Abstract
Gene expression is a quantitative trait that can be mapped genetically in structured populations to identify expression quantitative trait loci (eQTL). Genes and regulatory networks underlying complex traits can subsequently be inferred. Using a recently released genome sequence, we have defined cis- and trans-eQTL and their environmental response to low phosphorus (P) availability within a complex plant genome and found hotspots of trans-eQTL within the genome. Interval mapping, using P supply as a covariate, revealed 18,876 eQTL. trans-eQTL hotspots occurred on chromosomes A06 and A01 within Brassica rapa; these were enriched with P metabolism-related Gene Ontology terms (A06) as well as chloroplast- and photosynthesis-related terms (A01). We have also attributed heritability components to measures of gene expression across environments, allowing the identification of novel gene expression markers and gene expression changes associated with low P availability. Informative gene expression markers were used to map eQTL and P use efficiency-related QTL. Genes responsive to P supply had large environmental and heritable variance components. Regulatory loci and genes associated with P use efficiency identified through eQTL analysis are potential targets for further characterization and may have potential for crop improvement.
Expression quantitative trait loci (eQTL) are genetic regions associated with variation in gene expression among individuals (Kliebenstein, 2009). This variation can arise due to sequence polymorphisms in target genes, their cis-regulatory (proximal) or trans-regulatory (distal) regions, leading to phenotypic differences. Identifying variation in gene expression within a segregating mapping population is potentially of immense use, in particular within the plant and crop sciences (Druka et al., 2010). First, there is the opportunity to map the chromosomal positions of thousands of genes based on constitutive differences in expression between parents. Gene mapping at this scale is of great value, especially for organisms whose genomes have not been fully sequenced, and can contribute to efforts to integrate physical and genetic maps. Furthermore, in polyploids and/or where a genome contains ancestral duplications, a feature common among plant species, it will become feasible to map gene paralogues given sufficiently robust expression probes. Second, since a single gene could be associated with one or many eQTL, categorizing eQTL into cis- or trans-eQTL effects relative to the physical location of a gene can reveal regulatory networks controlling gene expression in the absence of a priori models. Third, by combining eQTL analysis with trait analysis, putative candidate genes associated with traits can be identified indirectly by correlation analysis. Candidate genes associated with important traits can then become targets for traditional genetic characterization (Druka et al., 2010).
Several eQTL studies have been reported in plants, including studies on Arabidopsis (Arabidopsis thaliana), barley (Hordeum vulgare), Populus, and maize (Zea mays; for review, see Kliebenstein, 2009; Druka et al., 2010). In studies where a physical map is available, eQTL have been categorized empirically as cis- and/or trans-eQTL based on assumptions about genetic linkage. The most significant eQTL tend to be cis-eQTL, which occur when a sequence polymorphism in a gene or promoter maps to quantitative variation in the gene transcript (Kliebenstein, 2009). This phenomenon, when combined with linkage, results in the standard “cis-diagonal” being seen when gene physical position is plotted against eQTL position (DeCook et al., 2006; Keurentjes et al., 2007; West et al., 2007; Kliebenstein, 2009). In contrast, sequence polymorphisms in regulatory genes such as transcription factors, which do not colocate with the physical position of the gene, can result in allelic variation at trans-eQTL. A cis-eQTL in a regulatory gene, therefore, can generate many trans-eQTL (Kliebenstein, 2009). Several trans-eQTL hotspots have been identified in plants, in which loci are associated with variation in the expression level of many hundreds or even thousands of genes, and these are thought to represent master regulatory loci with potentially pleiotropic effects (Kliebenstein, 2009). Typically, the statistical significance of associations between genetic loci and trans-eQTL are less than cis-eQTL, potentially due to the complex nature of regulatory gene networks.
The identification of cis- and trans-eQTL has already revealed information regarding the complex genetic architecture of variation in plant gene expression. However, relationships between classical phenotypic or trait QTL and variation in gene expression have not been widely reported, since most studies have been conducted under single experimental conditions. Notably, in barley, correlations between loci conferring resistance to rust pathogens and variation in gene expression have also been found using eQTL approaches (Druka et al., 2008; Chen et al., 2010). However, the identification of eQTL under abiotic stress has not yet been reported.
The aim of this study was to identify eQTL under altered soil phosphorus (P) supply. Brassica rapa was chosen since it is a close crop relative of Arabidopsis that retains a diploid genome structure, albeit including multiple paralogues. Recent hybridizations between the diploid A genome of B. rapa (vegetable and oil crops) and the C genome of Brassica oleracea (vegetable crops) have given rise to the widely grown amphidiploid Brassica napus (AC genome; canola/oilseed rape/colza, rutabaga/swede; Allender and King, 2010). Furthermore, extensive genetic and genomic resources for B. rapa have now been assembled. These include a rapid-cycling mapping population developed from highly inbred lines of rapid cycling (IMB211) and yellow sarson (R500) B. rapa (Iniguez-Luy et al., 2009), more than 2M Brassica GenBank sequences, and a B. rapa genome sequence made available in 2011 alongside other reference Brassica genome and resequencing projects (Multinational Brassica Genome Project; www.brassica.info/resource/sequencing.php), as well as oligonucleotide microarrays (Trick et al., 2009a; Love et al., 2010).
Identifying and characterizing eQTL under altered P supply will increase our understanding of P use efficiency (PUE) in plants. An improved understanding of the genetics of PUE at the individual gene level may provide new opportunities for crop improvement based on candidate gene and marker identification at a scale that is much more rapid than one based on trait QTL approaches alone (Hammond et al., 2009). There are pressing economical and environmental pressures to reduce our reliance on inorganic P fertilizers, including the development of crops that grow well under conditions of low soil P and that utilize P fertilizer inputs most efficiently.
RESULTS AND DISCUSSION
The transcriptional profiling of B. rapa recombinant inbred lines (RILs) grown in different environments has enabled us to characterize the genetic architecture of plant adaptation to low P availability. To our knowledge, this is the first study of its kind in which the heritability (H2) of global gene expression is estimated in response to a known abiotic stress (P availability) and the quantitative expression of genes is mapped across environments to identify eQTL associated with adaptation to P availability. To complete this study, we have utilized the BraIRRI mapping population (Iniguez-Luy et al., 2009). This population was derived from a cross between a highly inbred rapid-cycling B. rapa (IMB211, female) and a highly inbred annual yellow sarson (var trilocularis; R500, male), and the progeny selfed for eight generations to generate RILs.
