Abstract
We have identified two families with a previously undescribed lethal X-linked disorder of infancy; the disorder comprises a distinct combination of an aged appearance, craniofacial anomalies, hypotonia, global developmental delays, cryptorchidism, and cardiac arrhythmias. Using X chromosome exon sequencing and a recently developed probabilistic algorithm aimed at discovering disease-causing variants, we identified in one family a c.109T>C (p.Ser37Pro) variant in NAA10, a gene encoding the catalytic subunit of the major human N-terminal acetyltransferase (NAT). A parallel effort on a second unrelated family converged on the same variant. The absence of this variant in controls, the amino acid conservation of this region of the protein, the predicted disruptive change, and the co-occurrence in two unrelated families with the same rare disorder suggest that this is the pathogenic mutation. We confirmed this by demonstrating a significantly impaired biochemical activity of the mutant hNaa10p, and from this we conclude that a reduction in acetylation by hNaa10p causes this disease. Here we provide evidence of a human genetic disorder resulting from direct impairment of N-terminal acetylation, one of the most common protein modifications in humans.
Introduction
Researchers have used exon capture and high-throughput sequencing (exome sequencing) to identify the molecular etiology of several Mendelian disorders.1–5 It has been used in studies of a multigenerational pedigree6 and a de novo disorder,1 and it has been applied in molecular diagnostics.7 Most of these efforts have focused on characterizing previously described and recognizable genetic syndromes, such as Kabuki (MIM 147920),1 Miller (MIM 263750),2,8 and TARP (MIM 311900)9 syndromes. These efforts to search for the molecular etiology of known syndromes benefited from years of clinical evaluations, allowing researchers to combine unrelated individuals in the same cohort for analysis.
In contrast, exon capture and sequencing can help to identify previously unrecognized syndromes. We have characterized such a syndrome, in which the afflicted boys have an aged appearance, craniofacial anomalies, hypotonia, global developmental delays, cryptorchidism, and cardiac arrhythmias. To determine the genetic basis of this syndrome, we used X chromosome exon capture and sequencing and a recently developed probabilistic algorithm aimed at discovering disease-causing variants. We also demonstrate that this phenotype results from a decrease in the function of an enzyme involved in N-terminal acetylation of proteins.
Subjects and Methods
We describe two parallel genetic research efforts that converged on the same gene variant. The two groups working on this project became aware of each other's work after the genetic analyses had been completed, and therefore the methodologies used varied somewhat. When these methods differed, they are described separately.
The sample collection and analyses of families 1 and 2 were approved by the institutional review boards at the University of Utah and the National Human Genome Research Institute, respectively. Written informed consent was obtained from the parents of affected children for collected blood samples and extracting DNA from stored samples. Blood samples were collected and genomic DNA extracted with alkaline lysis and ethanol precipitation (Gentra Puregene, QIAGEN, USA). Two DNA samples from family 1 were extracted from stored formalin-fixed-paraffin-embedded (FFPE) tissues from the prior autopsies of two of the deceased boys. Slices from FFPE tissue blocks were digested overnight in proteinase K and then boiled for 10 min to inactivate the enzyme. The crude lysates were diluted 1:10 for PCR.
Genomic Microarrays and Copy-Number Variation Analysis
Copy-number variation in individual III-4 in family 1 was evaluated with two oligonucleotide-based genomic microarray platforms, the U-array Cyto6000 (manufactured by Agilent Technologies, Santa Clara, CA) and the SignatureChip Oligo Solution microarray (manufactured by Roche NimbleGen). Both platforms use a custom design with approximately 10 kb coverage for many clinically relevant syndromes and telomere and pericentromeric regions,; the genome-wide coverage is 75 kb for the U-array and 35 kb for the SignatureChipOS. The U-array data were analyzed with DNA Analytics 4.0.76 software from Agilent Technologies with the ADM-1 algorithm. The SignatureChipOS data were analyzed with Genoglyphix software (Signature Genomics, Spokane, WA).
X-Chromosome Exon Capture and Sequencing
Exon capture for family 1 was carried out with a commercially available Agilent in-solution method (SureSelect Human X chromosome kit, Agilent) as per the manufacturer guidelines with minor modifications. Randomly fragmenting the pure and high-molecular-weight genomic DNA samples with a Covaris S series Adaptive Focused Acoustics machine resulted in DNA fragments with a base pair peak of 150 to 200 bps. Adaptors were then ligated to both ends of the resulting fragments. The adaptor-ligated templates were purified by the Agencourt AMPure SPRI beads. Adaptor-ligated products were amplified by PCR with Phusion polymerase (six cycles) and subsequently purified with a QIAGEN QIAquick PCR purification kit. The quality and size distribution of the amplified product was assessed on an Agilent Bioanalyzer DNA 1000 chip. Amplified library (500 ng) with a size range of 200–400 bp was hybridized to SureSelect Biotinylated RNA Library (BAITS) for enrichment. Hybridized fragments were bound to the strepavidin beads whereas nonhybridized fragments were washed out after a 24 hr hybridization. The captured library was enriched by amplification by PCR (12 cycles), and the amplified product was subsequently qualified on an Agilent High Sensitivity DNA Bioanalyzer chip to verify the size range and quantity of the enriched library. Each captured library was sequenced with 76 bp single-end reads on one lane each of the Illumina GAIIx platform. Raw image files were processed by Illumina Pipeline v1.6 for base-calling with default parameters. All coordinates are based on the human genome build hg18. The pseudoautosomal regions were defined based on annotations within the UCSC genome browser. Specifically, these are the regions chrX:1–2709520, chrX:154584238–154913754, chrY:1–2709520, and chrY:57443438-57772954. For family 2, solution hybridization selection of the X exome (Sure Select, Agilent, Santa Clara, CA) was used to produce a paired-end sequencing library (Illumina, San Diego, CA) as previously described.9 Paired-end 75 bp reads were generated from the target-selected DNA library in one lane of an Illumina GAIIx. Coding changes were predicted with custom-designed software.9
Sanger Sequence-Confirmation Analysis
Amplification by PCR and Sanger sequencing were performed as described so that mutations and the cosegregation could be confirmed.10 Mutation numbering was performed according to Human Gene Variation Society nomenclature with reference NM_003491.2.
Sequence Alignment
For family 1, sequence reads were converted from Illumina fastq format to fastq files that conform to the Sanger specification for base-quality encoding with perl scripts. Burroughs Wheeler alignment (BWA)11 version 0.5.8 was used to align the sequencing reads, and the default parameters were used for fragment reads, to the human genome sequence build 36 downloaded from the UCSC Genome Browser or the 1000 Genomes Project websites.12 Alignments were converted from SAM format to sorted, indexed BAM files with SamTools.13 The picard tool was used to remove invalid alignments and remove duplicate reads from the BAM files. Regions surrounding potential indels were realigned with the GATK IndelRealigner tool.14 Variants were called with SamTools pileup command with the default parameters except that no upper limit was placed on the depth of coverage for calling variants. In addition, haploid chromosomes were called with the -N 1 options SamTools pileup. Variants were annotated with respect to their impact on coding features with perl scripts and the knownGenes and RefGenes tracks from the UCSC Genome Browser.15
Genotype Calling
Regions surrounding potential indels were realigned with the GATK IndelRealigner tool.14 Genotypes were called with both SamTools (pileup command) and the GATK UnifiedGenotypeCaller and IndelCaller.14 We then analyzed the union of single nucleotide variant (SNV) and indel variant calls from GATK and SamTools with the ANNOVAR program16 to identify exonic variants and to identify variants not previously reported in the 1000 Genomes Project and the dbSNP version 130. All variant calls not present on the X chromosome were removed given the X-linked nature of the disease. We analyzed the location and genotype of variants for each individual to locate the subset of variants on the X chromosome that were heterozygous in female carriers, hemizygous in individual III-4, and a hemizygous reference in the unaffected males. Each candidate variant was also screened for presence or absence in dbSNP 130,17 the 10Gen Dataset18 and variant data from the 1000 Genomes Project.12
We also used the read-mapping and SNV-calling algorithmGNUMAP19 independently to align the reads from the Illumina.qseq files to the X chromosome (human sequence build 36) and to simultaneously call SNVs. GNUMAP utilizes a probabilistic pair-hidden Markov model (PHMM) for base calling and SNV detection that incorporates base uncertainty on the basis of the quality scores from the sequencing run, as well as mapping uncertainty from multiple optimal and suboptimal alignments of the read to a given location to the genome. In addition, this approach applies a likelihood ratio test that provides researchers with straightforward SNV-calling cutoffs based on a p value cutoff or a false discovery control. Reads were aligned and SNVs called for the five samples. SNV calls for individual III-4, his brother, and his uncle were made assuming a haploid genome (because the calls are on the X chromosome), whereas heterozygous calls were allowed for the mother and grandmother. SNVs were selected based on a p value cutoff of 0.001. Because of the X-linked nature of the disease, candidate SNVs were selected that are heterozygous in the mother and grandmother and different between the uncle and brother and individual III-4.
