Abstract
Sequencing studies are increasingly being conducted to identify rare variants associated with complex traits. The limited power of classical single-marker association analysis for rare variants poses a central challenge in such studies. We propose the sequence kernel association test (SKAT), a supervised, flexible, computationally efficient regression method to test for association between genetic variants (common and rare) in a region and a continuous or dichotomous trait while easily adjusting for covariates. As a score-based variance-component test, SKAT can quickly calculate p values analytically by fitting the null model containing only the covariates, and so can easily be applied to genome-wide data. Using SKAT to analyze a genome-wide sequencing study of 1000 individuals, by segmenting the whole genome into 30 kb regions, requires only 7 hr on a laptop. Through analysis of simulated data across a wide range of practical scenarios and triglyceride data from the Dallas Heart Study, we show that SKAT can substantially outperform several alternative rare-variant association tests. We also provide analytic power and sample-size calculations to help design candidate-gene, whole-exome, and whole-genome sequence association studies.
Introduction
Genome-wide association studies (GWASs) have identified more than 1000 genetic loci associated with many human diseases and traits,1 yet common variants identified through GWASs often explain only a small proportion of trait heritability. The advent of massively parallel sequencing2 has transformed human genetics3,4 and has the potential to explain some of this missing heritability through identification of trait-associated rare variants.5 Although considerable resources have been devoted to sequence mapping and genotype calling,6–9 successful application of sequencing to the study of complex traits requires novel statistical methods that allow researchers to test efficiently for association given data on rare variants10 and to perform sample-size and power calculations to help design sequencing-based association studies.
Rare genetic variants, here defined as alleles with a frequency less than 1%–5%, can play key roles in influencing complex disease and traits.11 However, standard methods used to test for association with single common genetic variants are underpowered for rare variants unless sample sizes or effect sizes are very large.12,13 A logical alternative approach is to employ burden tests that assess the cumulative effects of multiple variants in a genomic region.12–18 Burden tests proposed to date are based on collapsing or summarizing the rare variants within a region by a single value, which is then tested for association with the trait of interest. For example, the cohort allelic sum test (CAST)14 collapses information on all rare variants within a region (e.g., the exons of a gene) into a single dichotomous variable for each subject by indicating whether or not the subject has any rare variants within the region and then applies a univariate test. Instead of collapsing by dichotomizing the number of rare variants within a region, collapsing by counting them is also possible.18 The combined multivariate and collapsing method12 extends CAST by collapsing rare variants within a region into subgroups on the basis of allele frequency, collapsing subgroups as in CAST, and applying a multivariate test to the subgroups. The weighted sum test (WST)13 specifically considers the case-control setting and collapses a set of SNPs into a single weighted average of the number of rare alleles for each individual. Numerous alternative methods are largely variations on these approaches.16,17,19
A limitation for all these burden tests is that they implicitly assume that all rare variants influence the phenotype in the same direction and with the same magnitude of effect (after incorporating known weights). However, one would expect most variants (common or rare) within a sequenced region to have little or no effect on phenotype, whereas some variants are protective and others deleterious, and the magnitude of each variant's effect is likely to vary (e.g., rarer variants might have larger effects). Hence, collapsing across all variants is likely to introduce substantial noise into the aggregated index, attenuate evidence for association, and result in power loss. Furthermore, burden tests require either specification of thresholds for collapsing or the use of permutation to estimate the threshold.16–20 Permutation tests are computationally expensive, especially on the whole-genome scale, and are difficult for covariate adjustment because permutation requires independence between the genotype and the covariates.
The recently proposed C-alpha test21 is a non-burden-based test and is hence robust to the direction and magnitude of effect. For case-control data, it compares the expected variance to the actual variance of the distribution of allele frequencies. These important advantages allow the C-alpha test to have improved power over burden-based tests, especially when the effects are in different directions. Despite these attractive features, the C-alpha test does not allow for easy covariate adjustment, such as for controlling population stratification, which is important in genetic association studies. The C-alpha test also uses permutation to obtain a p value when linkage disequilibrium is present among the variants, which is, as noted earlier, computationally expensive for whole-genome experiments. The approach has not been generalized to analysis of continuous phenotypes.
We propose in this paper the sequence kernel association test (SKAT), a flexible, computationally efficient, regression approach that tests for association between variants in a region (both common and rare) and a dichotomous (e.g., case-control) or continuous phenotype while adjusting for covariates, such as principal components, to account for population stratification.22 The kernel machine regression framework was previously considered for common variants.23,24 In this paper, we provide several essential methodological improvements necessary for testing rare variants. SKAT uses a multiple regression model to directly regress the phenotype on genetic variants in a region and on covariates, and so allows different variants to have different directions and magnitude of effects, including no effects; SKAT also avoids selection of thresholds. We develop a kernel association test to test the regression coefficients of the variants by using a variance-component score test in a mixed-model framework by accounting for rare variants.
SKAT is computationally efficient. This quality is especially important in genome-wide studies because SKAT only requires fitting the null model in which phenotypes are regressed on the covariates alone; p values are easily computed with simple analytic formulae. Additional features of SKAT include exploitation of local correlation structure, incorporation of flexible weights to boost power (e.g., by increasing the weight of rarer variants or incorporating functionality), and allowance for epistatic variant effects. As discussed in more detail below, under special cases, the SKAT, C-alpha test, and individual variant test statistics are closely related.
We demonstrate through simulation and analysis of resequencing data from the Dallas Heart Study that SKAT is often more powerful than existing tests across a broad range of models for both continuous and dichotomous data. We also investigate the factors that influence power for sequence association studies. Finally, we describe analytic tools to estimate statistical power and sample sizes to guide the design of new sequence association studies of rare variants with SKAT.
Material and Methods
Sequencing Kernel Association Test
SKAT is a supervised test for the joint effects of multiple variants in a region on a phenotype. Regions can be defined by genes (in candidate-gene or whole-exome studies) or moving windows across the genome (in whole-genome studies). For each region, SKAT analytically calculates a p value for association while adjusting for covariates. Adjustments for multiple comparisons are necessary for analyzing multiple regions, for example with the Bonferroni correction or FDR control.
