Orthophosphate (Pi) is an essential macronutrient that plays a central role in virtually all major metabolic processes in plants, particularly photosynthesis and respiration. Many metabolites are Pi monoesters, whereas the phosphoanhydride bonds of compounds such as ATP function to transfer energy from the energy-yielding process of photo-, oxidative, and substrate-level phosphorylation to the energy-dependent cellular processes of biosynthesis, ion pumping, and mechanical work. The massive use of Pi-containing fertilizers in agriculture demonstrates how the soluble Pi level of many soils is suboptimal for crop growth. Accessible reserves of rock phosphate—our major source of Pi fertilizers—are projected to be exhausted by the end of this century (Vance et al., 2003). The use of Pi fertilizers is also quite inefficient with less than 20% of applied Pi being absorbed by plants during their first growing season. The remaining Pi becomes immobile in the soil or leaches into and pollutes nearby surface waters. Agricultural Pi runoff is a primary factor in the eutrophication of lakes and marine estuaries, and has also resulted in blooms of toxic cyanobacteria. With the world’s population continuing its rapid increase, mankind faces a daunting challenge to produce sufficient food crops in the face of dwindling supplies of Pi fertilizers. A more comprehensive understanding of the biochemical and physiological mechanisms of plant Pi uptake and use is leading to the development of rational strategies and molecular tools for engineering nutrient-efficient cultivars needed to reduce agriculture’s overreliance on unsustainable Pi fertilizers. The aim of this Update article is to consider the influence of Pi nutrition on plant metabolism, with a focus on adaptive metabolic responses that serve to ameliorate the negative side effects of Pi deficiency. Examples of how metabolic Pi scavenging and recycling, and the unique flexibility of plant metabolism and bioenergetics may contribute to the survival of Pi-deficient (−Pi) plants are highlighted.
TRANSCRIPTIONAL VERSUS POSTTRANSCRIPTIONAL RESPONSES OF PLANT METABOLISM TO PHOSPHATE STARVATION
Plants have evolved the ability within species-dependent limits to acclimatize to extended periods of Pi deprivation by eliciting a complex array of morphological, physiological, and biochemical/metabolic adaptations collectively known as the Pi-starvation response (PSR). The PSR arises in part from the coordinated induction of hundreds of Pi-starvation inducible (PSI) genes encoding enzymes that reprioritize internal Pi use and maximize external Pi acquisition (Vance et al., 2003; Ticconi and Abel, 2004; Fang et al., 2009; Lin et al., 2009; Nilsson et al., 2010). As is well documented throughout this Focus Issue, many elements of the PSR are controlled at the transcriptional and translational level, and −Pi plants extensively remodel their transcriptome and proteome in ways that coordinate the requisite metabolic and morphological adaptations. Large collections of microarray data regarding plant, and particularly Arabidopsis (Arabidopsis thaliana), transcriptional responses to Pi starvation have: (1) shed light on the molecular identity and regulation underlying many classical biochemical and physiological adaptations to Pi deprivation, and (2) revealed that PSI gene expression is highly coordinated in a temporal and tissue-specific manner (Fang et al., 2009; Lin et al., 2009; Nilsson et al., 2010).
Recent studies are also emphasizing the importance of posttranscriptional mechanisms in the control of PSI enzyme expression and activity. This is reflected by proteomic profiling of −Pi rice (Oryza sativa), corn (Zea mays), and Arabidopsis, demonstrating that transcript abundance of various genes is not always indicative of protein accumulation during Pi deprivation (Fukuda et al., 2007; Li et al., 2008a; Tran and Plaxton, 2008). One of the best-characterized examples of posttranscriptional mechanisms in the Arabidopsis PSR is the regulatory module comprising the transcription factor PHR1, PHO2 that encodes the E2 ubiquitin conjugase UBC24, the microRNA399 (miR399), and the noncoding RNA At4 (Bari et al., 2006; Fang et al., 2009; Lin et al., 2009; Nilsson et al., 2010). miR399 regulates Pi homeostasis by controlling UBC24 expression. UBC24 functions during Pi sufficiency to promote the proteolytic turnover of PSI proteins, including high-affinity Pi transporters of the plasmalemma. Pi starvation induces PHR1 and activates expression of the phloem-mobile miR399. Binding of miR399 to complementary bases of UBC24 transcripts leads to the destruction of UBC24 mRNA, resulting in low levels of UBC24’s E2 ubiquitin conjugase activity, and the consequent accumulation of its downstream protein targets (Bari et al., 2006). Shoot-derived miR399 thus serves as a long-distance signal to suppress the expression of UBC24 in the roots. Expression of the ribo-regulator At4 is strongly induced during prolonged periods of Pi starvation (Fang et al., 2009; Lin et al., 2009; Nilsson et al., 2010). At4 binds to complementary bases of miR399, thereby inhibiting its silencing of UBC24 mRNA. This allows UBC24 levels to rapidly adjust to the dynamic balance of Pi supply and demand. High-throughput deep sequencing has identified additional miRNAs that control various genes involved in the Arabidopsis PSR (Hsieh et al., 2009). For example, miR165, miR778, miR827, and miRNA2111 are all highly induced in −Pi Arabidopsis, whereas miR169, miR395, and miR398 are repressed (Hsieh et al., 2009). Cross talk between these miRNAs may coordinate their expression under specific nutrient deficiencies, thereby facilitating plant survival in −Pi environments (Hsieh et al., 2009). Future challenges include the need to identify the specific targets of the various miRNAs, as well as to pinpoint the molecular mechanisms by which they control cellular Pi homeostasis.