Variant Annotation, Analysis, and Selection Tool Analysis
The SNV filtering was performed with the simple selection tool (SST) module in variant annotation, analysis, and selection tool (VAAST) on the basis of on the SNV position. Our analysis applied a disease model that did not require complete penetrance or locus homogeneity. We restricted the expected allele frequency of putative disease-causing variants within the control genomes to 0.1% or lower. The background file used in the analysis is composed of variants from dbSNP (version 130), 189 genomes from the 1000 Genomes Project,12 the 10Gen dataset,18 184 Danish exomes,20 and 40 whole genomes from the Complete Genomics Diversity Panel. VAAST candidate-gene prioritization analysis was performed with the likelihood ratio test under the dominant-inheritance model. An expected allele frequency of 0.1% or lower was assumed for the causal variant in the general population. After masking out loci of potentially low variant quality, SNVs in each gene were scored as a group. The significance level was assessed with individual permutation tests (the following VAAST analysis parameters were used “-m lrt -c X -g 4 -d 1.E8 -r 0.001 -x 35bp_se.wig -less_ram -inheritance d”).
Short-Tandem-Repeat Genotyping
Genotyping on family 1 was performed on DNA extracted from peripheral blood or FFPE tissues with a panel of 16 polymorphic short-tandem-repeat (STR) genotyping markers spanning the long arm of chromosome X (Applied Biosystems Linkage Mapping Set v2.5- MD10 and custom primers DXS8020, DXS1275, DXS6799, DXS1203, DXS8076, and DXS8037). Fluorescently labeled PCR products were separated with an Applied Biosystems 3130XL Genetic Analyzer and analyzed with GeneMapper Software version 3.7. Genotyping and haplotype analysis for family 2 was performed for a subset of markers on the X chromosome (DXS9900, DXS9896, DXS8016, DXS1003, DXS2132, DXS990, DXS6797, DXS8057, GATA31E08, DXS7127, and DXS1073) as described.21
Analysis of Haplotype Sharing
Using GNUMAP software and the X chromosome exon sequence data from family 1, we derived 1322 polymorphic markers on the mother's X chromosomes. We then determined the regions shared or not shared by individual III-4 and his unaffected brother.
Screening for Frequency of Causal Variants
We manually screened for the presence or absence of c.109T>C in NAA10 in dbSNP (version 130), 401 participants in the ClinSeq project,22 180 genomes from the 1000 Genomes Project,12 the 10Gen dataset,18 184 Danish exomes,20 and 40 whole genomes from the Complete Genomics Diversity Panel.
Sorting Intolerant From Tolerant Analysis
The sorting intolerant from tolerant (SIFT) sequence was used with the sequence of hNaa10p (NP_003482) as input. The following parameters were used: the database searched was the UniProt-SwissProt + TrEMBL 2010_09 and the median conservation of sequences was 3.00 (between 0 and 4.32, between 2.75 and 3.25 is recommended). The substitution at position 37 from S to P was predicted to affect protein function with a score of 0.00. The median sequence conservation was 3.02; 119 sequences were represented at this position. The threshold for intolerance is 0.05 (less is predicted not tolerated).
Plasmid Construction, Mutagenesis, Protein Production, Purification, and In Vitro Acetyltransferase Assays
The cDNA encoding hNaa10p WT was cloned into the pETM-41 vector (Maltose Binding Protein [MBP]/His-fusion) (from G. Stier, EMBL, Heidelberg, Germany) for expression in Escherichia coli. The plasmid encoding hNaa10p p.Ser37Pro was made by site-directed mutagenesis with the primers hNAA10 T109C F: 5′-CTTCTACCATGGCCTTCCCTGGCCCCAGCTC-3′ and hNAA10 T109C R: 5′-GAGCTGGGGCCAGGGAAGGCCATGGTAGAAG-3′ according to the instruction manual (QuikChange Site-Directed Mutagenesis Kit, Stratagene). Correct cloning and mutagenesis were verified by DNA sequencing and the plasmids were transformed into E. coli BL21 Star (DE3) cells (Invitrogen) by heat shock. The 200 ml cell cultures were grown in Luria Bertani (LB) medium to an OD600 nm of 0.6 at 37°C and subsequently transferred to 20°C. After 30 min of incubation, protein production was induced by adding IPTG to a final concentration of 0.5 mM. After 17 hr of incubation, the cells were harvested by centrifugation, and the bacterial pellets were stored at −20°C. E. coli pellets containing recombinant proteins were thawed at 4°C and lysed by sonication and French press in lysis buffer (1 mM DTT, 50 mM Tris-HCl [pH 7.4], 300 mM NaCl, and one tablet EDTA-free protease Inhibitor cocktail [Roche] per 50 ml). The cell extracts were applied on a metal affinity fast protein liquid chromatography column (HisTrap HP, GE Healthcare, Sweden). Fractions containing recombinant protein were pooled and subjected to size-exclusion chromatography (Superdex 75 16/60, GE Healthcare). Fractions containing the monomeric recombinant protein were pooled, and the protein purity was analyzed by SDS-PAGE gel electrophoresis. The protein concentrations were determined by OD280 nm measurements.
In the assays investigating the N-terminal acetyltransferase activity toward selected substrate peptides, purified MBP-hNaa10p WT or p.Ser37Pro were mixed with oligopeptides, acetyl-CoA and acetylation buffer. After incubation at 37°C, the acetylation reaction products were quantified with RP-HPLC as described previously.11,13 In the assays investigating the N-terminal acetyltransferase activity toward the acidic N termini of actins (DDDIA or EEEIA), 15 nM of purified MBP-hNaa10p WT or S37P were mixed with 250 μM of oligopeptides. In the assays where the AVFAD and SESSS based oligopeptides were tested, 250 nM of purified enzyme were mixed with 200 μM of mentioned oligopeptides. All samples were further mixed with 400 μM acetyl-CoA and acetylation buffer (1 mM DTT, 50 mM Tris-HCl [pH 8.5], 800 uM EDTA, 10% glycerol) in a total volume of 100 μl. The samples with actin-based oligopeptides as substrates were incubated at 37°C for 15 min, whereas the SESSS and AVFAD reactions were incubated at 37°C for 20 min. In the time-course acetylation experiments, the same conditions as mentioned above were used, and samples were collected at indicated times. For all aliquots collected, the enzyme activities were quenched by adding 5 μl of 10% TFA. The acetylation reaction products were quantified with RP-HPLC as described previously.11 All peptides were custom-made (Biogenes) to a purity of 80%–95%. All peptides (24-mers) used as substrates contain seven unique N-terminal amino acids because these are the major determinants influencing N-terminal acetylation. The next 17 amino acids are essentially identical to the ACTH peptide sequence (RWGRPVGRRRRPVRVYP); however, lysines were replaced by arginines so that any potential interference by Nɛ-acetylation would be minimized. The following oligopeptides were used, and proteins from which the seven N-terminal amino acids are derived are indicated: [H]AVFADLDRWGRPVGRRRRPVRVYP[OH], RNaseP protein p30 (P78346); [H]SESSSKSRWGRPVGRRRRPVRVYP[OH], high-mobility group protein A1 (P17096); [H]DDDIAALRWGRPVGRRRRPVRVYP[OH], β-actin (NP 001092); and [H]EEEIAALRWGRPVGRRRRPVRVYP[OH], γ-actin (NP 001092).
Clinical Reports
Family 1
Individual II-1 was born at 37½ weeks of gestation to a healthy 19-year-old G1P0→1. There had been concern for placental insufficiency. His birth weight was 2140 g (3rd−10th centile), and his length was 47 cm, (25th−50th centile); his orbitofrontal circumference (OFC) was 32 cm, (10th−25th centile), and his Apgar scores were 11 and 1.5 His perinatal course had been complicated by meconium aspiration leading to central depression. A number of distinctive features were noted at birth: large anterior and posterior fontanels, prominent eyes, large ears, flared nares, a narrow palate, a short neck, right cryptorchidism, fifth finger clinodactyly, relatively large great toes, metatarsus valgus, and very little subcutaneous fat.
At 6 months of age, his weight was 4.34 kg, (<< 5th centile), and his OFC was 39 cm, (<< 5th centile). He had severe global delays and had only achieved a social smile as a developmental milestone. His clinical course had been complicated by frequent apneic episodes, poor feeding, and eczema. Diagnostic studies had included karyotype analysis (46,XY), TORCH titers (which were normal), a computed tomography (CT) scan of the head demonstrating cerebral atrophy with enlarged ventricles and echocardiography suggesting peripheral pulmonary stenosis and possibly a septal defect.
At 11½ months old, he presented after 2–3 weeks of emesis and diarrhea. Upon admission he was noted to have an electrolyte imbalance, arrhythmias (premature ventricular complexes [PVCs], premature atrial contractions [PACs], supraventricular tachycardia [SVT], and ventricular tachycardia [Vtach]), and cardiomegaly, and went into cardiopulmonary arrest from which he could not be resuscitated. It should be noted that dysrhythmias occurred after the electrolyte imbalance had been corrected.
Individual II-6 was born at 381 weeks of gestation to a healthy 27-year-old G6P5→6. There had been concern for placental insufficiency, and his birth weight was 3065 g, (50th centile); his length was 47 cm, (25th−50th centile), and his OFC was 33.75 cm, (50th centile). His Apgar scores were 41 and 6.5 His perinatal course was complicated by respiratory distress and recognition of multiple minor anomalies: large anterior and posterior fontanels, prominent eyes, large ears, flared nares, a short neck, hypotonia, neurologic depression, and very little subcutaneous fat. Echocardiography was performed because of a murmur, which demonstrated persistence of the ductus arteriosus.
