Abstract
Site-directed mutagenesis via gene targeting (GT) based on homologous recombination is the ultimate mutation breeding technology because it enables useful information acquired from structural- and computational-based protein engineering to be applied directly to molecular breeding, including metabolic engineering, of crops. Here, we employed this rationale to introduce precise mutations in OASA2—an α-subunit of anthranilate synthase that is a key enzyme of tryptophan (Trp) biosynthesis in rice (Oryza sativa)—via GT, with subsequent selection of GT cells using a Trp analog. The expression level of OASA2 in plants homozygous and heterozygous for modified OASA2 was similar to that of nontransformants, suggesting that OASA2 transcription in GT plants was controlled in the same manner as endogenous OASA2, and that GT could lead to a lower risk of gene silencing than in conventional overexpression approaches. Moreover, we showed that enzymatic properties deduced from protein engineering or in vitro analysis could be reproduced in GT plants as evidenced by Trp accumulation levels. Interestingly, mature seeds of homozygous GT plants accumulated Trp levels 230-fold higher than in nontransformants without any apparent morphological or developmental changes. Thus, we have succeeded in producing a novel rice plant of great potential nutritional benefit for both man and livestock that could not have been selected using conventional mutagenesis approaches. Our results demonstrate the effectiveness of directed crop improvement by combining precision mutagenesis via GT with a knowledge of protein engineering.
Comparative genomics and structural- and computational-based protein engineering provide copious amounts of useful information that can be applied to crop improvement. In conventional mutation breeding technology using chemical mutagens and ionizing radiation, it remains quite difficult to select mutants harboring target genes modified exactly as required, because mutations occur randomly. Meanwhile, conventional transformation technology based on information from comparative genomics and protein engineering in crops remains a powerful tool in the field of molecular breeding. Agronomical traits such as herbicide tolerance, stress tolerance, and photosynthetic activity have been modified successfully in several crops using conventional transformation technologies that depend on overexpression of modified, highly functional enzymes (Rao, 2008). Moreover, conventional transformation technology is also essential for introducing exogenous genes that do not exist naturally in target plants, e.g. for the production of recombinant proteins. However, mutational approaches are considered better than conventional transformation approaches when introducing directed mutations into endogenous genes to confer novel traits to crops, because conventional transformation technologies cannot eliminate the endogenous targeted gene itself. Thus, a clear goal of current mutation breeding is to modify targeted endogenous genes directly to yield the desired mutations at high frequency.
Site-directed mutagenesis by gene targeting (GT) via homologous recombination (HR) is the ultimate mutation breeding technology. In our previous report, herbicide-tolerant rice (Oryza sativa) was produced successfully by site-directed mutagenesis of ACETOLACTATE SYNTHASE (ALS) via GT based on genome information from an already-discovered herbicide-tolerant rice-cultured cell line (Endo et al., 2007). However, to date, the successful production of plants exhibiting improved traits other than herbicide tolerance using pin-point mutagenesis via GT, to our knowledge, has not been reported (Lee et al., 1990; Hanin et al., 2001; Endo et al., 2006, 2007; Townsend et al., 2009). Moreover, to our knowledge, there are no reports of the production of a novel plant via precision engineering of an intrinsic gene via GT based on information from protein engineering.
Trp is one of the limiting essential amino acids in cereals, which play a huge role in human diets and livestock feed worldwide (Ishihara et al., 2007; Ufaz and Galili, 2008; Wakasa and Ishihara, 2009). Many secondary metabolites, such as serotonin, vitamin B3, and indole alkaloids, are derived from Trp. Trp deficiency can increase the risk of developing the vitamin-deficiency disease pellagra. Moreover, Trp supplementation to forage not only reduces sanitary and environmental risk by maintaining an adequate amino acid balance in the feed but also increases the growth rate of pigs and poultry (Van Cauwenberghe and Relandeau, 2000). However, synthetic Trp is expensive due to the relatively low efficiency of industrial Trp production (Ishihara et al., 2007). Thus, Trp fortification for human food and animal feed has great potential benefit.
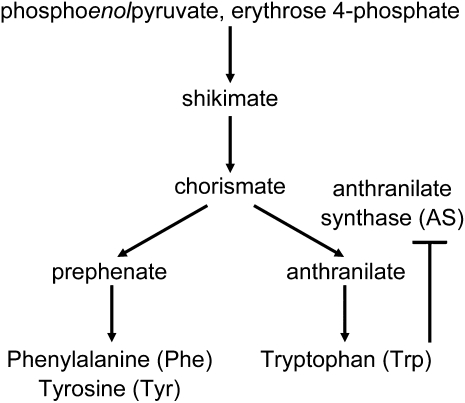
Anthranilate synthase (AS) plays a key role in the biosynthesis of Trp, and the α-subunit of AS is susceptible to feedback inhibition by Trp or its analogs (Fig. 1; Ishihara et al., 2007; Wakasa and Ishihara, 2009). To date, rice mutants insensitive to Trp analogs have been obtained in conventional mutation breeding programs (Wakasa and Widholm, 1987; Lee and Kameya, 1991; Kim et al., 2005). In addition, conventional transgenic approaches such as constitutive overexpression of the Trp-insensitive AS α-subunit have also been tried for the production of high-Trp plants (Tozawa et al., 2001; Wakasa et al., 2006). Mature seeds of rice plants produced by transgenic approaches accumulated 1,762 to 5,294 nmol/g Trp (Wakasa et al., 2006), while those produced by conventional mutation approaches accumulated 204 to 810 nmol/g Trp (Lee and Kameya, 1991; Kim et al., 2005).