Transcriptional Responses to Low P Availability Indicates Enhanced P Recycling
To determine conserved transcriptional responses to P availability, transcriptional profiles were determined across the BraIRRI mapping population within the treatments. To correct for multiple testing, a Benjamini and Hochberg multiple testing-corrected P < 0.05 (Benjamini and Hochberg, 1995) was used subject to greater than 2-fold difference in expression between low and optimal external P concentration ([P]ext). Analysis of transcriptional profiles between treatments identified 2,009 probes representing transcripts with significantly greater abundance at low [P]ext (Supplemental Table S2) and 1,217 probes representing transcripts with significantly less transcript abundance at low [P]ext (Supplemental Table S3). Probes with greater abundance included those with annotations to genes whose expression has been shown previously to change in response to low P availability (Hammond et al., 2005; White and Hammond, 2008; Fang et al., 2009; Nilsson et al., 2010). These included probes annotated as having phosphatase activity, such as a transcript with homology to Arabidopsis Purple Acid Phosphatase6 (At1g56360.1), which had a greater than 1,000-fold difference in expression between low and optimal [P]ext. In addition, six other transcripts with homology to Arabidopsis proteins containing phosphatase activity were in the top 50 transcripts with significantly greater abundance at low [P]ext (Supplemental Table S2). Proteins with acid phosphatase activity are known to be induced under P-limiting conditions and act to release P from various inorganic P monoesters to recycle inorganic P to essential metabolic processes (Li et al., 2002; Tran et al., 2010).
Several transcripts with significantly greater abundance at low [P]ext showed homology to proteins involved in the manipulation of membrane lipids. These included two transcripts with homology to glycerophosphoryl diester phosphodiesterases and a transcript with homology to Arabidopsis PLDP2, phospholipase D, which are involved in the catabolism of phospholipids, recycling P to essential metabolic processes and providing diacyglcerol for galactolipid biosynthesis (van der Rest et al., 2002; Li et al., 2006a, 2006b; Tjellström et al., 2008). Transcripts with homology to proteins involved in the biosynthesis of galactolipids and sulfolipids also had significantly greater abundance at low [P]ext, including two isoforms of 1,2-diacylglycerol 3-β-galactosyltransferase (MGDG and MGDC) (Supplemental Table S2). Under P-limiting conditions, plants can alter the composition of their membranes, reducing their phospholipid content by replacing phospholipids with galactolipids and sulfolipids (Essigmann et al., 1998; Andersson et al., 2003, 2005; Kobayashi et al., 2009), a process that may be regulated through cross talk between the auxin and cytokinin signaling pathways (Kobayashi et al., 2006), thus decreasing the structural requirement for P.
Transcripts with homology to proteins and transcripts involved in the regulation of plant responses to low P availability also had significantly greater abundance under low P availability (Supplemental Table S2). These included two transcripts with homology to two Arabidopsis SPX domain-containing proteins (At2g45130 and At5g20150) and a transcript with homology to Arabidopsis IPS1 (At3g09922). Proteins containing SPX domains have been implicated in the regulation of plant responses to low P availability, with the two proteins, SPX1 and SPX3, playing positive roles in plant adaptation to low P availability and acting in the PHR1 signaling cascade (Duan et al., 2008; Wang et al., 2009; Liu et al., 2010). The expression of the transcripts from the IPS1 family is induced rapidly and specifically in response to P starvation (Burleigh and Harrison, 1999; Martín et al., 2000; Hammond et al., 2003; Hou et al., 2005; Shin et al., 2006), and these noncoding transcripts sequester miR399 and serve to attenuate the miR399-mediated transcriptional responses to low P availability (Franco-Zorrilla et al., 2007).
Analysis of Gene Ontology (GO) terms associated with these transcripts was consistent with the role of these genes in plant adaptations to low P availability (Supplemental Tables S4 and S5). Transcripts with greater abundance under low P availability had an enrichment of GO terms attributed to flavonoid biosynthetic process (GO:0009813), sulfur compound biosynthetic process (GO:0044272), phosphoric ester hydrolase activity (GO:0042578), phosphatase activity (GO:0016791|GO:0016302), and response to nutrient levels (GO:0031667). The expression of these transcripts in the BraIRRI mapping population is thus consistent with previous observations of plant transcriptional responses to low P availability.
P-Responsive Transcripts Are Highly Heritable
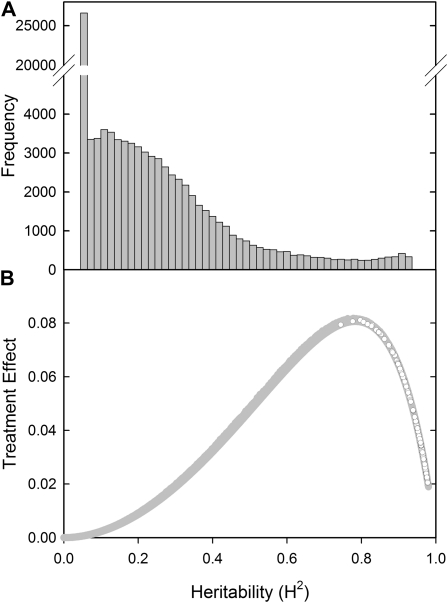
The H2 of gene expression was calculated for each probe on the array, using the log2-normalized signal values from all 78 genotypes at both low and optimal [P]ext. H2 was calculated as the genotype variance as a proportion of the genotype, genotype and environment ([P]ext) interaction, and a residual term using residual maximum likelihood procedures, with the effect of the [P]ext fixed (Fig. 1A; Supplemental Table S6). This provides a measure of how much variation in the expression of an individual gene was due to genotype relative to variation from environmental factors, such as experimental design and treatment. The transcript abundance for over 22% of probes had a H2 of zero, and 90% of the probes had a H2 of less than 0.5. Since the variance component model used to calculate H2 apportions the variance between genotype, environment, and their interaction, there is a mathematical interdependence between the H2 and the variance attributed to the environment component, [P]ext (Fig. 1B). This relationship identifies three groups of probes: (1) probes for which the transcript abundance is nonheritable and nonresponsive to [P]ext; (2) probes for which the transcript abundance is heritable and that are highly responsive to [P]ext; and (3) probes for which the transcript abundance is highly heritable but are not responsive to [P]ext (Fig. 1B).
Figure 1.
A, Frequency distribution for the H2 of gene expression for all probes on the Agilent Brassica array. B, H2 plotted against the treatment effect calculated in the residual maximum likelihood analysis. White circles represent GEMs identified from the array data and used to generate the GEMs map for the BraIRRI mapping population.