At 9 months of age, his weight was 6.38 kg, (<5th centile), his length was 68 cm, (5th centile), and his OFC was at 44.2 cm, (10th−25th centile). He had moderate to severe global developmental delays, achieving only a social smile and the ability to raise his head and roll over. He was described as being very fussy and irritable. His clinical course had been complicated by being a poor feeder, experiencing frequent otitis media, iron deficiency anemia, and mild eczema. His physical examination was notable because the anterior fontanel was open and he had down-slanting palpebral fissures; prominent eyes; long lashes; large ears (75th−97th centile); flared nares; a short columella; a short philtrum with an overhanging upper lip; a thin vermillion border of the lips; a narrow palate; microretrognathia; an umbilical hernia; small, “high riding” testes; long fingers; fullness to the dorsum of the feet; metatarsus valgus; capillary malformation over the glabella, eyelids, philtrum, and nape of neck; and hypertonia.
Diagnostic studies had included karyotype analysis (46, XY), very long-chain fatty acids (normal), glycerine kinase (normal), total carnitine (slightly elevated), urine organic acids (normal), biotinidase activity (normal), an EKG that was somewhat irregular, and a CT scan of the head demonstrating cerebral atrophy versus dysgenesis. At 9½ months of age, he died after presenting to the hospital with multiple apneic episodes.
Individual III-7 was born at 33 weeks of gestation to a 21-year-old G1P0→1, reported as having gestational diabetes; he was delivered by elective C-section because of concern that he suffered from polycystic kidneys, oligohydramnios, and pulmonary hypoplasia. His birth weight was 1559 g, (10th−25th centile), and he had a length of 39 cm, (10th centile); his OFC was 27 cm, (3th−10th centile), and his Apgar scores were 41 and 8.5 He was noted to have distinctive facial features and a large anterior fontanel and right-sided cryptorchidism and inguinal hernia. His newborn course was complicated by polycythemia, jaundice, low cortisol, and mild pulmonary hypoplasia. He spent approximately 5 weeks in the neonatal intensive care unit (NICU); he required assisted ventilation for 3 days with the rest of the time dedicated to feeding and growing. Diagnostic studies conducted during the neonatal period included renal ultrasonography (which was normal) and echocardiography, demonstrating a small persistent ductus arteriosus, a mildly decreased left ventricular systolic function, an abnormal appearing aortic valve, and an enlargement of the right ventricle, decreased right ventricular systolic function, and persistence of the foramen ovale.
At 5½ months of age, he was in his usual state of health until 2 days prior to admission, when he developed rhinorrhea; one day prior to admission he developed a fever. He presented to the hospital with increased irritability. At first he did not appear very ill, but he quickly progressed to shock and death.
At the autopsy he was noted to have a congenital hypotonia-lymphedema sequence; hypertelorism; a high, broad forehead with a frontal furrow; micrognathia; cardiomegaly (the right half of the heart was greater than left) and the persistence of the foramen ovale; adrenomegaly; and bilateral enlarged kidneys; and abnormal development, including fetal lobulations, glomerulocystic change, sclerotic glomeruli, micronodular distal tubule proliferation, hypoplastic or small testes, a right-sided inguinal hernia, and right-sided cryptorchidism.
Individual III-4 (Figures 1 and 2B) was born at 373 weeks of gestation to a 28-year-old G4P3→4, reported as having gestational diabetes. She had experienced preterm labor at 35 weeks; there had been decreased fetal movements, and concern for intrauterine growth retardation. His birth weight was 2410 g, (10th−25th centile), and he had a length of 44 cm (<10th centile), an OFC of 32 cm (10th−25th centile), and Apgar scores of 6,1 7,5 and 9.10 He was noted to have large anterior fontanels, prominent eyes, large ears, flared nares, a short neck, and very little subcutaneous fat. His newborn course had been complicated by hyperbilirubinemia, thrombocytopenia, and polycythemia.
Figure 1.
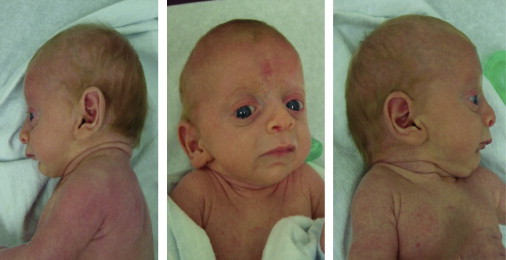
Triptych of Individual III-4 from Family 1
These pictures demonstrate the prominence of eyes, down-slanted palpebral fissures, thickened lids, large ears, flared nares, hypoplastic alae, short columella, protruding upper lip, and microretrognathia.
Figure 2.
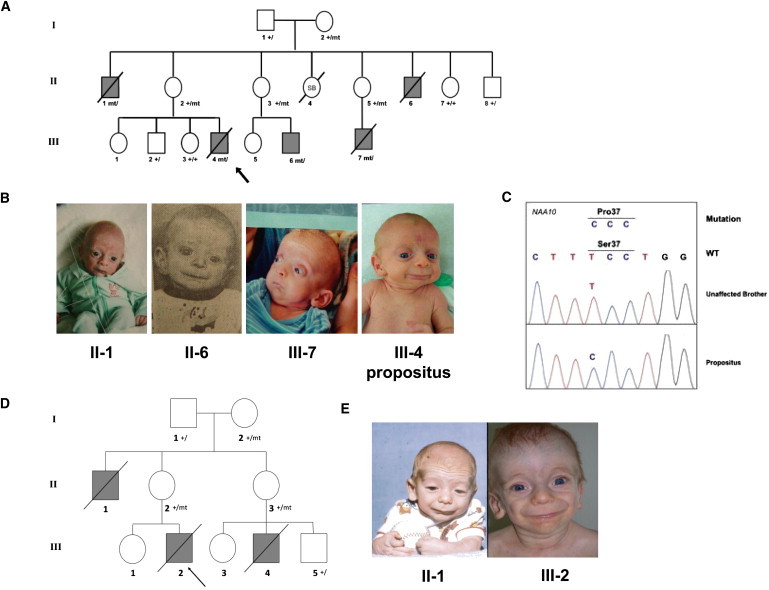
Pedigree Drawing and Pictures of Families 1 and 2
(A) Pedigree drawing for family 1. The most recent deceased individual, III-4, is the most well-studied subject in the family and is indicated by an arrow. Genotypes are marked for those in which DNA was available and tested. The following abbreviations are used: SB, stillborn; +, normal variant; mt, rare mutant variant.
(B) Pictures of four affected and deceased boys in this family, showing the aged appearance.
(C) Sanger sequencing results of NAA10 in individual III-4 from family 1.
(D) Pedigree for family 2. Individual III-2 is the most well-studied subject in the family and is indicated by an arrow.
(E) Picture of individuals II-I and III-2 in family 2 at ∼1 year of age.
At 3½ months of age his weight was 3.18 kg (<5th centile), and he had a length of 48 cm (<5th centile), and an OFC of 35 cm (2nd centile). He was able to smile. His clinical course was complicated by feeding difficulties and growth failure, but he had had no significant illness. He was noted to have a number of distinctive features, including prominent eyes, downslanted palpebral fissures, ocular hypertelorism, prominence of the cheeks, relatively large ears, a short nose, a longer philtrum, a narrow palate, microretrognathia, hypotonia, redundancy of the nuchal skin, skin laxity, and little subcutaneous fat.
At 15 months of age, his weight was 9.06 kg (<3rd centile), and he had a length of 78 cm (25th−50th centile) and an OFC of 43 cm (<3rd centile). He was able to smile. His clinical course had been complicated by severe scoliosis leading to restrictive lung disease, dysphagia, and eczema. He was unable to sit up, and he could not take food by mouth. He had been hospitalized on four separate occasions for apnea, a viral respiratory tract infection, an aspiration pneumonia, and a complication of surgery infection. He was noted to have a number of distinctive features including an anterior fontanel closed with fibrous tissue, a palpable metopic ridge, relative ocular hypertelorism (75th centile), deep-set eyes, downslanting palpebral fissures, long eyelashes, a bifid and depressed nasal tip, prominent nares, prominent ears, a short neck with excess nuchal skin, torticollis, pectus excavatum, scoliosis, and a small scrotal sack with left cryptorchidism. He had been able to build up subcutaneous fat stores, presumably because of G tube feeding,. He developed a very thick, dark head of hair by his first birthday.
Diagnostic studies had included urine organic acids (normal); serum immunoglobulins (normal); comparative genomic hybridization U-array 44 k platform (normal); and an MRI of the brain with spectroscopy that demonstrated bilateral symmetric globus pallidus T2 prolongation without diffusion restriction, a nonspecific elevation of choline and prominence of the Sylvian fissures, immature myelination of the splenium, and moderate lateral and third ventricular dilation without identified cause. An MRI of the spine demonstrated convex-right curvature reflecting compensation for moderately severe convex right C-shaped thoracic neuromuscular scoliosis without any noted congenital vertebral segmentation anomalies.
He had renal ultrasonography (normal), and echocardiography demonstrated a small perimembranous ventricular septal defect (VSD), patent foramen ovale or small secundum atrial septal defect, a mild left atrial enlargement, a trivial pulmonary stenosis, and a trivial bilateral branch pulmonary artery stenosis. Cranial radiography demonstrated a small sella turcica. Holter monitoring and the EKG had been entirely normal, but he eventually developed a nonspecific T-wave abnormality.
He died at 15 months of age after a protracted hospitalization during which he originally presented with multiple hypoxic episodes. He had a surgical correction of his severe scoliosis to optimize his pulmonary function. He developed a bradyarrhythmia and hypoxia and eventual pulseless electrical activity. After then suffering a full arrest and resuscitation, he was supported by mechanical ventilation until his parents chose to withdraw support.