Figure 1.
The biosynthesis pathway of aromatic amino acids in higher plants.
Structure-based protein engineering of OASA2—an α-subunit of AS in rice—showed that S126F and Y367A mutations confer Trp insensitivity, and the L530D mutation enhances the catalytic activity of AS (Tozawa et al., 2001; Kanno et al., 2005). Moreover, it has been shown that free Trp accumulates to levels 5- to 400-fold higher in transformed calli overexpressing modified OASA2 harboring S126F/L530D, Y367A/L530D, and Y367A than in nontransformants (Kanno et al., 2005; Wakasa and Ishihara, 2009). However, with a conventional mutagenesis approach, the simultaneous introduction of double mutations such as S126F/L530D and Y367A/L530D into OASA2 would be very difficult, if not impossible. Moreover, it would also be very difficult to introduce these mutations into OASA2 step by step by selection with different concentrations of 5-methyl-Trp (5MT) because the mutations confer different functions, i.e. Trp insensitivity, and enhancement of AS enzymatic activity of the OASA2 enzyme (Kanno et al., 2005). Thus, we consider site-directed mutagenesis via GT to be the best way to produce rice mutants harboring OASA2 with the desired double mutations.
In this study, we used a novel mutation-breeding strategy and produced Trp-accumulating rice by GT-mediated mutagenesis of OASA2. We successfully obtained mutant rice plants harboring OASA2 with S126F/L530D, Y367A/L530D, and Y367A mutations. Among them, we analyzed two kinds of GT plants harboring OASA2 with S126F/L530D and Y367A mutations, and showing different enzymatic kinetics from mutations analyzed in a previous study (i.e. 50% inhibition of initial activity values are >80- and 5-fold higher than the original enzyme, respectively; Kanno et al., 2005). Taking advantage of the intrinsic Trp translocation system, we successfully produced a novel high-Trp rice mutant via GT without any apparent morphological or developmental changes that could not have been selected using conventional mutagenesis approaches. To our knowledge, this study also provides the first demonstration of the effectiveness of directed crop improvement by combining precision mutagenesis via GT with knowledge of protein engineering.
RESULTS AND DISCUSSION
GT Experiments with Trp Analog
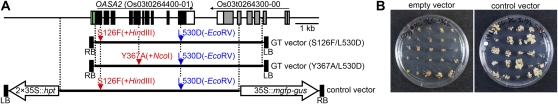
First, we established experimental conditions to introduce S126F/L530D mutations into OASA2 via GT. We constructed the GT vector (S126F/L530D) shown in Figure 2A. The GT vector for OASA2 harbors a deletion of the chloroplast-targeting signal corresponding to the first 49 amino acids of OASA2. This truncated version of OASA2 is expected to be nonfunctional, thus recovery of 5MT-tolerant callus and plants is expected only after HR between the GT vector and the chromosomal OASA2 locus. OASA2 exhibits sensitivity to both Trp and the Trp-analog 5MT (Tozawa et al., 2001; Kanno et al., 2004), and the latter was chosen as the selection agent for transformed calli and plants following introduction of the S126F/L530D mutations into OASA2 via GT. One potential problem for selecting GT products was that transcript levels of mutated OASA2 were expected to be relatively low in rice cells in which GT had occurred successfully because the mutated OASA2 is under the control of its own intrinsic promoter. Moreover, GT occurs at very low frequency compared to conventional transformation experiments. Thus, we optimized the concentration of 5MT required to select GT cells efficiently using a control vector containing a mutated OASA2 expression cassette driven by the intrinsic promoter and terminator (Fig. 2A); 500 μm 5MT was found to be optimal for selection of GT cells (Fig. 2B).
Figure 2.
GT experiments to introduce mutations into OASA2. A, Schematic representation of GT vectors and the control vector used in this study. As GT vectors, a 7.0-kb fragment containing a partial OASA2 fragment with S126F/L530D or Y367A/L530D mutations and silent mutations for CAPS markers was constructed. As a control vector, a 7.5-kb fragment containing a full OASA2 expression cassette with mutations was constructed. The S126F, Y367A, and L530D mutations (S126F; S [TCC] to F [TTC] at amino acid 126, Y367A; Y [TAC] to D [GCC] at amino acid 367, L530D; L [CTT] to D [GAC] at amino acid 530) and silent mutations for the CAPS markers (added HindIII site at 16-bp upstream of S126F; CAGCTT to AAGCTT, deleted EcoRV site at 22-bp upstream of L530D; GATATC to GATTTC) were introduced into the OASA2 fragment. The Y367A mutation generates additional recognition sites for NcoI; ACATGG to CCATGG. Left, The green, white, and black boxes show the putative chloroplast-targeting signal, untranslated region, and coding region of OASA2 (Os03t0264400-01), respectively. Right, The gray and white boxes show the putative gene structure of Os03t0264300-00, respectively. The horizontal arrows show the direction of transcription of each gene. B, Selection of rice calli transformed with empty vector (pPZP2028, left section) and control vector (right section) and selected on medium containing 500 μm 5MT.