Notation
Assume n subjects are sequenced in a region with p variant sites observed. Covariates might include age, gender, and top principal components of genetic variation for controlling population stratification.22 For the i-th subject, yi denotes the phenotype variable, Xi = (Xi1, Xi2, .., Xim) denotes the covariates, and Gi = (Gi1, Gi2, …, Gip) denotes the genotypes for the p variants within the region. Typically, we assume an additive genetic model and let Gij, = 0, 1, or 2 represent the number of copies of the minor allele. Dominant and recessive models can also be considered.
SKAT Model and Test for Linear SNP Effects
For a simple illustration of SKAT, we focus here on testing for a relationship between the variants and the phenotype by using classical multiple linear and logistic regression. We describe how the SKAT can incorporate epistatic effects later. To relate the sequence variants in a region to the phenotype, consider the linear model
| (Equation 1) |
when the phenotypes are continuous traits, and the logistic model
| (Equation 2) |
when the phenotypes are dichotomous (e.g., y = 0/1 for case or control). Here α0 is an intercept term, α = [α1,…, αm]' is the vector of regression coefficients for the m covariates, β = [β1,…,βp]' is the vector of regression coefficients for the p observed gene variants in the region, and for continuous phenotypes ɛi is an error term with a mean of zero and a variance of σ2. Under both linear and logistic models, and evaluating whether the gene variants influence the phenotype, adjusting for covariates, corresponds to testing the null hypothesis H0: β = 0, that is, β1 = β2 = … = βp = 0. The standard p-DF likelihood ratio test has little power, especially for rare variants. To increase the power, SKAT tests H0 by assuming each βj follows an arbitrary distribution with a mean of zero and a variance of wjτ, where τ is a variance component and wj is a prespecified weight for variant j. One can easily see that H0: β = 0 is equivalent to testing H0: τ = 0, which can be conveniently tested with a variance-component score test in the corresponding mixed model; this is known to be a locally most powerful test.25 A key advantage of the score test is that it only requires fitting the null model yi = α0 + α1'Xi + ɛi for continuous traits and the logit P(yi = 1) = α0 + α1'Xi for dichotomous traits.
Specifically, the variance-component score statistic is
| (Equation 3) |
where K = GWG', is the predicted mean of y under H0, that is for continuous traits and for dichotomous traits; and and are estimated under the null model by regressing y on only the covariates X. Here G is an n × p matrix with the (i, j)-th element being the genotype of variant j of subject i, and W = diag(w1,…, wp) contains the weights of the p variants.
In fact, K is an n × n matrix with the (i, i')-th element equal to . is called the kernel function, and measures the genetic similarity between subjects i and i' in the region via the p markers. This particular form of is called the weighted linear kernel function. We later discuss other choices of the kernel to model epistatic effects.
Good choices of weights can improve power. Each weight wj is prespecified, with only the genotypes, covariates and external biological information, that is estimated without using the outcome, and reflects the relative contribution of the j-th variant to the score statistic: if wj is close to zero, then the j-th variant makes only a small contribution to Q. Thus, decreasing the weight of noncausal variants and increasing the weight of causal variants can yield improved power. Because in practice we do not know which variants are causal, we propose to set , the beta distribution density function with prespecified parameters a1 and a2 evaluated at the sample minor-allele frequency (MAF) (across cases and controls combined) for the j-th variant in the data. The beta density is flexible and can accommodate a broad range of scenarios. For example, if rarer variants are expected to be more likely to have larger effects, then setting 0 < a1 ≤ 1 and a2 ≥ 1 allows for increasing the weight of rarer variants and decreasing the weight of common weights. We suggest setting a1 = 1 and a2 = 25 because it increases the weight of rare variants while still putting decent nonzero weights for variants with MAF 1%–5%. All simulations were conducted with this default choice unless stated otherwise. Note that a smaller a1 results in more strongly increasing the weight of rarer variants. Examples of weights across a range of a1 and a2 values are presented in Figure S1, available online. Note that a1 = a2 = 1 corresponds to wj = 1, that is all variants are weighted equally, and a1 = a2 = 0.5 corresponds to , that is wj is the inverse of the variance of the genotype of marker j, which puts almost zero weight for MAFs > 1% and can be used if one believes only variants with MAF < 1% are likely to be causal. Note that SKAT calculated with this weight is identical to the unweighted SKAT test with the standardized genotypes in Equations 1 and 2. Other forms of the weight as a function of MAF can also be used. Because SKAT is a score test, the type I error is protected for any choice of prechosen weights. Note that the weights used in the weighted sum test13 involve phenotype information and will therefore alter the null distribution of SKAT if such weights are used.
Under the null hypothesis, Q follows a mixture of chi-square distributions, which can be closely approximated with the computationally efficient Davies method.26 See Appendix A for details.
A special case of SKAT arises when the outcome is dichotomous, no covariates are included, and all wj = 1. Under these conditions, we show in Appendix A that the SKAT test statistic Q is equivalent to the C-alpha test statistic T. Hence, the C-alpha test can be seen as a special case of SKAT, or alternatively, SKAT can be seen as a generalized C-alpha test that does not require permutation but calculates the p value analytically, allows for covariate adjustment, and accommodates either dichotomous or continuous phenotypes. Because SKAT under flat weights is also equivalent to the kernel machine regression test23,24 and because the kernel machine regression test is in turn related to the SSU test,27 it follows transitively that SKAT under flat weights, the kernel machine regression test, the SSU test, and the C-alpha test are all equivalent and special cases of SKAT. Note that the null distribution is calculated differently via these methods, and SKAT gives more accurate analytic p values, especially in the extreme tail, when sample sizes are sufficient.
Relationship between Linear SKAT and Individual Variant Test Statistics
One can efficiently compute the test statistic Q by exploiting a close connection between the SKAT score test statistic Q and the individual variant test statistics. In particular, Q is a weighted sum of the individual score statistics for testing for individual variant effects. Hence, by letting gj = [G1j, G1j, …, Gnj]' denote the n × 1 vector containing the genotypes of the n subjects for variant j, it is straightforward to see that , where is the individual score statistic for testing the marginal effect of the j-th marker (H0: βj = 0) under the individual linear or logistic regression model of yi on Xi and only the j-th variant Gij:
for continuous phenotypes and
for dichotomous phenotypes. is estimated as for continuous traits and for dichotomous traits. As a score test, one needs to fit the null model only a single time to be able to compute the Sj for all individual variants j as well as all regions to be tested. Similarly, if multiple regions are under consideration, then the same can be used to compute the SKAT Q statistics for each region.
