Abstract
High throughput sequencing technology has opened a window into the vast communities of bacteria that live on and in humans, demonstrating tremendous variability, and that they play a large role in health and disease. The eukaryotic component of the human gut microbiome remains relatively unexplored with these methods, but turning these tools toward microbial eukaryotes in the gut will likely yield myriad insights into disease as well as the ecological and evolutionary principles that govern the gut microbiota. Microbial eukaryotes are common inhabitants of the human gut worldwide and parasitic taxa are a major source of morbidity and mortality, especially in developing countries, though there are also taxa that cause no harm or are beneficial. While the role microbial eukaryotes play in healthy individuals is much less clear, there are likely many complex interactions between the bacterial, archaeal, and eukaryotic microbiota that influence human health. Integrating eukaryotic microbes into a broad view of microbiome function requires an integrated ecological approach rather than one focused on specific, disease-causing taxa. Moving forward, we expect broad surveys of the eukaryotic microbiota and associated bacteria from geographically and socioeconomically diverse populations to paint a more complete picture of the human gut microbiome in health and disease.
Keywords: intestinal protozoa, host-associated communities, eukaryotic diversity
Introduction
Microbial eukaryotes are an important component of the human gut microbiome. Eukaryotes that reside in the human gut are distributed across the eukaryotic tree (Figure 1) and their relationship with the human host varies from parasitic to opportunistic to commensal to mutualistic. For the purposes of this review, we are focusing on the microbial eukaryotes and are not discussing metazoan parasites, which are thoroughly covered elsewhere (Stoll, 1947; Kassai, 1999; Muller, 2002; Bogitsh et al., 2005). We are also focusing on the gut microbiome as many eukaryotic microbes are found there, and it is the best-characterized human body site in terms of the bacterial communities.
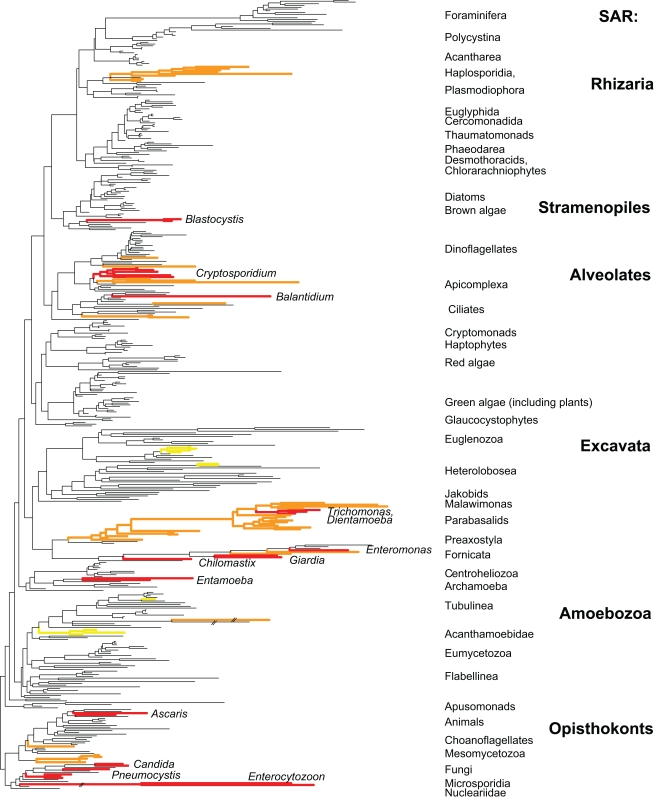
Figure 1.
Distribution of host-associated taxa across a broadly sampled eukaryotic tree of life. Red branches represent lineages found in the vertebrate gut with human-associated genera listed. Orange branches represent lineages found in invertebrates (some red lineages are also present in invertebrates). Yellow taxa are non-gut parasites of humans. Tree is based on Parfrey et al. (2010).