An extensive autopsy of individual III-4 revealed multiple minor external anomalies, severe scoliosis, an enlarged heart with a form fruste of a perimembranous VSD, and several findings that could be attributed to hypoxia and ischemia during individual III-4's terminal course, including slight waviness of myocardial fibers (early ischemic change), pulmonary congestion and intra-alveolar edema, serous effusions in all body cavities, mid- to central zone hepatocytic necrosis, and acute neuronal ischemia in the brain. Focal segmental and global glomerulosclerosis in the outer renal cortex suggested a more chronic or remote injury that might or might not be related to the underlying genetic disorder, but no other histologic or ultrastructural lesion was identified.
Individual III-6 was recently born at 354 weeks of gestation to a 28-year-old G2P1→2, reported as having gestational diabetes. His birth weight was 2604 g, (50th−75th centile), and he had a length of 48 cm, (75th centile), an OFC of 32.5 cm, (50th−75th centile), and an Apgar scores of 41 and 9.5 He was admitted directly to the NICU because of respiratory distress and remained there because of feeding difficulties and mild hyperbilirubinemia. He was noted to have a number of distinctive features including a relatively large anterior fontanel, prominent eyes, downslanted palpebral fissures, ocular hypertelorism, prominent cheeks, relatively large ears, a short nose, a longer philtrum, a narrow palate, microretrognathia, left cryptorchidism, a left inguinal hernia and a large right hydrocele, skin laxity, little subcutaneous fat, broad great toes, and hypotonia. Echocardiography demonstrated a thickened bicuspid aortic valve and mild pulmonary hypertension. A CT scan of the head was remarkable only for a cephalohematoma. His clinical course has been complicated thus far by continued feeding difficulties and growth failure.
Family 2
Family 2 presented originally in 1977 when individual II-1 (Figure 2D) was born at 43 weeks of gestation to his 25-year-old primigravid mother by Cesarean section because of cephalopelvic disproportion. He weighed 3.3 kg (25th−50th centile), measured 51 cm in length (50th−75th centile), and had a head circumference of 32 cm (<5th centile); he was assigned Apgar scores of 61 and 8.5 His testicles were undescended. He had poor feeding, jaundice with a peak bilirubin of 15 mg/dl, and vasomotor instability. He was transferred to a tertiary hospital at 8 days of age. At 7 months of age, he weighed 7.33 kg, measured 59 cm, and had a head circumference of 40.75 cm (all <5th centile). His ears and palpebral fissures were large for his chronological age, and he appeared to have bilateral ptosis. His had coarse facial features, horizontal wrinkles on his forehead, lax skin with irregular fat deposits beneath, and increased facial and body hair. His phallus was large at 4 cm, and his testes had descended. A skeletal survey showed delayed osseous development. His psychomotor development was delayed. A diagnosis of Donohue syndrome (MIM 246200) was suggested. At 8 months of age, he was admitted for traction of a congenitally dislocated left hip. Traction was insufficient to reduce the dislocation so a closed reduction under anesthesia was performed, and he was placed in a half-body cast. On the evening after the procedure, he apparently aspirated and had a respiratory arrest, and then had generalized seizures and an apparent posthypoxia encephalopathy. He recovered from this and was discharged but died at home several days later.
In 2005, individual III-2 (Figure 2D) was born at 39.7 weeks of gestation to a 25 year-old primigravida. The pregnancy was complicated by fetal supraventricular tachycardia, which occurred at 20 weeks of gestation and was controlled with maternal administration of flecainide. At birth he weighed 2.66 kg (5th centile), measured 46 cm (5th−10th centile), had a head circumference of 32 cm (5th−10th centile), and was assigned Apgar scores of 71 and 9.5 He had infraorbital creases, a horizontal crease across his chin, and prominent nasolabial folds. He had a glabellar vascular stain; his forehead was wrinkled, giving him a worried look, and he appeared aged. He had prominent areolae and nipples, which were normally spaced, but no palpable breast tissue. He had a generalized increase in fine body hair. He had bilateral inguinal hernias. The first and second toes appeared widely spaced. His karyotype was normal (500 bands). His copper and ceruloplasmin levels were low, but his plasma catecholamine levels were normal, ruling out Menkes syndrome (MIM 309400). His carbohydrate-deficient transferrin levels were normal. An echocardiogram revealed no structural or functional abnormalities. Inguinal herniorrhaphy was performed uneventfully at 6 weeks of age. At 5 months of age, he weighed 4.35 kg, measured 56.2 cm, and had a head circumference of 39 cm (all <5th centile). Despite hypertonia and hyperreflexia, his development was proceeding normally. He had eczema that was responding to topical corticosteroids. At 7.5 months, he weighed 5 kg, measured 61.3 cm and had a head circumference of 40 cm (all <5th centile), and his weight-for-length was 5%. He had little subcutaneous fat, which gave him a wizened look. His ears measured 4.6 cm (50th centile), appeared large for the size of his body, and had fleshy lobules. His philtrum measured 1.5 cm and appeared long and prominent. He was socially engaging, vocalized mostly with vowels, and could roll over, but could not sit independently. Subtelomeric fluorescence in situ hybridization analysis was normal. Eye evaluation showed only mild lagophthalmos. By 11 months of age, two incisors had erupted; a pincer grasp was emerging, and he could maintain a sitting position if placed into one. He vocalized with screeching and squealing but did not babble. At 11.5 months of age, he presented with symptoms of acute gastroenteritis and was admitted for rehydration. Clostridium difficile toxin testing was positive. He experienced three tonic-clonic seizures, and there was decerebrate posturing during the last. After loading with phenobarbital was performed, no further seizures occurred, and he returned to baseline. He continued to have excessive stool output, but electrolyte levels remained normal. He experienced increased difficulty in breathing and poor perfusion and was transferred to the prenatal intensive care unit, where he had a wide complex rhythm at 120–140 beats per minute. Ultimately, the rhythm became irregular, pulses were not palpable, and attempts to resuscitate him were unsuccessful. The autopsy showed severe micro- and macrovesicular steatosis. Because of the prenatal SVT and the rhythm disturbance without any causal electrolyte disturbance at the end of his life, his heart was evaluated postmortem for abnormalities of the conduction system. This analysis showed focal subendocardial scarring, but no cardiomyopathy or conduction system abnormalities were found. Rare inflammatory infiltrates suggested the possibility of a resolved infection.
In 2007, individual III-4 (Figure 2D) was born at 34 weeks of gestation to his 24-year-old mother, who had a 3-year-old daughter at the time. At 6.5 months of age, he had an episode of aspiration, after which gastrostomy and a tracheostomytubes were placed. At the age of 15 months, he was hospitalized for 5 weeks for an infection. The family was told a defibrillator was needed and surgery was done for this. Two days after his discharge, he died at home at the age of 16 months.
Results
Family 1
Individuals II-1 and II-6 from family 1 (Figure 1 and Figure 2) presented in the mid-1980s to the University of Utah Medical Center. These boys had striking similarity to each other with an array of shared manifestations (Table 1). Both subsequently died in infancy. At that time no specific diagnosis could be made, and the inheritance pattern was uncertain, though autosomal-recessive and X-linked inheritance modes were considered. X-linked inheritance was confirmed in the next generation, when individuals III-4 (Figure 1) and III-7 (Figure 2A) presented. The aged appearance was the most striking part of the disease.
Table 1.
Features of the Syndrome in Family 1
| Category | Features |
|---|---|
| Growth | postnatal growth failure |
| Development | global, severe delays |
| Facial | wrinkled foreheads; prominence of eyes, down-sloping palpebral fissures, thickened lids; large ears; flared nares, hypoplastic alae, short columella; protruding upper lip; microretrognathia |
| Skeletal | delayed closure of fontanels; broad great toes |
| Integument | redundancy/laxity of skin, minimal subcutaneous fat, cutaneous capillary malformations, very fine hair and eyebrows |
| Cardiaca | structural anomalies (ventricular septal defect, atrial level defect, pulmonary artery stenoses), arrhythmias (Torsade de points, PVCs, PACs, SVtach, Vtach), death usually associated with cardiogenic shock preceded by arrhythmia. |
| Genitala | inguinal hernia, hypo- or cryptorchidism |
| Neurologica | hypotonia progressing to hypertonia, cerebral atrophy neurogenic scoliosis |
Features of the syndrome demonstrating more variability. Though variable findings of the cardiac, genital and neurologic systems were observed, all affected individuals manifested some pathologic finding of each.
Family 1 X-Chromosome Exon Capture and Sequencing
Two genomic microarray analyses in individual III-4 did not show any likely causal copy-number variants. Accordingly, X chromosome exon capture and sequencing were used to screen for variants within coding regions. We established the following filtering criteria for five samples to determine the final set of variant calls: variants must be present on the X chromosome in individual III-4 (hemizygous), heterozygous in individual II-2 (the mother), heterozygous in individual I-2 (the grandmother), and absent in individuals III-2 and II-8 (the unaffected brother and unaffected uncle). Table 2 shows the coverage statistics of the X chromosome exon capture. The exon sequencing reads were processed by three independent variant-calling pipelines to increase the accuracy for SNVs and/or microindels (Table 3). These three variant-calling pipelines converged on a small list of candidate variants, which were annotated by ANNOVAR16 for functional importance. One mutation had not been seen previously in dbSNP or the 1000 Genomes Project database. It is missense mutation c.109T>C in NAA10, which predicts p.Ser37Pro (MIM 300013) and was confirmed by Sanger sequencing (Figure 2C). We confirmed that this mutation was not present in 401 participants (90% white and mixed European descent and 96% nonlatino) in the ClinSeq project22 nor was it seen in a combination of ∼6000 genomes or exomes (the majority of white and mixed European descent) collected in ongoing projects at Children's Hospital of Philadelpha, University of Utah, and/or BGI.