Accommodating Epistatic Effects and Prior Information under the SKAT
An attractive feature of SKAT is the ability to model the epistatic effects of sequence variants on the phenotype within the flexible kernel machine regression framework.28–30 To do so, we replace Gi'β by a more flexible function f(Gi) in the linear and logistic models (1) and (2) where f(Gi) allows for rare variant by rare variant and common variant by rare-variant interactions. Specifically, for continuous traits we use the semiparametric linear model23,29
| (Equation 4) |
and for dichotomous traits, we use the semiparametric logistic model24,30
| (Equation 5) |
Here the variants, Gi, are related to the phenotype through a possibly nonparametric function f(·), which is assumed to lie in a functional space generated by a positive semidefinite kernel function . Models (1) and (2) assume linear genetic effects and are specified by . By changing , one can allow for more complex models. Intuitively, is a function that measures genetic similarity between the i-th and i'-th subjects via the p variants in the region, and any positive semidefinite function can be used as a kernel function. We tailored several useful and commonly used kernels specifically for the purpose of rare-variant analysis: the weighted linear kernel, the weighted quadratic kernel, and the weighted identity by state (IBS) kernel.
The weighted linear kernel function implies that the trait depends on the variants in a linear fashion and is equivalent to the classical linear and logistic model presented in Equations 1 and 2. The weighted quadratic kernel implicitly assumes that the model depends on the main effects and quadratic terms for the gene variants and the first-order variant by variant interactions. The weighted IBS kernel , defines similarity between individuals as the number of alleles that share IBS. For additively coded autosomal genotype data, . The model implied by the weighted IBS kernel models the SNP effects nonparametrically.31 Consequently, this allows for epistatic effects because the function f(·) does not assume linearity or interactions of a particular order (e.g., the second order), Using the weighted IBS kernel removes the assumption of additivity because the number of alleles that are identical by state is a physical quantity that does not change on the basis of different genotype encodings.
We note that a kernel function that better captures both the similarity between individuals and the causal variant effects will increase power. In particular, if relationships are linear and no interactions are present, then the weighted linear kernel will have highest power. If interactions are present, the weighted quadratic and weighted IBS kernels can increase power. Our experience suggests using the IBS kernel when the number of interacting variants within the region is modest. As our understanding of genetic architecture improves so too will our knowledge of which kernel to use.
In each of the above kernels, wj is an allele specific weight that controls the relative importance of the jth variant and might be a function of factors such as allele frequency or anticipated functionality. Without prior information, we suggest the use of the suggested earlier. However, if prior information is available, for example some variants are predicted as functional or damaging via Polyphen32 or Sift,33 weights can be selected to increase the weight for likely functionality.
To test for the effects of gene variants in a region on a phenotype, one tests the null hypothesis H0: f(G) = 0. SKAT tests for this null hypothesis by assuming the n × 1 vector f = [f(G1), …, f(Gn)]' for the genetic effects of n subjects follows a distribution with mean zero and covariance τK, where τ is a variance component that indexes the effects of the variants.29,30 Hence, we can test the null hypothesis that corresponds to testing H0: τ = 0 by a variance-component score test. In particular, we simply replace K in Equation 3 by using the K discussed in this section, for example, the weighted IBS kernel, for epistatic effect. All subsequent calculations for computing a p value remain the same.
Because the SKAT evaluates significance via a score test, which operates under the null hypothesis, the SKAT is valid (in terms of protecting type I error) irrespective of the kernel and the weights used. Good choices of the kernel and the weights simply increase power.
Planning New Sequencing-Based Association Studies: Estimation of Power and Sample Size
Power and sample-size calculations are important in designing sequencing studies of complex traits. Using a modification of the higher-order moment-approximation method,34 we provide an analytic method to carry out efficiently such calculations for SKAT.35 Specifically, for a fixed sample size and α level, given a prior hypothesis on the genetic architecture of a particular region, the effect size, and the proportion and number of causal variants within a region, our method provides the power to detect the region as significant with SKAT. Similarly, if the desired power is fixed, the approach can be used to find the necessary sample size.
There are key differences between the power and sample-size estimation for single-variant- and region (set)-based tests. For a region (set)-based test, the power depends strongly on the underlying genetic architecture, and its estimation requires modeling this genetic architecture and the linkage disequilibrium (LD) between variants. Therefore, to estimate power to detect a particular region as associated with a phenotype requires specification of the significance level, sample size, which variants in the region are causal with corresponding effect size, and the LD structure of the variants in the region. Ideally, one could use prior data to assess the LD and MAF. Because prior data can be difficult to obtain, we currently recommend the use of either 1000 Genomes Project data36 or data simulated under a population genetics model.37 Relevant preliminary data will become increasingly available as sequencing studies become more common.
Our SKAT software uses simulated data based on the coalescent population genetic model (released with the software package) as a default in performing sample-size and power calculations, and instead of directly specifying the effects of any given variant, the user can input an MAF threshold for determining which variants are regarded as rare and also a proportion determining how many of the rare variants are causal. The causal variants are then randomly selected from the alleles with true MAF (based on simulated or preliminary data) less than the threshold. The magnitudes of the effects |βj| for causal variants are set to be equal to c × |log10 MAF| where c is determined on the basis of the maximum effect size the user would like to allow (described below in the power simulations section) at MAF = 10−4. This allows the effects of causal variants to decrease with MAFs. Because these parameters can be difficult to choose as a priori, power and sample size can be reasonably estimated by averaging results over a range of parameter values. Similarly, because the regional architecture can vary across different regions, for genome-wide studies, one can average over multiple randomly selected regions as currently implemented in the SKAT software.
Numerical Experiments and Simulations
To validate SKAT in terms of protecting type I error and to assess its power compared to burden tests and the accuracy of our power and sample-size tools, we carried out simulation studies under a range of configurations. For all simulations, we determined sequence genotypes by simulating 10,000 chromosomes for a 1 Mb region on the basis of a coalescent model that mimics the LD pattern local recombination rate and the population history for Europeans by using COSI.37
Type I Error Simulations
To investigate whether SKAT preserves the desired type I error rate at the near genome-wide threshold level, for example α = 10−6, it is necessary to conduct simulations with hundreds of millions of simulated datasets. Although SKAT is computationally efficient, generating such a large number of datasets is challenging. To reduce the computation burden, we took the following approach. Using 10,000 randomly selected sets of 30 kb subregions within a 1 Mb chromosome, we first generated 10,000 sets of genotypes G(n × p) from the coalescent model, with p variants on n subjects. Then, for each of the 10,000 simulated genotype data sets, we simulated 10,000 sets of continuous phenotypes such that we were able to obtain 108 individual genotype-phenotype data sets by using the model:
where X1 is a continuous covariate generated from a standard normal distribution, X2 is a dichotomous covariate taking values 0 and 1 with a probability of 0.5, and ɛ follows a standard normal distribution. Note that the continuous trait values are not related to the genotype so that the null model holds. The 30 kb regions on which the genotype values are based contained 605 variants on average, but the number of observed variants for any given data set was considerably less and depended on the sample size n, which we set to 500, 1000, 2500, and 5000.