Posttranslational mechanisms of metabolic control are another important feature of the PSR and include Pi’s role as an allosteric activator or inhibitor of many key control enzymes of intermediary plant metabolism (Plaxton and Podestá, 2006). This is illustrated by the major regulatory enzyme of starch biosynthesis, ADP-Glc pyrophosphorylase, which displays potent allosteric inhibition by Pi. Transgenic potato (Solanum tuberosum) plants expressing a Pi-insensitive bacterial ADP-Glc pyrophosphorylase overproduce starch (Stark et al., 1992). The results demonstrated that allosteric control of this plastidic enzyme, not the amount of its protein subunits, is the major physiological determinant of starch biosynthesis (Stark et al., 1992). Thus, the well-documented accumulation of starch by −Pi plant cells may largely arise from the release of ADP-Glc pyrophosphorylase from allosteric inhibition by Pi, owing to the large (up to 50-fold) reductions in cytoplasmic Pi pools that accompany long-term Pi deprivation (Duff et al., 1989; Vance et al., 2003). Similarly, PSI vacuolar acid phosphatase (APase) isozymes display potent product inhibition by Pi (Bozzo et al., 2004a; Veljanovski et al., 2006; Tran et al., 2010a). The vacuolar Pi concentration of Pi-sufficient plants (>10 mm) will exert significant feedback inhibition on their APase activity. Conversely, the depletion of vacuolar Pi pools that accompanies extended Pi deprivation (Fang et al., 2009) will effectively relieve the inhibition of PSI APases by Pi, thus contributing to their enhanced activity in vivo. It is evident that a complete description of the metabolic adjustments of −Pi plants not only requires the characterization of how Pi serves to control gene expression and protein turnover, but also how physiologically relevant changes in Pi levels modulate the activities of preexisting enzymes.
As discussed below, protein phosphorylation and glycosylation are emerging as essential posttranslational modifications that control the activity of and/or subcellular targeting of diverse enzymes up-regulated by −Pi plants (Gregory et al., 2009; Tran et al., 2010b). A challenging goal will be to document the functional consequences of posttranslational modifications such as reversible protein phosphorylation in the signaling and metabolic pathways involved in plant acclimation to nutritional Pi deprivation.
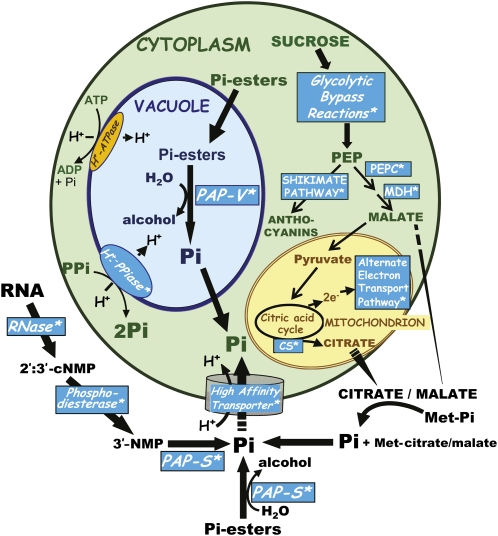
PHOSPHATE RECYCLING, MOBILIZATION, AND SCAVENGING DURING NUTRITIONAL PHOSPHATE DEPRIVATION
A common feature of the plant PSR is the development of dark-green or purple shoots due to anthocyanin accumulation brought about by PSI anthocyanin biosynthetic enzymes (Vance et al., 2003; Fang et al., 2009). Anthocyanins are a class of red/purple-colored flavonoids that can protect nucleic acids from UV damage and chloroplasts from photoinhibitory damage (Zeng et al., 2010). The up-regulation of high-affinity Pi transporters of the plasma membrane is another important component of the plant PSR (Fig. 1). These transporters are energized by ATP-dependent proton efflux, and actively assimilate Pi against a steep concentration gradient, as the soluble Pi concentration in the rhizosphere can be up to 10,000-fold lower than that of root cells. High-affinity Pi transporters of Arabidopsis belong to the nine-member PHT1 family and consist of H+/Pi symporters with 12 membrane-spanning domains (Fang et al., 2009; Lin et al., 2009). While all nine members are responsive to Pi starvation, each appears to exhibit a certain degree of tissue-specific expression, with some expressed in epidermal and root hair cells while others are expressed in stelar cells of the root. Consistent with this, Pht1;4 or Pht1;1 knockout plants had decreased Pi acquisition during Pi deficiency, while Pht1;1 (pho1) loss-of-function mutants fail to accumulate Pi in shoots during Pi sufficiency due to its role in xylem Pi loading (Fang et al., 2009; Lin et al., 2009).
Figure 1.