Table 2.
Coverage Statistics in Family 1 Based on GNUMAP
| Region | RefSeq Transcripts | Unique Exons | Percent Exon Coverage ≥ 1× | Percent Exon Coverage ≥ 10× | Unique Genes | Average Base Coverage | VAAST Candidate SNVs |
|---|---|---|---|---|---|---|---|
| X chromosomea | 1959 | 7486 | 97.8 | 95.6 | 913 | 214.6 | 1 (NAA10) |
| chrX:10054434–40666673 | 262 | 1259 | 98.1 | 95.9 | 134 | 213.5 | 0 |
| chrX:138927365–153331900 | 263 | 860 | 97.1 | 94.9 | 132 | 177.1 | 1 (NAA10) |
On the X chromosome, there are 8222 unique RefSeq exons. Of these exons, 736 were excluded from the SureSelect X-Chromosome Capture Kit because they were designated as pseudoautosomal or repetitive sequences (UCSC Genome Browser).
Table 3.
The SNV Count, Nonsynonymous Coding SNV Count, and Ti/Tv Ratio for Each Individual for Each Variant Analysis Pipeline in Family 1
| Sample | Pipeline | SNV Count | Nonsynonymous | Ti/Tv |
|---|---|---|---|---|
| III-4 | samtools | 1499 | 114 | 2.0 |
| GATK | 1546 + 236a | 146 + 6 (nonsynonymous + frame) | 2.0 | |
| GNUMAP | 2168 | 155 | 2.0 | |
| II-2 | samtools | 2512 | 219 | 1.6 |
| GATK | 1999 + 270a | 168 + 8 | 2.1 | |
| GNUMAP | 2893 | 183 | 2.0 | |
| III-2 | samtools | 1491 | 106 | 2.0 |
| GATK | 1509 + 252a | 134 + 10 | 2.0 | |
| GNUMAP | 2062 | 131 | 2.0 | |
| I-2 | samtools | 2637 | 229 | 1.5 |
| GATK | 2032 + 278a | 160 + 10 | 2.0 | |
| GNUMAP | 2920 | 183 | 1.9 | |
| II-8 | samtools | 1513 | 108 | 1.9 |
| GATK | 1572 + 243a | 136 + 8 | 1.9 | |
| GNUMAP | 1924 | 139 | 2.0 |
Microindels ascertained with GATK pipeline.
VAAST Analysis
We also used a recently developed tool (VAAST),23 which identifies disease-causing variants, to analyze the exon capture data from family 1. VAAST annotates the SNVs on the basis of their effect on coding sequences, selects the variants compatible with the pedigree, and performs a statistical analysis on the X chromosome genes to identify the variant(s) most likely to be disease-causing. In the candidate-gene identification step, VAAST uses a likelihood ratio test that incorporates both amino acid substitution frequencies and allele frequencies to prioritize candidate genes on the basis of SNVs present in those genes. The analyses by VAAST of the variant sets generated from the three variant-calling pipelines yielded similar results and identified the same causal mutation in NAA10. With SNVs from the affected child (individual III-4) alone, VAAST was able to narrow the candidate-gene list to fewer than five genes (Table 4). We then filtered the data by only selecting SNVs shared by the mother (individual II-2) and the affected child (individual III-4). This subset resulted in three, two, and two candidate genes in the GATK, Samtools, and GNUMAP datasets, respectively (Table 4). Next, we filtered the data by selecting SNVs shared by the mother (individual II-2), the maternal grandmother (individual I-2), and the affected child (individual III-4). This subset resulted in two, two, and one hits in the three datasets, respectively, and NAA10 ranked first in all three lists. When we further excluded the SNVs that were present in the unaffected brother and uncle, VAAST identified a single candidate disease-causing variant in NAA10 in the GATK and the Samtools datasets and two candidate disease-causing variants in NAA10 and RENBP in the GNUMAP dataset (Table 4).
Table 4.
Summary of the Filtering Procedure and Candidate Genes with VAAST
| SNV-Calling Pipeline | GATK | Samtools | GNUMAP |
|---|---|---|---|
| III-4 (total SNVs) | 1546 | 1499 | 2168 |
| III-4 (nsSNVs) | 146 | 114 | 155 |
| VAAST candidate genes (NAA10 ranking) | 4 (3) | 3 (2) | 5 (2) |
| Present in III-4 and mother II-2 (nsSNVs) | 122 | 107 | 116 |
| VAAST candidate genes (NAA10 ranking) | 3 (2) | 2 (1) | 2 (2) |
| Present in III-4, mother II-2, and grandmother I-2 (nsSNVs) | 115 | 95 | 104 |
| VAAST candidate genes (NAA10 ranking) | 2 (1) | 2 (1) | 1 (1) |
| Present in III-4, II-2, and I-2, absent in brother III-2 and uncle II-8 (nsSNVs) | 8 | 6 | 8 |
| VAAST candidate genes (NAA10 ranking) | 1 (1) | 1 (1) | 2 (1) |
Confirmation in Other Family Members
DNA samples from other members of family 1 were obtained from the medical examiner; the samples include DNA isolated from FFPE tissues from two of the deceased boys. Sanger sequencing in 14 DNA samples derived from blood or FFPE tissue from the family confirmed that the mutation in NAA10 cosegregated according to the deduced affection status or carrier status in all members of the family (Figure 2A and Figures S1–S5, available online). During the course of this work, another boy (individual III-6) was born to individual II-3 in family 1 (Figure S6). This boy was clinically diagnosed as having the syndrome and also was confirmed to have the mutation by Sanger sequencing.
Haplotype Analysis
Having identified a putative causative mutation from the X chromosome exon analysis, we performed haplotype analysis on family 1 to exclude regions of the X chromosome from which we could have possibly missed a mutation because of a lack of exon capture or a lack of sequencing coverage. We performed haplotype analysis by using two data sets: (1) the SNVs derived from X chromosome exon sequencing of individual III-4 and (2) STR genotyping of individual III-4, two other affected males (individuals II-6 and III-7) in the family, and carrier females I-2, II-2, and II-5. Haplotype analysis narrowed the possible candidate regions to X chromosome positions 10,054,434–40,666,673 (∼30 Mb) and 138,927,365–153,331,900 (∼14 Mb) (Table 5 and Table S1). The sequencing data showed that of the ∼155 Mb on the X chromosome, the affected brother (III-4) and unaffected brother (III-2) were identical at ∼71% and were recombinant at only 29%. The ∼14 Mb region is located on the telomeric end of the long arm of the X chromosome and includes NAA10. Subsequent addition of STR genotyping from the newborn affected male (III-6) further narrowed this interval to chrX:143,836,276–154,913,754 (telomere) (∼11 Mb), which still includes NAA10 (Table 5).
Table 5.
Haplotype Analysis in Affected Males from Family 1: FineMapChX_STR Results
| Sample | Allele | Markera | hg18 Position | Chromosome |
|---|---|---|---|---|
| III-4 | 219 | DXS1275 (AFM261ZH5) | 68,431,124 | Xq13.1 |
| III-6 | 225 | |||
| II-6 (FFPE tissue) | ? | |||
| III-7 (FFPE tissue) | 225 | |||
| III-4 | 253 | DXS8037 (AFMA285XG5) | 74,040,314 | Xq13.3 |
| III-6 | 257 | |||
| II-6 (FFPE tissue) | 242 | |||
| III-7 (FFPE tissue) | 257 | |||
| III-4 | 162 | DXS986 (AFM116XG1) | 79,267,784 | Xq21.1 |
| III-6 | 164 | |||
| II-6 (FFPE tissue) | 168 | |||
| III-7 (FFPE tissue) | 164 | |||
| III-4 | 106 | DXS8076 (AFMB357XE5) | 82,666,040 | Xq21.1 |
| III-6 | 98 | |||
| II-6 (FFPE tissue) | 98 | |||
| III-7 (FFPE tissue) | 98 | |||
| III-4 | 217 | DXS1203 (AFM262VG1) | 92,654,766 | Xq21.32 |
| III-6 | 217 | |||
| II-6 (FFPE tissue) | 217 | |||
| III-7 (FFPE tissue) | 217 | |||
| III-4 | 131 | DXS990 (AFM136YC7) | 92,887,320 | Xq21.32 |
| III-6 | 127 | |||
| II-6 (FFPE tissue) | 127 | |||
| III-7 (FFPE tissue) | 127 | |||
| III-4 | 253 | DXS6799 (GATA29G07) | 97,265,664 | Xq21.33 |
| III-6 | 257 | |||
| II-6 (FFPE tissue) | 257 | |||
| III-7 (FFPE tissue) | 257 | |||
| III-4 | 201 | DXS8020 (AFMA162TC1) | 99,455,440 | Xq22.1 |
| III-6 | 212 | |||
| II-6 (FFPE tissue) | 203 | |||
| III-7 (FFPE tissue) | 212 | |||
| III-4 | 128 | DXS1106 (AFM263WE1) | 102,618,641 | Xq22.2 |
| III-6 | 130 | |||
| II-6 (FFPE tissue) | 126 | |||
| III-7 (FFPE tissue) | 130 | |||
| III-4 | 317 | DXS8055 (AFMB291YE5) | 114,561,258 | Xq23 |
| III-6 | 315 | |||
| II-6 (FFPE tissue) | 317 | |||
| III-7 (FFPE tissue) | 315 | |||
| III-4 | 206 | DXS1001 (AFM248WE5) | 119,720,593 | Xq24 |
| III-6 | 199 | |||
| II-6 (FFPE tissue) | 206 | |||
| III-7 (FFPE tissue) | 199 | |||
| III-4 | 165 | DXS1047 (AFM150XF10) | 128,902,983 | Xq25 |
| III-6 | 165 | |||
| II-6 (FFPE tissue) | 165 | |||
| III-7 (FFPE tissue) | 160 | |||
| III-4 | 90 | DXS1227 (AFM317YE9) | 140,630,173 | Xq27.2 |
| III-6 | 82 | |||
| II-6 (FFPE tissue) | 90 | |||
| III-7 (FFPE tissue) | 82 | |||
| III-4 | 170 | DXS8043 (AFMB018WD9) | 143,836,276 | Xq27.3 |
| III-6 | 156 | |||
| II-6 (FFPE tissue) | 170 | |||
| III-7 (FFPE tissue) | 170 | |||
| III-4 | 83 | DXS8091 (AFM345WG9) | 147,410,588 | Xq28 |
| III-6 | 83 | |||
| II-6 (FFPE tissue) | 83 | |||
| III-7 (FFPE tissue) | 83 | |||
| c.109T>C in NAA10 | ∼152,850,921b | Xq28 | ||
| III-4 | 310 | DXS1073 (AFM276XH9) | 153,482,054 | Xq28 |
| III-6 | 310 | |||
| II-6 (FFPE tissue) | 310 | |||
| III-7 (FFPE tissue) | 310 |
Genomic position represents a single nucleotide within the STR.