We repeated the type I error simulations for dichotomous phenotypes as above, except the dichotomous outcomes were generated via the model:
where α0 was determined to set the prevalence to 1% and case-control sampling is used.
For both continuous and dichotomous simulations, we applied SKAT by using the default weighted linear kernel to each of the 108 data sets and estimated the empirical type I error rate as the proportion of p values less than α = 10−4, 10−5, or 10−6.
We note that the estimated type I error from this approach is not the same as the empirical type I error when genotypes are generated randomly for each simulation, because for each of the 10,000 genotype data sets, only the outcomes are resampled. However, our type I error estimator is still unbiased and results in very accurate type I error estimates. For larger α levels (0.05 and 0.01), we directly computed the empirical type I error rate by using data sets in which genotypes were randomly generated for each simulation.
Empirical Power Simulations
We simulated data sets in which 30 kb subregions were randomly selected from the generated 1 Mb chromosomes and used to create causal variants and a phenotype variable as well as additional simulated covariates. We generated continuous phenotypes by
where X1, X2, and ɛ are as defined for the type I error simulations, are the genotypes of the s causal rare variants (a randomly selected subset of the simulated rare variants, for example 5% of variants that have MAF < 3% in Figure 1), and the βs are effect sizes for the causal variants. Similarly, we generated dichotomous phenotypes for case-control data under the logistic model
where are again the genotypes for the causal rare variants and βs are log ORs for the causal variants. We controlled prevalence by α0 and set to it 1% unless otherwise stated. Under both models, we set the magnitude of each βj to such that rarer variants had larger effects. In the simulation studies, for continuous traits, c = 0.4, which gives the maximum effect size |βj| = 1.6 for variants with MAF = 10−4 and small effects |βj| = 0.28 for MAF = 0.2. For dichotomous traits, c = ln5/4 = 0.402, which gives the “maximum” OR = 5.0 (|βj| = ln5) for variants with MAF = 10−4 and smaller OR = 1.32 for MAF = 0.2. The effect size curves are given in Figure S2.
Figure 1.
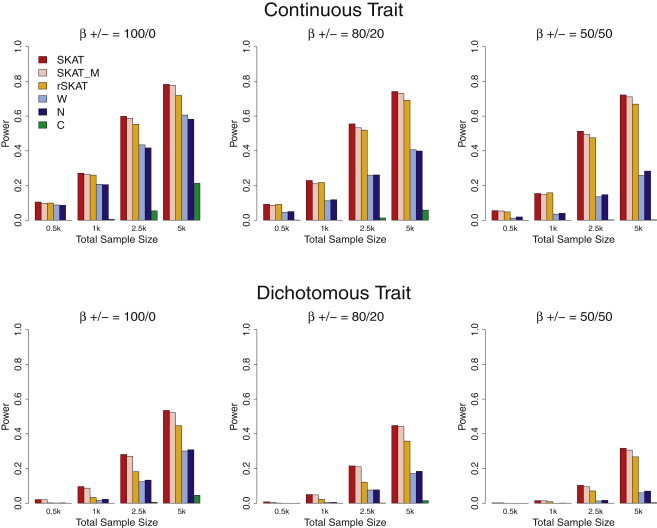
Simulation-Study-Based Power Comparisons of SKAT and Burden Tests
Empirical power at α = 10−6 under an assumption that 5% of the rare variants with MAF < 3% within random 30 kb regions were causal. Top panel: continuous phenotypes with maximum effect size (|β|) equal to 1.6 when MAF = 10−4; bottom panel: case-control studies with maximum OR = 5 when MAF = 10−4. Regression coefficients for the s causal variants were assumed to be a decreasing function of MAF as (j = 1,…,p [see Figure S2]), where c was chosen to result in these maximum effect sizes. From left to right, the plots consider settings in which the coefficients for the causal rare variants are 100% positive (0% negative), 80% positive (20% negative), and 50% positive (50% negative). Total sample sizes considered are 500, 1000, 2500, and 5000, with half being cases in case-control studies. For each setting, six methods are compared: SKAT, SKAT in which 10% of the genotypes were set to missing and then imputed (SKAT_M), restricted SKAT (rSKAT) in which unweighted SKAT is applied to variants with MAF < 3%, the weighted sum burden test (W) with the same weights as used by SKAT, counting-based burden test (N), and the CAST method (C). All the burden tests used MAF < 3% as the threshold. For each method, power was estimated as the proportion of p values < α among 1000 simulated data sets.
We compared SKAT, an unsupervised variation on the WST13 that uses weighted-count-based collapsing, counting-based collapsing,18 and CAST.14 For each of these tests, we considered variants with observed MAF < 3% as rare: whether CAST collapses depends on whether an individual exhibits any variants with allele frequency < 3%, the counting method counts the number variants with MAF < 3%, and the weighted count inflates the contribution of each rare variant by multiplying the genotype with the same beta-density-based weights as used in SKAT.
To accommodate missing genotypes commonly observed in sequence data, we considered the effect of imputing missing values by randomly setting 10% of the genotypes as missing, imputing genotypes on the basis of observed allele frequencies and Hardy-Weinberg equilibrium, and then applying SKAT to the imputed data. We also performed restricted SKAT (rSKAT) by applying unweighted SKAT to rare variants with MAF < 3%. Note that for dichotomous phenotypes, rSKAT is essentially equivalent to a covariate adjusted C-alpha test with the p value calculated analytically instead of via permutation. For each of the methods, power was estimated as the proportion of p values < α, where α = 10−6 to mimic genome-wide studies.