Eukaryotes are one of the three domains of life and are defined by the presence of nuclei. Animals, plants, and fungi are the most visible clades of eukaryotes, but these are just three of the 70+ lineages (Patterson, 1999), most of which are microbial. In recent decades, perspectives on the organization of eukaryotic diversity have shifted away from a Five Kingdom view emphasizing plants, animals, and fungi (Whittaker, 1969; Margulis et al., 1990). Molecular systematics and evolutionary studies clearly demonstrate that the bulk of the eukaryotic tree of life is microbial (Adl et al., 2005; Keeling et al., 2005; Parfrey et al., 2010), and eukaryotes are now divided into a small number of higher-level clades, although the structure of the eukaryotic tree is still stabilizing (Adl et al., 2005; Burki et al., 2010; Parfrey et al., 2010). Microbial eukaryotes are sometimes colloquially referred to as protists, although this term is not phylogenetically meaningful because microbial and macroscopic organisms intermingle in the eukaryotic tree of life (Adl et al., 2005; Parfrey et al., 2010). Similarly, terms such as protozoa, amebae, flagellates, and algae do not have phylogenetic meaning but are useful as morphological descriptors.
Approaches to Studying Host-Associated Microbial Eukaryotes
Microbial eukaryotes in the human gut have been studied primarily from a parasitological point of view and are generally considered to negatively impact human health. The methods typically used focus on elucidating the presence of specific parasitic taxa, traditionally with culture and microscopy-based approaches (Bogitsh et al., 2005; Church et al., 2010) and more recently with targeted molecular analyses such as PCR based assays (Stensvold et al., 2011). Broad surveys of the eukaryotic diversity in the gut microbiota have only been explored on a limited basis (Marchesi, 2010), but are likely to provide new insights into the role of microbial eukaryotes in the gut by providing data on the whole community, including uncultured organisms and potential interactions among taxa. Fewer than 1% of microbes are culturable, and culture independent (CI) studies of microbial communities have revealed abundant insights into the nature and function of bacterial and archaeal communities in diverse environments (Pace, 1997), including the human microbiome (Marchesi, 2010; Robinson et al., 2010). While studies on eukaryotes lag behind those of bacteria, CI approaches in environmental microbiology have also revealed hidden diversity and ecological interactions within eukaryotes (Caron et al., 2009; Steele et al., 2011).
In recent years barcoded high throughput sequencing of marker genes like small subunit ribosomal DNA (Hamady et al., 2008) coupled with analytical tools to assess patterns in the resulting data (Caporaso et al., 2010) have made it possible to assess the composition of bacterial communities and compare communities across many samples. These approaches have successfully been applied to bacteria and archaea, and have revolutionized our understanding of the human microbiome (Costello et al., 2009; Marchesi, 2010; Robinson et al., 2010). Comparing eukaryotic communities across a diverse collection of individuals from different geographic regions, economic status, and disease states are the next steps in characterizing the human gut microbiome.
There is growing body of theoretical and experimental literature on co-infection within the infectious disease community that stresses the importance of considering the action of disease agents within the context of other microbial players (e.g., Cox, 2001; Graham, 2008; Lafferty, 2010; Telfer et al., 2010). This may provide a framework for understanding the broader community context of the gut microbiome. Taking advantage of analytical tools such as co-occurrence networks, which detect positive or negative correlations among taxa (Steele et al., 2011), will also be crucial in detecting important interactions among community members. Together, these approaches will provide a baseline of expected microbial eukaryotes in healthy individuals and yield a better understanding of community changes associated with disease states.
Roles of Host-Associated Eukaryotes: Pathogen, Commensal, Beneficial
Understanding the prevalence and distribution of microbial eukaryotes in the human gut has large consequences for human health. This is especially true in the developing world, where microbial parasites represent a large source of morbidity and mortality (Kaplan et al., 2000; Haque, 2007). Major gastrointestinal pathogens include Entamoeba histolytica (amoebiasis), which infects millions of people and results in 40,000–100,000 deaths annually, and Giardia intestinalis (giardiasis), which is the most prevalent intestinal parasite (Haque, 2007). While intestinal protistan parasites are often considered a tropical malady, they are actually broadly distributed across the globe, and their prevalence within a population is often linked to poor sanitation of human waste (Bogitsh et al., 2005; Pritt and Clark, 2008).