Maximum Shared Region (hg18): chrX:143,836,276–154,913,754 (telomere) (11 Mb).
Coverage Analysis
Exon capture techniques cannot capture all exonic regions because of design issues with replicative regions; this can therefore result in an incomplete set of variants. The X chromosome Agilent design consists of over 3 Mb of exon intervals. We evaluated the capture and sequencing coverage for all the currently known 1959 chrX transcripts in RefSeq (hg18). On the X chromosome, there are 8222 unique RefSeq exons. Of these exons, 736 were excluded from the SureSelect X-Chromosome Capture Kit because they were designated as pseudoautosomal or repetitive sequences (UCSC Genome Browser). The remaining 7486 exons have an average length of 358.9 bases and are covered on average by 5.7 bait probes per exon (bait probes were 120 bases long). The average read coverage in these regions was 214.6 reads per base; in these regions an average of 97.8% of the bases were covered by one or more reads, and 95.6% were covered at 10× or better.
We also analyzed the average coverage for each gene (calculated as the total read bases within exons divided by the exon length) more specifically in the affected haplotype in the 14 Mb critical region derived from analysis of family 1 (Table S2). The average coverage was 185× among the 167 nonpseudoautosomal or duplicative RefSeq transcripts (110 unique genes) in the 14 Mb region. These 167 transcripts consist of 864 exons, of which 860 had some read coverage and 809 had reads covering more than 95% of the bases in the exon. Most of the exon portions in the 14 Mb region that were not covered by reads were from the 89 exons not included in the SureSelect X-Chromosome Capture Kit because they were in the pseudoautosomal or duplicative regions. There was some coverage on a handful of these exons as well as intronic and intergenic regions flanking covered exons as a consequence of the experimental protocol. For example, if an excluded exon or intronic sequence is close (∼500 bases) to an exon that is captured, the long fragment size (and the fact that we only sequence the end of the fragment) can lead to reads for the exon or intron. In the NAA10 haplotype region, we had greater than 10× coverage for 86% of the exons in the region for family 1; the corollary of this means that 97 exons do not have 10× coverage in the NAA10 region. However, of the 856 exons in the 14 Mb region that did have >10× coverage, the GNUMAP and VAAST approach found only two candidate SNVs that met our criteria (present in proband, not present in his brother and uncle, polymorphic in his mother and grandmother, nonsynonymous, not in the 1000 genomes or dbSNP), and in the exons in the 30 Mb region, no SNVs met our criteria. Theoretically, we could have nonetheless missed a crucially important variant, so we needed to use parallel approaches to prove causality of the variant in NAA10.
A Second Family
During preparation of a manuscript reporting the above results, another group (L.M.B., J.J.J, L.G.B.) communicated to us that they had also identified the NAA10 c.109T>C mutation in an apparently unrelated family (herein designated as family 2) with an indistinguishable phenotype (see Figure 2D and 2E). Haplotype analysis had been performed for family 2, and it identified a shared region of chrX:140,061,918–154,913,754 (∼14 Mb) in three carrier females (I-2, II-2, and II-3) that was not present in an unaffected male (III-5). The borders of the region were defined by recombinant marker GATA31E08 and the q terminus.
Massively parallel sequencing of X chromosome exons was done on DNA samples from an obligate carrier and her unaffected child as previously described.9 The entire X chromosome exon region target sequence was 2,784,426 bp, and the X chromosome exon region oligonucleotide library was designed to target 2,264,175 bp of this (81.3%). The target-selected DNA libraries from one female heterozygote (M87_4) and one unaffected male child (M87_5) were sequenced on one lane each of an Illumina GAIIx in paired-end 75 bp configuration, which yielded, respectively, 75,382,114 and 65,015,016 reads (separate results are given for each sample) or 5,729,040,664 and 4,941,141,216 bp of total sequence. Of the filtered aligning sequence, 20.9% and 11.9% could be uniquely aligned to the entire X chromosome exon region target (2,784,426 bases). This aligned sequence yielded a gross overall coverage of 2058× and 1775× of the entire X chromosome exon region target. The capture efficiency varied across the targets; 2,538,791 and 2,464,367 bp (89.8%) of the entire X chromosome exon region yielded ≥1× coverage, 2,305,571 and 2,251,868 bp (81.8%) yielded ≥10× coverage, and 2,254,446 and 2,183,075 bp (79.7%) yielded ≥20× coverage. The Most Probable Genotype (MPG) variant-calling software24 was able to make base calls on 2,295,223 and 2,378,985 bp of this sequence (82.4% and 85.4%).
The genotypes were filtered on the basis of several attributes that were felt to be appropriate for this disorder (Table 6). Heterozygosity was used because the test subject was an unaffected female carrier for an X-linked trait. We performed further filtering to include nonsynonymous, splice-site, frame-shifting, and nonsense variants. Filtering to exclude variants present in dbSNP or in the ClinSeq cohort (401 control individuals)22 was also performed. Finally, a filter that bounded the variants genomically within the shared 14 Mb haplotype region was applied. This left one variant, a single missense mutation, c.109T>C in NAA10. This variant was confirmed in the other individuals, and mutation status segregated with affection status and carrier status (Figure 2D).
Table 6.
Filtering Analysis of Family 2
| X chromosome exons | Exons in Shared Haplotype | |
|---|---|---|
| Total SNVs | 3441 | 585 |
| Heterozygous | 2381 | 418 |
| Stop/NonSyn/FS/Splicea | 136 | 35 |
| Not in dbSNP | 40 | 10 |
| Not in ClinSeq | 20 | 1 (NAA10) |
The following abbreviations are used: Stop, nonsense variants; NonSyn, nonsynonymous variants; FS, frame-shifting variants; Splice, splice-site variants.
Genome-wide Significance
We next assessed whether adding members of family 2 would improve the power of the VAAST analysis. We combined variants from individual III-4 in family 1 with the obligate carrier mother (II-2) in family 2 and performed VAAST analysis on the 216 coding SNVs present in either of the two individuals. After incorporating the mother from family 2, NAA10 was the only candidate gene, and the result was statistically significant (p value = 3.8 × 10−5; Bonferroni corrected p value = 3.8 × 10−5 × 729 = 0.028).
Lack of Relatedness of the Two Families
Both families are of mixed European ancestry. To evaluate whether the two families inherited the NAA10 variant from a recent shared founder, we examined SNVs surrounding this variant to test for evidence of identity-by-descent (IBD). Assuming no sequencing errors, the largest region consistent with IBD between the two probands is only 30 kb (Table 7). By relaxing this assumption to allow for multiple sequencing errors, we find the largest region consistent with IBD is 704 kb, which is approximately 1.5 centiMorgans (cM)25 (Figure 3). If the two probands were as distantly related as fourth cousins, the expected IBD segment size would be 20 cM (conditioned on ascertainment from the presence of a shared variant).26 Therefore, the variation between the two probands in this region is inconsistent with recent shared ancestry. If the two probands inherited the mutation from a more distant founder and if they share the entire 700 kb region, the estimated time to the founder would be around 3300 years ago. Because this variant is an X-linked recessive lethal mutation, the probability of its being eliminated in each transmission is 1/3, and the variant is therefore highly unlikely to have occurred 3300 years ago. We conclude that the disease-causing variant in the two families resulted from two independent mutational events.
Table 7.