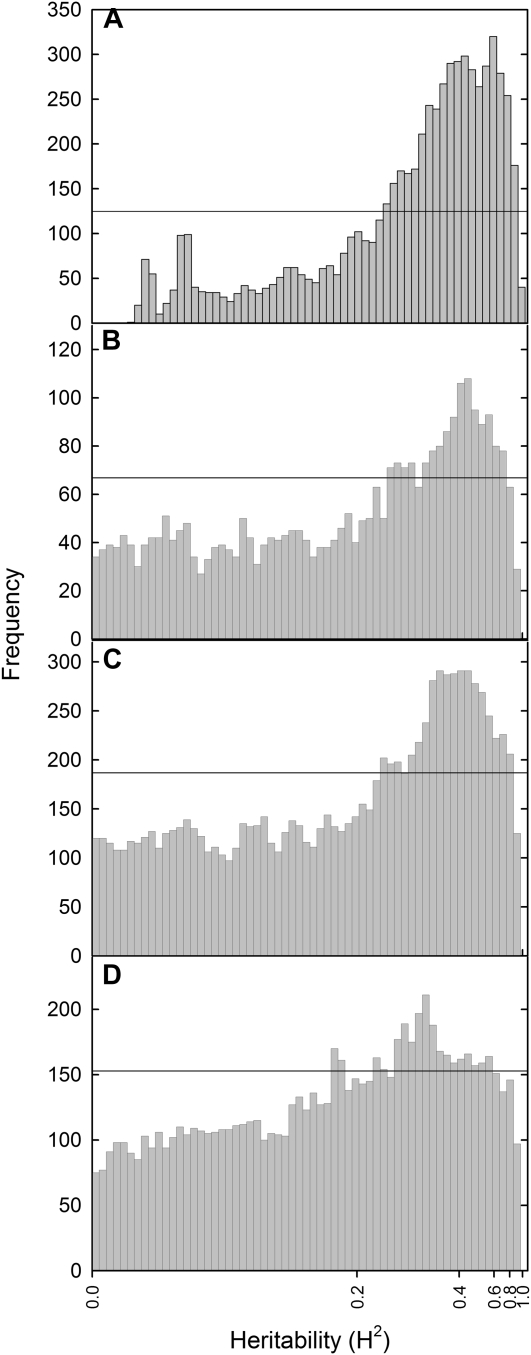
We hypothesized that the annotations for probes in group 2 will be enriched with genes known to respond to low P availability. Enrichment for genes identified as responding to low P availability (Supplemental Table S2) was determined in overlapping groups of 3,000 probes (e.g. 0–3,000, 1,500–4,500, 3,000–6,000, etc.) generated from a list of all probes ordered by H2 (e.g. probes in the group 0–3,000 had H2 = 0, probes in group 90,000–91,854 had H2 > 0.866). There was a significant (P < 0.05 for the χ2 distribution for a two-way contingency table) enrichment of probes identified as responding to low P availability (Supplemental Table S6) in groups of probes with H2 between 0.23 and 0.95, with an average treatment effect of 0.35 (Fig. 2A). GO term analysis of these significant groups also showed enrichment for GO terms associated with the chloroplast, ribosome, and carbohydrate metabolism. Groups of probes with H2 values less than 0.23 or greater than 0.95 showed no enrichment for any GO terms, with the exception of groups of probes with H2 = 0, which were enriched for a single GO term associated with the endomembrane system.
Figure 2.
Frequency distribution of genes differentially expressed between optimal and low P treatments identified in this paper (A), by Misson et al. (2005; B), by Morcuende et al. (2007; C), or by Wu et al. (2003; D) as a function of H2. Horizontal bars represent significance (P < 0.05) thresholds for overrepresentation of P-responsive genes within a bin of 3,000 genes.
Enrichment of genes responsive to low P availability in groups of genes with high H2 and treatment effects was also confirmed using independent data from transcriptional profiling studies of Arabidopsis responses to low P availability (Fig. 2, B–D; Wu et al., 2003; Misson et al., 2005; Morcuende et al., 2007). This suggests that transcriptional responses to P availability are highly heritable and provide potential targets for breeding crops with improved abilities to grow under low soil P conditions.
Gene Expression Markers Are Highly Heritable But Not Responsive to P Availability
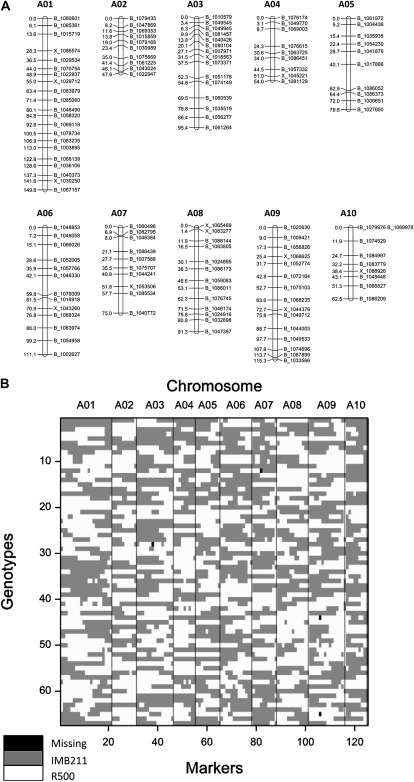
Using the gene expression data obtained for all the individual RILs in the BraIRRI mapping population, it was possible to identify gene expression markers (GEMs) in the genome suitable for mapping eQTL and QTL. In total, 125 robust GEMs were identified for 67 RILs (Fig. 3). The overall map length was 870.8 centimorgan (cM) across chromosomes A01 to A10, with an average distance between markers of 7.0 cM. The selection procedure used to generate the original 2,306 candidate GEMs was expected to enrich for single-feature polymorphisms (group 3 of the marker types described above). In practice, this was borne out, with all of the final selected markers showing more signal variation between parental alleles than due to [P]ext, with none of the GEMs present in transcripts responding to low P availability (Supplemental Tables S2 and S3), and an average H2 across the 125 markers of 0.93 (Fig. 1B). That the markers are largely nonresponsive to P indicates that their use for genetic map construction is justified, with the maps developed then being suitable for examining P-responsive transcripts (group 2 GEMs). The independence of marker types for different parts of the analysis gives confidence that the analysis is not tautological. Thus, the use of these spaced markers for eQTL and QTL analysis appears justified for the reasons described above. The segregation pattern shows a reasonable distribution of recombination break points across the 67 genotypes (Fig. 3B), although some degree of apparent local double recombination can be seen for a number of groups (e.g. lines 42 and 58 on A04; Supplemental Text S1).
Figure 3.
Distribution of 125 GEMs for the BraIRRI_04_2010a map (Supplemental Table S1) across the B. rapa A genome (A) and across the 67 genotypes of the BraIRRI mapping population used to identify eQTL and QTL associated with low and optimal P availability (B).