While the focus is generally on pathogens (in terms of study effort, genome sequencing, etc.), most microbial eukaryotes that reside in the gut do not cause harm, being either beneficial or commensal. Some eukaryotic microbes are considered probiotics, in particular the yeast Saccharomyces boulardii was originally isolated to combat Cholera, and it now marketed as a cure for diarrhea (McFarland and Bernasconi, 1993). Other eukaryotes have been shown to be beneficial in certain context, for example the hypothesized reduction in asthma and allergies in conjunction with hookworm infection (Falcone and Pritchard, 2005). Many other eukaryotic microbes are present in the gut but do no harm (they are commensals), including Pentatrichomonas and Entamoeba dispar (Bogitsh et al., 2005).
Human-associated microbial eukaryotes found in the gut are the focus of this article; however, eukaryotes are also part of the microbial community in other locations of the human microbiome. There is a low-diversity fungal community associated with human skin dominated by the genus Malassezia (Paulino et al., 2006). Fungi are also members of the oral microbiota; CI studies of western populations reveal diverse fungal lineages (Ghannoum et al., 2010). The mouth can harbor microbial eukaryotes that are closely related to taxa found in the gut such as Entamoeba gingivalis and Trichomonas tenax in individuals with poor oral hygiene (Bogitsh et al., 2005). Parasites that cause malaria (Plasmodium) and sleeping sickness (Trypanosoma brucei) reside in the liver and bloodstream, and other taxa cause problems when in brain tissue (e.g., Encephalitozoon, Toxoplasma, Naegleria; Bogitsh et al., 2005).
Current Surveys of the Eukaryotic Component of the Human Microbiome
Comprehensive study of the eukaryotic component of the human microbiome is just beginning and lags far behind our understanding of the bacterial communities. Initial CI surveys that were done with low-throughput methods of community finger printing and clone library sequencing are intriguing and suggest that there are numerous parallels between the bacterial and eukaryotic components of the microbiome. Fungi and Blastocystis are the dominant (and in many cases the only) eukaryotes in the gut microbiome of healthy individuals (Nam et al., 2008; Ott et al., 2008; Scanlan and Marchesi, 2008). The diversity of microbial eukaryotes within an individual is low, with fewer than 10 phylotypes recovered per individual (Ott et al., 2008; Scanlan and Marchesi, 2008). The limited data available thus far suggests that eukaryotic communities are stable across time and unique to individuals (Scanlan and Marchesi, 2008). The apparent diversity will likely rise as more individuals from diverse populations are sampled with next generation sequencing technologies by that enable rare taxa within the microbiota to be detected.
Evolutionary Context
The evolutionary patterns of microbial diversity within the vertebrate gut are similar across eukaryotes, bacteria, and archaea. Across all taxa only a small number of lineages have adapted to the gut environments, but those lineages are successful in colonizing a wide variety of hosts. Reduced diversity at deep phylogenetic levels (e.g., the Bacteroidetes and Firmicutes divisions of bacteria) is a hallmark of host-associated bacteria and archaea (Ley et al., 2006). This pattern is also evident in eukaryotes, where a small subset of eukaryotic lineages have host-associated representatives (Figure 1). Within the bacteria, the lineages present in the gut have flourished and produced extensive species and strain level diversity (Ley et al., 2006). The genetic diversity of microbial eukaryotes at shallow phylogenetic levels is lower than observed for bacteria, although there is still extensive strain (or species) level variation in isolates from different individuals (Parkar et al., 2010; Stensvold et al., 2011). The second major trend is that the taxa that have adapted to host-associated environments are broadly distributed as parasites and commensals of many different animal hosts. For example, commensal flagellate genera Chilomastix and Pentatrichomonas are found both in the bovine rumen and in the human gut (Williams and Coleman, 1992), and E. histolytica and E. invadens are parasitic in humans and snakes, respectively (Barnard and Upton, l994). This suggests that relatively few taxa have adapted to host-associated environments, but these taxa are successful in a variety of hosts and have diversified at shallow phylogenetic levels.