Relationship Inference of the Two Families
| Position |
First Family |
Second Family |
|||
|---|---|---|---|---|---|
| Inconsistent with IBD | Proband | Imputed Genotype of Proband | Unaffected Son | Mother | |
| 152802312 | X | G | A | G | AG |
| 152804123 | X | A | G | A | GA |
| 152821601 | X | A | G | A | GA |
| 152821643 | X | C | T | C | TC |
| 152825187 | X | G | A | G | AG |
| 152829448 | X | G | A | G | AG |
| 152853035 | G | G | A | GA | |
| 152860231 | X | T | G | G | GG |
| 152932023 | A | A | A | AA | |
| 152937386 | G | G | G | GG | |
| 152945374 | C | C | C | CC | |
| 153317692 | X | G | A | A | AA |
| 153321366 | G | G | G | GG | |
| 153534024 | X | C | A | C | AC |
| 153534719 | X | C | G | C | GC |
| 153556589 | X | A | G | A | GA |
| 153557591 | X | G | A | G | AG |
| 153557667 | X | T | C | T | CT |
Figure 3.
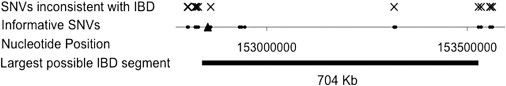
IBD Analysis of the Two Families in the Genomic Region Surrounding c.109T>C
Informative SNVs consist of two classes: rare SNVs present in the proband from family 1 in genomic regions with high coverage in the second family, and common SNVs present in the first proband and at least one member of the second family. Variant c.109T>C is indicated by the triangle. We imputed the genotype of the proband from family 2 from the genotypes of the mother and unaffected sibling of the second family (see Table 7). SNVs inconsistent with IBD, in which the imputed genotype of the second proband does not match the first proband, are indicated with an X. After allowing for multiple sequencing errors, the largest genomic segment consistent with IBD is around 700 kb in length.
Functional Analysis
N-terminal acetylation of proteins is catalyzed by N-terminal acetyltransferases (NATs). The primary NAT in terms of targeted substrates is the evolutionarily conserved NatA.27–29 The functional impact of N-terminal acetylation remains quite elusive, but recent data suggest a role as a destabilization signal for proteins.30 The human NatA complex is composed of the catalytic subunit hNaa10p (hARD1) and the auxiliary subunit hNaa15p (NATH/hNAT1), both essential for its activity.31 Increased NatA levels have been linked to tumor progression, and depletion of NatA subunits from cancer cells induces cell-cycle arrest and apoptosis.32 Human Naa10p is a protein of 235 amino acid residues, of which the first 178 residues compose a globular region, whereas the latter 57 residues are predicted to form an unstructured and flexible C-terminal tail.33 Thus, Ser37 is located in a structured part of hNaa10p. For many soluble globular proteins, Pro is known to be potentially disruptive for secondary structure elements such as the alpha-helix and beta-sheets.34 Thus, although the structure of hNaa10p is undetermined, the p.Ser37Pro mutation could indeed affect the structure of hNaa10p and thereby the catalytic activity. Ser37 and its surrounding residues are highly conserved among eukaryotes,31 suggesting an essential function. A SIFT analysis predicting whether an amino acid substitution affects protein function35 strongly suggested that a substitution from Ser to Pro at position 37 would affect protein function with a score of 0.00.
In order to directly assess the functional consequences of the hNaa10p p.Ser37Pro mutation, we analyzed the wild-type (WT) and the mutant proteins by a quantitative in vitro N-terminal acetylation assay. The enzyme activities of hNaa10p WT and hNaa10p p.Ser37Pro were determined with four unique peptides as substrate; these peptides having been previously shown to be acetylated by hNaa10p and NatA (Figure 4). Compared to hNaa10p WT, hNaa10p p.Ser37Pro displayed a 60%–80% reduction in NAT activity toward the in vivo substrate RNaseP protein p30 (AVFAD-) and toward β-actin (DDDIA-) and γ-actin (EEEIA-). In contrast, the activity toward the NatA substrate high-mobility group protein A1 (SESSS-) was only reduced by 20% (Figure 4). The oligopeptides AVFAD- and SESSS- represent classical cotranslational NatA substrates, being the N termini of proteins that are partially (AVFAD-) or fully (SESSS-) acetylated by NatA in HeLa cells.27 On the other hand, β-actin (DDDIA-) and γ-actin (EEEIA-) are nonclassical substrates recently shown to be acetylated more efficiently by hNaa10p/NatA than the classical substrates were, representing a posttranslational NAT activity.36
Figure 4.
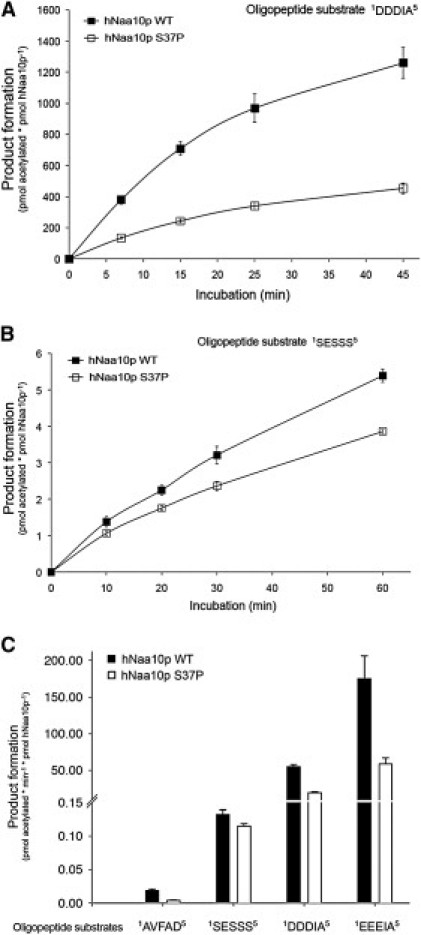
NAT Activity of Recombinant hNaa10p WT or p.Ser37Pro toward Synthetic N-Terminal Peptides
(A and B) Purified MBP-hNaa10p WT or p.Ser37Pro were mixed with the indicated oligopeptide substrates (200 μM for SESSS and 250 μM for DDDIA) and saturated levels of acetyl-CoA (400 μM). Aliquots were collected at indicated time points and the acetylation reactions were quantified with reverse phase HPLC peptide separation. Error bars indicate the standard deviation based on three independent experiments. The five first amino acids in the peptides are indicated, for further details see Subjects and Methods. Time-dependent acetylation reactions were performed to determine initial velocity conditions when comparing the WT and Ser37Pro NAT activities toward different oligopeptides.
(C) Purified MBP-hNaa10p WT or p.Ser37Pro were mixed with the indicated oligopeptide substrates (200 μM for SESSS and AVFAD and 250 μM for DDDIA and EEEIA) and saturated levels of acetyl-CoA (400 μM) and incubated for 15 min (DDDIA and EEEIA) or 20 min (SESSS and AVFAD) at 37°C in acetylation buffer. The acetylation activity was determined as above. Error bars indicate the standard deviation based on three independent experiments. Black bars indicate the acetylation capacity of the MBP-hNaa10p WT, whereas white bars indicate the acetylation capacity of the MBP-hNaa10p mutant p.Ser37Pro. The five first amino acids in the peptides are indicated.
Discussion
The data presented here show that a mutation in an enzyme involved in N-terminal acetylation of proteins leads to a distinct, previously undescribed X-linked phenotype in humans and that males who carry the hypomorphic hNaa10p p.Ser37Pro allele die in infancy with cardiac arrhythmias. N-terminal acetylation is one of the most common protein modifications in humans, occurring on approximately 80% of all human proteins27 It is catalyzed by several distinct NAT enzymes, of which the major one is NatA.29 The catalytic subunit of the NatA complex, hNaa10p, is essential for survival in the organisms Drosophila melanogaster,37 Trypanosoma brucei,38 and Caenorhabditis elegans.39 It is presumed that an amorphic NAA10 mutation would lead to embryonal lethality in humans, although one can only prove this by analyzing tissues from pregnancies that did not survive to term. The strong conservation of Naa10p Ser37, the predicted functional effect of p.Ser37Pro due to structural distortion, and the demonstrated disruption of catalytic activity by p.Ser37Pro (Figure 4) strongly imply that the hemizygous males with hNaa10p p.Ser37Pro have impaired NatA function. Thus, a variety of protein N termini, both those that are cotranslationally and those that are posttranslationally acetylated (e.g., actins), are likely to be insufficiently acetylated. Most likely, the serious consequences of the p.Ser37Pro mutation are caused by the lack of N-terminal acetylation for one or several proteins strictly requiring this modification for function or for maintenance of adequate amounts in the cell. Because hNaa10p has also been suggested to perform N-lysine (N-epsilon) acetylation of proteins such as beta-catenin,40 a lack of N-lysine acetylation of selected substrates could also cause the observed effects. Finally, proposed noncatalytic functions of hNaa10p41,42 could be affected in the p.Ser37Pro mutant and thereby also play a role in the observed phenotypes.
We have demonstrated herein that a probabilistic disease-causing variant discovery algorithm can readily identify and characterize the genetic basis of a previously unrecognized X-linked syndrome. We have also shown that this algorithm, when used in parallel with high-throughput sequencing, can identify variants with high prioritization for causing disease with as few as two individuals. In this instance, we screened ∼150 variants distributed among ∼2000 transcripts on the X chromosome in one sample from individual III-4 in family 1. With no prior filtering, we prioritized three to five possible candidate genes, and the mutation in NAA10 ranked second overall (Table 4). Including exon capture data from relatives increased NAA10's ranking to first overall in just one family. Furthermore, after combining variants from the proband in family 1 with the obligate carrier mother in family 2, VAAST identified NAA10 as the only statistically significant candidate.