The Expression QTL Hotspot on A06 Is Enriched with P-Responsive Transcripts
Using the genetic map based on GEMs (BraIRRI_04_2010a), data for individual probes across 67 lines and two [P]ext treatments were subjected to interval mapping, using [P]ext as a covariate. A total of 18,874 eQTL were identified across the genome, representing 15,910 unique probes, with log of the odds (LOD) scores ranging between 3.17 and 2,824.18 (Supplemental Table S7). Robust physical genomic locations were identified for 10,545 of the 15,910 probes associated with eQTL, allowing the positions of the genes to be compared with the positions of their associated eQTL and the nature of their regulation to be determined. Highly significant eQTL tend to be cis-eQTL, which occur when a sequence polymorphism in a gene or promoter maps to quantitative variation in the gene transcript (Kliebenstein, 2009). In contrast, sequence polymorphisms in regulatory genes such as transcription factors, which do not colocate with the physical position of the gene, can result in allelic variation at trans-eQTL. The midpoint between markers was used to define boundaries for the colocation of eQTL with their physical positions within the genome. Of the 14,257 eQTL that could be physically mapped, 6,304 colocated to the same chromosome as indicated by the eQTL mapping, with 3,236 colocating to the same physical position as the marker and defined as cis-eQTL (Supplemental Tables S8 and S9). This proportion of cis-eQTL to trans-eQTL is similar to that reported in the Arabidopsis Bay-0 × Sha RIL population (West et al., 2007) but lower than that observed in the Arabidopsis Landsberg erecta × Cape Verde Island RIL population (Keurentjes et al., 2007). The median LOD statistic associated with the 3,236 cis-eQTL was 8.4 and that for the 11,021 trans-eQTL was 4.0. This is consistent with previous eQTL studies, which have reported higher LOD scores associated with cis-eQTL compared with trans-eQTL (Keurentjes et al., 2007; West et al., 2007). The additive effect of the parental alleles was consistent between treatments, with 47.5% and 46.7% of eQTL resulting from a negative effect of the IMB211 allele under optimal [P]ext and low [P]ext, respectively, and 52.3% and 53.3% of eQTL resulting from a positive effect of the IMB211 allele under optimal [P]ext and low [P]ext, respectively.
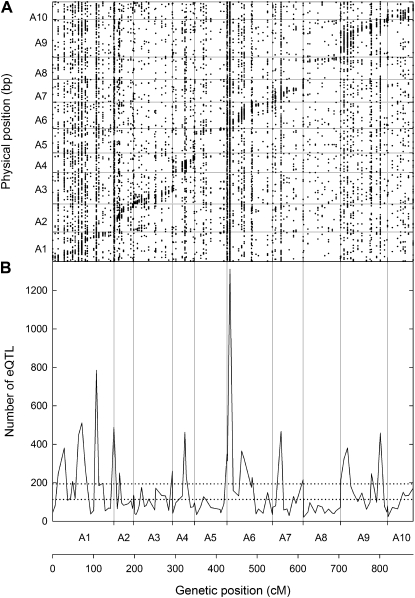
Using physical positions for the probes within the B. rapa genome and the number of eQTL associated with a specific marker above an empirical threshold, trans-eQTL hotspots were identified (Fig. 4). These are regions of the genome in which loci are associated with variation in the expression level of many hundreds or even thousands of genes. They are thought to represent master regulatory loci, with one or a few regulatory genes at this physical position controlling the expression of many genes associated with that position in the genome. We observed an average of 152.2 eQTL per marker, which is consistent with the expected number of eQTL per marker of 151.0, based on 125 markers and 18,876 eQTL. The 99.9% upper and lower confidence intervals of the Poisson distribution, which assumes that GEMs and eQTL are distributed equally, are 113 and 194. Thus, any marker associated with more than 194 eQTL is considered to be a regulatory hotspot, which is likely to contain many putative trans-eQTL. There are 27 markers with more than 194 eQTL (Fig. 4). These hotspots might represent gene-dense regions within the genome, and markers with less than 113 eQTL associated with them may represent regions of the genome with few genes, such as centromeres (Keurentjes et al., 2007). The most notable trans-eQTL hotspot occurs on chromosome A06, where marker 2 (B_1046058; 7.02 cM) is associated with 1,311 eQTL. Subsequent analysis revealed significant enrichment for GO terms associated with protein modification and P metabolism. Putative regulatory hotspots were also identified on chromosome A01 at marker 15 (B_1083235; 106.9 cM), which is associated with 785 eQTL, and is enriched with chloroplast and photosynthesis-related GO terms, on chromosome A04 at marker 5, which is enriched for His-related GO terms, and on chromosome A09 at marker 12 (B_1049533; 97.7 cM), which is enriched for cytoskeleton and nucleus-related GO terms.
Figure 4.
Comparison between genetic and physical positions of the 14,257 eQTL for which physical positions could be identified (A) and the distribution of eQTL associated with individual markers across the B. rapa A genome (B). Probe sequences were aligned to the concatenated scaffolds of the B. rapa Chiifu-401 genome sequence (version 1.0) to determine physical positions and compared with genetic locations determined through eQTL analyses.
Due to the low marker density, it is not possible to make direct comparisons to eQTL and QTL identified previously in Arabidopsis, but reanalyses of these data with a higher density map and improved genomic data will facilitate this and allow comparative genomics analyses of the eQTL and overlap of homologous genes between hotspots. Approaches that distinguish between paralogues within the genome, such as the use of high-density oligonucleotide arrays with exon-specific probes or RNAseq, will also increase the accuracy and depth of these analyses in the future (Trick et al., 2009b; Love et al., 2010).
