were 0.012 ± 0.004 μM and 0.009 ± 0.004 μM, respectively. As expected, the S96T/N142T replicon cells were less susceptible to 4′-azidocytidine compared to cells containing the wild-type replicon. The S282T replicon also remained as sensitive to inhibition by PSI-353661 (EC90 = 0.014 ± 0.003 μM) as wild-type (EC90 = 0.009 ± 0.004 μM). The S282T replicon was resistant to PSI-7851 and INX-08189.
Table 5.
Activity of PSI-353661 and other nucleoside or non-nucleoside analogs against replicons containing mutations that confer resistance to NS5B inhibitors. Values are reported as the mean ± S.D. from at least three independent experiments performed in duplicate.
| NS5B resistant mutations | HCV inhibitors | EC90 (μM) | EC90 fold change a | |
|---|---|---|---|---|
| WT replicon | Mutant replicon | |||
| S96T/ N142Tb | R1479 | 15.76 ± 3.67 | 74.12 ± 12.12 | 4.7 |
| PSI-7851 | 0.43 ± 0.23 | 0.39 ± 0.19 | 0.9 | |
| PSI-353661 | 0.012 ± 0.0040 | 0.0087 ± 0.0037 | 0.7 | |
| INX-08189 | 0.0054 ± 0.0036 | 0.0059 ± 0.0027 | 1.1 | |
|
| ||||
| S282Tc | R1479 | 22.35 ± 4.58 | 12.53 ± 3.78 | 0.6 |
| PSI-7851 | 0.44 ± 0.21 | 7.36 ± 1.02 | 16.7 | |
| PSI-353661 | 0.0093 ± 0.0041 | 0.014 ± 0.0026 | 1.5 | |
| INX-08189 | 0.018 ± 0.001 | 0.11 ± 0.004 | 6.1 | |
|
| ||||
| C316Yb | HCV-796 | 0.026 ± 0.017 | 1.92 ± 0.77 | 73.9 |
| PSI-353661 | 0.012 ± 0.0040 | 0.0098 ± 0.0035 | 0.8 | |
|
| ||||
| M414Tb | Benzothiadiazine | 1.23 ± 0.55 | 28.39 ± 13.70 | 23.1 |
| PSI-353661 | 0.012 ± 0.0040 | 0.012 ± 0.0022 | 1.0 | |
|
| ||||
| M423Tb | Thiophene | 3.65 ± 0.21 | 69.52 ± 9.24 | 19.1 |
| PSI-353661 | 0.012 ± 0.0040 | 0.0097 ± 0.0019 | 0.8 | |
|
| ||||
| P495Lb | 2-phenol indole | 1.17 ± 0.46 | 60.75 ± 19.90 | 51.9 |
| PSI-353661 | 0.012 ± 0.0040 | 0.012 ± 0.00034 | 1.0 | |
The fold change in EC90, the concentration at which 90% inhibition occurred, was calculated by normalizing the EC90 value in the mutant replicon with that of the wild-type.
Activity was determined by luciferase-based assay.
Activity was determined by RT-PCR assay.
We also tested PSI-353661 against replicons containing amino acid alterations that confer resistance to non-nucleoside inhibitors of NS5B: these included C316Y, which confers resistance to HCV-796; M414T, which confers resistance to benzothiadiazine compounds; M423T, which confers resistance to thiophene compounds; and P495L, which confers resistance to 2-phenol indol compounds (Howe et al., 2008; Le Pogam et al., 2006a; Le Pogam et al., 2006b; Nguyen et al., 2003; Tomei et al., 2003). Results showed that PSI-353661 remained fully active against HCV replicons harboring any one of these resistant mutations, which conferred various levels of resistance towards each of the corresponding reference compounds (Table 5).
3.4. PSI 353661 clears HCV replicon RNA and prevents viral rebound
Using the replicon system, PSI-353661 was evaluated for its ability to clear HCV replicon RNA from ET-Lunet cells. Replicon cells were incubated with PSI-353661, at approximately 2, 5, 10, and 20 times its EC50 value (4 nM), for 14-days in the absence of G418 and then for another 14-days in the presence of G418 and the absence of compound. Control cells incubated in the absence of PSI-353661 maintained a stable level of replicon RNA throughout the course of the experiment (Figure 2A). By the end of the first 14-days, a 1-log reduction of HCV replicon RNA was observed at 8 nM PSI-353661 (2× EC50) and a > 4-log decrease in HCV replicon RNA at the 20 nM dose. By day 10 HCV RNA was undetectable at the 40 and 80 nM doses (Figure 2A).
Figure 2. Rebound and clearance assay.

Replicon cells were treated with PSI-353661 for 2 weeks in the absence of G418. Fresh compound was added at every passage. After 2 weeks, cells were passaged into culture medium containing 0.25 mg/mL G418 without inhibitor and cultured for an additional 2 weeks without passaging. An aliquot of cells was harvested at each passage for replicon RNA analysis. Remaining cells were stained with crystal violet at the end of the experiment. (A) Effect of PSI-353661 on the level of HCV replicon RNA in the presence of 0 (□), 8 nM (▲), 20 nM (■), 40 nM (◆) and 80 nM (▲) PSI-353661. Values for the level of HCV RNA are the average of two separate assays. (B) Colony formation in the presence of PSI-353661.