After cocultivation of rice callus with Agrobacterium harboring the GT vector (S126F/L530D) and subsequent selection on 500 μm 5MT, we obtained a total of two regenerated plants that showed 5MT tolerance and were positive for cleaved amplified polymorphic sequences (CAPS) markers. Using the same strategy and the GT vector (Y367A/L530D) shown in Figure 2A, we succeeded in obtaining eight lines of candidate GT plants harboring OASA2 with Y367A/L530D mutations. Moreover, we also succeeded in obtaining six lines of candidate GT plants harboring OASA2 with the Y367A mutation using GT vector (Y367A/L530D). This is because the single mutation Y367A confers Trp insensitivity, and free Trp in transformed calli overexpressing modified OASA2 harboring Y367A accumulates to levels 5-fold higher than in nontransformants (Kanno et al., 2005; Wakasa and Ishihara, 2009). Thus, we succeeded in obtaining a total of 14 lines of candidate GT plants using GT vector (Y367A/L530D).
In this study, two regenerated GT candidate plants were obtained from 1,275 small pieces of calli (totally 20.4 g callus) for the S126F/L530D mutations, and 14 regenerated GT candidate plants were obtained from 3,475 small pieces of calli (totally 55.6 g callus) for the Y367A/L530D or Y367A mutations. In our previous GT experiments on the ALS locus, 72 GT plants were obtained from 1,500 small pieces of calli (Endo et al., 2007). Assuming a comparable transformation frequency in these GT experiments, the efficiency of isolating GT products was lower in this OASA2 GT system compared to the ALS GT system. This might be due to the short homologous sequence of GT vector used for the induction of the S126F mutation (described below) or the relatively inefficient 5MT selection system for GT products compared to the herbicide selection system used for GT products with the ALS GT system.
In further analyses, we used two lines (numbers 3-4 and 16-11) harboring OASA2 with S126F/L530D mutations, and two lines (numbers 5-11 and 4-4) harboring OASA2 with the Y367A mutation.
Molecular Analysis of GT Plants Harboring Mutated OASA2 with S126F/L530D
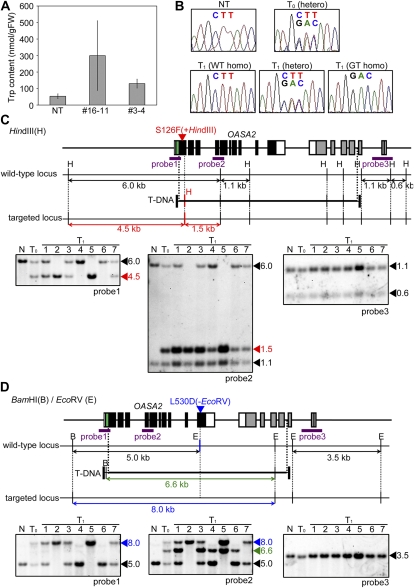
The amount of free Trp in calli from both numbers 3-4 and 16-11 was significantly higher than in nontransformants (Fig. 3A), suggesting that GT events had occurred successfully in these lines. However, subsequent genetic analyses were performed only with line number 3-4, since regenerated number 16-11 plants showed an albino phenotype (Supplemental Fig. S1A).
Figure 3.
Molecular analysis of GT plants harboring OASA2 with S126F/L530D mutations. A, Bar charts showing free Trp content in calli of nontransformant (NT), GT line 3-4 in the T2 generation (heterozygous), and GT line 16-11 in the T0 generation. Values are average ± sd (n = 3). B, Sequencing chromatograms of the mutation site L530D of number 3-4 in T0 and T1 generations. C, Southern-blot analysis of HindIII-digested number 3-4 in T0 and T1 generations using probes 1 to 3 (purple bars). The wild-type (6.0 kb) and targeted (4.5 kb) bands were detected using probe 1. Wild-type (1.1 and 6.0 kb) and targeted band or bands derived from random integrated T-DNA (1.5 kb) were detected using probe 2. Wild-type bands (1.1 and 0.6 kb) were detected using probe 3. N, Nontransformant. D, Southern-blot analysis of BamHI/EcoRV-digested number 3-4. Wild-type (5.0 kb) and targeted (8.0 kb) bands were detected using probe 1. Wild-type bands (5.0 kb), a band derived from random integrated T-DNA (6.6 kb), and the targeted band (8.0 kb) were detected using probe 2. A wild-type band (3.5 kb) was detected using probe 3.
To allow sequencing of OASA2 in regenerated number 3-4 plant (T0 generation) and its progeny (T1 generation), PCR was performed with primers that anneal to the region outside the GT construct (not present on the GT vector). Direct sequencing of PCR fragments confirmed that GT had occurred successfully (Fig. 3B). Southern-blot analysis of T0 and T1 generations of number 3-4 using HindIII with probes 1 and 2 (Fig. 3C) showed that a true GT event in which the wild-type OASA2 locus was modified as expected had indeed occurred because the wild-type 6.0-kb band was not observed in lines 2 and 5 of the T1 generation. In addition, additional genomic rearrangements in the OASA2 locus were not observed in Southern-blot analysis using probes 1 and 3 (Fig. 3C). These results were confirmed by Southern-blot analysis using BamHI/EcoRV with probe 2 (Fig. 3D), which showed that a 6.6-kb band derived from randomly integrated T-DNA was successfully segregated out in lines 2 and 7 of the T1 generation (Fig. 3D). Furthermore, genotyping of number 3-4 in the T2 generation showed that the ratio of wild-type, heterozygous, and homozygous alleles for the modified OASA2 locus was 11:32:15, which fits a 1:2:1 ratio (χ2 = 1.17; P = 0.56), suggesting that the introduced mutations were stably inherited in a Mendelian manner. Furthermore, Southern-blot analysis of the T0 generation of number 16-11 plants showed results similar to those obtained with number 3-4, suggesting that true GT had also occurred in number 16-11 (Supplemental Fig. S1B).