Power and Sample-Size Formulae
To demonstrate the utility and accuracy of our power and sample-size calculation method, we conducted several numerical experiments. We first illustrated the use of the methods by computing the sample size necessary to detect a 30 kb region with 5% of the variants with MAF < 3% being causal. We assume effect size (OR) increases with decreasing MAF, and seek 80% power at significance levels α = 10−6, 10−3, 10−2, corresponding to approximate genome-wide sequencing significance and candidate-gene-sequencing studies of 50 and five genes, respectively. We considered both continuous and dichotomous traits.
To show that the power estimated from our sample-size formula is accurate, we compared empirical power for SKAT under simulations to power estimated via our analytic method. Specifically, we simulated continuous and case-control data under the same setting as that used in the power simulations, and we estimated power as a function of the sample size by computing the proportion of p values < α = 10−6 and compared the empirical power curve to the power estimated by using our analytical method.
Results
Simulation of the Type I Error
The empirical type I error rates estimated for SKAT are presented in Table 1 for α = 10−4, 10−5, and 10−6 and suggest the type I error rate is protected for continuous phenotypes, though for smaller sample sizes the SKAT can be slightly conservative. For dichotomous phenotypes, SKAT is conservative for smaller sample sizes and very small α levels. Additional results from simulations of the type I error for SKAT and the competing methods are presented in Figure S3 for both continuous traits and dichotomous traits and show that at larger α levels, all of the considered tests correctly control at the α = 0.05 and 0.01 levels. These results show that SKAT is a valid method, and despite being conservative at low α levels, SKAT maintains good power relative to existing methods (see below). However, if sample sizes are small or sharp control of type I error is necessary, then standard permutation-based procedures can be used to generate a Monte Carlo p value for significance, though this can be computationally expensive and does not work in the presence of covariates, such as controlling for population stratification and require carful modifications.
Table 1.
Type I Error Estimates of SKAT Aimed at Testing an Association between Randomly Selected 30 kb Regions with a Continuous Trait at Type I Error Rates as Low as the Genome-wide α = 10−6 Level
| Total Sample Size (n) |
Continuous Phenotypes |
Dichotomous Phenotypes |
||||
|---|---|---|---|---|---|---|
| α = 10−4 | α = 10−5 | α = 10−6 | α = 10−4 | α = 10−5 | α = 10−6 | |
| 500 | 7.4 × 10−5 | 6.5 × 10−6 | 5.9 × 10−7 | 2.2 × 10−5 | 1.0 × 10−6 | 1.0 × 10−8 |
| 1000 | 8.5 × 10−5 | 8.2 × 10−6 | 8.0 × 10−7 | 5.0 × 10−5 | 3.5 × 10−6 | 2.3 × 10−7 |
| 2500 | 9.6 × 10−5 | 9.1 × 10−6 | 8.4 × 10−7 | 7.6 × 10−5 | 6.3 × 10−6 | 5.6 × 10−7 |
| 5000 | 9.8 × 10−5 | 9.6 × 10−6 | 8.8 × 10−7 | 8.9 × 10−5 | 7.8 × 10−6 | 7.0 × 10−7 |
Each entry represents type I error rate estimates as the proportion of p values α under the null hypothesis based on 108 simulated phenotypes.
Statistical Power of SKAT and Competing Methods
We compared the power of SKAT with three burden tests in a series of simulation studies for both continuous traits and dichotomous traits by generating sequence data in randomly selected 30 kb regions with a coalescent model.37 For our primary power simulation, within each region, 5% of variants with population MAF < 3% were randomly chosen as causal, the effect size of causal variants was a decreasing function of MAF, and 50%–100% of the causal variants being positively associated with the trait (See Materials and Methods and Figure S2). The simulated regions for our power analysis contained on average 605 variants (26 causal), of which 530.9 (88%), 502.9 (83%), and 422.8 (70%) had population MAF < 3%, < 1%, and < 0.1%, respectively. The average allele frequency spectrum across the samples is similar to that of the Dallas Heart Study data (Figure S4). Because the majority of variants have a low MAF, they might not be observed in any particular sample. The average number of observed variants (assuming no genotyping error) and the average number of observed causal variants are presented in Table 2.
Table 2.
Characteristics of the 30 kb Region Data Sets Used in the Simulation Studies
| Average Number of Observed Variants |
Sample Size (n) |
|||
|---|---|---|---|---|
| 500 | 1000 | 2500 | 5000 | |
| All traits∗ | 255 | 330 | 438 | 512 |
| Continuous trait∗∗ | 9.6 | 13.3 | 18.6 | 22.3 |
| Dichotomous trait (β ± = 100/0)∗∗ | 14.4 | 18.7 | 23.5 | 25.2 |
| Dichotomous trait (β± = 80/20)∗∗ | 13.3 | 17.1 | 22.0 | 24.3 |
| Dichotomous trait (β± = 50/50)∗∗ | 11.1 | 14.9 | 19.7 | 22.6 |
The number of observed variants∗ and the number of observed causal variants∗∗ within the region are averaged over the 1000 simulated data sets.
For continuous traits, SKAT had much higher power than all the burden tests, and the weighted count method tended to outperform the count and CAST methods (Figure 1). SKAT's power was robust to the proportion of causal variants that were positively associated with the trait, whereas the burden tests suffered substantial loss of power when causal variants had the opposite effects. The simulation results examining dichotomous traits were qualitatively similar in that SKAT dominated the competing methods. However, here the power of the SKAT decreased when both protective and harmful variants were present, although less so than for the burden tests. The difference in power for SKAT for different proportions of protective variants is due to the fact that given fixed population MAFs, protective variants imply negative log ORs and lower disease risk and hence lower MAFs in cases and more difficulties in observing rare variants in cases. The larger decrease in power for the competing methods is additionally driven by sensitivity to direction of effect due to aggregation of genotypes. Across all configurations, using imputed genotypes instead of the true genotype for 10% missing genotype data led to a very small reduction in power, despite the use of a very simple Hardy-Weinberg-based imputation strategy. This is true in part because most variants are rare.
Note that SKAT increases the weight of rare variants but does not require thresholding. To show that the superior performance of SKAT is intrinsic and is not driven by the particular choice of the weight used, we calculated rSKAT, which does not weight the rare variants but instead uses the same threshold as the burden tests. Our results, presented in Figure 1, show that rSKAT is still substantially more powerful than all three burden tests.
Power simulation results for other type I error rates (α = 0.01, 0.001), lower causal variant frequencies (population MAF < 1%), and other region sizes (10 kb and 60 kb) yielded the same conclusions (Figures S5–S8).