A model suggesting various adaptive metabolic processes (indicated by asterisks) that may help plants acclimatize to nutritional Pi deficiency. Alternative pathways of cytosolic glycolysis, mitochondrial electron transport, and tonoplast H+ pumping facilitate respiration and vacuolar pH maintenance by −Pi plant cells because they negate the dependence on adenylates and Pi, the levels of which become markedly depressed during severe Pi starvation. Large quantities of organic acids produced by PEPC, malate dehydrogenase (MDH), and citrate synthase (CS) may also be excreted by roots to increase the availability of: (1) mineral-bound Pi (by solubilizing calcium, iron, and aluminum phosphates = Met-Pi), and (2) organic P and its amenability to hydrolysis by secreted PAPs. During Pi deprivation vacuolar PAPs (PAP-V) are up-regulated to recycle Pi from nonessential intracellular Pi monoesters. Similarly, secreted PAPs (PAP-S) scavenge Pi from extracellular Pi monoester and nucleic acid fragment pools for its eventual uptake by PSI high-affinity Pi transporters of the plasma membrane.
Plants also increase the efficiency of Pi use during Pi starvation via up-regulation of a wide array of PSI hydrolases that scavenge and recycle Pi from intra- and extracellular organic phosphorus (P) compounds (Vance et al., 2003; Fang et al., 2009; Nilsson et al., 2010; Tran et al., 2010a). Thus, Arabidopsis plants cultivated on RNA as their sole source of exogenous Pi grow just as well as Pi-fertilized control plants, whereas a pdr1 mutant, which is defective in the induction of multiple PSI genes, requires Pi supplementation (Ticconi and Abel, 2004). This suggests that nucleic acids present in the soil’s decaying organic matter represent an important source of nutritional Pi that can be exploited by −Pi plants. The induction of secreted ribonucleases (RNases), nucleases, phosphodiesterases, and APases function in the systematic catabolism of soil-localized nucleic acids and their degradation products to mobilize Pi, which is made available for root uptake by high-affinity Pi transporters (Fig. 1; Ticconi and Abel, 2004; Fang et al., 2009). RNS1 is a PSI RNase isozyme that was the most abundant protein accumulating in the secretome of −Pi Arabidopsis suspension cells (Tran and Plaxton, 2008). In addition, PSI cyclic nucleotide phosphodiesterases function in concert with secreted RNases and APases to scavenge Pi from soil-localized nucleic acids (Ticconi and Abel, 2004).
Pi-starved plants also scavenge and conserve Pi by replacing their membrane phospholipids with amphipathic sulfolipids and galactolipids. A reduction in the phospholipid content of −Pi plant membranes coincides with increases in sulfolipid sulfoquinovosyldiacylglycerol and galactolipid digalactosyldiacylglycerol membrane lipids. SQD1 and SQD2 are PSI enzymes required for sulfolipid biosynthesis in −Pi Arabidopsis (Fang et al., 2009; Lin et al., 2009). An Arabidopsis sqd2 T-DNA insertional mutant showed reduced growth under −Pi conditions (Yu et al., 2002), which proves that sulfolipids are an important substitute for phospholipids during Pi deprivation. PLDζ1 and PLDζ2 are PSI phospholipases that catabolize phospholipids in −Pi Arabidopsis. Galactolipid digalactosyldiacylglycerol accumulation during Pi deprivation is reduced in the roots of −Pi pldζ1 single and pldζ1/pldζ2 double mutants (Li et al., 2006). PLDζ generates phosphatidic acid that can be dephosphorylated by an APase to release Pi and diacylglycerol. PLDζ1 and PLDζ2 also appear to be involved in regulating alterations in root architecture during Pi-limited growth. Phosphatidic acid was suggested to serve as a second messenger that activates a protein kinase-mediated protein phosphorylation cascade that controls root growth. In contrast to PLDζ function in roots, a nonspecific phospholipase C5 is responsible for phospholipid degradation in leaves during Pi starvation (Gaude et al., 2008).
A universal plant response to Pi deprivation is the up-regulation of intracellular (vacuolar) and secreted APases, enzymes that hydrolyze Pi from a broad and overlapping range of Pi monoesters with an acidic pH optimum (Fig. 1). As organic P typically constitutes about 50% of the total P in soil and is the predominant form of P found in soil solutions (Richardson, 2009), secreted APases are believed to function in Pi scavenging by roots of −Pi plants. This is reflected by the ability of Arabidopsis and other plants to scavenge and assimilate nutritional Pi from a broad range of exogenous organic P substrates, including RNA, DNA, ATP, 3-phosphoglycerate, and various hexose Ps such as Glc-6-P (Ticconi and Abel, 2004; Richardson, 2009; Liang et al., 2010). PSI APases are also secreted into the extracellular matrix and intercellular spaces (apoplast) of −Pi plant tissues (Barrett-Lennard et al., 1993; Kaida et al., 2009; Tran et al., 2010a). These APases have been hypothesized to function in Pi recycling from organic P compounds leaked from the −Pi cells (Barrett-Lennard et al., 1993). Similarly, intracellular (vacuolar) PSI APases scavenge and remobilize Pi from expendable intracellular Pi monoesters and anhydrides (Fig. 1). This is accompanied by marked reductions in levels of cytoplasmic P metabolites during extended Pi deprivation (Duff et al., 1989; Vance et al., 2003).