Southern-blot analysis (Fig. 3C) confirmed the successful introduction of the S126F mutation into the endogenous OASA2 gene by transforming with the GT vector shown in Figure 2A. In this vector, the S126F mutation is located adjacent to the right border of the T-DNA, which is protected from degradation due to the covalently bound pilot protein, virD2 (Dürrenberger et al., 1989), located 0.2-kb downstream from the start point of the truncated OASA2 sequence (Fig. 2A). A previous report has estimated the transfer frequency of point mutations in GT vector to the rice genome. The point mutation located 0.4-kb from the end of the T-DNA was successfully introduced to the rice genome in seven of eight independent lines, suggesting that one crossover event occurred within the 0.4-kb segment located at the end of the T-DNA in this GT experiment (Johzuka-Hisatomi et al., 2008). In our experiment, one crossover was thought to have occurred successfully within the 0.2-kb segment in OASA2 in number 3-4 and number 16-11. This result suggests that mutations located near the end of the T-DNA region can indeed be introduced via HR, and in fact it may be more efficient to place the mutation of interest at this position adjacent to the right border of the T-DNA.
Molecular Analysis of GT Plants Harboring Mutated OASA2 with Y367A
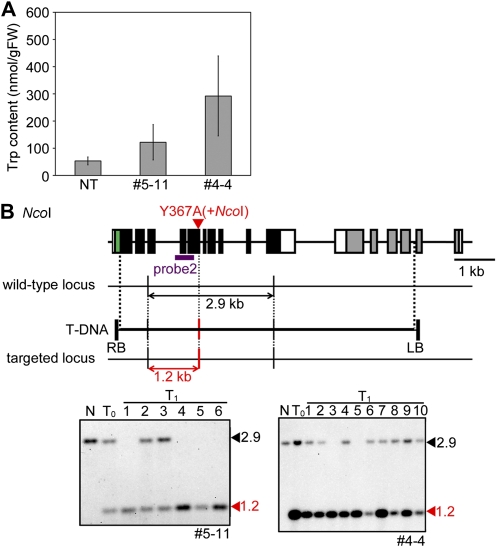
Amounts of free Trp in calli from both numbers 5-11 and 4-4 were significantly higher than in nontransformants (Fig. 4A). Southern-blot analysis showed that a true GT event had indeed occurred in both numbers 5-11 and 4-4 (Fig. 4B). In number 4-4, although the intensity of the 1.2-kb targeted band was similar to that of the wild-type 2.9-kb band in lines 6 and 10 of the T1 generation, band intensities of the targeted band were higher than that of the wild-type band in other lines (Fig. 4B). This result suggested that, in addition to true GT, random integration of T-DNA had occurred in number 4-4. Thus, further analyses were performed only with number 5-11, because extra genomic rearrangements due to random integration of T-DNA in number 4-4 could cause phenotypic changes. Genotyping analysis of 37 plants of number 5-11 in the T1 generation showed that the ratio of plants that were wild type, heterozygous, and homozygous for the modified OASA2 locus was 8:19:10, which fits a 1:2:1 ratio (χ2 = 0.24; P = 0.89), suggesting that mutations introduced into OASA2 are stably inherited to the T1 generation in a Mendelian manner.
Figure 4.
Molecular analyses of GT plant harboring OASA2 with Y367A mutation. A, Bar charts showing free Trp content in callus of nontransformant (NT), number 5-11 in the T2 generation (heterozygous), and number 4-4 in the T0 generation. Values are average ± sd (n = 3). B, Southern-blot analysis of NcoI-digested GT plants, number 5-11, and number 4-4 in the T0 and T1 generation with probe 2. Wild-type (2.9 kb) and targeted band or bands derived from random integrated T-DNA (1.2 kb) were detected using probe 2. N, Nontransformant.
Trp Hyperaccumulation in Mature Seeds of GT Plants
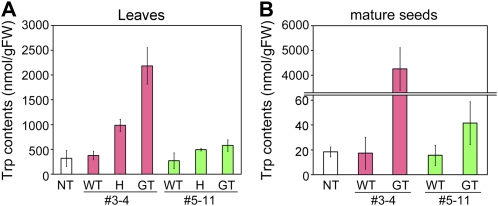
We determined the free Trp content in leaves of both lines 3-4 and 5-11. Leaves of number 3-4 plants heterozygous and homozygous for the GT locus accumulated 985 and 2,184 nmol Trp/g fresh weight (FW), i.e. up to 3.1- and 6.8-fold more than in nontransformants, respectively (Fig. 5A). Similarly, the Trp content in homozygous leaves, in which the Trp-sensitive wild-type OASA2 was completely eliminated, was higher than that in heterozygous leaves of number 5-11 plants (Fig. 5A), suggesting that Trp accumulation in leaves was correlated with the copy number of mutated OASA2 due to a gene dosage effect of the mutated OASA2.
Figure 5.
Free Trp contents in numbers 3-4 and 5-11. Bar charts show Trp contents in leaves (A) and mature seeds (B) of nontransformant (NT), and wild-type (WT) plants, and plants homozygous (GT) and heterozygous (H) in numbers 3-4 and 5-11. Values are average ± sd (n = 3).