In the 30 kb genomic regions considered, reflecting analysis of genome-wide sequencing data, it is unlikely that a large proportion of the rare variants are all causal. However, for exome-scale sequencing, the number of observed rare variants can be considerably smaller and the proportion of causal rare variants can be greater. Hence, we also conducted power simulations for smaller region sizes (3 kb and 5 kb) and larger proportions of causal variants (10%, 20%, and 50%). Results for both continuous and dichotomous phenotypes are presented in Figures S9–S12 and show that if 50% of the rare variants are causal and that all of the causal variants have effects in the same direction, then SKAT and rSKAT are less powerful compared to collapsing methods, with count-based collapsing having the greatest power. This result held for both 3 kb and 5 kb regions and is expected because the collapsing methods implicitly assume that all of the variants are causal and have unidirectional effects. In all other settings we considered, SKAT was the most powerful method.
Power and Sample-Size Estimation
To illustrate our power and sample-size calculation method, in Figure 2 we present the estimated sample-size curves as a function of maximum effect sizes (ORs for dichotomous traits) necessary to detect a 30 kb region with 5% of the variants with MAF < 3% being causal. Table 3 presents estimated sample sizes for several configurations of practical interest. Additional sample-size curves when causal variants are rarer (MAF < 1%) or occur more frequently (10% of variants are causal) or when prevalence is varied (5%, 0.1%) can be found in Figures S13–S15. These results show that, for a given region, one will have more power (and a lower required sample size) to detect rare causal variants if the percentage of variants that are causal is higher, the causal rare variants have higher MAFs and/or larger effect sizes (e.g., odds ratios [ORs]), and the effects are more consistently in the same direction. For case-control designs, lower prevalence yields higher power because given the same OR and population MAF, the lower prevalence results in enrichment of more harmful (ORs > 1) variants, that is higher MAFs, across both cases and controls, that is for rarer diseases harmful rare variants are more likely to be observed. Conversely, if the prevalence is low, fewer protective variants (ORs < 1), that is lower MAFs, are likely to be observed in the sample.
Figure 2.
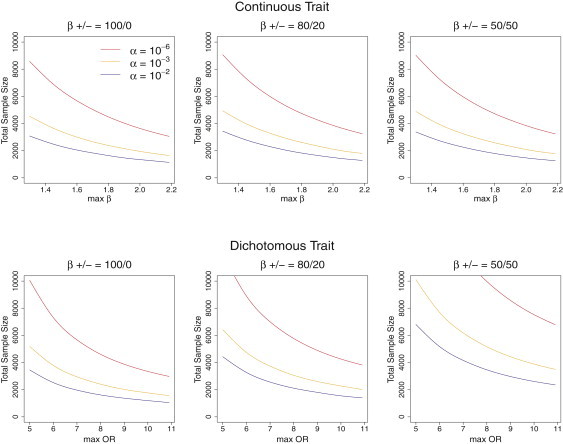
Sample Sizes Required for Reaching 80% Power
Analytically estimated sample sizes required for reaching 80% power to detect rare variants associated with a continuous (top panel) or dichotomous phenotype in case-control studies (half are cases) (bottom panel) at the α = 10−6, 10−3, and 10−2 levels, under the assumption that 5% of rare variants with MAF < 3% within the 30 kb regions are causal. Plots correspond to 100%, 80%, and 50% of the causal variants associated with increase in the continuous phenotype or risk of the dichotomous phenotype. Regression coefficients for the s causal variants were assumed to be the same decreasing function of MAF as that in Figure 1. The absolute values of Required total sample sizes are plotted again the maximum effect sizes (ORs) when MAF = 10−4. Estimated total sample sizes were averaged over 100 random 30 kb regions.
Table 3.
Required Total Sample Size to Achieve 80% Power to Detect Rare Variants Associated with a Continuous or Dichotomous Case-Control Phenotype at the Genome-wide Level α = 10−6
| Total Sample Size |
Maximum β = 1.6/ Maximum OR = 5 |
Maximum β = 1.9/ Maximum OR = 7 |
||
|---|---|---|---|---|
| 5% Causal | 10% Causal | 5% Causal | 10% Causal | |
| Continuous trait | 5,990 | 1,800 | 4,260 | 1,290 |
| Dichotomous trait with prevalence 10% | 15,120 | 4,810 | 9,650 | 3,120 |
| Dichotomous trait with prevalence 1% | 12,030 | 3,870 | 7,010 | 2,290 |
Power was estimated via the analytical formulae assuming 5% or 10% of variants with MAF < 3% are causal. Regression coefficients for the s causal variants were assumed to be a decreasing function of MAF, (j = 1,…,s), where 80% of βj's are positive and 20% are negative; see Figure S2. Required total sample sizes (cases and controls) are given for different “maximum” effect sizes (or ORs) when MAF = 10−4 and different prevalences for case-control studies. Estimated sample sizes were averaged over 100 random 30 kb regions.
We also compared the power and sample-size formulae estimates to the empirical, simulation-based power estimates for both continuous and dichotomous traits. The curves plotted in Figure 3 show that the empirical power is accurately approximated by our analytical formula.
Figure 3.
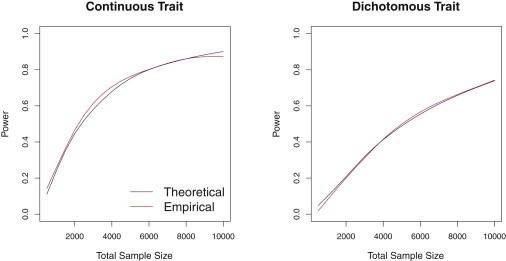
Power Comparisons Based on Simulation and Analytic Estimation
Power as a function of total sample size estimated by simulation with 1000 replicates and by the proposed power formula for continuous and dichotomous case-control traits. Simulation configurations correspond to those used in Figure 1, in which 80% of the regression coefficients for the causal rare variants were positive.
Application to Dallas Heart Study Data
We analyzed sequence data on 93 variants in ANGPTL3 (MIM 604774), ANGPTL4 (MIM 605910), and ANGPTL5 (MIM 607666) in 3476 individuals from the Dallas Heart Study38 to test for association between log-transformed serum triglyceride (logTG) levels and rare variants in these genes. We adjusted for sex and ethnicity (black, Hispanic, or white) but did not adjust for age as a large number of subjects have missing ages. In addition to testing for association via SKAT and the three burden tests considered earlier, we also applied the permutation-based varying-threshold method (VT) and the Polyphen-score-adjusted VT (VTP),16 which are based on the residuals obtained from regressing the phenotype on the covariates and assume gene-covariate independence. Because VT and VTP require permutation, they are computationally expensive when applied genome wide. For VTP, we used the Polyphen score for rare variants (MAF < 0.01) and assigned a constant score of 0.5 to all other variants. We also analyzed a dichotomized phenotype on the highest and lowest quartiles of each of the six sex-ethnicity groups (Table 4).