Trait QTL Colocalize with eQTL
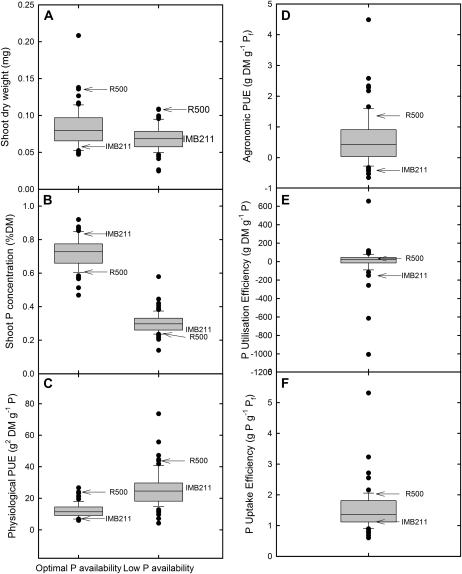
Transgressive segregation of trait values beyond the parental values was observed for all traits measured in the BraIRRI mapping population (Fig. 5). Shoot P concentration (shoot [P]) varied 1.97-fold at optimal P availability and 4.17-fold at low P availability, with population mean shoot [P] of 0.72% for lines grown at optimal P availability and 0.30% for lines grown at low P availability. Measures of PUE also varied among the 67 RILs grown at optimal and low P availability (Fig. 5). Genetic loci associated with the responsiveness to [P]ext were subsequently mapped using these data (Table I). Of the 13 trait QTL mapped, six colocalize with eQTL, including QTL associated with P utilization efficiency (g dry matter g−1 tissue P) at marker 2 (B_1046058; 7.02 cM) on A06, QTL associated with physiological PUE (g2 dry matter g−1 tissue P) at optimal [P]ext at marker 7 (B_1022937; 48.85 cM) on A01, and marker 5 (B_1057766; 35.87 cM) on A06. A QTL associated with agronomic PUE (g dry matter g−1 fertilizer P applied) also colocalized with marker 5 on A06.
Figure 5.
Shoot dry weight (A), shoot P concentration (B), physiological PUE (C), agronomic PUE (D), P utilization efficiency (E), and P uptake efficiency (F) for the B. rapa BraIRRI mapping population. DM, Dry matter; P, shoot P concentration; Pf, fertilizer P applied. Data are residual maximum likelihood-estimated means for plants grown in compost under CE conditions at low and optimal [P]ext. The boundaries of the box closest to and farthest from zero indicate the 25th and 75th percentiles, respectively. The solid line within the box indicatee the median. Error bars indicate the 10th and 90th percentiles. Circles indicate outliers. Parents of the mapping population are indicated.
Table I. Significant (P < 0.05) QTL associated with shoot P and measures of PUE in B. rapa.
Shoot dry matter, shoot [P], and measures of PUE were determined in 67 RILs of the BraIRRI mapping population. Plants were grown under CE conditions in compost containing 9 mg L−1 (low) or 30 mg L−1 (optimal) Olsen extractable P. Dry weight and shoot [P] were determined after 18 d of growth, and measures of PUE were calculated according to Hammond et al. (2009). QTL positions were estimated in QTL Cartographer 2.0 using the zmapQTL model 6 composite interval mapping option. DM, Dry matter; P, shoot [P]; Pf, fertilizer P applied.
| Trait | Chromosome | Marker | Position | LOD Score | Additive Effect | LOD Threshold (P < 0.05) |
| cM | ||||||
| Dry weight at optimal [P]ext (mg) | A01 | 3 | 13.6 | 3.632 | −0.010 | 2.352 |
| Dry weight at optimal [P]ext (mg) | A06 | 6 | 43.1 | 2.646 | −0.010 | 2.352 |
| Dry weight at low [P]ext (mg) | A01 | 6 | 47.0 | 3.186 | −0.007 | 2.758 |
| Shoot [P] at low [P]ext (% DM) | A09 | 8 | 63.6 | 2.529 | −0.027 | 2.409 |
| Physiological P use at optimal [P]ext (g2 DM g−1 P) | A01 | 7 | 48.9 | 2.811 | −1.564 | 2.693 |
| Physiological P use at optimal [P]ext (g2 DM g−1 P) | A06 | 5 | 35.9 | 5.178 | −2.344 | 2.693 |
| Physiological P use at low [P]ext (g2 DM g−1 P) | A01 | 6 | 45.0 | 2.585 | −4.118 | 2.500 |
| P efficiency ratio at low [P]ext (g DM g−1 P) | A06 | 4 | 32.6 | 4.243 | −55.303 | 2.107 |
| P utilization efficiency (g DM g−1 P) | A06 | 2 | 14.2 | 1.684 | 51.696 | 1.536 |
| P utilization efficiency (g DM g−1 P) | A09 | 11 | 92.7 | 3.116 | 125.712 | 1.536 |
| P utilization efficiency (g DM g−1 P) | A09 | 13 | 107.8 | 3.995 | −103.788 | 1.536 |
| Agronomic PUE (g DM g−1 Pf) | A06 | 5 | 35.9 | 3.473 | −0.348 | 2.276 |
| P uptake efficiency (g P g−1 Pf) | A05 | 6 | 40.1 | 2.903 | −0.342 | 2.101 |
Previously, QTL for shoot [P] and PUE traits have been identified in Arabidopsis, B. oleracea, B. rapa, and B. napus mapping populations (Bentsink et al., 2003; Loudet et al., 2003; Lisec et al., 2008; Wu et al., 2008; Zhao et al., 2008; Hammond et al., 2009; Liu et al., 2009; Ding et al., 2010; Yang et al., 2010). The high colinearity and synteny between the Arabidopsis, B. oleracea, and B. rapa genomes enables the identification of conserved loci between these species (Parkin et al., 2005; Schranz et al., 2006). QTL for leaf [P] have been located in B. rapa on chromosomes A01, A03, and A08 at 27, 40, and 47 cM, respectively (Zhao et al., 2008) and for shoot [P] in B. oleracea on C3 at 30 and 107 cM and on C7 at 33 cM (Hammond et al., 2009). A QTL associated with shoot [P] at low [P]ext was identified on A9 in this study and did not colocate with previous studies. QTL associated with dry weight at low and optimal [P]ext and physiological PUE at optimal and low [P]ext colocalize with QTL associated with physiological PUE at high and low [P]ext, P uptake efficiency (g tissue P g−1 fertilizer P applied), and shoot dry weight identified on chromosome 1 in B. oleracea (Hammond et al., 2009). Alignment with QTL identified in Arabidopsis for shoot [P] reveals some colocalization between loci. QTL mapped to the top of A06 in this study colocalize with QTL for shoot [P] identified previously (Bentsink et al., 2003; Loudet et al., 2003) on the top of Arabidopsis chromosome 1. This suggests that loci for shoot [P] may be conserved in the Brassicaceae, but further work, including identification of the genes responsible for these QTL, is required to confirm this. Interestingly, several of the QTL associated with dry weight at optimal [P]ext on A01 and A06 and physiological PUE at optimal [P]ext, QTL associated with P utilization efficiency, and agronomic PUE on A06 all colocalize with eQTL hotspots. Further research is required to confirm the genetic nature of any links between these QTL.