FEEDING HUNGRY PLANTS: THE ROLE OF PURPLE ACID PHOSPHATASES IN PHOSPHATE NUTRITION
Purple APases (PAPs) comprise the largest class of plant APases. They exhibit a distinctive purple or pink color in solution that results from a charge transfer transition at about 560 nm from the metal-coordinating Tyr to the metal ligand Fe(III) (Tran et al., 2010a). Most PAPs are nonspecific APases that hydrolyze Pi from a broad spectrum of Pi monoesters over a wide pH range. However, mammalian PAPs expressed in macrophages and spleen cells after phagocytosis likely function in the generation of reactive oxygen species via a Fenton reaction involving the Fe(II) of the active site (Tran et al., 2010a). Similarly, (1) several plant PAPs that exhibit significant APase activity also have alkaline peroxidase activity, and (2) overexpression of a soybean (Glycine max) PAP, GmPAP3, increased tolerance to oxidative damage imposed during salinity stress (Li et al., 2008b). Mammalian and bacterial PAPs also function as phosphotyrosyl phosphatases, implying a role in signal transduction (Tran et al., 2010a). Likewise, a PAP (NtPAP12) of tobacco (Nicotiana tabacum) cell walls is highly active against phosphotyrosylated peptides (Kaida et al., 2009), whereas a variety of APases from other plant sources have significant activity with phosphotyrosine or other phosphoamino acids as substrates (Bozzo et al., 2002, 2004a; Veljanovski et al., 2006). Interestingly, transgenic expression of NtPAP12 resulted in altered cell wall composition and enhanced β-glucan synthase activity, suggesting that this tobacco PAP isozyme might function as a protein phosphatase involved in the control of cell wall biosynthesis (Kaida et al., 2009).
Many studies have focused on the role that individual PAP isozymes play in intra- and/or extracellular Pi scavenging and recycling during Pi starvation. The transcription factors PHR1, WRKY75, and ZAT6 have been implicated in the control of PAP expression in −Pi Arabidopsis, while other studies have revealed PSI PAPs whose synthesis is controlled by posttranscriptional mechanisms (Veljanovski et al., 2006; Tran and Plaxton, 2008; Lin et al., 2009; Tran et al., 2010b). In contrast, Pi resupply to −Pi plants quickly represses PSI PAP genes while inducing specific proteases that target intracellular and secreted PSI PAPs (Bozzo et al., 2004b; Nilsson et al., 2010; Tran et al., 2010a). Identification and characterization of PSI PAPs is required to define the molecular mechanisms underlying this biochemical hallmark of the plant PSR, as well as to identify suitable targets for improving crop Pi acquisition. PSI PAPs have been biochemically characterized from several species including tomato (Solanum lycopersicum), lupin (Lupinus albus), bean (Phaseolus vulgaris), tobacco, and Arabidopsis (Bozzo et al., 2002, 2004a, 2006; Vance et al., 2003; Veljanovski et al., 2006; Tran et al., 2010a, 2010b). White lupin secretes abundant amounts of an APase known as LaSAP2 from its proteoid cluster roots when subjected to Pi starvation (Wasaki et al., 2000). LaSAP2 shares 65% amino acid sequence identity with AtPAP12, a secreted PSI Arabidopsis PAP isozyme (Tran et al., 2010a, 2010b). This similarity extends to its promoter region, where binding by PHR1 can direct enhanced LaSAP2 expression under −Pi conditions. By contrast, a secreted APase of −Pi yellow lupin roots is orthologous to AtPAP26, a dual-targeted (vacuolar and secreted) PAP up-regulated by −Pi Arabidopsis (Veljanovski et al., 2006; Tran et al., 2010a, 2010b). As with AtPAP26 (see below), the lupin AtPAP26 ortholog is constitutively transcribed irrespective of the plant’s nutritional Pi status.
The Arabidopsis genome encodes 29 putative PAP isozymes whose transcriptional expression is dependent upon various developmental and environmental factors (Tran et al., 2010a). In contrast to the abundance of PAP genomic and transcript expression data, comparatively little information is available on the identity and biochemical features of specific PAP isozymes that contribute to intra- versus extracellular Pi scavenging by −Pi plants. AtPAP17 was one of the first PSI PAPs to be purified and characterized from −Pi Arabidopsis. It exists as a low-molecular-mass (34 kD) monomeric PAP and is transcriptionally induced in roots and leaves of −Pi Arabidopsis (del Pozo et al., 1999). This suggests that AtPAP17 might be involved in the metabolism of reactive oxygen species during general stress, rather than playing significant Pi recycling or scavenging roles in −Pi Arabidopsis. The determination of AtPAP17’s subcellular localization and the phenotypic impact that modifying its expression has on transgenic Arabidopsis will help to fully establish its function(s) during Pi deprivation or senescence.
Recent biochemical and functional genomic studies have established the dual-targeted AtPAP26 as the predominant intracellular (vacuolar) APase, as well as a major secreted APase isozyme up-regulated by −Pi Arabidopsis (Veljanovski et al., 2006; Hurley et al., 2010; Tran et al., 2010b). Transient expression of an AtPAP26-GFP fusion construct coupled with imaging via epifuorescence microscopy confirmed AtPAP26’s vacuolar localization (Hurley et al., 2010). A homozygous atpap26 T-DNA insertional mutant exhibited a large reduction in extractable shoot and root, and root-secreted APase activities, as well as impaired development when subjected to Pi deficiency (Hurley et al., 2010). The results demonstrated that AtPAP26 is a principal contributor to PSI APase activity, and that it plays an important role in the Pi metabolism of −Pi Arabidopsis.
