Abstract
Like all plants, potato has evolved a surveillance system consisting of a large array of genes encoding for immune receptors that confer resistance to pathogens and pests. The majority of these so-called resistance or R proteins belong to the super-family that harbour a nucleotide binding and a leucine-rich-repeat domain (NB-LRR). Here, sequence information of the conserved NB domain was used to investigate the genome-wide genetic distribution of the NB-LRR resistance gene loci in potato. We analysed the sequences of 288 unique BAC clones selected using filter hybridisation screening of a BAC library of the diploid potato clone RH89-039-16 (S. tuberosum ssp. tuberosum) and a physical map of this BAC library. This resulted in the identification of 738 partial and full-length NB-LRR sequences. Based on homology of these sequences with known resistance genes, 280 and 448 sequences were classified as TIR-NB-LRR (TNL) and CC-NB-LRR (CNL) sequences, respectively. Genetic mapping revealed the presence of 15 TNL and 32 CNL loci. Thirty-six are novel, while three TNL loci and eight CNL loci are syntenic with previously identified functional resistance genes. The genetic map was complemented with 68 universal CAPS markers and 82 disease resistance trait loci described in literature, providing an excellent template for genetic studies and applied research in potato.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-011-1602-z) contains supplementary material, which is available to authorized users.
Introduction
Potato (Solanum tuberosum ssp. tuberosum L.) is an important food crop in temperate climates. Worldwide more than 19 million ha. of potatoes are grown with a total economic value higher than 31 billion US$ (www.potato2008.org/en/world/index.html). Potato was brought to Europe by Spanish conquistadors in the sixteenth century from its centre of origin, the Andean region of South America where it was domesticated about 8,000 years ago (Bradshaw and Ramsay 2005). However, with the introduction of potato as a crop a range of pathogens and pests were introduced as well, causing devastating yield losses, including the Great Irish Famine by the oomycete Phytophthora infestans (Bradshaw and Ramsay 2005). Therefore, breeding for resistance is central for disease control in potato.
To defend themselves against pathogens and pest, plants have evolved an innate surveillance system encoded by a large set of resistance genes. Most resistance genes are single dominant and confer resistance in a gene-for-gene specific manner (Flor 1971). Most known resistance genes belong to the class of genes encoding a nucleotide binding (NB) and leucine rich repeats (LRR) domains (reviewed by Tameling and Takken 2008). R genes that belong to this super-family can be subdivided into genes encoding proteins with an N-terminal coiled-coil (CC) domain or a Toll-like Interleukin receptor (TIR) domain. They confer resistance to completely unrelated taxonomic groups like bacteria, fungi, viruses and nematodes by the activation of a defence response that prevents the pathogen from spreading (Bendahmane et al. 1999; Lawrence et al. 1995; Milligan et al. 1998; Mindrinos et al. 1994; van der Vossen et al. 2000; Vos et al. 1998; Whitham et al. 1994).
To date, thirteen functional NB-LRR resistance genes have been identified in potato i.e. R1 (Ballvora et al. 2002), Rpi-blb2 (van der Vossen et al. 2005), Gro1 (Paal et al. 2004), R3a (Huang et al. 2005), Gpa2 (van der Vossen et al. 2000), Rx1 (Bendahmane et al. 1999), Rx2 (Bendahmane et al. 2000), Rpi-blb3, Rpi-abpt, R2, R2-like, and Rpi-mcd1 (Lokossou et al. 2009), and Rpi-blb1 (van der Vossen et al. 2003). However, the potato genome harbours many more functional NB-LRR genes. In twenty or more regions in the potato genome, one or more resistance traits have been mapped, but none of the underlying genes have been identified yet (Bakari et al. 2006; Caromel et al. 2005; Caromel et al. 2003; Celebi-Toprak et al. 2002; Flis et al. 2005; Gebhardt and Valkonen 2001; Grube et al. 2000; Hein et al. 2009; Marczewski et al. 2001; Marczewski et al. 2002; Marczewski et al. 2006; Sato et al. 2006; Song et al. 2005; Szajko et al. 2008).
To facilitate the cloning and characterisation of resistance genes, various genetic maps have been constructed for the potato genome. The first genetic map was published in 1988 (Bonierbale et al. 1988). In 2001, Gebhardt and Valkonen published a genetic map of potato showing the location of all resistance traits known at that time. From this study, they could conclude that the distribution of R genes and QTLs is not random, but that they often reside in clusters or so-called ‘hotspots of resistance’ in the genome. Later on, an ultra high dense (UHD) genetic map with 10,365 AFLP loci in 1,118 bins was constructed (van Os et al. 2006), providing genome-wide marker saturation that facilitated the construction of a genome-wide physical map of potato (Borm 2008).
In this study, the UHD map (van Os et al. 2006) and the physical map (Borm 2008) of the diploid potato clone RH89-039-16 (RH) were used to construct a genetic map showing the genome-wide distribution of NB-LRR resistance gene homologs in the potato genome. In total, a set of 288 selected BAC clones derived from 47 NB-LRR resistance gene loci has been sequenced, which resulted in the detection of 738 partial and full length RGHs using BLASTp (Gish and States 1993). The RGHs can be subdivided into 448 CC-NB-LRR encoding sequences located at 32 different loci, whereas 280 sequences belong to the TIR-NB-LRR subclass derived from 15 different loci. Positional information of 82 resistance traits previously described was retrieved from literature and included in this study, resulting in a comprehensive integrated genome-wide genetic map of NB-LRR encoding RGHs and disease resistance loci in potato. Potential applications of the results obtained in this study to enhance the identification of genes underlying important disease resistance traits in potato and other solanaceous species will be discussed.
Materials and methods
BAC library screening
A BAC library of the diploid S. tuberosum spp. tuberosum clone RH89-039-16 (RH) consisting of 78,336 clones and an estimated coverage of five times the diploid genome was available (Borm 2008). The BAC clones were spotted in duplo on three macro-array filters by RZPD (Berlin, Germany).
Degenerate primers were designed based on P-loop and GLPL motifs of the Nucleotide Binding (NB) domains found in 54 known potato, tomato and pepper NB-LRR resistance gene sequences obtained from Genbank (data not shown). Identical thermal cycling conditions, using the primer combinations in Table 1, were used for all PCR amplifications: 1′ 94 C -(30″ 94C – 30″ 52C – 1′ 72C) 35× – 5′ 72C.
Table 1.
Degenerate primer combinations used for the amplification of NBS sequences from potato
| Forward primer | Forward primer sequence | Reverse primer | Reverse primer sequence |
|---|---|---|---|
| Ploop1 | ggi ggi ntr ggi aar acr ac | GLPL1 | cci ray ggr rat cgy sai tty ig |
| GLPL3B | cct gar ggr ray cri gaa sai ca | ||
| GLPL5 | cck gar ggi rat cgk rri ttt ca | ||
| GLPL7A | ccy iaa ggi aat cgw gar ytt ca | ||
| GLPL7B | cct way ggi gaw cgw rat iyt ca | ||
| GLPL8 | cct aac ggt gat cgc aac | ||
| Ploop2 | ggi ggi nty ggi aar acr ac | GLPL2 | cct aac ggt gat cgc aac aay yg |
| GLPL3A | ccy aat ggw gat ygt taa tga ma |
The resulting PCR products were purified directly using the QIAquick PCR purification kit (QIAGEN) according to the manufacturer’s instructions. Purified PCR products were ligated in the pGEMTeasy vector (QIAGEN) and transformed to DH5α (Invitrogen) cells according to the manufacturer’s instructions. For each primer combination, 96 clones were re-amplified using the original primers and clones representative of each product size were sent for Sanger sequencing (Greenomics, Wageningen, The Netherlands). Probes for screening the BAC library were selected from the resulting sequences as described in the results. Probe construction and filter hybridisation was performed by Greenomics (Wageningen, The Netherlands), using 15 different probe pools based on the 96 NB sequences.
BAC analysis
For BACs selected for sequencing, the insert size was determined using CHEF gel electrophoresis on a 1% agarose gel (Seakem® Gold, FMC, Philadelphia, PA, USA) in 0.5×TBE buffer at 4°C using a BIORAD CHEF DR II system (Bio-Rad laboratories, Hercules, CA, USA) at 200 V with a pulse time of 5–15 s for 18 h.