The common patterns of diversity in the gut microbiome seen across the three domains of life are likely driven by the fact that host-associated environments, and especially the gut, are unique microbial habitats that are difficult to colonize because the stable, warm, low-oxygen, and eutrophic conditions represent an extreme environment (Ley et al., 2008). Eukaryotes that live in the gut are anaerobic or microaerophilic, and in most cases have highly reduced mitochondria (Hjort et al., 2010). Global analysis of bacterial communities demonstrate that host-associated communities are quite distinct from their free-living counterparts (Ley et al., 2008). While global patterns of eukaryotic diversity have yet not been examined quantitatively, it is likely that similar patterns to bacteria will emerge, as most gut taxa are known only from host-associated environments and are found across a range of hosts (Kreier and Baker, 1987). However, extensive CI studies of eukaryotes may reveal additional hidden diversity either within known lineages or in lineages not previously found within the microbiome.
Eukaryotic Communities in Other Hosts
The eukaryotic microbiota has been extensively studied in animals where eukaryotes play a more central role, such as ruminants and termites, and these studies may provide expectations for the role of eukaryotes in the human microbiome. Culture independent studies have shown that mice harbor fungal communities that are much more diverse than those found in humans (Scupham et al., 2006; Marchesi, 2010), and animals that break down cellulose may generally harbor higher eukaryotic diversity. Eukaryotes make up the bulk of microbiota residing in the rumen and hindgut of many herbivores. Yet, even here the high-level phylogenetic diversity is limited: for example, the multitude of rumen ciliates all belong to a single order, the Litostomatea (Williams and Coleman, 1992). The human commensal Balantidium also belongs to this order. One of the best-characterized examples of mutualism occurs between symbiotic flagellates that reside in the hindgut of basal termites and the wood-eating cockroach Cryptocercus and break down cellulose (Radek, 1999). These insects rely on the parabasalid and oxymonad symbionts to break down cellulose and release energy, and these flagellates can constitute 15–30% of the body weight of the termite (Radek, 1999). Going a step further, parabasalids of the genus Trichonympha that inhabit Cryptocercus are themselves intimately associated with bacterial symbionts (Carpenter et al., 2009). There are also complex endosymbioses between rumen ciliates, methanogenic Archaea (related to Methanobrevibacter smithii) and proteobacteria (Irbis and Ushida, 2004).
Examining the eukaryotic microbiota associated with diverse animals has the potential to reveal new lineages of eukaryotes as several clades are known only from the microbiome of specific taxa. For example opalinids, a clade of stramenopiles, are only found in the hindgut of amphibians and hypermastigid parabasalids in the hindguts of termites and cockroaches. Parasites of invertebrate animals are more broadly distributed across the eukaryotic tree (Orange branches in Figure 1), and several novel lineages have been recently discovered (e.g., Hertel et al., 2002). This pattern likely stems from two factors: “invertebrates” are a far more diverse category phylogenetically than are vertebrates, and invertebrate-associated environments span a far broader range of conditions.
The Ecological Context of the Gut Microbiome
The species composition and general patterns of diversity found in the intestinal microbiota are best understood from an ecological perspective (reviewed in Ley et al., 2006; Robinson et al., 2010). Eukaryotes in the gut are not acting in isolation but are part of a complex community of microorganisms that includes bacteria, archaea, and viruses that interact through predation (Wildschutte et al., 2004), competition (Graham, 2008; Lafferty, 2010), and mutualism (Radek, 1999). The microbiota are also interacting and responding to the host immune system. Thus, it is important to determine changes in the bacterial and archaeal communities that correspond to the presence or absence of eukaryotic microbes. Next-generation sequencing of the total microbiota across diverse samples will enable this type of investigation. Placing eukaryotic microbes within the ecological context of the microbiome may be important for determining factors that influence pathogenicity of those that reside in the human gut. An ecological perspective will be particularly useful in understanding asymptomatic infections, opportunistic infections, and complex phenotypes such as IBS and malnutrition.