On the other hand, mature seeds of homozygous numbers 3-4 and 5-11 plants accumulated 42 and 4,256 nmol Trp/g FW, i.e. up to 230- and 2.3-fold more than in nontransformants, respectively (Fig. 5B). In particular, mature seeds of homozygous number 3-4 plants accumulated 14.13 ± 0.91 μg Trp per seed, suggesting that an amount of 280 g of rice grains in a day could meet the daily requirement for Trp in an adult human weighing 60 kg (4 mg/kg/d [Joint WHO/FAO/UNU Expert Consultation, 2007]). This Trp level in mature seeds of homozygous number 3-4 plants compares favorably with that of plants generated by conventional transgenic approaches (Wakasa et al., 2006). Furthermore, we were interested to find that mature seeds of homozygous number 3-4 plants accumulated Trp to levels up to 230-fold higher than in nontransformants (Fig. 5B) despite the fact that leaves of the same lines accumulated Trp up to only 6.8-fold higher than in nontransformants (Fig. 5A). Since Trp in old tissues has been reported to translocate to young tissues (Matsuda et al., 2010), it is possible that free Trp is synthesized and accumulated gradually and, consequently, additional Trp in source organs translocates to seeds in GT plants.
To evaluate the effect of Trp hyperaccumulation in rice, we measured the content of other amino acids in mature seeds of nontransformed and homozygous number 3-4 plants. The contents of all amino acids, including Phe and Tyr—synthesized from chorismate in the branched pathway of Trp biosynthesis (Fig. 1)—in mature seeds of homozygous number 3-4 plants were higher compared to nontransformants (Table I). However, the ratio of amino acids other than Trp in mature seeds of homozygous number 3-4 plants to that in nontransformants was comparable to that of the overall amount of total amino acids (Table I), suggesting that Trp can be accumulated specifically using this strategy. Many secondary metabolites such as indole alkaloids are derived from Trp. In previous reports, indole compounds including indole acetic acid were accumulated in transgenic rice plants in which Trp accumulated to a high level (Wakasa et al., 2006; Matsuda et al., 2010), suggesting that various kinds of Trp-derived metabolites could accumulate to high levels in our GT plants. Thus, to evaluate the additional effects of Trp fortification via GT-mediated mutagenesis, nontargeted metabolome analysis should be performed using GT plants in further studies.
Table I. Free amino acid content in mature seed.
Free amino acid content in mature seed (nmol/g FW; average ± sd, n = 3). NT, Nontransformant.
| Amino Acid | NT | GT (no. 3-4) | GT/NT |
| Gly | 12.3 ± 8.1 | 32.0 ± 15.5 | 2.6 |
| Ala | 67.0 ± 15.8 | 352.8 ± 89.8 | 5.3 |
| Ser | 26.6 ± 2.0 | 121.2 ± 72.4 | 4.6 |
| Pro | 52.1 ± 20.4 | 168.9 ± 112.0 | 3.2 |
| Val | 9.1 ± 2.6 | 45.4 ± 20.5 | 5.0 |
| Thr | 1.0 ± 0.3 | 2.1 ± 0.7 | 2.1 |
| Leu | 1.5 ± 0.5 | 7.6 ± 3.9 | 5.1 |
| Ile | 1.8 ± 0.6 | 8.9 ± 4.5 | 4.9 |
| Asn | 137.1 ± 47.4 | 739.2 ± 174.0 | 5.4 |
| Asp | 214.5 ± 106.1 | 461.0 ± 172.2 | 2.1 |
| Gln | 3.7 ± 0.6 | 39.8 ± 24.3 | 10.8 |
| Lys | 5.6 ± 2.1 | 54.9 ± 19.4 | 9.8 |
| Glu | 254.2 ± 42.5 | 364.2 ± 161.8 | 1.4 |
| Met | 1.1 ± 0.3 | 4.3 ± 2.6 | 3.9 |
| His | 20.9 ± 7.9 | 70.6 ± 50.9 | 3.4 |
| Phe | 1.5 ± 0.6 | 12.5 ± 6.1 | 8.3 |
| Arg | 21.6 ± 8.7 | 175.0 ± 83.8 | 8.1 |
| Tyr | 6.6 ± 2.0 | 55.7 ± 27.1 | 8.4 |
| Trp | 18.3 ± 3.9 | 4,255.6 ± 855.0 | 232.5 |
| Total amino acids | 856.4 ± 244.3 | 6,990.6 ± 1,787.6 | 8.2 |
It was suggested that Trp fortification and the concomitant alteration of the amount of Trp-derived metabolites may have some negative effect on yield (Wakasa et al., 2006). In this study, there were no noticeable morphological or developmental changes in GT plants grown in the greenhouse, although the field performance of these GT plants should be evaluated in detail and very carefully in further analyses (Supplemental Fig. S2). Additional Trp in old tissues can translocate to young tissues (Matsuda et al., 2010); this Trp translocation system could minimize the risk of metabolic load due to excess Trp biosynthesis in GT plants, although it requires the highest energy input of all the amino acids (Kanno et al., 2005; Ishihara et al., 2007; Wakasa and Ishihara, 2009). Moreover, mutated OASA2 expression under the control of a vegetative organ-specific promoter could be sufficient to accumulate high Trp in seeds using the Trp translocation system (Matsuda et al., 2010), suggesting that modification of the intrinsic OASA2 promoter via GT-mediated mutagenesis might be one approach to reduce the negative effects of Trp accumulation on field performance in GT plants in further study.