To determine whether PSI-353661 had cleared the HCV replicon after 14-days of treatment, the compound was withdrawn and the cells were incubated in the presence of G418 for an additional 14-days. Cells that contained replicon RNA grew in the presence of G418 whereas cells that were cleared of the replicon did not survive G418 treatment. As can be seen in Figure 2B, partial clearance of replicon RNA was achieved with 20 nM PSI-353661 whereas 40 and 80 nM PSI-353661 completely eliminated the replicon, which resulted in the clearance of cells from those cultures and prevented rebound in HCV replicon RNA.
3.5. Metabolic profile of PSI-353661 in primary human hepatocytes
Cellular extracts from primary human hepatocytes incubated with 5 μM [3H]-PSI-353661 were analyzed by ion exchange HPLC and the metabolites identified by comparing the retention time of the radiolabeled metabolites to the retention time of known, chemically synthesized standards (Figure 1). As shown in Figure 3A, incubating primary human hepatocytes for 30 minutes with 5 μM [3H]-PSI-353661 led to the formation of the following intracellular metabolites: PSI-6567, PSI-352878, PSI-353131, PSI-353224, PSI-353222, PSI-353579 and PSI-352666. After 4 hours incubation, the major metabolites were PSI-6567, PSI-353222, PSI-353579 and PSI-352666 with PSI-352666 being the predominant metabolite (Figure 3B)
Figure 3. Identification of metabolites.
Primary human hepatocytes were incubated with 5 μM [3H]-labeled PSI-353661 for 30 minutes (A) or 4 hours (B) and formation of the intracellular metabolites were monitored by an anion exchange HPLC chromatography.
A time course experiment was performed to assess the effect of time on the metabolism of PSI-353661 during continuous exposure of primary human hepatocytes to the compound (Figure 4A and 4B). Throughout the course of the experiment, PSI-353661 was undetectable. The predominant metabolite formed when incubating PSI-353661 with primary human hepatocytes was the 5′-triphosphate, PSI-352666. PSI-352666 reached its maximum concentration at about 4 hours and then declined. The monophosphate, PSI-353222, and the diphosphate, PSI-353579, appeared to have reached their maximum concentration after about 1-2 hours of exposure. PSI-353131 reached its maximum concentration after 0.25 hours and declined rapidly thereafter. The concentration of the 6-methoxypurine monophosphate metabolite, PSI-353224, declined rapidly after reaching its maximum concentration after 0.5-1 hour of exposure. Both nucleoside analogs, PSI-352878 and PSI-6567, reached peak concentration after about 0.5-1h of exposure.
Figure 4.
Metabolic profile of PSI-353661. Time course of formation of PSI-353661 metabolites was monitored in primary human hepatocytes treated with 5 μM [3H]-labeled PSI-353661 up to 24 hours (A). The earlier time points were shown by plotting the same graph up to 4 hours (B). Formation of the metabolites was followed in primary hepatocytes treated with varying concentrations of exogenous PSI-353661 for 4 hours and the amount on intracellular metabolites were plotted against concentration of exogenously added PSI-353661 (C).
In order to determine whether increasing the exposure of hepatocytes to higher concentrations of PSI-353661 would lead to increased formation of PSI-352666, primary human hepatocytes were incubated with PSI-353661 at different concentrations up to a maximal concentration of 100 μM for 4 hr. PSI-353222, PSI-353579, and PSI-352666 increased with increasing concentrations of exogenous PSI-353661, but the rate of increase declined when the extracellular concentration of PSI-353661 was higher than 10 μM (Figure 4C). However, the concentration of PSI-6567, PSI-353131 and PSI-353224 appeared to continue to increase as the extracellular concentration of PSI-353661 increased.
3.6. Identification of enzymes involved in metabolism of PSI-353661 to 2′-fluoro-2′-C-methylguanosine-5′-triphosphate
For PSI-353661 to be metabolized to PSI-352666 (2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-triphosphate), it must first be converted to 2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-monophosphate (PSI-353222), which involves a 4-step process. The first step is hydrolysis of the carboxyester by cellular enzymes. The second step is facilitated by the free carboxyl group performing a putative nucleophilic attack on the phosphorous releasing the phenol to produce the intermediate metabolite PSI-353131. The third step is removal of the amino acid moiety to form PSI-353224. The last step requires demethylation at the O6 position of PSI-353224 to give PSI-353222.
We have previously demonstrated that the hydrolysis of the carboxylester linkage between the carboxylic acid and the alaninyl moiety isopropyl group of the phosphoramidate prodrug PSI-7977 is hydrolyzed by carboxylesterase 1 (CES1) and cathepsin A (Cat A) (Murakami et al., 2010). Here we demonstrate that PSI-353661 was hydrolyzed to PSI-353131 by CatA in a time-dependent manner with a specific activity of 0.80 μmole min-1mg-1 (Figure 5A). CES1 also hydrolyzed PSI-353661 but at a lower rate (estimated to be 0.034 μmole min-1mg-1) and the reaction did not progress in a time-dependent manner (Figure 5A).