Table 4.
Analysis of the Dallas Heart Study Sequencing Data
| SKAT | C | N | W | VTa | VTPa | |
|---|---|---|---|---|---|---|
| Continuous TG level | 9.5 × 10−5 | 1.9 × 10−3 | 7.2 × 10−5 | 2.3 × 10−4 | 3.5 × 10−4 | 2.0 × 10−5 |
| Dichotomized TG level | 1.3 × 10−4 | 3.2 × 10−2 | 2.2 × 10−3 | 3.1 × 10−3 | 8.6 × 10−3 | 2.1 × 10−3 |
Analysis of the Dallas Heart Study sequencing data with SKAT, the weighted sum burden test (W), the counting-based burden test (N), the CAST method (C), the varying-threshold method (VT), and the Polyphen-score adjusted VT (VTP) method. Beta (1, 25) is used as the weight in the SKAT and the weighted sum test.
p values are estimated on the basis of 106 permutations.
SKAT was by far the most powerful test for the dichotomous trait. For continuous traits, SKAT has much smaller p values than two burden methods (CAST and WST) and VT, and has a slightly higher p value than the counting-based burden test (N) and VTP. Note that SKAT was easier to apply because it did not require prior functional information (available for only a subset of variants) or permutation, and it adjusted for covariates without assuming gene-covariate independence.
Computation Time
The computation time for the SKAT depends on the sample size and the number of markers. To analyze a 30 kb region sequenced on 1000, 2500, or 5000 individuals, SKAT required 0.21, 0.73, and 2.3 s, respectively, for continuous traits and ∼20% longer for dichotomous traits, on a 2.33 GHz laptop with 6 Gb memory. Analyzing 300 kb, 3 Mb, or 3 Gb (the entire genome) on 1000 individuals requires 2.5 s, 25 s, and 7 hr, respectively.
Discussion
We propose SKAT as a supervised, flexible, and computationally efficient statistical method that tests for association between a continuous or dichotomous phenotype and rare and common genetic variants in sequencing-based association studies. We demonstrate that SKAT's power is greater than that of several burden tests over a range of genetic models. Furthermore, we have developed analytical power and sample-size calculations for SKAT that assist in designing sequencing-based association studies.
Like burden tests, SKAT performs region-based testing. However, SKAT has several major advantages over the existing tests. As a supervised method, SKAT directly performs multiple regressions of a phenotype on genotypes for all variants in the region, adjusting for covariates. Hence, as with conventional multiple regression models, neither directionality nor magnitudes of the associations are assumed a priori but are instead estimated from the data. To test efficiently for the joint effects of rare variants in the region on the phenotype, SKAT assumes a distribution for the regression coefficients of the markers, whose variances depend on flexible weights. SKAT performs a score-based variance-component test, whose calculation only requires fitting the null model by regressing phenotypes on covariates alone and computing p values analytically. The flexible regression framework also allows us to allow for epistatic effects.
Besides region-based analysis, SKAT can also be applied to any biologically meaningful SNP set. As SKAT is a regression-based method, it can be easily extended to survival, and longitudinal and multivariate phenotypes and hence provides a comprehensive framework for a wide variety of sequencing-based association studies.
The ability to obtain a p value directly without the need for permutation is an attractive feature of SKAT, and allows for rapid estimation of p values in exome and genome-wide sequencing studies. Our simulations showed that for continuous phenotype, the p values are accurate when the sample size is moderate or large; for dichotomous phenotypes, the p values are conservative at lower α levels (e.g., < 10−4) if the sample size is modest or small. Permutation can be used to obtain a more accurate estimate in the absence of covariates. In the presence of covariates, for example population stratification, standard permutations fail and require careful modifications. Despite the conservative nature of the score test, SKAT often still has higher power than competing methods at small α levels.
SKAT can be combined with collapsing strategies to form a hybrid testing approach. If most of the variants within a range of allele frequencies are causal and have the same directionality (i.e., under settings that are optimal for burden-based tests), collapsing these variants and then applying SKAT to the collapsed variants can improve power. For example, because singletons are common in sequencing studies (57 of 93 variants in the Dallas Heart Study data), a possible hybrid strategy is to first collapse all of the singletons into a single value and then apply SKAT to the collapsed value and the other 36 variants. Compared to the original SKAT, this strategy gives a slightly lower p value, 3.1 × 10−5, for the continuous trait and a slightly higher p value, 1.6 × 10−4, for the dichotomous trait. Simulation studies showed that the two methods are of similar power under the settings we used to generate Figure 1.
An important feature of SKAT is that it allows for incorporation of flexible weight functions to boost analysis power, for example by increasing the weight of variants with lower MAFs and decreasing the weight of information from variants inferred with lower confidence. Good choices of weights are likely to improve the power of the association test with SKAT, although simulations show that even equal weights can yield high power when combined with thresholding. In our simulation studies, we employed a class of flexible continuous weights as a function of MAF by using the beta function, which increases the weight of rare variants and does not require thresholding. Users can define other types of weight functions. To further improve analysis power, one can estimate weights by incorporating information besides MAF, for example by using the Polyphen score or integrating other annotation information, which will become increasingly available as our understanding of genome variation improves. Therefore, because of its flexibility, SKAT has the capacity to mature, and its power to increase, as the field progresses.
Acknowledgments
This work was supported by grants P30 ES010126 (to M.C.W.), DMS 0854970 and R01 GM079330 (to T.C.), R01 HG000376 (to M.B.), and R37 CA076404 and P01 CA134294 (to S.L. and X.L.). We thank Jonathan Cohen, Alkes Price, and Shamil Sunyaev for providing the Dallas Heart Study data and Larisa Miropolsky for help with the software development.