Sequence analysis
Sequencing to six-fold coverage and assembly of BAC clones to phase 1 was done by Greenomics (Wageningen, The Netherlands). Genomic BAC sequences were translated into 6 frames using a PERL (www.perl.com) script. BLASTp (Gish and States 1993) was used to match a set of 59 known functional resistance genes, separated into different domains based on literature (Table 2), against the database of translated BACs. BLASTp output took the form of a list of homologous BACs for each R gene domain, which was parsed using R2BAC (Goto et al. 2010) to identify the location of matching R gene domains inside the BACs. BAC sequences have been submitted to Genbank: AC238121, AC238183, AC232102, AC237998, AC238257, AC238084, AC238124, AC238122, AC238122, AC238122, AC238122, AC238122, AC238122, AC238122, AC238122, AC238261, AC238307, AC237896, AC237892, AC238268, AC237880, AC238396, AC238310, AC238277, AC238378, AC238011, AC238356, AC238396, AC234535, AC237944, AC238154, AC237844, AC238274, AC237846, AC238200, AC2382831, AC237953, AC237983, AC237843, AC238108, AC238211, AC238221, AC238176, AC238227, AC238336, AC238074, AC238299, AC238304, AC238344, AC238372, AC238222, AC237902, AC237930, AC232101, AC237980, AC237842, AC238190, AC232105, AC237826, AC237833, AC237875, AC234558, AC237927, AC238292, AC237906, AC237966, AC238063, AC238128, AC238172, AC238197, AC237841, AC237847, AC237855, AC238340, AC238022, AC237958, AC237973, AC234529, AC237970, AC237837, AC238099, AC237996, AC237865, AC237914, AC238314, AC238386, AC238387, AC237832, AC237840, AC238065, AC237848, AC237853, AC237857, AC237859, AC236625, AC237861, AC237862, AC237866, AC239995, AC238021, AC238132, AC239974, AC238178, AC238279, AC240056, AC239998, AC240052, AC239956, AC239979, AC239982, AC239958, AC240012, AC240037, AC240057, AC238125, AC240042, AC239955, AC238020, AC240025, AC239996, AC240059, AC240062, AC240070, AC240041, AC240072, AC240016, AC239971, AC238036, AC240047, AC239983, AC237905, AC240023, AC240063, AC240065, AC239989, AC240010, AC240048, AC239981, AC239980, AC240000, AC239980, AC240066, AC239987, AC240004, AC237886, AC240014, AC240007, AC240024, AC240040, AC240022, AC240073, AC240011, AC240014, AC240015, AC239967, AC239976, AC240003, AC240064, AC240069, AC239965, AC239975, AC237835, AC239991, AC239992, AC240020, AC240029, AC240044, AC240055, AC240071, AC240005, AC240006, AC232094, AC240028, AC240038, AC238291, AC239966, AC240001, AC240009, AC240013, AC240019, AC232092, AC240124, AC238273, AC237872, AC240032, AC232110, AC232116, AC240050, AC237883, AC237889 and AC23789.
Table 2.
Functional NB-LRR resistance genes from various plant species
| R protein | Origin | References |
|---|---|---|
| Bs2 | Capsicum annuum | Tai et al. (1999) |
| Bs4 | Lycopersicon esculentum | Schornack et al. (2004) |
| CaMi | Capsicum annuum | Chen et al. (2007) |
| Dm3 | Bremia lactucae | Meyers et al. (1998) |
| Gpa2 | Solanum tuberosum ssp. andigena | van der Vossen et al. (2000) |
| Gro1-4 | Solanum spegazzinii | Paal et al. (2004) |
| Hero | Lycopersicon esculentum | Ernst et al. (2002) |
| HRT | Arabidopsis thaliana | Cooley et al. (2000) |
| I-2 | Lycopersicon pimpinellifolium | Ori et al. (1997) |
| L10 | Linum usitatissimum | Ellis et al. (1999) |
| L6 | Linum usitatissimum | Lawrence et al. (1995) |
| Lr1 | Triticum aestivum | Cloutier et al. (2007) |
| Lr10 | Triticum aestivum | Feuillet et al. (2003) |
| Lr21 | Triticum aestivum | Huang et al. (2003) |
| M | Linum usitatissimum | Anderson et al. (1997) |
| Mi-1 | Lycopersicon peruvianum | Milligan et al. (1998) |
| Mla1 | Hordeum vulgare | Zhou et al. (2001) |
| N | Nicotiana tabacum | Whitham et al. (1994) |
| N | Linum usitatissimum | Dodds et al. (2001a) |
| Nrc1 | Lycopersicon esculentum | Gabriëls et al. (2007) |
| Nrg1 | Nicotiana benthamiana | Peart et al. (2005) |
| P2 | Linum usitatissimum | Dodds et al. (2001b) |
| Pi-ta | Oryza sativa | Bryan et al. (2000) |
| Pi36 | Oryza sativa | Liu et al. (2007) |
| Pi37 | Oryza sativa | Lin et al. (2007) |
| Pib | Oryza sativa | Wang et al. (1999) |
| Pm3b | Triticum aestivum | Yahiaoui et al. (2004) |
| Prf | Lycopersicon esculentum | Salmeron et al. (1996) |
| R1 | Solanum demissum | Ballvora et al. (2002) |
| R2 | Solanum demissum | Lokossou et al. (2009) |
| R2-like | Solanum spp. | Lokossou et al. (2009) |
| R3a | Solanum demissum | Huang et al. (2005) |
| Rcy1 | Arabidopsis thaliana | Sekine et al. (2006) |
| Rp1D | Zea mays | Collins et al. (1999) |
| Rpg1-b | Glycine max | Ashfield et al. (2004) |
| Rpi-abpt | Solanum bulbocastanum | Lokossou et al. (2009) |
| Rpi-blb1 | Solanum bulbocastanum | van der Vossen et al. (2003) |
| Rpi-blb2 | Solanum bulbocastanum | van der Vossen et al. (2005) |
| Rpi-blb3 | Solanum bulbocastanum | Lokossou et al. (2009) |
| Rpm1 | Arabidopsis thaliana | Grant et al. (1995) |
| Rpp1 | Arabidopsis thaliana | Botella et al. (1998) |
| Rpp13 | Arabidopsis thaliana | Bittner-Eddy et al. (2000) |
| Rpp2A/B | Arabidopsis thaliana | Sinapidou et al. (2004) |
| Rpp4 | Arabidopsis thaliana | van der Biezen et al. (2002) |
| Rpp5 | Arabidopsis thaliana | Parker et al. (1997) |
| Rpp8 | Arabidopsis thaliana | McDowell et al. (1998) |
| Rps2 | Arabidopsis thaliana | Bent et al. (1994) |
| Rps4 | Arabidopsis thaliana | Gassmann et al. (1999) |
| Rps5 | Arabidopsis thaliana | Warren et al. (1998) |
| RRS-1 | Arabidopsis thaliana | Deslandes et al. (2002) |
| Rx1 | Solanum tuberosum ssp. andigena | Bendahmane et al. (1999) |
| Rx2 | Solanum demissum | Bendahmane et al. (2000) |
| SSI4 | Arabidopsis thaliana | Shirano et al. (2002) |
| Sw-5 | Lycopersicon peruvianum | Spassova et al. (2001) |
| Tm-2(2) | Lycopersicon esculentum | Lanfermeijer et al. (2003) |
| WRR4 | Arabidopsis thaliana | Borhan et al. (2008) |
| Xa1 | Oryza sativa | Yoshimura et al. (1998) |
| Y-1 | Solanum tuberosum ssp. andigena | Vidal et al. (2002) |
SSR analysis
Primers (Supplemental Table 1) for Simple Sequence Repeat (SSR) markers (Goldstein et al. 1995) were based on BAC sequences obtained in this study and were used on BAC DNA and genomic DNA using the following thermal cycling conditions: 5′ 94C – (30″ 94C – 30″ 56C – 30″ 72C)25× – 7″ 72C. SSR markers were visualised using a Li-cor sequencer (Li-cor, Lincoln, NB, USA) according to the manufacturer’s instructions.
CAPS analysis
Cleaved Amplified Polymorphic Sequences (CAPS) analysis (Konieczny and Ausubel 1993) was performed using the following thermal cycling conditions: 5′ 94 C – (30″ 94C – 30″ Tm – 30″ 72C)30× – 7″ 72C. Primers were either designed on BAC sequences within this study, or on sequence information derived from the PoMaMo database (http://gabi.rzpd.de/projects/Pomamo/; Meyer et al. 2005) or the SOL Genomics Network (http://solgenomics.net/; Mueller et al. 2005), or obtained from literature. Primers, annealing temperature (Tm), the appropriate endonuclease for the detection of polymorphism and the source are listed in Supplemental Table 2. DNA fragments of the CAPS markers were separated on 1% agarose in 1× TAE buffer at 120 V.
NBS profiling
NBS profiling (van der Linden et al. 2004), was performed on the RH × SH mapping population to create a map of NBS specific markers in potato (van der Linden, unpublished results). Markers segregating from RH were expected to (partly) resemble bands derived from RGHs identified in this study. To anchor RGH containing BACs with these NBS specific markers, a similar NBS profiling study was performed on the RH BAC library that was pooled (Borm 2008) for the direct identification of single NB-LRR containing BACs. For this, NB-site specific primers NB1, NB2, NB5a6, and NB9 (van der Linden et al. 2004) were used to screen these complex pooled-BAC-pools from the RH BAC library in an NBS profiling assay. NBS profiling was performed essentially as described previously (van der Linden et al. 2004). Adaptors were ligated to the restrictions sites of AluI, HaeIII, MseI, RsaI and TaqI. The NB-site specific primers in combination with adaptor primers were used for PCR, and fragments were fractionated using denaturing polyacrylamide gel electrophoresis. Bands derived from RGH containing RH BACs were compared to the NBS profiling markers derived from RH (van der Linden, unpublished results). Co-migrating bands in the complex BAC pools and the genomic DNA of RH that were constructed with the same enzyme/primer combination were assumed to be derived from the same locus. This method was used to anchor RGH containing BACs to the bin signatures of the UHD map (van Os et al. 2006).
Genetic mapping
A mapping population of 136 F1 genotypes from the cross between the diploid potato clones RH x SH83-92-488 (SH) was available (Rouppe van der Voort et al. 1997). For genetic mapping in this study, a subset of 45 offspring genotypes was selected using the software package MapPop (Vision et al. 2000) that identifies the most informative genotypes from a mapping population based on the maximum number of recombination events distributed over the genome. The consensus bin signatures of the UHD map of potato (van Os et al. 2006) were used as input for MapPop. Segregating bands were mapped with the software package BINmap+ (Borm 2008), that uses the bin signatures of the UHD map to match segregation patterns. Genomic DNA from SH, RH and progeny was extracted as described (van Os et al. 2006). BAC DNA was isolated using a high-throughput protocol, adapted from (Sambrook et al. 1989) as described (Borm 2008).