The range of the host response to many individual microbial eukaryotes varies greatly from asymptomatic to causing morbidity or mortality. For example, many people infected with known parasites, such as E. histolytica and G. intestinalis, are asymptomatic (Prado et al., 2005; Pritt and Clark, 2008). In the case of E. histolytica, only 10% of infections are invasive and result in amoebic dysentery, but these cases often lead to death (Pritt and Clark, 2008). Conversely, a number of microbial eukaryotes that are normally commensal or absent from the human microbiota can occasionally cause disease when they become abundant or infect immunocompromised people. Many factors underlie the differential manifestations of pathology, including host immune response, prior exposure to the pathogen, host genetics, and nutritional status, and co-infection with multiple parasites (e.g., Pritt and Clark, 2008). Differences in the bacterial and archaeal communities may be a further factor determining whether or not an individual manifests symptoms when their intestinal tract is colonized by protistan parasites. Building a large database of co-occurring eukaryotic and bacterial communities will allow one to test the hypothesis that changes in the bacterial community are responsible for the variance in the pathogenicity of eukaryotic parasites.
Opportunistic parasites are a significant source of morbidity in immune compromised patients, although these same taxa may be present in healthy people without apparent consequence. These emerging opportunistic pathogens include Cryptosporidium parvum, Pneumocystis carnii, and microsporidia, and all generally cause severe diarrhea (Kaplan et al., 2000). This shift in pathogenicity between healthy and immunocompromised individuals might result from changes in the microbial community in addition to changes in the immune system. These parasitic infections may be similar to Clostridium difficile induced diarrhea, which has been shown to result from disruption of the gut microbial community (Walk and Young, 2008). A better understanding of the microbial ecology of opportunistic infections may lead to more effective treatment options, as it has with C. difficile (Walk and Young, 2008). There are also other organisms that may be present in a healthy microbiome community, but become problematic when their numbers increase, including the flagellate Chilomastix (Bogitsh et al., 2005) and the fungus Candida albicans (Schulze and Sonnenborn, 2009). In these cases it may be very important to look simultaneously at the bacterial and eukaryotic communities to determine whether a shift in the normal (bacteria dominated) microbiota is correlated with a rise in opportunistic microbial eukaryotes.
Eukaryotes have also been implicated as causative agents of diseases such as IBS (Blastocystis), IBD (fungi), and “leaky gut” (Candida; Boorom et al., 2008; Ott et al., 2008; Schulze and Sonnenborn, 2009). These diseases, which have become prevalent in western populations, are due to complex changes in the microbial community. Elucidating community-wide changes, rather than presence or absence of specific taxa, will be crucial to understanding the cause and potential treatment of in these complex polymicrobial diseases (Swidsinski et al., 2009). Samples taken from along the gastrointestinal tract may also be important to determine whether community changes associated with IBS or IBD are restricted to the site of disease or are systemic changes in the overall gut community, as has been the observed in bacterial communities (Peterson et al., 2008).
Several eukaryotic parasites are thought to interact with malnutrition and increase morbidity in parts of the developing world. For example, children infected with Giardia may experience reduced growth even when the infection is asymptomatic (Prado et al., 2005). Other eukaryotes that inhabit the gut are only pathogenic when their host is malnourished or otherwise stressed, as in the ciliate Balantidium (Schuster and Ramirez-Avila, 2008). These eukaryotes may interact with the bacterial community to impact nutrient cycling and absorption in the gut. Given the patchy distribution of these parasitic taxa in human populations it should be possible to assess whether the presence of these taxa correlate with differences in the bacterial community host malnourishment.