Two PAP isozymes secreted into the culture media by −Pi Arabidopsis suspension cells were purified, identified by mass spectrometry, and characterized as an AtPAP12 homodimer and AtPAP26 monomer composed of glycosylated 60- and 55-kD subunits, respectively (Tran et al., 2010b). AtPAP12 and AtPAP26 both exhibited a high specific APase activity, broad substrate selectivity, and overlapping, but nonidentical pH-activity profiles. Their combined activities endow −Pi Arabidopsis with an efficient biochemical machinery for scavenging Pi from external Pi monoesters located in the rhizosphere or apoplastic environment. A surprising outcome of this research was that secreted AtPAP26 of −Pi Arabidopsis exists as a pair of kinetically distinct glycoforms that differentially bind to lectins such as jack bean (Canavalia ensiformis) Concanavalin-A and snow drop (Galanthus nivalis) agglutinin (Tran et al., 2010b). Glycosylation is an important posttranslational modification that influences enzyme localization, stability, and/or kinetic properties. The determination of glycan structures and linkages at each N-linked glycosylation site is required to test the hypothesis that the dual targeting and kinetic differences of the vacuolar and secreted AtPAP26 glycoforms of −Pi Arabidopsis arise from differential glycosylation. Although vacuolar and secreted AtPAP26 polypeptides are markedly up-regulated by −Pi Arabidopsis, AtPAP26 transcripts are relatively abundant and invariant irrespective of nutritional Pi status (Veljanovski et al., 2006; Hurley et al., 2010; Tran et al., 2010b). Several proteomic studies subsequently documented a variety of intracellular and secreted proteins that are also controlled posttranscriptionally mainly at the level of protein accumulation in plants responding to changes in environmental Pi availability (Fukuda et al., 2007; Li et al., 2008a; Tran and Plaxton, 2008). This highlights the need to integrate transcript profiling with parallel biochemical and proteomic analyses of the plant PSR, as the combined datasets will provide a more robust depiction of how alterations in gene expression may be linked to adaptive changes in the metabolism of −Pi plants.
Analysis of atpap12 and atpap26 T-DNA single mutants, and an atpap12/atpap26 double mutant has confirmed that AtPAP12 and AtPAP26 account for most of the root-secreted APase activity of −Pi wild-type seedlings (Tran et al., 2010b), and that they play an essential role in Pi scavenging during cultivation on nucleic acids and Pi monoesters as the sole source of exogenous P (W.D. Robinson, H.T. Tran, and W.C. Plaxton, unpublished data). Although secreted AtPAP12 or AtPAP26 of −Pi Arabidopsis cannot hydrolyze Pi from phytic acid (inositol hexaphosphate), products of Pi hydrolysis from phytic acid (e.g. inositol P) were not tested and should be assessed in future studies. Phytase activity has been reported to constitute less than 1% of the total APase activity of Arabidopsis root extracts, and Arabidopsis is unable to acquire P from exogenous phytate owing to the absence of an extracellular phytase (Richardson, 2009). AtPAP15 is the only member of the AtPAP family that has been shown to possess both APase and phytase activity, and appears to play an important role in mobilizing Pi from phytate reserves during seed or pollen germination (Wang et al., 2009). Since its expression is unresponsive to Pi deprivation and does not occur in root hair or epidermal cells, AtPAP15 does not appear to function in extracellular Pi scavenging. However, overexpression of AtPAP15 containing a carrot (Daucus carota) extracellular-targeting peptide significantly improved the growth and Pi use efficiency of transgenic soybean plants cultivated on sand containing phytate as their sole source of P (Wang et al., 2009). By contrast, transgenic plants overexpressing secreted phytases showed no advantage in their growth or Pi nutrition when cultivated in various agricultural soils (Richardson, 2009; Tran et al., 2010a). This indicates that the bulk of soil organic P accessible to secreted PAPs may not be an effective substrate for transgenic plants that secrete phytase.
The biochemical characterization of intra- and extracellular PSI tomato PAP isozymes indicated their probable role in Pi scavenging and recycling by −Pi tomato (Bozzo et al., 2002, 2004a). Decreased cytoplasmic Pi, a consequence of prolonged Pi starvation is met by a highly specific response that involves temporal and tissue-specific synthesis of several PSI tomato PAP isozymes (Bozzo et al., 2006). An APase recently identified as PvPAP3 by mass spectrometry was highly induced in both leaves and roots of −Pi bean plants (Liang et al., 2010). The transient expression of 35S:PvPAP3-GFP constructs in onion (Allium cepa) epidermal cells indicated that it is secreted into the apoplast (Liang et al., 2010). PvPAP3 was most active with ATP as a substrate, suggesting that it might function in acclimation to Pi starvation through the use of extracellular ATP as a Pi source from the environment. Both animal and plant cells secrete ATP into the extracellular matrix, and extracellular ATP is essential for maintaining plant cell viability (Tran et al., 2010a).
PAPs are obvious targets for engineering Pi-efficient crops, as they play a crucial role in Pi recycling and scavenging by −Pi plants. Owing to microbial activity, the application of Pi fertilizers increases both the inorganic and organic P content of agricultural soils, thereby influencing the amount of organic P that is available for PAP hydrolysis (Richardson, 2009). It is therefore of considerable interest to determine whether the efficiency of Pi fertilizer application can be improved by the overexpression of secreted PAPs such as AtPAP12 and AtPAP26 in genetically engineered crop plants. Indeed, the secreted AtPAP12 ortholog of −Pi white lupin (LaSAP2) was recently overexpressed in tobacco (Wasaki et al., 2009). The transgenic plants exhibited enhanced Pi uptake and growth during cultivation on −Pi soils. This was attributed to LaSAP2 having several advantages in soil, including stability over a broad range of pH values and temperatures, broad substrate specificity, and a long half-life in soil suspension (Wasaki et al., 2009).