Figure 5. Hydrolysis of PSI-353661 by CatA and CES1.
CatA and CES1 reactions were performed in the presence of 100 μM PSI-353661 (A). Error bars represent standard deviation from three independent experiments. Metabolism of PSI-353661 was studied in primary human hepatocytes from different donors by following formation of the triphosphate, PSI-352666 in the presence of a CatA inhibitor, telaprevir, (Panel B; Donors A,B, and C) or a CES1 inhibitor, BNPP (Panel C; Donors D, E, and F).
To further substantiate that CatA and CES-1 were involved in the metabolism of PSI-353661, we studied the effect of telaprevir, an inhibitor of CatA and BNPP, a known inhibitor of CES-1 (Murakami et al., 2010), on the metabolism of PSI-353661 in primary human hepatocytes by following formation of the triphosphate, PSI-352666, in the presence of these inhibitors (Figure 5B and C). BNPP inhibited the formation of PSI-352666 in hepatocytes from three different donors in a dose-dependent manner. However, as was observed with the phosphoramidate prodrug PSI-7977, the metabolism of PSI-353661 was not inhibited by telaprevir in hepatocytes from two of the three donors (Murakami et al., 2010).
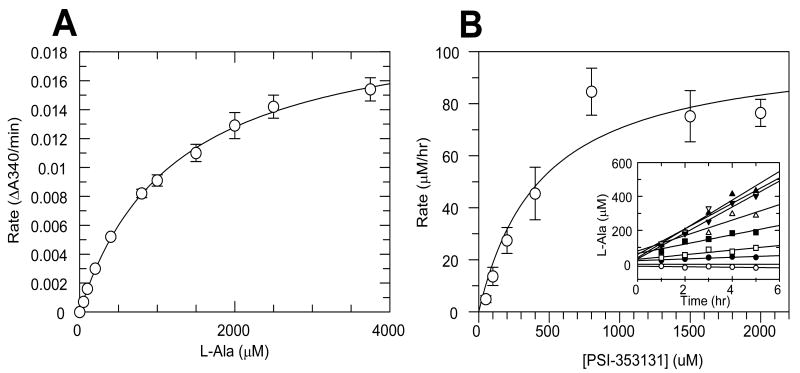
The second enzymatic step in the activation of PSI-353661 can be catalyzed by histidine triad nucleotide-binding protein 1 (Hint1). The ability of Hint1 to catalyze the hydrolytic removal of L-alanine from PSI-353131 was tested using a two-step reaction. First, steady-state parameters for L-alanine with Ala-DH were determined as shown in Figure 6A. The Michaelis constant (Km) for alanine was 1227 ± 65 μM and the Vmax was 0.0206 ± 0.0005 ΔA340/min. This Ala-DH rate versus concentration curve was used to determine the concentration of alanine generated in the Hint1 reactions. After Hint1 reactions were performed, the concentration of the L-Ala product formed was quantified by performing the Ala-DH reaction and measuring the initial rate. The amount of L-Ala product was quantified at different incubation times to determine the rate of PSI-353224 formation in the Hint1 reaction (Figure 6B inset). The steady-state kinetic parameters for PSI-353131 were determined by plotting the reaction rates against PSI-353131 concentration (Figure 6B). The Km value for PSI-353131 was 445 ± 182 μM and kcat was 3.44 min-1.
Figure 6. Hint1 activity assay.
A standard curve for L-alanine dehydrogenase (L-AlaDH) was generated by plotting the initial rates against the L-Ala concentrations (A). L-Ala formation was monitored for Hint1-mediated deamination of PSI-353131 (B). Rates were calculated from the individual time courses (inset). Bars in the graphs represent the standard error of the linear regression analysis from the individual time courses.
It has been reported that an adenosine deaminase-like enzyme (ADAL1, N6-methyl-AMP aminohydrolase, abacavir monophosphate deaminase) is capable of catalyzing the hydrolytic deamination of natural substrates N6-methyl AMP, N6,N6-dimethyl-AMP and N6-methyl dAMP and N6 substituted purine-5′-monophosphate analogs (Schinkmanova et al., 2006; Schinkmanova et al., 2008). Because removal of the methyl group at the O6 position of the guanine base is required during activation of PSI-353661 we examined the ability of ADAL1 to catalyze this reaction. PSI-353661, PSI-353131, and PSI-353224, along with N6-methyl AMP and O6-methyl GMP, were tested for their ability to serve as substrates for purified recombinant human ADAL1. Of the five compounds tested, only N6-methyl AMP, O6-methyl GMP and PSI-353224 were substrates for ADAL1 (Table 6). The Km and kcat values for PSI-353224 were 9.0 ± 2.1 μM and 5.84 ± 0.40 s-1, respectively, and a kcat/Km of 0.65 μM-1sec-1. Km and kcat values for O6-methyl GMP were 5.2 ± 0.9 μM and 2.33 ± 0.07 s-1, respectively, and a kcat/Km value similar to that obtained for PSI-353224. N6-methyl AMP appeared to be a slightly less efficient substrate for ADAL1 with a kcat/Km of 0.13 μM-1sec-1. Recombinant human adenylate deaminase (AMPD) was also evaluated for its ability to use PSI-353224 as a substrate. PSI-353224 (100 μM) was incubated with AMPD for up to 72 hours and the reaction mixture analyzed by HPLC. AMPD failed to convert PSI-353224 to PSI-353222, indicating that PSI-353224 was not a substrate for AMPD.