Prospects for the Future
Incorporation of eukaryotes into human microbiome studies is just beginning, and will benefit from broad CI surveys of the eukaryotic communities in healthy and diseased individuals. These surveys should encompass diverse populations of healthy and diseased individuals, and should exploit within-subject time-series designs (to control for inter-individual diversity of the microbiome), and use identical twins (to control for genetic variability). Analyzing these data with emerging computational tools, such as co-occurrence networks, and pipelines for comparing large numbers of samples (Caporaso et al., 2010) will enable the patterns in these data to be elucidated. In-depth sequencing of the meta-transcriptome of eukaryotes in the gut is another technique that will offer a window into the functioning of eukaryotes in the gut microbiome (Gianoulis et al., 2009). This could be targeted toward eukaryotes by selecting Poly(A) + RNAs for sequencing, although depletion of host transcripts in an unbiased way will be a major issue. Genomic surveys of uncultured host-associated eukaryotes will soon be possible using techniques such as single-cell genomics and transcriptomics (Heywood et al., 2011; Yoon et al., 2011). By connecting single-cell techniques, improved phylogenetic tree construction, broad surveys, and the parameters of host phenotypes, these studies have great potential to improve our understanding of the links between microbial eukaryotes and human health, and our understanding of the eukaryotic component of the tree of life.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many thanks to Valerie McKenzie and members of the Knight lab for comments on the manuscript, especially Catherine Lozupone, Jesse Zaneveld, Justin Kuczynski, and Daniel McDonald. This manuscript was improved following comments from two reviewers. Support from the Bill and Melinda Gates Foundation, the Crohn’s and Colitis Foundation of America, the National institutes of Health, and the Howard Hughes Medical Institute is acknowledged.
References
- Adl S. M., Simpson A. G. B., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., McCourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 10.1111/j.1550-7408.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Barnard S., Upton S. (l994). A Vetrinary Guide to the Parasites of Reptiles. Malabar, FL: Krieger Publishing Company [Google Scholar]
- Bogitsh B., Carter C., Oeltmann T. (2005). Human Parasitology. Amsterdam: Elsevier [Google Scholar]
- Boorom K. F., Smith H., Nimri L., Viscogliosi E., Spanakos G., Parkar U., Li L. H., Zhou X. N., Ok U. Z., Leelayoova S., Jones M. S. (2008). Oh my aching gut: irritable bowel syndrome, Blastocystis, and asymptomatic infection. Parasit. Vectors 1, 40. 10.1186/1756-3305-1-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F., Kudryavtsev A., Matz M. V., Aglyamova G. V., Bulman S., Fiers M., Keeling P. J., Pawlowski J. (2010). Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol. Biol. 10, 377. 10.1186/1471-2148-10-377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Tumbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. (2010). Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron D. A., Worden A. Z., Countway P. D., Demir E., Heidelberg K. B. (2009). Protists are microbes too: a perspective. ISME J. 3, 4–12 10.1038/ismej.2008.101 [DOI] [PubMed] [Google Scholar]
- Carpenter K. J., Chow L., Keeling P. J. (2009). Morphology, phylogeny, and diversity of Trichonympha (Parabasalia: Hypermastigida) of the wood-feeding cockroach Cryptocercus punctulatus. J. Eukaryot. Microbiol. 56, 305–313 10.1038/ismej.2008.101 [DOI] [PubMed] [Google Scholar]
- Church C., Neill A., Schotthoefer A. M. (2010). Intestinal infections in humans in the rocky mountain region, United States. J. Parasitol. 96, 194–196 10.1645/GE-2229 [DOI] [PubMed] [Google Scholar]
- Costello E. K., Lauber C. L., Hamady M., Fierer N., Gordon J. I., Knight R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox F. E. (2001). Concomitant infections, parasites and immune responses. Parasitology 122(Suppl.), S23–S38 10.1017/S003118200001698X [DOI] [PubMed] [Google Scholar]