In summary, based on information from structure-based protein engineering, we have succeeded in producing, via GT, novel high-Trp rice mutants that could never have been selected with conventional mutagenesis approaches.
Mutation Breeding via GT in Crop Improvement
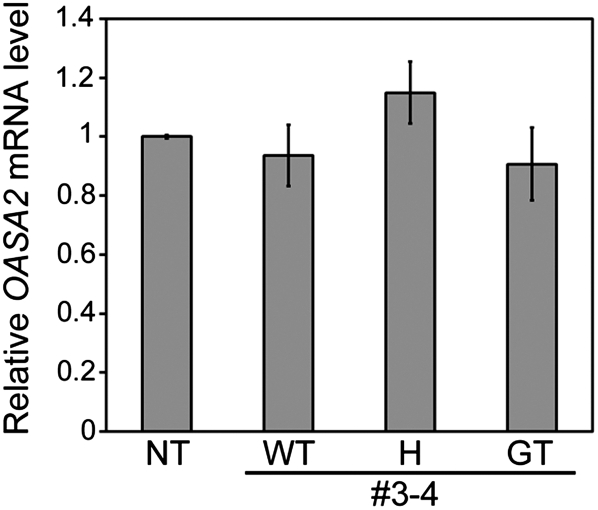
In this study, we also showed the following advantages of site-directed mutagenesis via GT in crop improvement. (1) OASA2 mRNA levels in plants homozygous or heterozygous for modified OASA2 were similar to those in wild-type plants, as expected (Fig. 6). This result is similar to that presented in our previous report (Endo et al., 2007). We suggest that site-directed mutagenesis via GT leads to a lower risk of gene silencing due to the use of intrinsic cis-acting elements to regulate transcription of the mutated target gene at the original locus, and due to the lack of additional copies of the target gene as would be present in conventional transgenic approaches. (2) In both leaves and mature seeds of GT plants, the Trp content of line number 3-4 was higher than that in number 5-11 (Fig. 5). This result was attributed to the level of Trp insensitivity and enzymatic activity; the 50% inhibition of initial activity values of OASA2 carrying the S126F/L530D mutation are >16-fold higher than those of Y367A (Kanno et al., 2005). A similar tendency was reported in transformed rice overexpressing mutated OASA2 (Kanno et al., 2005; Wakasa and Ishihara, 2009). Thus, enzymatic properties that had been discovered from in vitro analysis of mutant type of enzymes could be reproduced in plants produced by GT-mediated mutagenesis.
Figure 6.
OASA2 expression level in number 3-4 quantitative RT-PCR of OASA2 in leaves of nontransformant (NT), wild type (WT), and homozygous (GT) and heterozygous (H) for the modified OASA2. OASA2 mRNA levels were normalized to the OsActin1 mRNA level as a control and are presented as a ratio of that observed in NT. Values are average ± sd (n = 3).
We also demonstrated the practical potential of site-directed mutagenesis in plants via GT and the usefulness of selection using a metabolite analog that inhibits growth of wild-type cells or supports growth of gene-targeted cells, information obtained from structure-based protein engineering, and a knowledge of metabolite translocation systems for metabolic engineering via GT. We believe that this system could be used widely to produce high-value-added crops, such as fortification of particular metabolites, enhancement of stress tolerance, and the modification of the sensitivity to plant growth regulators. However, at least two difficulties with GT in plants remain to be solved: frequency and versatility. (1) In this study, GT frequency was estimated at 0.16% and 0.40% (two GT lines from 1,275 pieces of calli for the S126F/L530D mutation and 14 lines from 3,475 pieces of calli for the Y367A/L530D or Y367A mutation, respectively), which we believe represents the baseline of the endogenous HR system in rice as described above. Recently, target-site-specific chimeric nucleases, such as zinc-finger nuclease have been shown to improve GT frequencies when modifying an endogenous target gene in maize (Zea mays; Shukla et al., 2009) and tobacco (Nicotiana tabacum; Townsend et al., 2009), with GT frequencies estimated at 10- to 1,000-fold higher as a result of zinc-finger nuclease expression (de Pater et al., 2009). Thus, highly efficient GT systems exploiting chimeric nucleases have great potential to produce specifically designed plants. (2) Our GT system without the use of exogenous positive selection markers is a powerful technology for site-directed mutagenesis. Alternatively, site-directed mutagenesis can be achieved together with the use of exogenous selection markers (Terada et al., 2007; Johzuka-Hisatomi et al., 2008). GT systems using exogenous selection markers should be used with target genes or traits that are less easily selectable, because they allow modification of any gene in principle. However, exogenous sequences including exogenous markers should be completely eliminated to produce mutants in which only the desired mutations are introduced in the target gene, because such markers remain in the rice genome in this system. Although successful GT using exogenous markers and subsequent application of site-specific recombination using Cre-loxP has been reported, the efficiency of Cre-loxP-mediated recombination is still low and the footprints of loxP sites remain at the targeted genome locus (Terada et al., 2010). Thus, the next step is to establish a GT system using exogenous markers, with the subsequent complete elimination of exogenous positive selection markers leaving no footprint using an HR system (Hare and Chua, 2002).
MATERIALS AND METHODS
Primers
Primers used in this study are listed in Supplemental Table S1.