In silico anchoring
To determine overlap between some RGH containing BACs and previously anchored BAC contigs from the physical map of potato (Borm 2008) and thereby providing a genetic anchor for these RGH containing BACs, the BACend tool was used. The BACend tool uses a high stringency (98% nucleotide identity) BLAST to compare a query sequence to the BAC-end sequences of the RH BAC library (Borm 2008), displaying results in their physical map context. The BAC-end tool also filters BLAST results to ensure that BAST hits are structurally sound (ignoring non full-length BLAST hits in the middle of sequence- fragments) and non-repetitive (as determined from the BAC-end sequences) and of sufficient length (>100 base-pairs).
BLASTn (Altschul et al. 1990) was performed using R gene sequences [Mi-1 (Milligan et al. 1998) and Rpi-blb2 (van der Vossen et al. 2005)] or marker sequences of a known genomic location. The marker sequences were derived from SOL Genomics Network (http://solgenomics.net/; Mueller et al. 2005), or kindly provided by G. van der Linden (NBS blast). For marker sequences, a threshold of >98% nucleotide identity over the complete length of the marker sequence was used. For the R gene BLAST, the threshold was lower (>80%), based on the average similarity within an R gene cluster, but matching the complete ORF of the R gene. With these stringent criteria, it was assumed that when a match in an RGH containing BAC is found, such BAC has the same genetic location.
Results
Identification of BACs harbouring NB-LRR Resistance Gene Homologs (RGHs)
To assess the NB-LRR resistance gene super-family in the potato genome, sequence information of the conserved NB domain was used to design probes for the screening of a BAC library derived from the diploid potato genotype RH89-039-16 (RH). In total, eight primer combinations were designed based on the conserved P-loop and the GLPL motifs in the NB domain and used to amplify and sequence 150 fragments from genomic DNA of RH. Using Neighbour-Joining analysis, the 150 sequences could be divided into 32 groups with >85% similarity (data not shown). For each group, three representative probes were selected that ranged in size from 450 to 600 base pairs. The resulting collection of 96 probes was divided into 15 probe-pools to screen a BAC library of RH. This resulted in the detection of 1,535 BAC clones potentially harbouring resistance gene homologs (RGHs) encoding NB-LRR proteins.
Selection of BACs for sequencing
RGH sequences are often located in arrays of homologous sequences in so-called R gene clusters in the genome of plants, including potato [e.g. GroI (Paal et al. 2004), R1 (Ballvora et al. 2002) and Gpa2 (van der Vossen et al. 2000)]. Therefore, it is likely that members of a single cluster will be present in multiple overlapping BACs, forming a physical map contig. To identify overlap between the positive BACs and to remove redundant BACs, the physical map of RH (Borm 2008) was used to locate 1,402 of the 1,533 positive BAC clones in 502 physical map contigs. Many physical map contigs harboured only one out of several BACs that hybridised with an NBS probe. This was considered to be the result of a falsely positive signal. Therefore, 310 BACs were omitted from further analysis. The remaining, 192 physical map contigs were selected for further analysis as they contained at least two positive overlapping BACs.
To obtain a genome-wide collection of RGH sequences, one representative BAC from each contig was selected for Sanger sequencing (6× coverage). Of the 131 positive BAC clones that could not be assigned to a physical map contig, but may represent small clusters or even single R genes, 96 were randomly selected for sequencing too. In total, 33 million base pairs of sequence data were obtained for 288 BACs, divided into 2,958 sequence contigs (on average 10.27 sequence contigs per BAC).
Identification of resistance gene homologs
To identify the RGHs for each BAC, a high-throughput method was developed to search the complete set of BAC sequences that we obtained. A local BLASTp search was performed on six translated frames of the complete collection of BAC sequences with the sequences of the CC/TIR, NB and LRR domains of a set of 59 known resistance proteins (Table 2). This resulted in 2,184 significant BLAST hits (E value ≤0.05), which are presented in Supplemental Table 3. If a sequenced BAC fragment harbours different R protein domains in the correct order on a DNA sequence read of about 5 kb, it is assumed that they represent one single gene. In this way, we could reconstruct a total of 738 partial and full-length RGH sequences derived from 195 BACs, which could be subdivided in 280 RGHs that harbour a TIR domain and 448 that harbour a CC domain (Supplemental Table 3).
Anchoring the RGH containing BACs to the genetic map of potato
To determine the genomic locations of the RGHs, we first compared our set of corresponding BACs to the physical map of potato (Borm 2008) that was constructed from the same BAC library. In the latest version of the physical map, 53 of the RGH containing BACs were genetically anchored or merged to contigs with other RGH containing BACs (de Boer et al., unpublished data). In addition, a variety of anchoring methods were used (Table 3). For SSR, CAPS and NBS profiling, a subset of 45 most informative genotypes of the SHxRH mapping population was selected. This combination of genetic mapping and in silico anchoring enabled us to determine the genetic location of 169 of the 195 RGH containing BACs (87%). Twenty-three BACs could only be anchored to a linkage group of the genetic map and not to a bin (range). Eighteen of them could be added to a locus based on their RGH content. The remaining 5 BACs are anchored to chromosome X (4) and XII (1). Based on their RGH content, the four BACs on chromosome X could be combined into one locus. A complete overview of the anchoring of the BACs to the linkage groups is depicted in Supplemental Fig. 1. Thirty-eight BACs were mapped by more than one independent method, confirming their map locations. Specific information on the map positions per BAC is provided in Supplemental Table 3.
Table 3.
Overview of the methods used to anchor the sequenced BAC clones to the genetic map of potato
| Mapping method | Number of BACs mapped |
|---|---|
| Potato physical map | 53 |
| SSR | 56 |
| CAPS | 16 |
| NBS profiling | 13 |
| BACend tool | 34 |
| BLAST | 35 |
Identification of 47 RGH loci in the potato genome
Our results show that the majority of RGH containing BACs map in clusters across the potato genome, except for LGII and LGIII (Supplemental Fig. 1). A total of 151 BACs mapped to a position with at least one other BAC harbouring highly homologous RGHs, suggesting they are derived from complex loci. Eighteen BACs were mapped alone, or did not harbour RGHs with similar BLAST results as those mapping at the same genetic position. Eight of these BACs harbour more than one RGH, suggesting they are representing relatively small clusters, while ten harbour only one RGH that may be a simple locus (Supplemental Table 3).
Sometimes, the BACs mapping to the same genetic position all harbour highly homologous RGHs, like for example the locus at the distal end of the long arm of chromosome V, where all BACs harbour only Rpi-blb2 homologs (Supplemental Fig. 1). However, BACs harbouring different types of RGH are not always genetically separated from each other, although they might have some physical distance. For example, on the short arm of chromosome VI (Supplemental Fig. 1) nine BACs harbour Rpi-blb2 homologs, six BACs harbour Bs4 homologs, and one BAC harbours both and the genetic map positions of the BACs are partially overlapping.
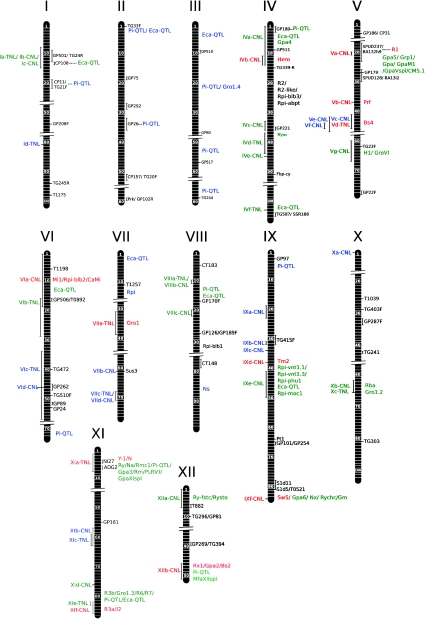
On the basis of these observations, we developed an overall strategy for assigning BACs to discrete RGH loci whereby we assumed that RGHs resulting in similar BLAST hits and present on BAC clones with the same genetic map location are derived from the same locus. Using this definition, 47 RGH loci could be identified (Fig. 1 and Table 4). The RGH locus names are composed of the chromosome number, a letter for each RGH locus on that chromosome and an indication whether they represent TNL or CNL RGHs.
Fig. 1.
Schematic representation of the genome-wide integrated genetic map of RGHs and resistance trait loci in potato. Chromosomes or linkage groups are represented by broad vertical bars with the bin signatures (van Os et al. 2006) separated by white horizontal bars. To the right of each linkage group, genetic markers are indicated in black. Previously described R genes which reside at syntenic loci are indicated in red and previously described R genes for which no syntenic loci is shown are indicated in black. Resistance trait loci that map approximately to an RGH locus are indicated in green and otherwise in blue. Horizontal dashed bars indicate that the marker used to map a previously described resistance trait locus is also mapped in the UHD map. To the left of each linkage group, RGH (TNL and CNL) loci are indicated in red if they are syntenic to previously described R genes, in green if they map approximately at the same location as a resistance trait locus and in blue if they do not. Thin vertical bars represent genetic intervals and thin horizontal bars a genetic map position
Table 4.