- Falcone F. H., Pritchard D. I. (2005). Parasite role reversal: worms on trial. Trends Parasitol. 21, 157–160 10.1016/j.pt.2005.02.002 [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A., Jurevic R. J., Mukherjee P. K., Cui F., Sikaroodi M., Naqvi A., Gillevet P. M. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6, e1000713. 10.1371/journal.ppat.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianoulis T. A., Raes J., Patel P. V., Bjornson R., Korbel J. O., Letunic I., Yamada T., Paccanaro A., Jensen L. J., Snyder M., Bork P., Gerstein M. B. (2009). Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl. Acad. Sci. U.S.A. 106, 1374–1379 10.1073/pnas.0808022106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A. (2008). Ecological rules governing helminth–microparasite coinfection. Proc. Natl. Acad. Sci. U.S.A. 105, 566–570 10.1073/pnas.0707221105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Walker J. J., Harris J. K., Gold N. J., Knight R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5, 235–237 10.1038/nmeth.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque R. (2007). Human intestinal parasites. J. Health Popul. Nutr. 25, 387–391 [PMC free article] [PubMed] [Google Scholar]
- Hertel L. A., Bayne C. J., Loker E. S. (2002). The symbiont Capsaspora owczarzaki, nov gen. Nov sp., isolated from three strains of the pulmonate snail biomphalaria glabrata is related to members of the mesomycetozoea. Int. J. Parasit. 32, 1183–1191 10.1016/S0020-7519(02)00066-8 [DOI] [PubMed] [Google Scholar]
- Heywood J., Sieracki M., Bellows W., Poulton N., Stepanauskas R. (2011). Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 2011, 674–684 10.1038/ismej.2010.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjort K., Goldberg A. V., Tsaousis A. D., Hirt R. P., Embley T. M. (2010). Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos. Trans. R. Soc. B Biol. Sci. 365, 713–727 10.1098/rstb.2009.0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irbis C., Ushida K. (2004). Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50, 203–212 10.2323/jgam.50.203 [DOI] [PubMed] [Google Scholar]
- Kaplan J. E., Jones J. L., Dykewicz C. A. (2000). Protists as opportunistic pathogens: public health impact in the 1990s and beyond. J. Eukaryot. Microbiol. 47, 15–20 10.1111/j.1550-7408.2000.tb00004.x [DOI] [PubMed] [Google Scholar]
- Kassai T. (1999). Veterinary Helminthology. Oxford: Butterworth-Heinemann [Google Scholar]
- Keeling P. J., Burger G., Durnford D. G., Lang B. F., Lee R. W., Pearlman R. E., Roger A. J., Gray M. W. (2005). The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676 10.1016/j.tree.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Kreier J., Baker J. (1987). Parasitic Protozoa. Boston: Allen and Unwin, Inc [Google Scholar]
- Lafferty K. D. (2010). Interacting parasites. Science 330, 187–188 10.1126/science.1196915 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. (2008). Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Peterson D. A., Gordon J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Marchesi J. R. (2010). Prokaryotic and eukaryotic diversity of the human gut. Adv. Appl. Microbiol. 72, 43–62 [DOI] [PubMed] [Google Scholar]
- Margulis L., Corliss J. O., Melkonian M., Chapman D. J. (eds.) (1990). Handbook of Protoctista. The Jones and Bartlett Series in Life Sciences. Boston: Jones and Bartlett [Google Scholar]
- McFarland L., Bernasconi P. (1993). Saccharomyces boulardii: a review of an innovative biotherapeutic agent. Microb. Ecol. Health Dis. 6, 157–171 [Google Scholar]
- Muller R. (2002). Worms and Human Disease. New York, NY: CABI Publishing [Google Scholar]
- Nam Y. D., Chang H. W., Kim K. H., Roh S. W., Kim M. S., Jung M. J., Lee S. W., Kim J. Y., Yoon J. H., Bae J. W. (2008). Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J. Microbiol. 46, 491–501 10.1007/s12275-008-0199-7 [DOI] [PubMed] [Google Scholar]
- Ott S. J., Kuhbacher T., Musfeldt M., Rosenstiel P., Hellmig S., Rehman A., Drews O., Weichert W., Timmis K. N., Schreiber S. (2008). Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43, 831–841 10.1080/00365520801935434 [DOI] [PubMed] [Google Scholar]
- Pace N. R. (1997). A molecular view of microbial diversity and the biosphere. Science 276, 734–740 10.1126/science.276.5313.734 [DOI] [PubMed] [Google Scholar]