INDUCED SYNTHESIS AND EXCRETION OF ORGANIC ACIDS
An important component of the plant PSR is root excretion of organic acids into the rhizosphere (Fig. 1), which results in rhizosphere acidification. The mechanism responsible for the release of organic acids (which are dissociated into their anionic forms at cytoplasmic pH) was unknown until plasma membrane H+-pumping ATPases were shown to be involved in plant adaptation to Pi starvation (Yan et al., 2002). This has been correlated with the up-regulation of novel membrane channels needed to transport anions such as citrate and malate from root cells into the rhizosphere (Diatloff et al., 2004). Thus, the current model of organic acid excretion comprises two separate transport processes, one involving an active H+ efflux driven by a plasma membrane H+-ATPase, and the second entailing channel-like transporters that allow the passive efflux of organic anions (Yan et al., 2002; Diatloff et al., 2004). Organic acid excretion results in the chelation of metal cations that immobilize Pi (e.g. Al3+, Ca2+, Fe2/3+), thus increasing soil solution Pi concentrations by up to 1,000-fold. Organic acid excretion also: (1) mobilizes Pi bound in humic-metal complexes, (2) enhances both the solubility of organic P and its amenability to dephosphorylation by secreted PAPs, and (3) may also promote the growth of symbiotic rhizosphere microbes that facilitate root Pi acquisition (Vance et al., 2003; Ticconi and Abel, 2004; Fang et al., 2009; Richardson, 2009). The amount of carbon (C) exuded as organic acids can be enormous, ranging from 10% to greater than 25% of the total plant dry weight (Vance et al., 2003). Enhanced synthesis of organic acids by −Pi plants has been correlated with up-regulation of phosphoenolpyruvate (PEP) carboxylase (PEPC; and its activation by reversible phosphorylation), malate dehydrogenase, and citrate synthase (Fig. 1), and elevated rates of dark CO2 fixation (Vance et al., 2003; Gregory et al., 2009). Overexpression of a maize PEPC gene increased organic acid synthesis and excretion in rice (Fang et al., 2009). Similarly, overexpression of mitochondrial citrate synthase in either carrot or Arabidopsis plants resulted in enhanced levels of citrate excretion from roots and improved growth of plants cultivated in media containing insoluble aluminum Pi or in −Pi acidic soil (Fang et al., 2009).
METABOLIC FLEXIBILITY OF HUNGRY PLANTS
A striking feature of plant metabolism is that the same step in a metabolic pathway can frequently be accomplished in a variety of different ways. This metabolic flexibility is reflected by genetic engineering experiments in which gene-silencing technologies were used to partially or fully eliminate an enzyme traditionally considered to be essential and yet the resulting transgenic plants were able to grow and develop more or less normally (Plaxton and Podestá, 2006). Metabolic redundancy represents an essential component of the biochemical adaptations of plants and allows them to respond dynamically to their constantly changing and stressful environment. As a consequence of the large decline in cytoplasmic Pi levels that follows severe Pi stress, large (up to 80%) reductions in intracellular levels of ATP, ADP, and related nucleoside Ps also occur (Duff et al., 1989; Plaxton and Podestá, 2006). This is expected to inhibit C flux through the enzymes of classical glycolysis that are dependent upon adenylates or Pi as cosubstrates (Fig. 2). Despite depleted intracellular Pi and adenylate pools, −Pi plants must continue to maintain respiratory C flux to generate energy and C skeletons required for key anabolic processes.
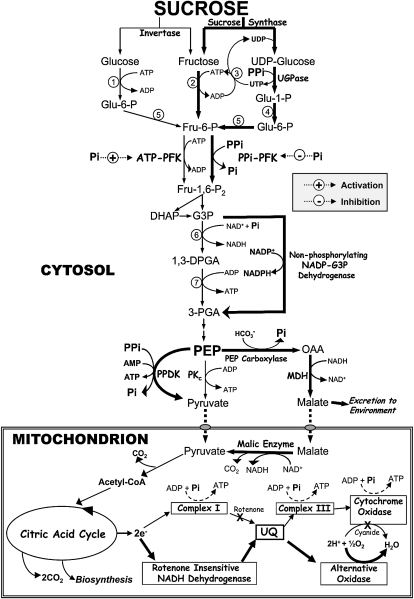
Figure 2.
Alternative pathways of cytosolic glycolysis and mitochondrial electron transport (indicated by bold arrows) that may promote the survival of Pi-deprived plants. A key component of this model is the critical role played by PPi-dependent glycolytic bypass enzymes and metabolic Pi recycling systems during Pi deprivation. Enzymes that catalyze the numbered reactions are as follows: 1, hexokinase; 2, fructokinase; 3, nucleoside diphosphate kinase; 4, phosphoglucose mutase; 5, phosphoglucose isomerase; 6, NAD-dependent glyceraldehyde-3-P dehydrogenase (phosphorylating); 7, 3-phosphoglycerate kinase. Abbreviations are as described in the text or as follows: DHAP, dihydroxyacetone-P; G3P, glyceraldehyde-3-P; 1,3-DPGA, 1,3-P2-glycerate; 3-PGA, 3-phosphoglycerate; MDH, malate dehydrogenase; OAA, oxaloacetate; UGPase, UDP-Glc pyrophosphorylase; UQ, ubiquinone.