List of RGH loci, their map position in the UHD map of potato and the sequence homology to the closest known R protein for each domain (based on BLASTp results)
| RGH locus | BIN | Best BLAST result per domain (e-value) | ||
|---|---|---|---|---|
| TIR/CC | NB | LRR | ||
| Ia-TNL | 8-15 | BS4 (2.4e-76) | N-Nt (2.6e-138) | N-Nt (2.9e-15) |
| Ib-CNL | 8-15 | Mi-1 (5.8e-68) | Rpi-blb2 (2.7e-116) | CaMi (3.2e-68) |
| Ic-CNL | 8-15 | – | Dm3 (5.5e-13) | Dm3 (2.9e-07) |
| Id-TNL | 81 | Gro1-4 (1.3e-25) | N-Nt (3.4e-59) | N-Nt (2.9e-17) |
| IVa-CNL | 1-14 | Bs2 (1.9e-22) | Dm3 (3.3e-48) | Dm3 (9.2e-11) |
| IVb-CNL | 11-14 | Hero (0.0) | Hero (1.1e-155) | Hero (2.9e-124) |
| IVc-CNL | 33-36 | – | Dm3 (3.3e-51) | Dm3 (2.7e-12) |
| IVd-TNL | 37-44 | Bs4 (5.8e-73) | Bs4 (1.5e-97) | Bs2 (3.7e-12) |
| IVe-CNL | 37-45 | – | Dm3 (9.9e-51) | Dm3 (1.1e-08) |
| IVf-TNL | 103 | – | – | Rpp5 (0.004) |
| Va-CNL | 20-21 | R1 (0.0) | R1 (2.6e-116) | R1 (0.0) |
| Vb-CNL | 37 | Prf (1.1e-109) | Prf (3.2e-107) | Prf (1.9e-165) |
| Vc-CNL | 41-46 | Rpi-blb2 (5.7e-119) | Rpi-blb2 (6.3e-120) | CaMi (1.8e-75) |
| Vd-TNL | 41-46 | Bs4 (3.8e-74) | Bs4 (4.8e-155) | Y1 (1.4e-110) |
| Ve-CNL | 42-47 | I2 (1.6e-52) | R3a (7.9e-158) | R3a (0.0) |
| Vf-CNL | 44-46 | – | Dm3 (7.5e-53) | Dm3 (4.7e-08) |
| Vg-CNL | 65-67 | Rpi-blb2 (4.4e-117) | Rpi-blb2 (1.4e-115) | CaMi (7.3e-68) |
| VIa-CNL | 11-19 | CaMi (0.0) | Rpi-blb2 (0.0) | Rpi-blb2 (3.9e-162) |
| VIb-TNL | 16-29 | Bs4 (6.1e-81) | Bs4 (7.7e-139) | Y1 (5.5e-116) |
| VIc-TNL | 34-57 | Gro1 (2.4e-42) | Bs4 (1.3e-69) | Gro1 (1.5e-53) |
| VId-CNL | 46 | Rx1 (3.5e-31) | Gpa2 (1.8e-141) | Rx1 (4.6e-98) |
| VIIa-TNL | 31-39 | Gro1-4 (2.9e-169) | Gro1-4 (1.3e-92) | Gro1-4 (2.9e-169) |
| VIIb-CNL | 61 | – | Rps5 (3.6e-38) | Dm3 (7.7e-07) |
| VIIc-TNL | 68-71 | Y1 (5.9e-42) | Bs4 (8.3e-35) | N-Nt (1.1e-79) |
| VIId-CNL | 68-71 | – | Gpa2 (4.4e-91) | Bs2 (5.9e-44) |
| VIIIa-TNL | 10-11 | Bs4 (6.1e-74) | Y1 (3.7e-06) | Y1 (1.1e-64) |
| VIIIb-CNL | 10-11 | Bs2 (6.6e-86) | Gpa2 (1.2e-151) | Gpa2 (1.6e-66) |
| VIIIc-CNL | 20-22 | Rpi-blb2 (3.8e-133) | Rpi-blb2 (1.1e-44) | Rpi-blb2 3.6e-77) |
| IXa-CNL | 19-23 | Rx1 (2.2e-33) | Gpa2 (3.7e-153) | Rx1 (2.0e-51) |
| IXb-CNL | 31-32 | R3a (8.6e-46) | R3a (1.5e-154) | I2 (1.3e-138) |
| IXc-CNL | 35 | Rpi-blb2 (4.8e-51) | Rpi-blb2 (4.6e-94) | Mi1 (7.3e-34) |
| IXd-CNL | 48 | – | Tm2 (1.0e-56) | Tm2 (3.6e-15) |
| IXe-CNL | 41-50 | – | Sw5 (3.9e-58) | Sw5 (1.1e-17) |
| IXf-CNL | 84 | Sw5 (0.0) | Sw5 (0.0) | Sw5 (5.4e-131) |
| Xa-CNL | 1 | Bs2 (6.7e-24) | Dm3 (1.4e-46) | Dm3 (1.2e-07) |
| Xb-CNL | 62-66 | – | Rps5 (2.0e-23) | – |
| Xc-TNL | 66 | – | – | Bs4 (0.0002) |
| Xd-CNL | – | I2 (9.2e-63) | I2 (0.0) | I2 (0.0) |
| XIa-TNL | 4-8 | Bs4 (2.1e-82) | Y1 (1.2e-159) | Y1 (4.8e-159) |
| XIb-CNL | 57-62 | I2 (8.7e-58) | R3a (4.7e-174) | I2 (0.0) |
| XIc-TNL | 59-63 | Bs4 (8.7e-60) | Bs4 (4.8e-143) | Y-1 (4.4e-36) |
| XId-CNL | 77 | CaMi (0.0) | Rpi-blb2 (1.6e-164) | Rpi-blb2 (1.5e-148) |
| XIe-TNL | 82-83 | Bs4 (2.9e-68) | Bs4 (2.5e-79) | Y-1 (8.2e-70) |
| XIf-CNL | 77-86 | CaMi (0.0) | R3a (0.0) | I2 (0.0) |
| XIIa-CNL | 1-7 | R1 (7.9e-105) | R1 (3.1e-87) | R1 (1.2e-96) |
| XIIb-CNL | 46-51 | Bs2 (0.0) | Gpa2 (0.0) | Rx1 (0.0) |
| XIIc-TNL | – | Bs4 (1.7e-77) | Bs4 (1.6e-155) | N-Nt (5.7e-98) |
Loci are highlighted in bold if they are syntenic to known functional R genes as shown in Fig. 1
To see if the RGH loci identified in this study co-localise to R gene loci described previously, the approximate genetic map positions of a set of 21 known solanaceous R genes (Table 5) were included in the RGH map. From this, we could conclude that eleven of these RGH loci co-localise with known functional R genes like for instance the Rpi-blb2 locus at the short arm of chromosome VI and the Gpa2/Rx1 locus at the distal end of the short arm of chromosome XII (Fig. 1). However, no RGH loci with homology to the potato R genes R2 (and homologs) and Rpi-blb1 or the tomato genes Nrc1 and Nrg1 were found. Thirty-six RGH loci are not syntenic to existing R genes and considered to be novel RGH loci harbouring TNL (12) and CNL (24) encoding R gene homologs.
Table 5.
List of known functional solanaceous R genes, the genomic location in the genotype in which they were identified and the corresponding location in the UHD map of RH
| R gene | Genomic location | Orthologous location in RH | References |
|---|---|---|---|
| Hero | LG4 short arm | LGIV bin10-14 | Ernst et al. (2002) and Ganal et al. (1995) |
| R2 | LGIV short arm, centromeric | Unknown | Li et al. (1998 and Lokossou et al. (2009) |
| R2-like | LGIV short arm, centromeric | Unknown | Lokossou et al. (2009) and Park et al. (2005a) |
| Rpi-blb3 | LGIV short arm, centromeric | Unknown | Lokossou et al. (2009) |
| Rpi-abpt | LGIV short arm, centromeric | Unknown | Lokossou et al. (2009) and Park et al. (2005b) |
| R1 | LGV short arm | LGV bin 20-21 | Ballvora et al. (2002) and Leonards-Schippers et al. (1992) |
| Prf | LG5 short arm | LGV bin37 | Martin et al. (1991) Salmeron et al. (1994) Salmeron et al. (1996) |
| Bs4 | LGV short arm | LGV bin41-46 | Ballvora et al. (2001) and Schornack et al. (2004) |
| Mi-1 | LG6 short arm | LGVI bin1-19 | Kaloshian et al. (1998) and Milligan et al. (1998) |
| Rpi-blb2 | LGVI short arm | LGVI bin1-19 | van der Vossen et al. (2005) |
| Gro1 | LGVII | LGVII bin31-39 | Barone et al. (1990) Paal et al. (2004) |
| Rpi-blb1 | LGVIII long arm | Unknown | van der Vossen et al. (2003) |
| Tm2 | LG9 long arm, centromeric | LGIX bin38 | Lanfermeijer et al. (2003) |
| Sw5 | LG9 long arm, telomeric | LGIX bin84 | Spassova et al. (2001) and Stevens et al. 1995) |
| Y1 | LGXI long arm, telomeric | LGXI bin1-9 | Hamalainen et al. (1998) and Vidal et al. (2002) |
| N | A region syntenic to potato LGXI long arm, telomeric | LGXI bin1-9 | Hehl et al. (1999) and Whitham et al. (1994) |
| R3a | LGXI short arm, telomeric | LGXI bin82-86 | Huang et al. (2005) and Huang et al. (2004) |
| I2 | LG11, telomeric | LGXI bin82-86 | Ori et al. (1997) and Segal et al. (1992) |
| Rx1 | LGXII, short arm, telomeric | LGXII bin46-51 | Bendahmane et al. (1999) and Ritter et al. (1991) |
| Gpa2 | LGXII, short arm, telomeric | LGXII bin46-51 | Rouppe van der Voort et al. (1997) and van der Vossen et al. (2000) |
| Bs2 | LG9 (a region syntenic to potato LGXII, short arm, telomeric) | LGXII bin46-51 | Mazourek et al. (2009) and Tai et al. (1999) |
Integrating disease resistance trait loci in the RGH genetic map
For the integration of potato disease resistance trait loci in the RGH map, 68 universal CAPS markers were first mapped in RH (Fig. 1) to create anchor points for each linkage group to facilitate the comparison of genetic maps obtained for different Solanum genotypes. For two resistance QTLs [i.e. Eca1a (Zimnoch-Guzowska et al. 2000) and K31_T1-11 (Oberhagemann et al. 1999)], the CAPS markers could be used directly, because the same marker was used to map the resistance trait in the original mapping population. In most cases, however, the tomato-EXPEN 2000 map (Fulton et al. 2002) on the Sol Genomics Network (Mueller et al. 2005), or the PoMaMo database (Meyer et al. 2005) were used to estimate the genomic position by comparing the location of the marker used to map the resistance trait and the location of the markers mapped in this study. In this way, 58 more trait loci for which no functional R gene has been identified yet could be integrated in the UHD genetic map of RH. The loci that have been mapped were as reviewed (Gebhardt and Valkonen 2001; Hein et al. 2009). In addition, fifteen loci were integrated that were not described in these reviews i.e. Ny-1 (Szajko et al. 2008), Ry-f sto (Flis et al. 2005), Ry sto (Song et al. 2005), Ry chc (Sato et al. 2006), Gm and Rm (Marczewski et al. 2006), Ns (Marczewski et al. 2002), Ny tbr (Celebi-Toprak et al. 2002), PLRV.I (Marczewski et al. 2001), PLRV.4 (Marczewski et al. 2004), Rl adg (Velásquez et al. 2007), MfaXII spl (Bakari et al. 2006), GpaM1 (Caromel et al. 2003), GpaV sspl and GpaXI sspl (Caromel et al. 2005). After integration of this information in the RGH map, we could observe that twenty-one RGH loci may be linked to forty-six disease traits for which no R gene sequence information is available yet. In addition, thirteen remaining RGH loci were still not linked to any disease resistance trait.