Although we have noted that the affected infants have an aged appearance, we have not established any direct link with progeria or other progeroid syndromes. The autopsies did not reveal any premature arteriosclerosis or degeneration of vascular smooth muscle cells, as is seen in Hutchinson-Gilford progeria syndrome (MIM 176670).43,44 Cell lines now being derived from family 1 and possibly future animal models will provide important insights about the pathophysiology underlying this previously unrecognized syndrome.
Acknowledgments
We express our gratitude to the families for their extraordinary cooperation and assistance. We also thank David Nix, Nina Glomnes, and Whitney Fitts for advice and/or technical assistance with family 1. Exon capture and sequencing for family 1 was paid for by the Department of Psychiatry, University of Utah (to G.J.L.). Collection of DNA and phenotyping for family 1 was supported by the Clinical Genetics Research Program: Phenotyping Core, under CCTS grant UL1RR025764 at the University of Utah. The University of Utah Microarray and Genomic Analysis core facility was supported by award number P30CA042014 from the National Cancer Institute. Functional analyses were supported by the Research Council of Norway (grant 197136 to T.A.) and the Norwegian Cancer Society (to J.R.L. and T.A.). B.M. and M.Y. were supported by National Human Genome Research Institute (NHGRI) 1RC2HG005619, K.W. by a pilot/methodological study award from NIH/National Center for Research Resources grant UL1 RR025774, J.X. by NIH/NHGRI K99HG005846, and W.E.J. by NHGRI 5R01HG5692. The research on family 2 was supported by Intramural Funds of the NHGRI, NIH (L.G.B.). These authors thank Danielle Brinkman for the initial consenting and records gathering for family 2 and Caitlin Krause and Jamie Teer for technical support.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
ANNOVAR Software, http://www.openbioinformatics.org/annovar/
Complete Genomics Diversity Panel, http://www.completegenomics.com/sequence-data/download-data/
GATK Software, http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit
GNUMAP, http://dna.cs.byu.edu/gnumap/
Online Mendelian Inheritance in Man(OMIM), http://www.omim.org
SIFT (Sorting Intolerant From Tolerant) Analysis, http://sift.bii.a-star.edu.sg/www/SIFT_seq_submit2.html
UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloğlu A., Ozen S., Sanjad S. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierce S.B., Walsh T., Chisholm K.M., Lee M.K., Thornton A.M., Fiumara A., Opitz J.M., Levy-Lahad E., Klevit R.E., King M.C. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am. J. Hum. Genet. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilgüvar K., Oztürk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A.O., Gökben S. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedges D.J., Burges D., Powell E., Almonte C., Huang J., Young S., Boese B., Schmidt M., Pericak-Vance M.A., Martin E. Exome sequencing of a multigenerational human pedigree. PLoS ONE. 2009;4:e8232. doi: 10.1371/journal.pone.0008232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnefond A., Durand E., Sand O., De Graeve F., Gallina S., Busiah K., Lobbens S., Simon A., Bellanné-Chantelot C., Létourneau L. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS ONE. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston J.J., Teer J.K., Cherukuri P.F., Hansen N.F., Loftus S.K., Chong K., Mullikin J.C., Biesecker L.G., NIH Intramural Sequencing Center (NISC) Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. Am. J. Hum. Genet. 2010;86:743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston J.J., Olivos-Glander I., Killoran C., Elson E., Turner J.T., Peters K.F., Abbott M.H., Aughton D.J., Aylsworth A.S., Bamshad M.J. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: Robust phenotype prediction from the type and position of GLI3 mutations. Am. J. Hum. Genet. 2005;76:609–622. doi: 10.1086/429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evjenth R., Hole K., Ziegler M., Lillehaug J.R. Application of reverse-phase HPLC to quantify oligopeptide acetylation eliminates interference from unspecific acetyl CoA hydrolysis. BMC Proc. 2009;3(Suppl 6):S5. doi: 10.1186/1753-6561-3-S6-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durbin R.M., Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evjenth R., Hole K., Karlsen O.A., Ziegler M., Arnesen T., Lillehaug J.R. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J. Biol. Chem. 2009;284:31122–31129. doi: 10.1074/jbc.M109.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39(Database issue):D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., Dicuccio M., Federhen S. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reese M.G., Moore B., Batchelor C., Salas F., Cunningham F., Marth G.T., Stein L., Flicek P., Yandell M., Eilbeck K. A standard variation file format for human genome sequences. Genome Biol. 2010;11:R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clement N.L., Snell Q., Clement M.J., Hollenhorst P.C., Purwar J., Graves B.J., Cairns B.R., Johnson W.E. The GNUMAP algorithm: Unbiased probabilistic mapping of oligonucleotides from next-generation sequencing. Bioinformatics. 2010;26:38–45. doi: 10.1093/bioinformatics/btp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Vinckenbosch N., Tian G., Huerta-Sanchez E., Jiang T., Jiang H., Albrechtsen A., Andersen G., Cao H., Korneliussen T. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 21.Kurpinski K.T., Magyari P.A., Gorlin R.J., Ng D., Biesecker L.G. Designation of the TARP syndrome and linkage to Xp11.23-q13.3 without samples from affected patients. Am. J. Med. Genet. A. 2003;120A:1–4. doi: 10.1002/ajmg.a.10201. [DOI] [PubMed] [Google Scholar]
- 22.Biesecker L.G., Mullikin J.C., Facio F.M., Turner C., Cherukuri P.F., Blakesley R.W., Bouffard G.G., Chines P.S., Cruz P., Hansen N.F., NISC Comparative Sequencing Program The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yandell M., Huff C.D., Hu H., Singleton M., Moore B., Xing J., Jorde L.B., Reese M.G. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21 doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teer J.K., Bonnycastle L.L., Chines P.S., Hansen N.F., Aoyama N., Swift A.J., Abaan H.O., Albert T.J., Margulies E.H., Green E.D., NISC Comparative Sequencing Program Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 26.Huff C.D., Witherspoon D.J., Simonson T.S., Xing J., Watkins W.S., Zhang Y., Tuohy T.M., Neklason D.W., Burt R.W., Guthery S.L. Maximum-likelihood estimation of recent shared ancestry (ERSA) Genome Res. 2011;21:768–774. doi: 10.1101/gr.115972.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J.E., Vandekerckhove J., Lillehaug J.R., Sherman F., Gevaert K. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen J.R., Kayne P.S., Moerschell R.P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polevoda B., Arnesen T., Sherman F. A synopsis of eukaryotic Nalpha-terminal acetyltransferases: Nomenclature, subunits and substrates. BMC Proc. 2009;3(Suppl 6):S2. doi: 10.1186/1753-6561-3-S6-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang C.S., Shemorry A., Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnesen T., Anderson D., Baldersheim C., Lanotte M., Varhaug J.E., Lillehaug J.R. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 2005;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gromyko D., Arnesen T., Ryningen A., Varhaug J.E., Lillehaug J.R. Depletion of the human Nα-terminal acetyltransferase A induces p53-dependent apoptosis and p53-independent growth inhibition. Int. J. Cancer. 2010;127:2777–2789. doi: 10.1002/ijc.25275. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez-Puig N., Fersht A.R. Characterization of the native and fibrillar conformation of the human Nalpha-acetyltransferase ARD1. Protein Sci. 2006;15:1968–1976. doi: 10.1110/ps.062264006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou P.Y., Fasman G.D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 35.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Damme P., Evjenth R., Foyn H., Demeyer K., De Bock P.J., Lillehaug J.R., Vandekerckhove J., Arnesen T., Gevaert K. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N{alpha}-acetyltransferases and point to hNaa10p as the post-translational actin N{alpha}-acetyltransferase. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.004580. M110, 004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Mijares M., Gall M.D., Turan T., Javier A., Bornemann D.J., Manage K., Warrior R. Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev. Dyn. 2010;239:2813–2827. doi: 10.1002/dvdy.22418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingram A.K., Cross G.A., Horn D. Genetic manipulation indicates that ARD1 is an essential N(infinity)-acetyltransferase in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;111:309–317. doi: 10.1016/s0166-6851(00)00322-4. [DOI] [PubMed] [Google Scholar]
- 39.Sönnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 40.Lim J.H., Park J.W., Chun Y.S. Human arrest defective 1 acetylates and activates beta-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–10682. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- 41.Hua K.T., Tan C.T., Johansson G., Lee J.M., Yang P.W., Lu H.Y., Chen C.K., Su J.L., Chen P.B., Wu Y.L. N-α-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218–231. doi: 10.1016/j.ccr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Lee C.F., Ou D.S., Lee S.B., Chang L.H., Lin R.K., Li Y.S., Upadhyay A.K., Cheng X., Wang Y.C., Hsu H.S. hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capell B.C., Tlougan B.E., Orlow S.J. From the rarest to the most common: Insights from progeroid syndromes into skin cancer and aging. J. Invest. Dermatol. 2009;129:2340–2350. doi: 10.1038/jid.2009.103. [DOI] [PubMed] [Google Scholar]
- 44.Liu G.H., Barkho B.Z., Ruiz S., Diep D., Qu J., Yang S.L., Panopoulos A.D., Suzuki K., Kurian L., Walsh C. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