Table 6. Kinetic parameters for ADAL1-mediated demethylation.
| Km (μM) | kcat (s-1) | kcat / Km (μM-1s-1) | |
|---|---|---|---|
| N6-meAMP | 12.5 ± 1.6 | 1.58 ± 0.12 | 0.13 |
| O6-meGMP | 5.2 ± 0.9 | 2.33 ± 0.07 | 0.45 |
| PSI-353661 | No activity | ||
| PSI-353131 | No activity | ||
| PSI-353224 | 9.0 ± 2.1 | 5.84 ± 0.40 | 0.65 |
Values are reported as the mean ± S.D. from three independent experiments.
PSI-353222 was effectively phosphorylated to the corresponding 5′-diphosphate analog, PSI-353579, by recombinant hGUK1. Phosphorylation of PSI-353222 by hGUK1 was measured using a coupled spectrophotometric reaction. PSI-353222 was a substrate for purified hGUK1 with Km and kcat values of 64.9 ± 1.6 μM and 0.31 ± 0.03 s-1, respectively. The natural substrate, guanosine-5′-monophosphate (GMP) was phosphorylated efficiently by hGUK1 with a kcat/Km value of 3.9 μM-1s-1 (Table 7). However, compared with GMP, PSI-353222 was significantly less efficient as a substrate. The Km for PSI-353222 indicated that PSI-353222 did not bind as tightly to the enzyme compared to GMP (3.4-fold weaker) and the kcat value for PSI-353222 was significantly reduced, approximately 240-fold lower than that for GMP. No activity was observed when PSI-353224 was used as substrate at a concentration of 500 μM, the highest concentration tested. These results strongly suggest that the hydrolysis of the O6-methyl group of PSI-353224 occurs before phosphorylation to the diphosphate takes place.
Table 7. Kinetic parameters for hGUK1-mediated phosphorylation.
| Km (μM) | kcat (s-1) | kcat / Km (μM-1s-1) | |
|---|---|---|---|
| GMP | 19.1 ± 4.5 | 74.7 ±17.6 | 3.9 |
| PSI-353222 | 64.9 ± 1.6 | 0.31 ± 0.03 | 0.005 |
| PSI-353224 | No activity | ||
Values are reported as the mean ± S.D. from three independent experiments.
Phosphorylation of PSI-353579 (2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′- diphosphate) to PSI-352666 can be carried out by at least three enzymes: nucleoside diphosphate kinase (NDPK), pyruvate kinase (PK), and 3-phosphoglycerate kinase (3-PGK). Of the three enzymes NDPK was the most efficient in catalyzing the reaction. The apparent steady-state parameters for PSI-353579 and the natural substrate, guanosine-5′-diphosphate (GDP), with NDPK are summarized in Table 8. The Km for PSI-353579 was slightly higher compared to the Km for GDP, while the kcat for both was similar. Overall the catalytic efficiency (kcat/Km) for PSI-353579 was similar to that of GDP. Phosphorylation of PSI-353579 was also observed with PK and 3-PGK; however, the kinetic parameters were unable to be determined due to the poor affinity (Km too high) of PSI-353579 for these enzymes.
Table 8. Apparent kinetic parameters for NDPK-mediated phosphorylationa.
| Km (μM) | kcat (s-1) | kcat / Km (μM-1s-1) | |
|---|---|---|---|
| GDP | 56.2 ± 4.9 | 38.8 ± 5.0 | 0.69 |
| PSI-353579 | 66.3 ± 2.2 | 44.6 ± 4.9 | 0.67 |
Kinetic parameters are apparent as PK contributed in the phosphorylation.
Values are reported as the mean ± S.D. from three independent experiments.
3.7. Inhibition of HCV NS5B polymerase
The active triphosphate form, PSI-352666, acts as an alternate substrate for HCV RdRp to inhibit RNA synthesis. The ability of PSI-352666 to inhibit NS5B polymerase from different genotypes was tested in the presence of 1 μM NTPs. PSI-352666 was found to be active against recombinant NS5B from genotypes 1b, 2a, 3a, and 4a. with average IC50 values of 1.0 ± 0.2, 4.7 ± 0.6, 1.3 ± 0.5, and 4.2 ± 0.8 μM, respectively.