ALTERNATIVE REACTIONS OF CYTOSOLIC GLYCOLYSIS AND TONOPLAST PROTON PUMPING CONTRIBUTE TO THE METABOLIC FLEXIBILITY OF PHOSPHATE-STARVED PLANTS
At least six Pi- and adenylate-independent glycolytic bypass enzymes in addition to the inorganic pyrophosphate (PPi)-dependent H+-pump (H+-PPiase) of the tonoplast membrane have been reported to be significantly up-regulated by −Pi plant cells (Figs. 1 and 2; Duff et al., 1989; Palma et al., 2000; Vance et al., 2003; Plaxton and Podestá, 2006). These PSI bypasses facilitate glycolytic flux and vacuolar pH maintenance during severe Pi stress when the intracellular levels of adenylates and Pi are very low. Several of these bypasses employ PPi to perform cellular work, while simultaneously conserving ATP and recycling Pi (Figs. 1 and 2). Bypass enzymes such as PPi-dependent phosphofructokinase (PPi-PFK), PEPC, pyruvate Pi dikinase (PPDK), and the tonoplast H+-PPiase also promote intracellular Pi recycling, as Pi is a by-product of their reaction.
The PEPC-malate dehydrogenase-malic enzyme glycolytic bypass allows −Pi plants to maintain PEP to pyruvate flux when the activity of cytosolic pyruvate kinase (PKc) becomes ADP limited (Fig. 2). Metabolite determinations and kinetic studies of PKc and PEPC in Catharanthus roseus suggested that the contribution of PEPC to the metabolism of PEP increased in −Pi cells in vivo (Nagano et al., 1994). Further evidence for the operation of this PKc bypass in −Pi C. roseus was provided by the rapid release of 14CO2 from organic compounds derived from [14C]malate and fixed H14CO3 (Nagano et al., 1994). Levels of PEPC, malate dehydrogenase, and malic enzyme transcripts, protein, and/or enzymatic activity are markedly up-regulated in response to Pi deprivation in a wide range of plant species (Vance et al., 2003; Plaxton and Podestá, 2006). Recent developments have clarified the molecular basis of the PEPC response to Pi stress in Arabidopsis cell cultures and seedlings, rice leaves, and white lupin proteoid roots, which involves the up-regulation of specific PEPC and PEPC protein kinase isozymes under the control of multiple transcription factors. The simultaneous induction and in vivo phosphorylation activation of the PEPC isozyme AtPPC1 at its conserved Ser-11 phosphorylation site was recently confirmed to be an integral component of the Arabidopsis PSR (Gregory et al., 2009). To our knowledge, this study provided the first unequivocal evidence that regulatory enzyme phosphorylation contributes to the plant PSR, and is consistent with the observation that the most prominent PSI genes of Arabidopsis include the PEPC protein kinase-encoding genes AtPPCK1 and AtPPCK2 (Fang et al., 2009; Gregory et al., 2009; Lin et al., 2009; Nilsson et al., 2010). BHLH32 is a transcription factor that functions as a negative regulator of several PSI genes in Arabidopsis, including AtPPCK1 and AtPPCK2 (Chen et al., 2007). Characterization of additional components of the signal transduction pathways that link nutritional Pi status with the control of AtPPC1, AtPPCK1, and AtPPCK2 transcription and translation, as well as AtPPC1, and AtPPCK1, and AtPPCK2 turnover will be a fruitful area for future research.
THE USE OF INORGANIC PYROPHOSPHATE CONSERVES ATP DURING PHOSPHATE STARVATION
PPi is a by-product of a host of biosynthetic reactions, including the polymerization reactions involved in the final steps of macromolecule synthesis. The large amounts of PPi produced during biosynthesis are not always wasted, but can be employed by various anaerobic microorganisms as well as the plant cytosol to enhance the energetic efficiency of several cellular processes. In contrast to animals, the plant cytosol lacks soluble PPiase and thus contains PPi concentrations of up to 0.5 mm. Furthermore, plant cytosolic PPi levels are remarkably insensitive to abiotic stress such as anoxia or Pi starvation that elicit significant reductions in cellular ATP pools (Duff et al., 1989; Plaxton and Podestá, 2006; Huang et al., 2008). The mechanisms of cytosolic PPi homeostasis in stressed versus nonstressed plant cells remain elusive. Anabolism, and hence the rate of PPi production, would be reduced under stressed conditions such as anoxia or Pi starvation. However, PPi would continue to be generated during the biosynthesis of essential macromolecules needed to support diminished growth and/or to replace those that have become damaged.
That PPi-powered processes may be a crucial facet of the metabolic adaptations of plants to environmental extremes that lead to depressed ATP (but not PPi) pools is indicated by the significant up-regulation of Suc synthase, UDP-Glc pyrophosphorylase, PPi-PFK, PPDK, and the tonoplast H+-PPiase by anoxia or extended Pi starvation (Duff et al., 1989; Palma et al., 2000; Plaxton and Podestá, 2006; Huang et al., 2008). As Pi exerts reciprocal allosteric effects on the activity of ATP-PFK (potent activator) and PPi-PFK (potent inhibitor; Fig. 2; Plaxton and Podestá, 2006), the large reduction in cytoplasmic Pi pools of −Pi plants should promote the in vivo activity of PPi-PFK while curtailing that of ATP-PFK. This has been corroborated by a study of transgenic tobacco plants overexpressing 6-phosphofructo-2-kinase that concluded that the glycolytic contribution of PPi-PFK increases and ATP-PFK decreases under conditions of Pi deficiency (Fernie et al., 2002).