GT Vector Construction
To construct GT vectors, a 4.4-kb fragment containing OASA2 was amplified by PCR from rice (Oryza sativa) genomic DNA using the high-fidelity DNA polymerase KOD -plus- (TOYOBO) with the primer set OASA2 f1/r1. The resultant PCR fragments were cloned into the vector pBlueScript using EcoRI/XhoI. The introduction of S126F, Y367A, and L530D mutations into OASA2 was performed using a QuickChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol with primer sets OASA2 f2/r2, OASA2 f3/r3, and OASA2 f4/r4, respectively. We also amplified a 9.7-kb fragment containing OASA2 by PCR from rice genomic DNA using KOD -plus- (TOYOBO) with primer set OASA2 f5/r5. To construct the GT vector for S126F/L530D, the resultant PCR fragments were cloned into the vector pPZP2028 using KpnI/PacI (a derivative of pPZP202 [Hajdukiewicz et al., 1994], with the addition of rare restriction [AscI/PacI] sites to both ends of the multicloning site of pPZP202). The 3.8-kb fragment containing wild-type OASA2 was replaced with OASA2 containing the introduced S126F and L530D mutations on a SacII/XhoI fragment. Finally, the 7.5-kb fragment was amplified by PCR from this vector using KOD -plus- with primer set f5/r6. The resultant PCR fragments were cloned into the vector pPZP201 using KpnI/EcoRI. To construct the GT vector for Y367A/L530D, the 2.0-kb fragment containing mutated OASA2 harboring Y367A was replaced with wild-type OASA2 using SacI/BstXI. Finally, the 1.4-kb fragment containing mutated OASA2 harboring the S126F mutation was replaced with wild-type OASA2 amplified using OASA2 f6/r3 with KpnI/SacI. To construct the positive control vector, the 7.5-kb fragment containing mutated OASA2 was digested from this vector using SpeI and cloned into the vector pCAMBIA1304 digested with XbaI.
Plant Materials and Transformation
Rice (‘Nipponbare’) was used in this study. The binary plasmids described above were transferred into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by electroporation. Rice callus was transformed as described (Toki et al., 2006). Dehulled mature seeds were inoculated on callus induction (N6D) medium solidified with 0.4% gelrite (Wako Pure Chemical Industries) after sterilization with 70% ethanol, and 2.5% sodium hypochlorite, and cultured for 3 to 4 weeks at 33°C under constant light. After a 3-d cocultivation, callus was washed and cultured on N6D medium containing 500 μm 5MT (Sigma-Aldrich) and 400 mg L−1 carbenicillin (Nakalai tesque) or 12.5 mg L−1 meropenem (Wako Pure Chemical Industries) for 1 month at 33°C. For regeneration, calli growing vigorously on 5MT were transferred to regeneration medium with 300 μm 5MT, 200 mg L−1 carbenicillin, or 12.5 mg L−1 meropenem, and cultured for 1 month at 30°C under constant light. Shoots arising from callus on regeneration medium were transferred to Murashige and Skoog medium without phytohormones (Murashige and Skoog, 1962) to promote vigorous root growth.
Extraction of Rice Genomic DNA and Southern-Blot Analysis
For extraction of DNA, rice seedlings were harvested, immediately frozen in liquid N2, and stored at −80°C. For Southern-blot analysis, genomic DNA was extracted from leaves using Nucleon PhytoPure (GE Healthcare) according to the manufacturer’s protocol. Southern-blot analysis was performed according to a standard protocol. Specific DNA probes were prepared using the PCR digoxigenin probe synthesis kit (Roche Diagnostics) according to the manufacturer’s protocol with the primer set: OASA2 f1/r7 (probe 1), OASA2 f7/r8 (probe 2), and OASA2 f8/r9 (probe 3). Signals in Southern-blot hybridization were detected and analyzed using Lumivision PRO (TAITEC).
Extraction of Total RNA and Quantitative Reverse Transcription-PCR
For extraction of RNA, rice leaf blades were harvested, immediately frozen in liquid N2, and stored at −80°C. Total RNA was extracted from leaves using an RNeasy Plant mini kit (QIAGEN) according to the manufacturer’s protocol. Total RNA was used for reverse transcription (RT) using the Oligo(dT)20 primer with ReverTra Ace (TOYOBO). Reverse-transcribed cDNAs were prepared from three different lines. Transcript levels of each gene were measured by real-time PCR using an ABI7300 (Applied Biosystems) and the Power SYBR green PCR master mix (Applied Biosystems) according to the manufacturer’s protocol, with the primer set RT-f/RT-r. A PCR fragment of each gene was used to prepare standard curves for quantification.
Sequencing Analysis
PCR was performed using the primer set OASA2 f1/r6 with KOD -plus-. Sequences of purified PCR fragments were read with an ABI3130 sequencer (Applied Biosystems) and analyzed with Sequence Scanner.
Amino Acid Analysis
Callus cultured on N6D medium (Toki et al., 2006) without Gly and Pro for 2 weeks, leaves (the aerial part of the seedlings) grown on Murashige and Skoog medium without phytohormones (Murashige and Skoog, 1962) (Toki et al., 2006) for 2 weeks, and dehulled mature seeds grown in the greenhouse were used for amino acid analysis. Sample preparation and amino acid quantification by capillary electrophoresis time-of-flight mass spectrometry followed the method of Ohkama-Ohtsu (Ohkama-Ohtsu et al., 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Molecular analysis of line 16-11.