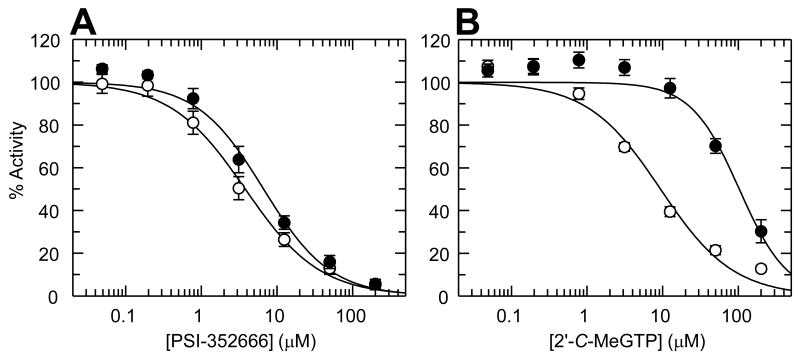
Additionally, we compared the ability of PSI-353666 and 2′-α-OH-2′-β-C-meGTP to inhibit wild-type and S282T mutant genotype 1b RdRp in the presence of 5 μM NTPs (Figure 7A and B). With PSI-352666, there was approximately a 2-fold difference in IC50 values between wild-type (IC50 = 4.0 ± 1.1 μM) and S282T mutant (IC50 = 9.2 ± 1.4 μM) (Figure 7A). When the reaction was performed with 2′-α-OH-2′-β-C-meGTP as the inhibitor, the S282T mutant RdRp showed a more than 10-fold reduction in sensitivity when compared with wild-type NS5B (Figure 7B). The IC50 values for 2′-α-OH-2′-β-C-meGTP with the wild-type and the S282T RdRp were 6.8 ± 1.7 μM and 103 ± 18 μM, respectively.
Figure 7. Inhibition of wild type and S282T HCV NS5B polymerase with PSI-352666 and 2′-α-OH-2′-β-C-methylGTP.
Activity of NS5B polymerase activity was plotted against concentrations of PSI-352666 (A) or 2′-α-OH-2′-β-C-methylGTP (B). The experiments were performed using wild type (open circles) and S282T mutant (closed circles) enzymes. Bars represent the standard deviation from at least three independent assays performed in duplicate.
4. Discussion
PSI-353661 is a single diastereoisomer of a phosphoramidate prodrug of 2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-monophosphate currently undergoing preclinical evaluation. In addition to the phosphoramidate motif, PSI-353661 contains an O6-methyl modification on the guanine base. Our results demonstrated that PSI-353661 has potent activity against HCV in both the replicon assay and the infectious virus assay. We compared the activity of PSI-353661 with that of the recently reported phosphoramidate prodrug INX-08189 (McGuigan et al., 2010). Both PSI-353661 and INX-08189 showed similar activity in the replicon assay. However, INX-08189 was significantly more cytotoxic than PSI-353661.
Intracellular phosphorylation studies with PSI-353661 and primary human hepatocytes clearly show that PSI-353661 was transported into cells and phosphorylated to the active triphosphate form, PSI-352666. PSI-352666 was formed rapidly and achieved a high concentration (>50 μM) after 4 hours of exposure before declining. The first step in the metabolism of PSI-353661 involves hydrolysis of the carboxyl ester bond between the alaninyl moiety and the isopropyl alcohol (McGuigan et al., 1998a; McGuigan et al., 1998b). Recently, we demonstrated that PSI-7977 was a substrate for both CatA and CES1 (Murakami et al., 2010). Since PSI-353661 and PSI-7977 are both Sp isomers and have the identical phosphoramidate moiety in common, we tested CatA and CES1 for their ability to catalyze this hydrolytic reaction. As was observed with PSI-7977, PSI-353661 was a substrate for both enzymes. However, as we previously reported for PSI-7977 (Murakami et al., 2010), incubating CES1 with PSI-353661 resulted in unusual kinetics. Only a small fraction of PSI-353661 (∼10%) was converted to product, PSI-353131. Although we do not completely understand the mechanism of this behavior, it is our hypothesis that PSI-353661 bind in an orientation that is essentially nonproductive. Further studies are ongoing to better understand this mechanism.
The effect of telaprevir and BNPP on the formation of the active triphosphate, PSI-352666, provides further evidence supporting the role of CatA and CES1 in the metabolism of PSI-353661. While telaprevir was an inhibitor of CatA, inhibition of the formation PSI-352666 by telaprevir was observed in hepatocytes from only one of three different donors. This may be due to differences in the level of expression of CatA in the hepatocytes from the different donors. It was previously demonstrated by Western blot analysis that the expression level of both CatA and CES1 varies substantially from one donor to another (Murakami et al., 2010). Therefore the lack of inhibition of PSI-353661 metabolism by telaprevir in hepatocytes from two of the three donors may be because CES1 is predominantly contributing to PSI-353661 hydrolysis in the cells from these two donors due to the differential expression of CatA and CES1
Once the carboxyl ester is hydrolyzed, the phenol on the phosphate moiety is released via a non-enzymatic chemical reaction that involves a nucleophilic attack of the carboxyl group at the phosphate affording the alaninyl phosphate intermediate, PSI-353131. As has been demonstrated by others, this chemical step is followed by the enzymatic removal of the amino acid moiety from the phosphate by Hint 1 (Chou et al., 2007; Murakami et al., 2010). While other enzymes may be involved in removing the amino acid moiety, our studies show that Hint1 is capable of converting PSI-353131 to PSI-353224.