Discussion
Here, we present a comprehensive overview of the genome-wide distribution of NB-LRR loci harbouring resistance gene homologs (RGHs) in potato. RGH sequences were retrieved after analysing the sequences of 288 unique BAC clones, which were selected from a BAC library of the diploid potato clone RH89-039-16 (S. tuberosum ssp. tuberosum). This resulted in the identification of 738 partial and full-length NB-LRR encoding genes. Based on homology of the RGH sequences with known resistance gene proteins, the complete set of RGH could be subdivided in 280 TIR-NB-LRR (TNL) encoding sequences and 448 sequences encoding CC-NB-LRR (CNL). The RGH containing BACs were anchored to the genetic and physical map of potato, which allowed us to determine the location of R gene loci in the potato genome. A total of 47 NB-LRR loci were detected in the potato genome, including 36 novel loci and 11 loci syntenic with previously identified functional resistance gene loci (Hero, R1, Bs4, Prf, Rpi-blb2/Mi1/CaMi, Gro1, Sw5, Y-1/N, R3a/I2 and Gpa2/Rx1/Bs2). A literature survey was conducted to integrate the resistance trait loci described for potato. This showed that they often co-localise with the TNL and CNL loci obtained in this study. Hence, the integrated genome-wide genetic map of potato presented in this paper provides an excellent template for the development of markers for marker-assisted selection and for candidate gene approaches for the identification of functional R genes.
Leister et al. (1996) produced a genetic map in potato using (amongst others) NB-domain targeting RFLP markers (St markers). As expected, the majority of the 27 St markers map in an area where NB-LRR sequences were detected in this study. In addition, we identified at least 27 loci that do harbour RGHs but which were not detected by an St marker. However, nine St markers present on chromosomes II, III, IV, VIII, X and XII map to a genomic location where no RGHs have been detected. This discrepancy may indicate that our NB-based screening of the RH BAC library was not saturated, which is supported by the observation that we were not able to identify a number of R genes, including R2, R2-like, Rpi-blb3/Rpi-abpt on chromosome IV (Lokossou et al. 2009) and Rpi-blb1 on chromosome VIII (van der Vossen et al. 2003). This bias in our collection can be explained by the lack in sequence homology between the NB regions used to design the probes to screen the BAC library and the corresponding region in the NB domain of this particular set of R genes (data not shown). By using a BAC library covering 5× the genome, it is very well possible that not all RGH loci present in the potato genome were represented in the library. It is very unlikely that some RGH loci are not present in RH, because unpublished results for the Rx1/Gpa2 locus and for RGH locus Vg-CNL show that they are present in over 500 genotypes derived from different Solanum species.
The data presented in this study show that the genomic distribution of RGH loci is not random as illustrated by the low number of RGHs present on chromosome I compared to for instance the large number of RGHs identified on chromosome XI that harbours six RGH-loci, corresponding to 6 St markers. This non-random distribution of RGH loci was anticipated by Gebhardt and Valkonen (2001) based on the non-random distribution of resistance trait loci. Therefore, the genome-wide NB-LRR map presented in this study was integrated with resistance trait loci as reviewed by Gebhardt and Valkonen (2001) and Hein et al. (2009). The map has been supplemented with 15 loci that have not been described by these two papers. Thirty-eight resistance trait loci approximately co-localise with TNL and CNL loci (Fig. 1), providing a template for a candidate-gene approach. Interestingly, many of these are quantitative resistance traits, suggesting NB-LRR resistance genes underlie these quantitative loci. At least ten RGH loci do not co-localise with known resistance trait loci, which could potentially facilitate a candidate gene approach for the identification of genes underlying as yet unmapped resistance traits.
In compliance with previously analysed plant species like Arabidopsis (Meyers et al. 2003) poplar (Kohler et al. 2008) and rice (Zhou et al. 2004), this study shows that the potato genome harbours a substantial and highly diverse super-family of NB-LRR genes, which can be divided in a TIR and a CC class of RGHs. The ratio between TNL and CNL as observed in this study is 5:8, which is in the range of the ratio observed in other dicot species like Arabidopsis, lettuce, poplar and grapevine, where the ratio TIR:CC is 2:1, 1:1, 3:7 and 1:2, respectively (Kohler et al. 2008; McHale et al. 2009; Meyers et al. 2003; Yang et al. 2008). Remarkably, in monocot species the TIR RGHs are almost absent. Rice contains only three TIR-NBS sequences, but they lack the LRR domain and have very deviant NBS sequences (Bai et al. 2002; Zhou et al. 2004).
Most RGHs are present on complex loci (Michelmore and Meyers 1998) and the findings in this study comply with this. However, in eleven cases only one single RGH could be allocated to a locus. They may represent simple loci, but it can also be the result of incomplete sequencing. Interestingly, many loci, both complex and simple, reside in hotspots of resistance genes. In these regions (for instance on chromosome V between bins 40 and 50) the distinction between different clusters could not be based on genetic segregation and is purely based on sequence divergence (Supplemental Fig. 1). On the basis of this genetic map, it is impossible to determine whether these RGH clusters are still physically separated or completely intermingled. However, in three cases, two different types of RGH are even present on the same BAC. In the Arabidopsis genome sequence, mixed RGH clusters have been identified (Meyers et al. 2003) and therefore it is possible that the RGH clusters are not physically discrete. If they are, the fact that two types of RGH are present on one BAC proves that they can be physically very close in potato (i.e. less than 100 kb).
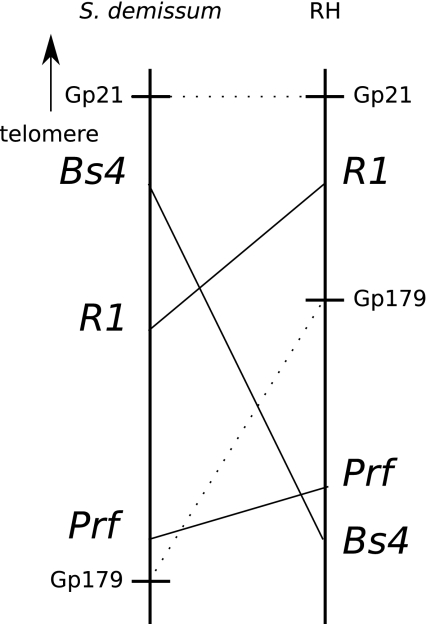
Grube et al. (2000) showed that R gene loci in the solanaceous crop genera tomato, potato and pepper are present on corresponding positions and that homologs of previously identified R genes derived from tomato and tobacco could be identified in syntenious positions in pepper. In this study, it was shown that RGH sequences co-localise with resistance gene loci in potato. Synteny of R gene loci between different potato genotypes or even other solanaceous species is illustrated by the fact that NB-LRR sequences homologous to previously identified R genes in potato, tomato, pepper and tobacco have been found at corresponding genomic locations. However, it is also known that resistance gene loci are very dynamic regions (Ballvora et al. 2007; Kuang et al. 2005) and that micro-synteny seldom exists. At a larger scale, synteny can also be partly lost between different genotypes. This is shown on chromosome V, where three functional R genes have previously been identified, namely R1, Prf and Bs4 (Ballvora et al. 2002; Salmeron et al. 1996; Schornack et al. 2004). Although two of these genes (Prf and Bs4) are derived from tomato, a partly sequenced physical map of this region in S. demissum revealed that homologs of all three genes are present in potato (Kuang et al. 2005). Indeed, in RH, the homologs of all three genes have been identified and mapped as well (this study). In S. demissum, the order of the three clusters, including the two markers Gp21 and Gp179 and coming from the telomere, is Gp21, Bs4, R1, Prf, Gp179 (Kuang et al. 2005). Interestingly, in the genetic map of RH the order is Gp21, R1, Gp179, Prf, Bs4 (Fig. 2). The markers in RH are fitted to the BIN signatures of the UHD map, which is the most parsimonious marker order based on 10,000 markers and considered to be very robust (Isidore et al. 2003; van Os et al. 2006). Nevertheless, an incorrect marker order can never be completely ruled out. This lack of synteny, genuine or artificial, can be a drawback for applications.