CONCLUSION
Using high-density microarrays, we have identified robust GEMs within the BraIRRI mapping population. Combined with the recently released B. rapa genome sequence, we have defined cis- and trans-eQTL and their environmental responses to low P availability within a complex plant genome. Putative hotspots of trans-eQTL within the genome were also identified. Genes responsive to P supply had large environmental and heritable variance components. This suggests that transcriptional responses to P availability are highly heritable and provide potential targets for breeding crops with improved abilities to grow under low soil P conditions. Further analyses are required to identify the major regulators underlying trans-eQTL hotspots and the biochemical, morphological, and physiological processes they regulate in the plant adaptations to low soil P availability.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of 78 informative lines from the BraIRRI mapping population of Brassica rapa (2n = 2x = 10; A genome) and the two parent lines were selected for study. The establishment of the BraIRRI population is described by Iniguez-Luy et al. (2009). Briefly, the population was derived from a cross between a highly inbred rapid-cycling B. rapa (IMB211, female), derived from the WI Fast Plant population, produced from 10 generations of selection for early flowering and rapid generation time (Williams and Hill, 1986), and a highly inbred annual yellow sarson (var trilocularis; R500, male). An initial F1 plant was self-pollinated, and 160 RILs were obtained by single seed descent to the S4 generation (Iniguez-Luy et al., 2009) and subsequently to S8 (C. Hopkins, unpublished data, Rothamsted Research, UK).
Plants were grown from seed in individual pots (9 cm × 9 cm × 8 cm; Desch Plantpak) containing a moistened peat-based compost under controlled-environment conditions. The potting mix consisted of 25% sand and 75% (v/v) compost (Shamrock medium grade sphagnum peat; Scotts UK). Unfertilized, the potting mix had an Olsen extractable P (Olsen et al., 1954) of 8 mg L−1. Two [P]ext treatments of 9 mg L−1 (low) or 30 mg L−1 (optimal) Olsen extractable P were produced by incorporating 0.075 and 0.45 g of sieved (mesh, 500 μm) single superphosphate (7% P) per liter of compost, and used to induce a growth response based on previous studies with Brassica (Hammond et al., 2009). Both compost treatments contained the following nutrients in sufficient amounts to prevent deficiencies: NH4NO3 (0.4 g L−1), KNO3 (0.75 g L−1), ground limestone (2.25 g L−1), magnesium limestone (2.25 g L−1), and a fritted trace elements mixture containing boron, copper, iron, manganese, molybdenum, and zinc (WM255; Fargro Ltd.) at 0.4 g L−1. Potting mixes were made in bulk using a paddle mixer (model 156; St. Moritz). The controlled environment (CE) was a walk-in high-specification growth room (Weiss-Gallenkamp) set to a 16-h photoperiod using metal halide lamps, giving a photon flux density between 400 and 700 nm (photosynthetically active radiation) of 250 μmol photons m−2 s−1 at plant height. The day/night settings for temperature and relative humidity were 18°C/15°C and 76%/71%.
The experimental design was based on an α-design (John and Williams, 1995). The experiment was done in three runs, with all 78 RILs plus parents grown in each run. Each run comprised 12 blocks. Each block contained two subblocks: one for low [P]ext and one for optimal [P]ext treatments. Each subblock was placed on an independent tray, with capillary matting, and irrigated through automated drippers with deionized water. Within a block, the same set of lines was sown in the same (randomized) order in each subblock. Each subblock contained up to 42 pots, comprising six individual plants per RIL for six or seven RILs.
All plants were harvested 21 d after sowing. For each line at each treatment, fully expanded leaves from three plants (six-leaf stage, not flowering) were sampled, snap frozen in liquid nitrogen, freeze dried, ground, and stored at −80°C. For the remaining three plants, leaf, stem, and cotyledon fresh weight and dry weight were determined. Leaf samples were digested by the addition of 2 mL of nitric acid to 0.3 g of dried, ground material and processed in a closed vessel acid digestion microwave (MARSXpress; CEM). Digested samples were diluted with 23 mL of deionized water and analyzed using inductively coupled plasma emission spectrometry (JY Ultima 2; Jobin Yvon) to determine shoot [P].
RNA Extraction and Array Processing
RNA was extracted from leaf samples from one experimental run (run 2; 78 lines at low and optimal [P]ext, with one line duplicated [i.e. 158 samples]) and from leaf samples from the parent lines at low and optimal P in all three experimental runs (12 samples) using a modified TRIzol extraction method (Hammond et al., 2006). Extracted total RNA was purified using the RNA Cleanup protocol for RNeasy columns with on-column DNase digestion to remove residual genomic DNA (Qiagen). Samples of total RNA were checked for integrity and quality using an Agilent Bioanalyzer (Agilent Technologies). RNA samples were labeled with the QuickAmp Labeling Kit (Agilent Technologies) and hybridized to Agilent Brassica 95k 60-mer arrays (Trick et al., 2009a) for 17 h at 65°C at 10 rpm. The arrays were washed, then scanned on an Agilent G2565CA scanner, according to the manufacturer’s instructions. Data files were generated using Agilent Feature Extraction Software (version 10.7.3.1). Array files have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/; accession no. GSE27052).
Initial Data Analysis
Data files (n = 170) were imported into GeneSpring GX (version 11.0.2; Agilent Technologies). Signal values within arrays were normalized to the 50th percentile. Normalized signal values for individual probes were standardized to the median signal value for the probe across all arrays. The log2-normalized signal values were exported as a data matrix with 170 columns (unique RNA samples) and 91,854 (92k) rows (array probes; data available from Gene Expression Omnibus accession no. GSE27052). GO enrichment was performed using the GO category analysis tool in Gene Spring GX with a P value, adjusted for multiple testing using the method of Benjamini and Hochberg (1995), of 0.05. GO terms were assigned to individual probes based on their homology to Arabidopsis (Arabidopsis thaliana) gene sequences.
Custom GenStat scripts (VSN International; available on request from the corresponding author) were used to batch process the log2-normalized signal values in groups of up to 1,500 genes. Genotypic and environmental variance components were estimated for each probe, where environment represents low and optimal [P]ext and genotype represents the effect of line within the population. A variance components model was used to allocate sources of variation, with environment defined as a fixed factor and [genotype + (genotype.environment) + residual] defining the random term. The model was fitted using residual maximum likelihood procedures (Patterson and Thompson, 1971; Robinson, 1987). H2 was calculated using the method of Cullis et al. (2006) as H2 = 1 – mean(pev)/sG, where pev is the vector prediction error variance for the line effects and sG is the line variance component. This can be interpreted as broad-sense mean line H2, which is approximately equal to H2 = sG/(sG + sGE + s/2), where sGE is the genotype environment variance component and s is the residual variance (Cullis et al., 2006).