Based on our metabolism and enzyme studies, the next step in the metabolism of PSI-353661 involved the demethylation of the compound resulting in the formation of 2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-monophosphate (PSI-353222). ADAL1 was identified as an enzyme capable of catalyzing this reaction. The last two steps in the metabolic pathway of PSI-353661 were catalyzed by hGUK1 and NDPK (the enzyme predominately involved in converting the diphosphate to the triphosphate), PK and 3-PGK.
The 5′-triphosphate, PSI-352666, was found to be an inhibitor of recombinant NS5B RdRp from genotype 1b, 2a, 3a, and 4a with IC50 values ranging from 1.0 ± 0.2 to 4.7 ± 0.6 μM. Given that PSI-352666 was a potent inhibitor of the NS5B of HCV genotypes 1-4 and that PSI-353661 demonstrated potent activity against genotype 1a, 1b, and 2a replicons and in the H77 (genotype 1a) and JFH1 (genotype 2a) infectious virus assays suggests that PSI-353661 would provide broad genotype coverage.
We assessed the activity profile of PSI-353661 against replicons containing amino acid alterations in NS5B known to confer resistance to nucleoside analogs, S282T and S96T/N142T. Replicons containing either the S282T or the S96T/N142T amino acid alteration showed no change in sensitivity to PSI-353661 compared to the wild-type control replicon. As yet we have been unable to select for a mutation(s) that confer resistance to PSI-353661. However the S282T amino acid alteration did confer resistance to INX-08189. This was not surprising since the S282T amino acid alteration also confers resistance to the parent nucleoside, 2′-C-methylguanosine (Migliaccio et al., 2003). The active form of both prodrugs, the corresponding 5′-triphosphate, was tested against both recombinant wild-type NS5B and NS5B containing the S282T amino acid alteration. It is clear that there is a marked distinction between a purine having a 2′-β-methyl substitution with a 2′-α-fluoro versus a 2′-α-hydroxy substitution. Unlike the pyrimidines where the S282T amino acid alteration confers resistance to both the 2′-α-fluoro and the 2′-α-hydroxy substituted 2′-β-methyl analogs, the combination of the 2′-β-methyl group and the 2′-α-fluorine substitution seems to impart unique characteristics to the guanine nucleoside, which clearly has a dramatic effect on the resistance profile. It is not yet clear what it is about the 2′-α-fluorine relative to the α-hydroxyl substitution that imparts this differentiating characteristic. The unique effect conveyed by the 2′-α-fluorine group could be the result of an electronic effect imparted on the nucleoside or some as yet unknown interaction with the polymerase.
Overall, these studies characterize the in vitro activity and safety and the mechanism of action of PSI-353661. Based on its preclinical in vitro activity and safety profile, PSI-353661 warrants further investigation.
Acknowledgments
Dr. Nigel Bourne and Ronald L. Veselenak have no conflict and their work was performed at the University of Texas Medical Branch was supported in part by a contract (AI-25488) from the National Institute of Allergy and Infectious Diseases. We thank Dr. David Frick (University of Wisconsin, Milwaukee, WI) and Dr. Julie Heck (New York Medical College, Valhalla, NY) for providing the recombinant NS5B polymerase derived from the genotype 2a JFH-1 strain.