Appendix A
Estimating the Null Distribution for Q
Under the null hypothesis, Q follows a mixture of chi-square distributions.29,30 More specifically, we define where is the n × (p + 1) matrix equal to [1, X]. For continuous phenotypes, where is the estimator of σ under the null model where f(G) = 0, and I is an n × n identity matrix. For dichotomous phenotypes, where is the estimated probability that the i-th subject is a case under the null model. Then under the null model
| (Equation 6) |
where (λ1, λ2, …, λn) are the eigenvalues of , and are independent random variables.
Several approximation and exact methods have been suggested to obtain the distribution of Q.39 Among these, the Davies exact method,26 based on inverting the characteristic function of Equation 6, appears to work well in practice and is used here.
SKAT Is a Generalization of the C-Alpha Test
The recently proposed the C-alpha test has advantages over burden tests in that it explicitly models the possibility that minor alleles can be deleterious or protective. However, it does not currently allow for the analysis of quantitative outcomes or the inclusion of covariates and p value calculation requires permutation. We demonstrate that for a dichotomous trait in the absence of covariates, the C-alpha test statistic is equivalent to the SKAT statistic with unweighted linear kernel, which is the same as the kernel machine test in Wu et al.24
Suppose the j-th variant is observed times in the cases, out of times total in cases and controls, and that . For a dichotomous trait and no covariates, the C-alpha test statistic
| (Equation 7) |
Denote . Because is the mean of under the null hypothesis of no association, is the C-alpha test statistic without mean centering. Because and , where is the j-th column of the genotype matrix G and , it can be easily shown that
| (Equation 8) |
Note that under the unweighted linear kernel, K = GG' and if no covariates are present. Hence, Equation 8 is identical to Equation 3, that is is equivalent to the SKAT test statistic with unweighted linear kernel.
Although the SKAT statistic with unweighted linear kernel and the C-alpha test statistic are equivalent, SKAT and C-alpha test use different null distributions to assess significance: C-alpha test uses a normal approximation, whereas we use a mixture of chi-squares. The normal approximation gives a valid p value when the tested rare variants are independent and sample sizes are large, and so requires an assumption of linkage equilibrium. In the presence of LD, permutation is used by the C-alpha test for significance testing. One can easily see that the test statistic takes a quadratic form of y, which follows a mixture of chi-square distributions. SKAT approximates this distribution directly with the Davies method and hence gives accurate estimation of significance regardless of the LD structure when sample size is sufficient.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org/
Online Mendelian Inhereitance in Man (OMIM), http://www.omim.org
SKAT software, http://www.hsph.harvard.edu/∼xlin/software.html
References
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mardis E.R. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 4.Ansorge W.J. Next-generation DNA sequencing techniques. New Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Eichler E.E., Flint J., Gibson G., Kong A., Leal S.M., Moore J.H., Nadeau J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R.Q., Li Y.R., Fang X.D., Yang H.M., Wang J., Kristiansen K., Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bansal V., Harismendy O., Tewhey R., Murray S.S., Schork N.J., Topol E.J., Frazer K.A. Accurate detection and genotyping of SNPs utilizing population sequencing data. Genome Res. 2010;20:537–545. doi: 10.1101/gr.100040.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvajal-Carmona L.G. Challenges in the identification and use of rare disease-associated predisposition variants. Curr. Opin. Genet. Dev. 2010;20:277–281. doi: 10.1016/j.gde.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Schork N.J., Murray S.S., Frazer K.A., Topol E.J. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen B.E., Browning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgenthaler S., Thilly W.G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Li B., Leal S.M. Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 2009;5:e1000481. doi: 10.1371/journal.pgen.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han F., Pan W. A data-adaptive sum test for disease association with multiple common or rare variants. Hum. Hered. 2010;70:42–54. doi: 10.1159/000288704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris A.P., Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zawistowski M., Gopalakrishnan S., Ding J., Li Y., Grimm S., Zöllner S. Extending rare-variant testing strategies: analysis of noncoding sequence and imputed genotypes. Am. J. Hum. Genet. 2010;87:604–617. doi: 10.1016/j.ajhg.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asimit J., Zeggini E. Rare variant association analysis methods for complex traits. Annu. Rev. Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 21.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Kwee L.C., Liu D., Lin X., Ghosh D., Epstein M.P. A powerful and flexible multilocus association test for quantitative traits. Am. J. Hum. Genet. 2008;82:386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M.C., Kraft P., Epstein M.P., Taylor D.M., Chanock S.J., Hunter D.J., Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin X. Variance component testing in generalised linear models with random effects. Biometrika. 1997;84:309–326. [Google Scholar]
- 26.Davies R. The distribution of a linear combination of chi-square random variables. J. R. Stat. Soc. Ser. C Appl. Stat. 1980;29:323–333. [Google Scholar]
- 27.Pan W. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genet. Epidemiol. 2009;33:497–507. doi: 10.1002/gepi.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristianini N., Shawe-Taylor J. Cambridge Univ Press; Cambridge: 2000. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. [Google Scholar]
- 29.Liu D., Lin X., Ghosh D. Semiparametric regression of multidimensional genetic pathway data: least-squares kernel machines and linear mixed models. Biometrics. 2007;63:1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D., Ghosh D., Lin X. Estimation and testing for the effect of a genetic pathway on a disease outcome using logistic kernel machine regression via logistic mixed models. BMC Bioinformatics. 2008;9:292. doi: 10.1186/1471-2105-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleuret, F., and Sahbi, H. (2003). Scale-invariance of support vector machines based on the triangular kernel. In 3rd International Workshop on Statistical and Computational Theories of Vision. (ftp://ftp.inria.fr/INRIA/publication/publi-pdf/RR/RR-4601.pdf).
- 32.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 34.Liu H., Tang Y., Zhang H. A new chi-square approximation to the distribution of non-negative definite quadratic forms in non-central normal variables. Comput. Stat. Data Anal. 2009;53:853–856. [Google Scholar]
- 35.Lee, S., Wu, M.C., Cai, T., Li, Y., Boehnke, M., and Lin, X. (2011). Power and sample size calculations for designing rare variant sequencing association studies. In Harvard University Technical Report. (http://www.hsph.harvard.edu/∼xlin).
- 36.Durbin R.M., Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romeo S., Yin W., Kozlitina J., Pennacchio L.A., Boerwinkle E., Hobbs H.H., Cohen J.C. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duchesne P., Lafaye De Micheaux P. Computing the distribution of quadratic forms: Further comparisons between the Liu-Tang-Zhang approximation and exact methods. Comput. Stat. Data Anal. 2010;54:858–862. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