Development of GEMs
To enable mapping of eQTL and trait QTL, a robust set of 125 GEMs were selected for a subset of 67 genotypes (BraIRRI_04 subpopulation). First, the mean log2-normalized expression for each of the 92k transcripts was calculated for each of the two parents at both [P]ext treatments (i.e. IMB211 [n = 6] and R500 [n = 6]). Transcripts were then ranked following a two-tailed Student’s t test in ascending order of significance. Based on a parental segregation threshold of P < 0.001, a subset of 2,306 transcripts was selected for exploration as suitable GEMs. For GEMs mapping, a reiterative procedure was followed in JoinMap4 (Kyazma; van Ooijen, 2006) using a combination of maximum likelihood mapping (MLM) and regression mapping (RM) approaches. A putative marker data set was identified through selection from the original 2,306 transcripts, minimizing missing data. Selecting for good separation of IMB211 and R500 allele scores within the 78 lines produced a data set of 838 putative GEM loci, and these were combined with 224 RFLP and simple sequence repeat markers from the S4 BraIRRI map (Iniguez-Luy et al., 2009) for initial group determination before the S4 BraIRRI markers were removed before map construction. Grouping used the “independence LOD” option ranging from LOD 2 to 15 in two LOD steps, with the appropriate level for defining a linkage group being determined by visual inspection of the groupings produced. Generally, the final LOD stringency selected was between LOD = 3 and LOD = 5, with the exception of A02 and A03, as described below. Results from MLM and RM (maps 1 and 2 only, with a “jump” value of 3) were used to create initial linkage group maps, using default settings unless otherwise stated. The comparison of results from MLM and RM iterations was used to test the robustness of observed GEM order. The final set of 125 markers was generated through a reiterative process of removing GEMs through examining “fit and stress” parameters for MLM and also through visual inspection of graphical genotypes for MLM and RM for each map iteration (for detailed discussion, see JoinMap4 manual). GEM markers containing clear miscalls were removed, focusing on double recombination events in small genetic distances (1–5 cM). From the initial “grouping” derived from 838 GEM markers, a “minimal” map was created by continuing to remove markers to try to produce a framework at approximately 10 cM resolution for optimal QTL analysis. For both BraIRRI and GEM data, A02 and A03 were difficult to separate at the “group” stage, so very high stringency was applied, suggesting a genuine (but unknown) genetic effect. The resulting GEMs-based map (Fig. 3; Supplemental Table S1; Supplemental Text S1; BraIRRI_04_2010a), containing 125 GEMs mapped across 67 individuals from the BraIRRI mapping population, was subsequently used for QTL and eQTL analyses.
Identifying eQTL and Trait QTL
eQTL associated with individual probes were identified using a two-stage process. GenStat procedure QIBDPROBABILITIES (Boer and Thissen, 2009) was used to calculate a set of genetic predictors at the marker positions on the GEMs genetic map (BraIRRI_04_2010a) for 67 RILs (BraIRRI_04 subpopulation). Initially, a parallel regression model was used to identify possible QTL effects across 1,500 genes simultaneously, by testing for a combined QTL + (QTL × environment) effect, where QTL indicates a genetic predictor used as a covariate. This model was fitted for each genetic predictor in turn. For probes with a significant result in the first step, simple interval mapping, using genetic predictors within the mixed model framework, was performed using QTL procedures implemented in GenStat (Boer et al., 2007). Briefly, a mixed model was fitted with fixed environment effects and QTL × environment interaction for each genetic predictor in turn and random terms comprising genotype, genotype × environment, and residual terms. A compound symmetry structure was used to account for genetic covariance across environments.
We aligned individual probes on the 95k oligonucleotide array to prepublication concatenated scaffolds of the B. rapa Chiifu-401 genome sequence (version 1.0, 255.9 Mb, representing 90% of the assembled sequences), kindly provided by Xiaowu Wang (Institute of Vegetables and Flowers-Chinese Academy of Agricultural Sciences). Probes that aligned with three or fewer locations with a match of 98% or greater within the genome sequence were then assigned to specific chromosome sequence coordinates. Therefore, we were able to distinguish cis- and trans-eQTL. cis-eQTL were defined as those whose physical position was in the same region as the marker associated with it. The region for defining cis-eQTL was taken as the midpoint between the marker associated the eQTL and the adjacent markers or the end of the chromosome for markers at the ends of chromosomes. eQTL that fell outside of this region were defined as trans-eQTL. eQTL hotspots were identified when the number of eQTL associated with a specific marker exceeded an empirical threshold. Here, the threshold was defined as the upper 99% confidence interval for the Poisson distribution, assuming an equal distribution of GEMs and eQTL.
Trait QTL associated with shoot [P], dry weight, and fresh weight at optimal and low [P]ext and measures of PUE (Hammond et al., 2009) were identified using the same 67 RILs (BraIRRI_04 subpopulation) and 125 GEMs (BraIRRI_04_2010a map). Marker mean and QTL positions were estimated in QTL Cartographer 2.0 (S. Wang, C.J. Basten, and Z.-B. Zeng, 2001–2004, Windows QTL Cartographer 2.0, Department of Statistics, North Carolina State University, Raleigh) using the zmapQTL model 6 composite interval mapping option. Five background cofactors were determined by forward stepwise regression and a 10-cM window, with genome scanning at 2 cM. This procedure estimated the LOD score and additive effect every 2 cM along each chromosome. Significant LOD thresholds were determined empirically based on 1,000 permutations and ranged from 1.54 to 2.81. The additive effect of each QTL was reported relative to the contribution of alleles from the female IMB211 parent.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. GEM map BraIRRI_04_2010a.
Supplemental Table S2. Transcripts with significantly greater abundance at low [P]ext.
Supplemental Table S3. Transcripts with significantly less abundance at low [P]ext.
Supplemental Table S4. Enrichment of GO terms in transcripts with significantly greater abundance at low [P]ext.
Supplemental Table S5. Enrichment of GO terms in transcripts with significantly less abundance at low [P]ext.
Supplemental Table S6. Variance components and H2 of Brassica transcripts.
Supplemental Table S7. Expression QTL associated with low and optimal P availability.
Supplemental Table S8. cis-eQTL associated with low and optimal P availability.
Supplemental Table S9. trans-eQTL associated with low and optimal P availability.
Supplemental Text S1. Further information on the generation of the BraIRRI_04_2010a map.
Supplementary Material
Acknowledgments
We are grateful to Xiaowu Wang, who kindly provided us with prepublication access to B. rapa Chiifu-401 genome scaffolds, and to Johan van Ooijen for discussions on mapping strategy.