Abbreviations
- HCV
hepatitis C virus
- HPLC
high pressure liquid chromatography
- PCR
polymerase chain reaction
- ddC
dideoxycytidine
- GMP
guanosine-5′ -monophosphate
- GDP
guanosine-5′ -diphosphate
- RdRp
HCV RNA-dependent RNA polymerase
- NS5B
HCV non-structural protein 5B
- Cat A
cathepsin A
- CES1
carboxylesterase 1
- Hint 1
histidine triad nucleotide-binding protein 1
- ADAL1
adenosine deaminase-like protein 1
- hGUK1
human guanylate kinase
- NDPK
human nucleoside diphosphate kinase
- PK
pyruvate kinase
- 3-PGK
3-phosphoglycerate kinase
- Km
Michaelis constant
- kcat
turnover number
- kcat/Km
catalytic efficiency
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afdahl N, Godofsky E, Dienstag J, Rustgi VK, Schick L, McEniry D, Zhou XJ, Chao G, Fang C, Fielman B, Myers M, Brown N. Final phase I/II trial results for NM283, a new polymerase inhibitor for hepatitis C: antiviral efficacy and tolerance in patients with HCV-1 infection, including previous interferon failures. 55th Annual Meeting of the American Association for the Study of Liver Diseases; Boston, MA, USA. 2004. p. 726A. [Google Scholar]
- Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J Virol. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Bao D, Chun BK, Naduthambi D, Nagaratham D, Rachakonda S, Reddy PG, Ross BS, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Niu C, Micholochick Steuer HM, Furman PA, Otto MJ, Sofia MJ. Discovery of PSI-353661, a novel purine nucleotide prodrug for the treatment of HCV infection. ACS Med Chem Lett. 2010;2:130–135. doi: 10.1021/ml100209f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Baraniak J, Kaczmarek R, Zhou X, Cheng J, Ghosh B, Wagner CR. Phosphoramidate pronucleotides: a comparison of the phosphoramidase substrate specificity of human and Escherichia coli histidine triad nucleotide binding proteins. Mol Pharm. 2007;4:208–217. doi: 10.1021/mp060070y. [DOI] [PubMed] [Google Scholar]
- Clark JL, Mason JC, Hollecker L, Stuyver LJ, Tharnish PM, McBrayer TR, Otto MJ, Furman PA, Schinazi RF, Watanabe KA. Synthesis and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methyl purine nucleosides as inhibitors of hepatitis C virus RNA replication. Bioorg Med Chem Lett. 2006;16:1712–1715. doi: 10.1016/j.bmcl.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Duarte MI, Andrade HF, Jr, Mariano ON, Corbett CE, Sesso A. Baseline volume data of human liver parenchymal cell. J Submicrosc Cytol Pathol. 1989;21:275–279. [PubMed] [Google Scholar]
- Fleischer RD, Lok AS. Myopathy and neuropathy associated with nucleos(t)ide analog therapy for hepatitis. B J Hepatol. 2009;51:787–791. doi: 10.1016/j.jhep.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Herlihy KJ, Graham JP, Kumpf R, Patick AK, Duggal R, Shi ST. Development of intergenotypic chimeric replicons to determine the broad-spectrum antiviral activities of hepatitis C virus polymerase inhibitors. Antimicrob Agents Chemother. 2008;52:3523–3531. doi: 10.1128/AAC.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe AY, Cheng H, Johann S, Mullen S, Chunduru SK, Young DC, Bard J, Chopra R, Krishnamurthy G, Mansour T, O'Connell J. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob Agents Chemother. 2008;52:3327–3338. doi: 10.1128/AAC.00238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan P, Fu Q, Lam W, Liou JY, Dutschman G, Cheng YC. Phosphorylation of pyrimidine deoxynucleoside analog diphosphates: selective phosphorylation of L-nucleoside analog diphosphates by 3-phosphoglycerate kinase. J Biol Chem. 2002;277:5453–5459. doi: 10.1074/jbc.M109025200. [DOI] [PubMed] [Google Scholar]
- Lam AM, Murakami E, Espiritu C, Steuer HM, Niu C, Keilman M, Bao H, Zennou V, Bourne N, Julander JG, Morrey JD, Smee DF, Frick DN, Heck JA, Wang P, Nagarathnam D, Ross BS, Sofia MJ, Otto MJ, Furman PA. PSI-7851, a pronucleotide of beta-D-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob Agents Chemother. 2010;54:3187–3196. doi: 10.1128/AAC.00399-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pogam S, Jiang WR, Leveque V, Rajyaguru S, Ma H, Kang H, Jiang S, Singer M, Ali S, Klumpp K, Smith D, Symons J, Cammack N, Najera I. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology. 2006a;351:349–359. doi: 10.1016/j.virol.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Le Pogam S, Kang H, Harris SF, Leveque V, Giannetti AM, Ali S, Jiang WR, Rajyaguru S, Tavares G, Oshiro C, Hendricks T, Klumpp K, Symons J, Browner MF, Cammack N, Najera I. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J Virol. 2006b;80:6146–6154. doi: 10.1128/JVI.02628-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Copeland WC. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003;2:812–822. doi: 10.1038/nrd1201. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown MF, Rajyaguru S, Le Pogam S, Ali S, Jiang WR, Kang H, Symons J, Cammack N, Najera I. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob Agents Chemother. 2008;52:1604–1612. doi: 10.1128/AAC.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuigan C, Madela K, Aljarah M, Gilles A, Brancale A, Zonta N, Chamberlain S, Vernachio J, Hutchins J, Hall A, Ames B, Gorovits E, Ganguly B, Kolykhalov A, Wang J, Muhammad J, Patti JM, Henson G. Design, synthesis and evaluation of a novel double pro-drug: INX-08189. A new clinical candidate for hepatitis C virus. Bioorg Med Chem Lett. 2010;20:4850–4854. doi: 10.1016/j.bmcl.2010.06.094. [DOI] [PubMed] [Google Scholar]