Fig. 2.
Comparative map representing the short arm of chromosome V of S. demissum and S. tuberosum (RH). The vertical bars represent the genetic maps. Genetic positions of markers Gp21 and Gp179 are indicated with a horizontal bar and their comparison between the two maps with a dashed line. Comparison of the RGH loci syntenic to Bs4, R1 and Prf are indicated with lines. The direction of the telomere is indicated with an arrow
Breeders want to incorporate agronomically interesting resistance traits in their breeding material. Marker assisted selection is a technique that can facilitate this process. However, for marker assisted selection, it is essential that markers are available that are diagnostic for the trait of interest (Moloney et al. 2010). It was shown that markers that are genetically very close in a potato mapping population are not always diagnostic in breeding material (Moloney et al. 2010). A specific marker developed on the resistance gene itself has the highest change of being diagnostic. It was shown that cluster specific markers can be developed in the LRR region, which is the most variable part of the resistance gene (Bakker et al. 2003); (Finkers-Tomczak et al. 2011). It is also possible to develop gene specific markers for each member of a resistance gene cluster (Finkers-Tomczak et al. 2011). The genome-wide integrated map presented in this study, links resistance trait loci with NB-LRR loci, and will provide a template to design cluster-specific markers and closely linked markers for the benefit of marker-assisted selection and candidate gene approaches. The potato genome sequence will be available soon (Visser et al. 2009), and this will allow us to fill the gaps and complete the collection of NB-LRR sequences.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1 SSR forward (F) and reverse (R) primer sequences and simple sequence repeats (PDF 73 kb)
Supplemental Table 2 CAPS primer sequences, their annealing temperature (Tm) in °C and their appropriate endonuclease (enzyme). (PDF 96 kb)
Supplemental Table 3 A list of RGH loci per linkage group as identified in this study. Each row represents a significant RGH domain BLASTp hit, with in the columns from left to right: The name of the RGH locus, the BAC fragment on which the BLASTp hit was found, the Genbank accession number of the BAC sequence, the linkage group (LG) and the bin range (bin) on which the BAC is mapped in the UHD map, the translational frame where the BLASTp hit was found and the amino acid position of the BLASTp hit (start – stop) on the BAC fragment. The significance of the BLASTp hit (Expected) and the method that was used to map the BAC (anchor). The last column indicates the number of (partial) RGHs that were detected. In the first columns, the R genes syntenic to the RGH loci are indicated in italics. (PDF 335 kb)
Supplemental Fig. 1 Schematic representation of the genetically mapped BACs harbouring RGHs and the defined RGH loci The broad black vertical bar represents the genetic map with the bin signatures (van Os et al. 2006) separated by white horizontal bars. To the right of the genetic map, the BAC clones that are mapped at this location are presented, with a black horizontal bar to indicate their genetic interval. On the left side, the cluster names, the universal markers and linked cloned R genes are indicated, with the interval based on the genetic location of the BAC clones or the bin range of the markers. The red BAC names indicate the presence of CNL RGHs and the absence of TNL RGHs. The green BAC names indicate the presence of TNL RGHs and the absence of CNL RGHs and the blue BAC names indicate the presence of both TNL and CNL RGHs. (PDF 194 kb)
Acknowledgments
We would like to thank Gerda Sabatino for her help with the NBS profiling work. This research was financed by the Centre for BioSystems Genomics (CBSG), which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research, The Dutch Technology Foundation and the EU Integrated Projects APOPHYS (QLRT-2001-01849) BioExploit (FOOD-CT-2005-513959).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG. Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002;12:1871–1884. doi: 10.1101/gr.454902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakari KA, Marie-Claire K, Bernard C, Jean-Paul D, Didier F, Maria MD, Daniel E, Didier M. A major gene mapped on chromosome XII is the main factor of a quantitatively inherited resistance to Meloidogyne fallax in Solanum sparsipilum. Theor Appl Genet. 2006;112:699–707. doi: 10.1007/s00122-005-0173-2. [DOI] [PubMed] [Google Scholar]
- Bakker E, Butterbach P, Rouppe van der Voort J, Van der Vossen E, Van Vliet J, Bakker J, Goverse A. Genetic and physical mapping of homologs of the virus resistance gene Rx1 and the cyst nematode resistance gene Gpa2 in potato. Theor Appl Genet. 2003;106:1524–1531. doi: 10.1007/s00122-003-1213-4. [DOI] [PubMed] [Google Scholar]
- Ballvora A, Schornack S, Baker BJ, Ganal M, Bonas U, Lahaye T. Chromosome landing at the tomato Bs4 locus. Mol Gen Genomics. 2001;266:639–645. doi: 10.1007/s004380100583. [DOI] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313x.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- Ballvora A, Jocker A, Viehover P, Ishihara H, Paal J, Meksem K, Bruggmann R, Schoof H, Weisshaar B, Gebhardt C (2007) Comparative sequence analysis of Solanum and Arabidopsis in a hot spot for pathogen resistance on potato chromosome V reveals a patchwork of conserved and rapidly evolving genome segments. BMC Genomics 8:art. no. 112 [DOI] [PMC free article] [PubMed]
- Barone A, Ritter E, Schachtschabel U, Debener T, Salamini F, Gebhardt C. Localization by Restriction-Fragment-Length-Polymorphism mapping in potato of a major dominant gene conferring resistance to the potato cyst nematode Globodera rostochiensis. Mol Gen Genet. 1990;224:177–182. doi: 10.1007/BF00271550. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 2000;21:73–81. doi: 10.1046/j.1365-313x.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. Rps2 of Arabidopsis-Thaliana—a leucine-rich repeat class of plant-disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000;21:177–188. doi: 10.1046/j.1365-313x.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- Bonierbale MW, Plaisted RL, Tanksley SD. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan MH, Gunn N, Cooper A, Gulden S, Tor M, Rimmer SR, Holub EB. WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol Plant Microbe Interact. 2008;21:757–768. doi: 10.1094/MPMI-21-6-0757. [DOI] [PubMed] [Google Scholar]
- Borm TJA. Construction and use of a physical map of potato. Wageningen: Wageningen University; 2008. p. 136. [Google Scholar]
- Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JDG. Three genes of the arabidopsis RPP1 complex resistance locus recognize distinct peronospora parasitica avirulence determinants. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw JE, Ramsay G. Utilisation of the Commonwealth Potato Collection in potato breeding. Euphytica. 2005;146:9–19. [Google Scholar]
- Bryan GT, Wu KS, Farrall L, Jia YL, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2033–2045. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caromel B, Mugniéry D, Lefebvre V, Andrzejewski S, Ellissèche D, Kerlan MC, Rousselle P, Rousselle-Bourgeois F. Mapping QTLs for resistance against Globodera pallida (Stone) Pa2/3 in a diploid potato progeny originating from Solanum spegazzinii. Theor Appl Genet. 2003;106:1517–1523. doi: 10.1007/s00122-003-1211-6. [DOI] [PubMed] [Google Scholar]
- Caromel B, Mugniery D, Kerlan MC, Andrzejewski S, Palloix A, Ellisseche D, Rousselle-Bourgeois F, Lefebvre V. Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ratio of Globodera pallida and together for developing a necrotic reaction. Mol Plant Microbe Interact. 2005;18:1186–1194. doi: 10.1094/MPMI-18-1186. [DOI] [PubMed] [Google Scholar]
- Celebi-Toprak F, Slack SA, Jahn MM. A new gene, Nytbr, for hypersensitivity to Potato virus Y from Solanum tuberosum maps to chromosome IV. Theor Appl Genet. 2002;104:669–674. doi: 10.1007/s001220100749. [DOI] [PubMed] [Google Scholar]
- Chen R, Li H, Zhang L, Zhang J, Xiao J, Ye Z. CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annuum L.) confers nematode resistance in tomato. Plant Cell Rep. 2007;26:895–905. doi: 10.1007/s00299-007-0304-0. [DOI] [PubMed] [Google Scholar]
- Cloutier S, McCallum BD, Loutre C, Banks TW, Wicker T, Feuillet C, Keller B, Jordan MC. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol Biol. 2007;65:93–106. doi: 10.1007/s11103-007-9201-8. [DOI] [PubMed] [Google Scholar]
- Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley MB, Pathirana S, Wu H-J, Kachroo P, Klessig DF. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 2000;12:663–676. doi: 10.1105/tpc.12.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 2001;27:439–453. doi: 10.1046/j.1365-313x.2001.01114.x. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst K, Kumar A, Kriseleit D, Kloos D-U, Phillips MS, Ganal MW (2002) The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J 31:127–136 [DOI] [PubMed]
- Feuillet C, Travella S, Stein N, Albar L, Nublat A, Keller B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA. 2003;100:15253–15258. doi: 10.1073/pnas.2435133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers-Tomczak A, Bakker E, de Boer J, van der Vossen E, Achenbach U, Golas T, Suryaningrat S, Smant G, Bakker J, Goverse A. Comparative sequence analysis of the potato cyst nematode resistance locus H1 reveals a major lack of co-linearity between three haplotypes in potato (Solanum tuberosum ssp.) Theor Appl Genet. 2011;122:595–608. doi: 10.1007/s00122-010-1472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flis B, Hennig J, Strzelczyk-Zyta D, Gebhardt C, Marczewski W. The Ry-fsto gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122718 in PVY resistant potato cultivars. Mol Breed. 2005;15:95–101. [Google Scholar]