References
- Allender C, King GJ. (2010) Clarifying the origins of the amphiploid species Brassica napus L. using chloroplast and nuclear molecular markers. BMC Plant Biol 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS. (2005) Phosphate-limited oat: the plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Bentsink L, Yuan K, Koornneef M, Vreugdenhil D. (2003) The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor Appl Genet 106: 1234–1243 [DOI] [PubMed] [Google Scholar]
- Boer MP, Thissen JTM. (2009) Procedure QIBDPROBABILITIES. GenStat Release 12 Reference Manual, Part 3: Procedure Library PL20. VSN International, Hemel, UK [Google Scholar]
- Boer MP, Wright D, Feng L, Podlich DW, Luo L, Cooper M, van Eeuwijk FA. (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177: 1801–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hackett CA, Niks RE, Hedley PE, Booth C, Druka A, Marcel TC, Vels A, Bayer M, Milne I, et al. (2010) An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE 5: e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis BR, Smith AB, Coombes N. (2006) On the design of early generation variety trials with correlated data. J Agric Biol Environ Stat 11: 381–393 [Google Scholar]
- DeCook R, Lall S, Nettleton D, Howell SH. (2006) Genetic regulation of gene expression during shoot development in Arabidopsis. Genetics 172: 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Yang M, Hu Y, Liao Y, Shi L, Xu F, Meng J. (2010) Quantitative trait loci affecting seed mineral concentrations in Brassica napus grown with contrasting phosphorus supplies. Ann Bot (Lond) 105: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Bonar N, Druka I, Zhang L, Marshall DF, Steffenson BJ, Close TJ, Wise RP, et al. (2008) Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp. tritici in barley. Theor Appl Genet 117: 261–272 [DOI] [PubMed] [Google Scholar]
- Druka A, Potokina E, Luo Z, Jiang N, Chen X, Kearsey M, Waugh R. (2010) Expression quantitative trait loci analysis in plants. Plant Biotechnol J 8: 10–27 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZY, Shao C, Meng YJ, Wu P, Chen M. (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci 176: 170–180 [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Bowen HC, White PJ, Mills V, Pyke KA, Baker AJM, Whiting SN, May ST, Broadley MR. (2006) A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol 170: 239–260 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, Craigon DJ, Higgins J, Emmerson ZF, Townsend HJ, White PJ, May ST. (2005) Using genomic DNA-based probe-selection to improve the sensitivity of high-density oligonucleotide arrays when applied to heterologous species. Plant Methods 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ, King GJ, Bowen HC, Hayden R, Meacham MC, Mead A, Overs T, Spracklen WP, et al. (2009) Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. J Exp Bot 60: 1953–1968 [DOI] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK. (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28: 353–364 [Google Scholar]
- Iniguez-Luy FL, Lukens L, Farnham MW, Amasino RM, Osborn TC. (2009) Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor Appl Genet 120: 31–43 [DOI] [PubMed] [Google Scholar]
- John ER, Williams ER. (1995) Cyclic and Computer Generated Designs. Chapman & Hall, London [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJ, Vreugdenhil D, Koornneef M, Jansen RC. (2007) Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. (2009) Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu Rev Plant Biol 60: 93–114 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Awai K, Nakamura M, Nagatani A, Masuda T, Ohta H. (2009) Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T, Takamiya K-I, Ohta H. (2006) Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J 47: 238–248 [DOI] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D. (2002) Purple acid phosphatases of Arabidopsis thaliana: comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X. (2006a) Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Welti R, Wang X. (2006b) Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation: roles of phospholipases D ζ1 and D ζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol 142: 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, Willmitzer L. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling H-Q, Wu C, Lian X, Wu P. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu J, Yang J, Li R, Shi L, Zhang C, Long Y, Xu F, Meng J. (2009) Analysis of genetic factors that control shoot mineral concentrations in rapeseed (Brassica napus) in different boron environments. Plant Soil 320: 255–266 [Google Scholar]
- Loudet O, Chaillou S, Krapp A, Daniel-Vedele F. (2003) Quantitative trait loci analysis of water and anion contents in interaction with nitrogen availability in Arabidopsis thaliana. Genetics 163: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love CG, Graham NS, O Lochlainn S, Bowen HC, May ST, White PJ, Broadley MR, Hammond JP, King GJ. (2010) A Brassica exon array for whole-transcript gene expression profiling. PLoS ONE 5: e12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrera-Estrella L, Nussaume L, Thibaud MC. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng WM, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Olsen SR, Cole CV, Watanabe FS, Dean LA. (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA Circular 939. Gov. Printing Office, Washington DC [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson HD, Thompson R. (1971) Recovery of inter-block information when block sizes are unequal. Biometrika 58: 545–554 [Google Scholar]
- Robinson DL. (1987) Estimation and use of variance components. Statistician 36: 3–14 [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. (2006) The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11: 535–542 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ. (2006) Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Tjellström H, Andersson MX, Larsson KE, Sandelius AS. (2008) Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ 31: 1388–1398 [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. (2010) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Trick M, Cheung F, Drou N, Fraser F, Lobenhofer EK, Hurban P, Magusin A, Town CD, Bancroft I. (2009a) A newly-developed community microarray resource for transcriptome profiling in Brassica species enables the confirmation of Brassica-specific expressed sequences. BMC Plant Biol 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Long Y, Meng J, Bancroft I. (2009b) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotech J 7: 334–346 [DOI] [PubMed] [Google Scholar]
- van der Rest B, Boisson AM, Gout E, Bligny R, Douce R. (2002) Glycerophosphocholine metabolism in higher plant cells: evidence of a new glyceryl-phosphodiester phosphodiesterase. Plant Physiol 130: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen (2006) JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma, Wageningen, The Netherlands [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. (2009) Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol 51: 663–674 [DOI] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA. (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Hammond JP. (2008) Phosphorus nutrition of terrestrial plants. White PJ, Hammond JP, , The Ecophysiology of Plant-Phosphorus Interactions. Springer, Dordrecht, The Netherlands, pp 51–81 [Google Scholar]
- Williams PH, Hill CB. (1986) Rapid-cycling populations of Brassica. Science 232: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Wu J, Yuan YX, Zhang XW, Zhao J, Song X, Li Y, Li X, Sun R, Koornneef M, Aarts MGM, Wang XW. (2008) Mapping QTLs for mineral accumulation and shoot dry biomass under different Zn nutritional conditions in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Soil 310: 25–40 [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Ding GD, Shi L, Feng J, Xu F, Meng J. (2010) Quantitative trait loci for root morphology in response to low phosphorus stress in Brassica napus. Theor Appl Genet 121: 181–193 [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Jamar DCL, Lou P, Wang YH, Wu J, Wang XW, Bonnema G, Koornneef M, Vreugdenhil D. (2008) Quantitative trait loci analysis of phytate and phosphate concentrations in seeds and leaves of Brassica rapa. Plant Cell Environ 31: 887–900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.