- McGuigan C, Sutton PW, Cahard D, Turner K, O'Leary G, Wang Y, Gumbleton M, De Clercq E, Balzarini J. Synthesis, anti-human immunodeficiency virus activity and esterase lability of some novel carboxylic ester-modified phosphoramidate derivatives of stavudine (d4T) Antivir Chem Chemother. 1998a;9:473–479. doi: 10.1177/095632029800900603. [DOI] [PubMed] [Google Scholar]
- McGuigan C, Tsang HW, Sutton PW, De Clercq E, Balzarini J. Synthesis and anti-HIV activity of some novel chain-extended phosphoramidate derivatives of d4T (stavudine): esterase hydrolysis as a rapid predictive test for antiviral potency. Antivir Chem Chemother. 1998b;9:109–115. doi: 10.1177/095632029800900202. [DOI] [PubMed] [Google Scholar]
- Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- Moyle G. Toxicity of antiretroviral nucleoside and nucleotide analogues: is mitochondrial toxicity the only mechanism? Drug Saf. 2000;23:467–481. doi: 10.2165/00002018-200023060-00001. [DOI] [PubMed] [Google Scholar]
- Murakami E, Bao H, Ramesh M, McBrayer TR, Whitaker T, Micolochick Steuer HM, Schinazi RF, Stuyver LJ, Obikhod A, Otto MJ, Furman PA. Mechanism of activation of beta-D-2′-deoxy-2′-fluoro-2′-c-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob Agents Chemother. 2007;51:503–509. doi: 10.1128/AAC.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E, Niu C, Bao H, Micolochick Steuer HM, Whitaker T, Nachman T, Sofia MA, Wang P, Otto MJ, Furman PA. The mechanism of action of beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine involves a second metabolic pathway leading to beta-D-2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-triphosphate, a potent inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother. 2008;52:458–464. doi: 10.1128/AAC.01184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E, Tolstykh T, Bao H, Niu C, Micolochick Steuer HM, Bao D, Chang W, Espiritu C, Bansal S, Lam AM, Otto MJ, Sofia MJ, Furman PA. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J Biol Chem. 2010;285:34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Pockors P, Godofsky E, Rodriguez-Torres M, Everson G, Fried M, G R, Harrison S, Nyberg L, Shiffman M, Chan A, Hill G. High end-of-treatment response (84%) after 4 weeks of R1626, peginterferon alfa-2A (40 kDa) and ribavirin followed by a further 44 weeks of peginterferon alfa-2A and ribavirin. 43rd Annual Meeting of the European Association for the Study of the Liver (EASL); Milan, Italy. 2008. [Google Scholar]
- Nguyen TT, Gates AT, Gutshall LL, Johnston VK, Gu B, Duffy KJ, Sarisky RT. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob Agents Chemother. 2003;47:3525–3530. doi: 10.1128/AAC.47.11.3525-3530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkmanova M, Votruba I, Holý A. N6-methyl-AMP aminohydrolase activates N6-substituted purine acyclic nucleoside phosphonates. Biochem Pharmacol. 2006;71:1370–1376. doi: 10.1016/j.bcp.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Schinkmanova M, Votruba I, Shibata R, Han B, Liu X, Cihlar T, Holý A. Human N6-methyl-AMP/DAMP aminohydrolase (Abacavir 5′-monophosphate deaminase) is capable of metabolizing N6-substituted purine acyclic nucleoside phosphonates. Collect Czech Chem Commun. 2008;73:275–291. [Google Scholar]
- Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- Stuyver LJ, Lostia S, Adams M, Mathew JS, Pai BS, Grier J, Tharnish PM, Choi Y, Chong Y, Choo H, Chu CK, Otto MJ, Schinazi RF. Antiviral activities and cellular toxicities of modified 2′,3′-dideoxy-2′,3′-didehydrocytidine analogues. Antimicrob Agents Chemother. 2002;46:3854–3860. doi: 10.1128/AAC.46.12.3854-3860.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver LJ, McBrayer TR, Tharnish PM, Clark J, Hollecker L, Lostia S, Nachman T, Grier J, Bennett MA, Xie MY, Schinazi RF, Morrey JD, Julander JL, Furman PA, Otto MJ. Inhibition of hepatitis C replicon RNA synthesis by beta-D-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir Chem Chemother. 2006;17:79–87. doi: 10.1177/095632020601700203. [DOI] [PubMed] [Google Scholar]
- Stuyver LJ, McBrayer TR, Tharnish PM, Hassan AE, Chu CK, Pankiewicz KW, Watanabe KA, Schinazi RF, Otto MJ. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J Virol. 2003;77:10689–10694. doi: 10.1128/JVI.77.19.10689-10694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji N, Tanji K, Kambham N, Markowitz GS, Bell A, D'Agati VD. Adefovir nephrotoxicity: possible role of mitochondrial DNA depletion. Hum Pathol. 2001;32:734–740. doi: 10.1053/hupa.2001.25586. [DOI] [PubMed] [Google Scholar]
- Tomei L, Altamura S, Bartholomew L, Biroccio A, Ceccacci A, Pacini L, Narjes F, Gennari N, Bisbocci M, Incitti I, Orsatti L, Harper S, Stansfield I, Rowley M, De Francesco R, Migliaccio G. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2003;77:13225–13231. doi: 10.1128/JVI.77.24.13225-13231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]