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell. 2002;14:1457–1467. doi: 10.1105/tpc.010479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen, Jack H, Ekengren, Sophia K, van Ooijen, Gerben, Abd-El-Haliem, Ahmed M, van den Berg, Grardy CM, Rainey, Daphne Y, Martin, Gregory B, Takken, Frank LW, de Wit, Pierre JGM, Joosten, Matthieu HAJ. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 2007;50:14–28. doi: 10.1111/j.1365-313X.2007.03027.x. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Simon R, Brommonschenkel SH, Arndt M, Phillips MS, Tanksley SD, Kumar A. Genetic mapping of a wide spectrum nematode resistance gene (Hero) against Globodera rostochiensis in tomato. Mol Plant Microbe Interact. 1995;8:886–891. doi: 10.1094/mpmi-8-0886. [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JPT. Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Linares AR, Cavallisforza LL, Feldman MW. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Prins P, Nakao M, Bonnal R, Aerts J, Katayama T. BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26:2617–2619. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis Rpm1 gene enabling dual- specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Grube RC, Radwanski ER, Jahn M. Comparative genetics of disease resistance within the Solanaceae. Genetics. 2000;155:873–887. doi: 10.1093/genetics/155.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalainen JH, Sorri VA, Watanabe KN, Gebhardt C, Valkonen JPT. Molecular examination of a chromosome region that controls resistance to potato Y and A potyviruses in potato. Theor Appl Genet. 1998;96:1036–1043. [Google Scholar]
- Hehl R, Faurie E, Hesselbach J, Salamini F, Whitham S, Baker B, Gebhardt C. TMV resistance gene N homologs are linked to Synchytrium endobioticum resistance in potato. Theor Appl Genet. 1999;98:379–386. [Google Scholar]
- Hein I, Birch PRJ, Danan S, Lefebvre V, Achieng Odeny D, Gebhardt C, Trognitz F, Bryan GJ (2009) Progress in mapping and cloning qualitative and quantitative resistance against phytophthora infestans in potato and its wild relatives. Potato Res 1–13
- Huang L, Brooks SA, Li WL, Fellers JP, Trick HN, Gill BS. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics. 2003;164:655–664. doi: 10.1093/genetics/164.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Vleeshouwers V, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E. The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant Microbe Interact. 2004;17:428–435. doi: 10.1094/MPMI.2004.17.4.428. [DOI] [PubMed] [Google Scholar]
- Huang SW, van der Vossen EAG, Kuang HH, Vleeshouwers V, Zhang NW, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- Isidore E, van Os H, Andrzejewski S, Bakker J, Barrena I, Bryan GJ, Caromel B, van Eck H, Ghareeb B, de Jong W, van Koert P, Lefebvre V, Milbourne D, Ritter E, van der Voort JR, Rousselle-Bourgeois F, van Vliet J, Waugh R. Toward a marker-dense meiotic map of the potato genome: lessons from linkage group I. Genetics. 2003;165:2107–2116. doi: 10.1093/genetics/165.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloshian I, Yaghoobi J, Liharska T, Hontelez J, Hanson D, Hogan P, Jesse T, Wijbrandi J, Simons G, Vos P, Zabel P, Williamson VM. Genetic and physical localization of the root-knot nematode resistance locus Mi in tomato. Mol Gen Genet. 1998;257:376–385. doi: 10.1007/s004380050660. [DOI] [PubMed] [Google Scholar]
- Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kuang HH, Wei FS, Marano MR, Wirtz U, Wang XX, Liu J, Shum WP, Zaborsky J, Tallon LJ, Rensink W, Lobst S, Zhang PF, Tornqvist CE, Tek A, Bamberg J, Helgeson J, Fry W, You F, Luo MC, Jiang JM, Buell CR, Baker B. The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologs and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 2005;44:37–51. doi: 10.1111/j.1365-313X.2005.02506.x. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Dijkhuis J, Sturre MJG, de Haan P, Hille J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol Biol. 2003;52:1039–1051. doi: 10.1023/a:1025434519282. [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG. The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene Rps2 and the tobacco viral resistance gene N. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Ballvora A, Salamini F, Gebhardt C. A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet. 1996;14:421–429. doi: 10.1038/ng1296-421. [DOI] [PubMed] [Google Scholar]
- Leonards-Schippers C, Gieffers W, Salamini F, Gebhardt C. The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Mol Gen Genet. 1992;233:278–283. doi: 10.1007/BF00587589. [DOI] [PubMed] [Google Scholar]
- Li X, Van Eck HJ, Rouppe Van Der Voort JNAM, Huigen DJ, Stam P, Jacobsen E. Autotetraploids and genetic mapping using common AFLP markers: the R2 allele conferring resistance to Phytophthora infestans mapped on potato chromosome 4. Theor Appl Genet. 1998;96:1121–1128. [Google Scholar]
- Lin F, Chen S, Que ZQ, Wang L, Liu XQ, Pan QH. The blast resistance gene Pi37 encodes a nucleotide binding site-leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics. 2007;177:1871–1880. doi: 10.1534/genetics.107.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Lin F, Wang L, Pan QH. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics. 2007;176:2541–2549. doi: 10.1534/genetics.107.075465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PR, Visser RG, Jacobsen E, van der Vossen EA. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant-microbe Interact MPMI. 2009;22:630–641. doi: 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- Marczewski W, Flis B, Syller J, Schäfer-Pregl R, Gebhardt C (2001) A major quantitative trait locus for resistance to Potato leafroll virus is located in a resistance hotspot on potato chromosome XI and is tightly linked to N-gene-like markers. Mol Plant-Microbe Inter 14:1420–1425 [DOI] [PubMed]
- Marczewski W, Hennig J, Gebhardt C. The Potato virus S resistance gene Ns maps to potato chromosome VIII. Theor Appl Genet. 2002;105:564–567. doi: 10.1007/s00122-002-0976-3. [DOI] [PubMed] [Google Scholar]
- Marczewski W, Flis B, Syller J, Strzelczyk-Zyta D, Hennig J, Gebhardt C. Two allelic or tightly linked genetic factors at the PLRV.4 locus on potato chromosome XI control resistance to potato leafroll virus accumulation. Theor Appl Genet. 2004;109:1604–1609. doi: 10.1007/s00122-004-1780-z. [DOI] [PubMed] [Google Scholar]
- Marczewski W, Strzelczyk-Zyta D, Hennig J, Witek K, Gebhardt C. Potato chromosomes IX and XI carry genes for resistance to potato virus M. Theor Appl Genet. 2006;112:1232–1238. doi: 10.1007/s00122-006-0224-3. [DOI] [PubMed] [Google Scholar]
- Martin GB, Williams JGK, Tanksley SD. Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci USA. 1991;88:2336–2340. doi: 10.1073/pnas.88.6.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazourek M, Cirulli ET, Collier SM, Landry LG, Kang BC, Quirin EA, Bradeen JM, Moffett P, Jahn MM. The fractionated orthology of Bs2 and Rx/Gpa2 supports shared synteny of disease resistance in the solanaceae. Genetics. 2009;182:1351–1364. doi: 10.1534/genetics.109.101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Dhandaydham M, Long TA, Aarts MGM, Goff S, Holub EB, Dangl JL. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale LK, Truco MJ, Kozik A, Wroblewski T, Ochoa OE, Lahre KA, Knapp SJ, Michelmore RW. The genomic architecture of disease resistance in lettuce. Theor Appl Genet. 2009;118:565–580. doi: 10.1007/s00122-008-0921-1. [DOI] [PubMed] [Google Scholar]
- Meyer S, Nagel A, Gebhardt C (2005) PoMaMo—a comprehensive database for potato genome data. Nucleic Acids Res 33:D666–D670 [DOI] [PMC free article] [PubMed]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell. 1998;10:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson V. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell. 1998;10:1307–1319. doi: 10.1105/tpc.10.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene Rps2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Moloney C, Griffin D, Jones PW, Bryan GJ, McLean K, Bradshaw JE, Milbourne D. Development of diagnostic markers for use in breeding potatoes resistant to Globodera pallida pathotype Pa2/3 using germplasm derived from Solanum tuberosum ssp. andigena CPC 2802. Theor Appl Genet. 2010;120:679–689. doi: 10.1007/s00122-009-1185-0. [DOI] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD. The SOL Genomics Network. A comparative resource for Solanaceae biology and beyond. Plant Physiol. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhagemann P, Chatot-Balandras C, Schäfer-Pregl R, Wegener D, Palomino C, Salamini F, Bonnel E, Gebhardt C (1999) A genetic analysis of quantitative resistance to late blight in potato: towards marker-assisted selection. Mol Breed 5:399–415
- Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. The l2C family from the wilt disease resistance locus l2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell. 1997;9:521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paal J, Henselewski H, Muth J, Meksem K, Menendez CM, Salamini F, Ballvora A, Gebhardt C. Molecular cloning of the potato Gro1–4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004;38:285–297. doi: 10.1111/j.1365-313X.2004.02047.x. [DOI] [PubMed] [Google Scholar]
- Park TH, Vleeshouwers V, Huigen DJ, van der Vossen EAG, van Eck HJ, Visser RGF. Characterization and high-resolution mapping of a late blight resistance locus similar to R2 in potato. Theor Appl Genet. 2005;111:591–597. doi: 10.1007/s00122-005-2050-4. [DOI] [PubMed] [Google Scholar]