Supplemental Figure S2. Growth of line 3-4 in the greenhouse.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank K. Wakasa for stimulating discussion and H. Rothnie for editing. We also thank K. Osakabe, K. Abe, M. Endo, S. Nonaka, A. Yokoi, and N. Ohtsuki for discussion and K. Amagai, R. Aoto, C. Furusawa, A. Nagashii, E. Ozawa, and F. Suzuki for technical assistance.
References
- de Pater S, Neuteboom LW, Pinas JE, Hooykaas PJJ, van der Zaal BJ. (2009) ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol J 7: 821–835 [DOI] [PubMed] [Google Scholar]
- Dürrenberger F, Crameri A, Hohn B, Koukolíková-Nicola Z. (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci USA 86: 9154–9158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Osakabe K, Ichikawa H, Toki S. (2006) Molecular characterization of true and ectopic gene targeting events at the acetolactate synthase gene in Arabidopsis. Plant Cell Physiol 47: 372–379 [DOI] [PubMed] [Google Scholar]
- Endo M, Osakabe K, Ono K, Handa H, Shimizu T, Toki S. (2007) Molecular breeding of a novel herbicide-tolerant rice by gene targeting. Plant J 52: 157–166 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J. (2001) Gene targeting in Arabidopsis. Plant J 28: 671–677 [DOI] [PubMed] [Google Scholar]
- Hare PD, Chua NH. (2002) Excision of selectable marker genes from transgenic plants. Nat Biotechnol 20: 575–580 [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. (1993) New Agrobacterium helper plasmids for gene-transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Ishihara A, Matsuda F, Miyagawa H, Wakasa K. (2007) Metabolomics for metabolically manipulated plants: effects of tryptophan overproduction. Metabolomics 3: 319–334 [Google Scholar]
- Johzuka-Hisatomi Y, Terada R, Iida S. (2008) Efficient transfer of base changes from a vector to the rice genome by homologous recombination: involvement of heteroduplex formation and mismatch correction. Nucleic Acids Res 36: 4727–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint WHO/FAO/UNU Expert Consultation (2007) Protein and amino acid requirements in human nutrition. World Health Organ Tech Rep Ser 935: 1–265 [PubMed] [Google Scholar]
- Kanno T, Kasai K, Ikejiri-Kanno Y, Wakasa K, Tozawa Y. (2004) In vitro reconstitution of rice anthranilate synthase: distinct functional properties of the alpha subunits OASA1 and OASA2. Plant Mol Biol 54: 11–22 [DOI] [PubMed] [Google Scholar]
- Kanno T, Komatsu A, Kasai K, Dubouzet JG, Sakurai M, Ikejiri-Kanno Y, Wakasa K, Tozawa Y. (2005) Structure-based in vitro engineering of the anthranilate synthase, a metabolic key enzyme in the plant tryptophan pathway. Plant Physiol 138: 2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Lee IS, Jang CS, Kang SY, Seo YW. (2005) Characterization of the altered anthranilate synthase in 5-methyltryptophan-resistant rice mutants. Plant Cell Rep 24: 357–365 [DOI] [PubMed] [Google Scholar]
- Lee HY, Kameya T. (1991) Selection and characterization of a rice mutant resistant to 5-methyltryptophan. Theor Appl Genet 82: 405–408 [DOI] [PubMed] [Google Scholar]
- Lee KY, Lund P, Lowe K, Dunsmuir P. (1990) Homologous recombination in plant cells after Agrobacterium-mediated transformation. Plant Cell 2: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Ishihara A, Takanashi K, Morino K, Miyazawa H, Wakasa K, Miyagawa H. (2010) Metabolic profiling analysis of genetically modified rice seedlings that overproduce tryptophan reveals the occurrence of its inter-tissue translocation. Plant Biotechnol 27: 17–27 [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ohkama-Ohtsu N, Oikawa A, Zhao P, Xiang C, Saito K, Oliver DJ. (2008) A gamma-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol 148: 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AG. (2008) The outlook for protein engineering in crop improvement. Plant Physiol 147: 6–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng XD, et al. (2009) Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. (2007) Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol 144: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada R, Nagahara M, Furukawa K, Shimamoto M, Yamaguchi K, Iida S. (2010) Cre-loxP mediated marker elimination and gene reactivation at the waxy locus created in rice genome based on strong positive-negative selection. Plant Biotechnol 27: 29–37 [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu FL, Maeder ML, Joung JK, Voytas DF. (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozawa Y, Hasegawa H, Terakawa T, Wakasa K. (2001) Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2: tryptophan accumulation in transgenic rice expressing mutant of OASA1. Plant Physiol 126: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufaz S, Galili G. (2008) Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol 147: 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenberghe S, Relandeau C. (2000) L-tryptophan supplementation to enhance piglet growth. Ajinomoto Tech Bull 23: 1–12 [Google Scholar]
- Wakasa K, Hasegawa H, Nemoto H, Matsuda F, Miyazawa H, Tozawa Y, Morino K, Komatsu A, Yamada T, Terakawa T, et al. (2006) High-level tryptophan accumulation in seeds of transgenic rice and its limited effects on agronomic traits and seed metabolite profile. J Exp Bot 57: 3069–3078 [DOI] [PubMed] [Google Scholar]
- Wakasa K, Ishihara A. (2009) Metabolic engineering of the tryptophan and phenylalanine biosynthetic pathways in rice. Plant Biotechnol 26: 523–533 [Google Scholar]
- Wakasa K, Widholm JM. (1987) A 5-methyltryptophan resistant rice mutant, MTR1, selected in tissue-culture. Theor Appl Genet 74: 49–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.