- Parfrey L. W., Grant J., Tekle Y. I., Lasek-Nesselquist E., Morrison H. G., Sogin M. L., Patterson D. J., Katz L. A. (2010). Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 59, 518–533 10.1093/sysbio/syq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkar U., Traub R. J., Vitali S., Elliot A., Levecke B., Robertson I., Geurden T., Steele J., Drake B., Thompson R. C. A. (2010). Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet. Parasitol. 169, 8–17 10.1016/j.vetpar.2009.12.032 [DOI] [PubMed] [Google Scholar]
- Patterson D. J. (1999). The diversity of eukaryotes. Am. Nat. 154, S96–S124 10.1086/303287 [DOI] [PubMed] [Google Scholar]
- Paulino L. C., Tseng C. H., Strober B. E., Blaser M. J. (2006). Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 44, 2933–2941 10.1128/JCM.00785-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D. A., Frank D. N., Pace N. R., Gordon J. I. (2008). Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 10.1016/j.chom.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado M. S., Cairncross S., Strina A., Barreto M. L., Oliveira-Assis A. M., Rego S. (2005). Asymptomatic giardiasis and growth in young children; a longitudinal study in Salvador, Brazil. Parasitology 131, 51–56 10.1017/S0031182005007353 [DOI] [PubMed] [Google Scholar]
- Pritt B., Clark C. G. (2008). Amebiasis. Mayo Clin. Proc. 83, 1154–1160 10.4065/83.10.1154 [DOI] [PubMed] [Google Scholar]
- Radek R. (1999). Flagellates, bacteria, and fungi associated with termites: diversity and function in nutrition - a review. Ecotropica 5, 183–196 [Google Scholar]
- Robinson C. J., Bohannan B. J. M., Young V. B. (2010). From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74, 453–476 10.1128/MMBR.00014-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan P. D., Marchesi J. R. (2008). Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2, 1183–1193 10.1038/ismej.2008.76 [DOI] [PubMed] [Google Scholar]
- Schulze J., Sonnenborn U. (2009). Yeasts in the gut: from commensals to infectious agents. Dtsch. Arztebl. Int. 106, 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster F. L., Ramirez-Avila L. (2008). Current world status of Balantidium coli. Clin. Microbiol. Rev. 21, 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham A. J., Presley L. L., Wei B., Bent E., Griffith N., McPherson M., Zhu F. L., Oluwadara O., Rao N., Braun J., Borneman J. (2006). Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72, 793–801 10.1128/AEM.72.1.793-801.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J., Countway P. D., Xia L., Vigil P., Beman J., Kim D., Chow C., Sachdeva R., Jones A., Schwalbach M., Rose J., Hewson I., Patel A., Sun F., Caron D., Fuhrman J. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 1–12 10.1128/AEM.72.1.793-801.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensvold C. R., Lebbad M., Verweij J. J. (2011). The impact of genetic diversity in protozoa on molecular diagnostics. Trends Parasitol. 27, 53–58 10.1016/j.pt.2010.11.005 [DOI] [PubMed] [Google Scholar]
- Stoll N. R. (1947). This wormy world. J. Parasitol. 33, 1–18 10.2307/3273478 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Herber A. (2009). Mucosal flora in crohn’s disease and ulcerative colitis – an overview. J. Physiol. Pharmacol. 60, 61–71 [PubMed] [Google Scholar]
- Telfer S., Lambin X., Birtles R., Beldomenico P., Burthe S., Paterson S., Begon M. (2010). Species interactions in a parasite community drive infection risk in a wildlife population. Science 330, 243–246 10.1126/science.1190333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk S., Young V. (2008). Emerging insights into antibiotic-associated diarrhea and Clostridium difficile infection through the lens of microbial ecology. Interdiscip. Perspect. Infect. Dis. 2008, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker R. H. (1969). New concepts of kingdoms of organisms. Science 163, 150–160 10.1126/science.163.3863.150 [DOI] [PubMed] [Google Scholar]
- Wildschutte H., Wolfe D. M., Tamewitz A., Lawrence J. G. (2004). Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. U.S.A. 101, 10644–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A., Coleman G. (1992). The Rumen Protozoa. New York: Springer-Verlag [Google Scholar]
- Yoon H. S., Price D. C., Stepanauskas R., Rajah V. D., Sieracki M. E., Wilson W. H., Yang E. C., Duffy S., Bhattacharya D. (2011). Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717 10.1126/science.1203163 [DOI] [PubMed] [Google Scholar]