- Park TH, Vleeshouwers VGAA, Hutten RCB, Van Eck HJ, Van Der Vossen E, Jacobsen E, Visser RGF. High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol Breed. 2005;16:33–43. [Google Scholar]
- Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, vanderBiezen EA, Moores T, Dean C, Daniels MJ, Jones JDG. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR Protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol. 2005;15:968–973. doi: 10.1016/j.cub.2005.04.053. [DOI] [PubMed] [Google Scholar]
- Ritter E, Debener T, Barone A, Salamini F, Gebhardt C. RFLP mapping on potato chromosomes of 2 genes controlling extreme resistance to potato virus X (PVX) Mol Gen Genet. 1991;227:81–85. doi: 10.1007/BF00260710. [DOI] [PubMed] [Google Scholar]
- Rouppe van der Voort J, Wolters P, Folkertsma R, Hutten R, Van Zandvoort P, Vinke H, Kanyuka K, Bendahmane A, Jacobsen E, Janssen R, Bakker J. Mapping of the cyst nematode resistance locus Gpa2 in potato using a strategy based on comigrating AFLP markers. Theor Appl Genet. 1997;95:874–880. [Google Scholar]
- Salmeron JM, Barker SJ, Carland FM, Mehta AY, Staskawicz BJ. Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell. 1994;6:511–520. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GED, Rommens CMT, Scofield S, Kim H-S, Lavelle DT, Dahlbeck D, Staskawicz B. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Moleculal cloning: a loboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato M, Nishikawa K, Komura K, Hosaka K. Potato virus Y resistance gene, Rychc, mapped to the distal end of potato chromosome 9. Euphytica. 2006;149:367–372. [Google Scholar]
- Schornack S, Ballvora A, Gurlebeck D, Peart J, Baulcombe D, Ganal M, Baker B, Bonas U, Lahaye T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severly truncated derivatives of AvrBs4 and overexpressed AvrBs3 (vol 37, pg 46, 2004) Plant J. 2004;37:787. doi: 10.1046/j.1365-313x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- Segal G, Sarfatti M, Schaffer MA, Ori N, Zamir D, Fluhr R. Correlation of genetic and physical structure in the region surrounding the I2Fusarium oxysporum resistance locus in tomato. Mol Gen Genet. 1992;231:179–185. doi: 10.1007/BF00279789. [DOI] [PubMed] [Google Scholar]
- Sekine K-T, Ishihara T, Hase S, Kusano T, Shah J, Takahashi H. Single amino acid alterations in Arabidopsis thaliana RCY1 compromise resistance to Cucumber mosaic virus, but differentially suppress hypersensitive response-like cell death. Plant Mol Biol. 2006;62:669–682. doi: 10.1007/s11103-006-9048-4. [DOI] [PubMed] [Google Scholar]
- Shirano Y, Kachroo P, Shah J, Klessig DF. A gain-of-function mutation in an arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell. 2002;14:3149–3162. doi: 10.1105/tpc.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tör M, Crute I, Bittner-Eddy P, Beynon J. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Song YS, Hepting L, Schweizer G, Hartl L, Wenzel G, Schwarzfischer A. Mapping of extreme resistance to PVY (Rysto) on chromosome XII using anther-culture-derived primary dihaploid potato lines. Theor Appl Genet. 2005;111:879–887. doi: 10.1007/s00122-005-0010-7. [DOI] [PubMed] [Google Scholar]
- Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, Prins M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol Breed. 2001;7:151–161. [Google Scholar]
- Stevens MR, Lamb EM, Rhoads DD. Mapping the Sw-5 locus for tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor Appl Genet. 1995;90:451–456. doi: 10.1007/BF00221989. [DOI] [PubMed] [Google Scholar]
- Szajko K, Chrzanowska M, Witek K, Strzelczyk-Zyta D, Zagórska H, Gebhardt C, Hennig J, Marczewski W (2008) The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor Appl Genetics 116:297–303 [DOI] [PMC free article] [PubMed]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Takken FLW. Resistance proteins: Scouts of the plant innate immune system. Eur J Plant Pathol. 2008;121:243–255. [Google Scholar]
- van der Biezen E, Freddie CT, Kahn K, Parker JE, Jones JDG (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29:439–451 [DOI] [PubMed]
- van der Linden CG, Wouters D, Mihalka V, Kochieva EZ, Smulders MJM, Vosman B. Efficient targeting of plant disease resistance loci using NBS profiling. Theor Appl Genet. 2004;109:384–393. doi: 10.1007/s00122-004-1642-8. [DOI] [PubMed] [Google Scholar]
- van der Vossen E, Rouppe van der Voort J, Kanyuka K, Bendahmane A, Sandbrink H, Baulcombe D, Bakker J, Stiekema W, Klein-Lankhorst R. Homologs of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J. 2000;23:567–576. doi: 10.1046/j.1365-313x.2000.00814.x. [DOI] [PubMed] [Google Scholar]
- van der Vossen E, Sikkema A, Hekkert BTL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313x.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- van Os H, Andrzejewski S, Bakker E, Barrena I, Bryan GJ, Caromel B, Ghareeb B, Isidore E, de Jong W, van Koert P, Lefebvre V, Milbourne D, Ritter E, van der Voort JNAMR, Rousselle-Bourgeois F, van Vliet J, Waugh R, Visser RGF, Bakker J, van Eck HJ. Construction of a 10, 000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics. 2006;173:1075–1087. doi: 10.1534/genetics.106.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez AC, Mihovilovich E, Bonierbale M (2007) Genetic characterization and mapping of major gene resistance to potato leafroll virus in Solanum tuberosum ssp. andigena. Theor Appl Genetics 114:1051–1058 [DOI] [PubMed]
- Vidal S, Cabrera H, Andersson RA, Fredriksson A, Valkonen JPT. Potato gene Y-1 is an N gene homolog that confers cell death upon infection with potato virus Y. Mol Plant Microbe Interact. 2002;15:717–727. doi: 10.1094/MPMI.2002.15.7.717. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Shmoys DB, Durrett RT, Tanksley SD. Selective mapping: a strategy for optimizing the construction of high-density linkage maps. Genetics. 2000;155:407–420. doi: 10.1093/genetics/155.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser RGF, Bachem CWB, de Boer JM, Bryan GJ, Chakrabati SK, Feingold S, Gromadka R, van Ham RCHJ, Huang S, Jacobs JME, Kuznetsov B, de Melo PE, Milbourne D, Orjeda G, Sagredo B, Tang X. Sequencing the Potato genome: outline and first results to come from the Elucidation of the sequence of the world’s third most important food crop. Am J Potato Res. 2009;86:417–429. [Google Scholar]
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Osterenk J, De Both M, Peleman J, Liharska T, Hontelez J, Zabeau M. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol. 1998;16:1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999;19:55–64. doi: 10.1046/j.1365-313x.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S, Dineshkumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic-virus resistance gene-N—similarity to Toll and the Interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Yahiaoui N, Srichumpa P, Dudler R, Keller B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004;37:528–538. doi: 10.1046/j.1365-313x.2003.01977.x. [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang X, Yue JX, Tian D, Chen JQ. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Gen Genomics. 2008;280:187–198. doi: 10.1007/s00438-008-0355-0. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang Z-X, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci USA. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Vale G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P. Cell-autonomous expression of barley mla1 confers race-specific resistance to the powdery mildew fungus via a rar1-independent signaling pathway. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Gen Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- Zimnoch-Guzowska E, Marczewski W, Lebecka R, Flis B, Schäfer-Pregl R, Salamini F, Gebhardt C (2000) QTL analysis of new sources of resistance to Erwinia carotovora ssp. atroseptica in Potato done by AFLP, RFLP, and resistance-gene-like markers. Crop Sci 40:1156–1167
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 SSR forward (F) and reverse (R) primer sequences and simple sequence repeats (PDF 73 kb)
Supplemental Table 2 CAPS primer sequences, their annealing temperature (Tm) in °C and their appropriate endonuclease (enzyme). (PDF 96 kb)
Supplemental Table 3 A list of RGH loci per linkage group as identified in this study. Each row represents a significant RGH domain BLASTp hit, with in the columns from left to right: The name of the RGH locus, the BAC fragment on which the BLASTp hit was found, the Genbank accession number of the BAC sequence, the linkage group (LG) and the bin range (bin) on which the BAC is mapped in the UHD map, the translational frame where the BLASTp hit was found and the amino acid position of the BLASTp hit (start – stop) on the BAC fragment. The significance of the BLASTp hit (Expected) and the method that was used to map the BAC (anchor). The last column indicates the number of (partial) RGHs that were detected. In the first columns, the R genes syntenic to the RGH loci are indicated in italics. (PDF 335 kb)
Supplemental Fig. 1 Schematic representation of the genetically mapped BACs harbouring RGHs and the defined RGH loci The broad black vertical bar represents the genetic map with the bin signatures (van Os et al. 2006) separated by white horizontal bars. To the right of the genetic map, the BAC clones that are mapped at this location are presented, with a black horizontal bar to indicate their genetic interval. On the left side, the cluster names, the universal markers and linked cloned R genes are indicated, with the interval based on the genetic location of the BAC clones or the bin range of the markers. The red BAC names indicate the presence of CNL RGHs and the absence of TNL RGHs. The green BAC names indicate the presence of TNL RGHs and the absence of CNL RGHs and the blue BAC names indicate the presence of both TNL and CNL RGHs. (PDF 194 kb)