Alternative PPi-dependent cytosolic reactions of glycolysis and tonoplast H+ pumping confer a considerable bioenergetic advantage that can extend the survival time of plant cells that have become ATP depleted owing to environmental stresses such as Pi starvation or anoxia. This has been corroborated by studies of mutant or transgenic plants exhibiting altered levels of PPi-dependent enzymes and pathways. For example: (1) a 60% reduction in root PPi levels of transgenic potato plants overexpressing a bacterial soluble PPiase resulted in decreased plant growth and overall vitality during hypoxia stress, lower ATP levels, and an impaired ability to resume aerobic growth following 4 d of hypoxia (Mustroph et al., 2005), (2) a crucial role for Suc synthase in maintaining glycolysis in hypoxic maize roots was established by examination of mutants deficient in Suc synthase activity (Ricard et al., 1998), and (3) overexpression of the tonoplast H+-PPiase resulted in transgenic plants that outperformed controls when subjected to nutritional Pi deprivation (Yang et al., 2007). As outlined in Figures 1 and 2, the induction of PPi-dependent bypass enzymes may help plants acclimate to nutritional Pi deprivation by: (1) circumventing ATP-limited reactions, (2) conserving limited cellular pools of ATP, while (3) recycling precious Pi from PPi.
MITOCHONDRIAL ELECTRON TRANSPORT CHAIN BYPASSES OF PHOSPHATE-STARVED PLANTS
The significant reductions in intracellular Pi and ADP levels that follow extended Pi deprivation will impede respiratory electron flow through the cytochrome pathway at the sites of coupled ATP synthesis. However, the presence of nonenergy conserving pathways of mitochondrial electron transport provides a mechanism whereby respiratory flux can be maintained under conditions when the availability of ADP and/or Pi are restrictive (e.g. during severe Pi deprivation). Plants acclimate to Pi stress by the up-regulation and/or increased engagement of the nonenergy conserving (e.g. rotenone- and/or cyanide-insensitive) pathways of the mitochondrial electron transport chain (Figs. 1 and 2; Rychter and Mikulska, 1990; Plaxton and Podestá, 2006). This allows continued functioning of the mitochondrial citric acid cycle and electron transport chain with limited ATP production and may thereby contribute to the survival of −Pi plants. This has been corroborated by the impaired growth and metabolism of −Pi transgenic tobacco unable to synthesize a functional alternative oxidase (AOX; Sieger et al., 2005). Lack of AOX under Pi limitation was correlated with increased levels of proteins commonly associated with oxidative stress. It was concluded that AOX respiration provides a crucial adaptive mechanism by which plant cells can modulate their growth response to Pi availability and that AOX also has nutrient-specific roles in maintaining cellular redox and C balance (Sieger et al., 2005).
HIGH-THROUGHPUT TRANSCRIPT AND PROTEOME PROFILING INDICATES THAT PHOSPHATE STARVATION INDUCIBILITY OF GLYCOLYTIC AND MITOCHONDRIAL ELECTRON TRANSPORT BYPASS PROTEINS IS WIDESPREAD IN PLANTS
The application of high-throughput transcript and proteome profiling technologies has allowed researchers to simultaneously catalog the effects of Pi deficiency on the expression of many genes and proteins in plants such as Arabidopsis, rice, maize, bean, and white lupin. Interestingly, the enhanced expression of alternative glycolytic enzymes (such as Suc synthase, UDP-Glc pyrophosphorylase, PPi-PFK, and PPDK) and mitochondrial electron transport proteins (such as AOX) that do not require Pi or adenylates as cosubstrates has been frequently observed (Plaxton and Podestá, 2006; Fukuda et al., 2007; Li et al., 2008a; Fang et al., 2009; Nilsson et al., 2010). The differential regulation of genes involved in primary metabolism demonstrates the activation of genes involved in bypassing the ATP- and Pi-dependent enzymes, and changing respiratory metabolism required to generate ATP and C skeletons during Pi deficiency.
CONCLUDING REMARKS
Important insights into the plant PSR have been acquired through extensive transcriptomic analyses that have revealed hundreds of PSI genes encoding proteins that are believed to help −Pi plants reprioritize internal Pi use and maximize Pi acquisition from the soil (Vance et al., 2003; Ticconi and Abel, 2004; Fang et al., 2009; Lin et al., 2009; Nilsson et al., 2010). However, these results must be balanced by the fact that transcript abundance does not necessarily reflect cognate protein levels, and that transcript profiling provides no information about either the subcellular location of gene products nor posttranslational modifications that may be essential for their function, transport, and activation. Thus, a thorough understanding of metabolic aspects of the PSR will require the effective integration of transcript profiling and related genomic tools with proteomic, enzymological/biochemical, and metabolomic approaches. In particular, a better understanding of the extent to which PPi and changes in flux through alternative enzymes and pathways influence plant stress tolerance is relevant to the applied goal of engineering transgenic crops having improved resistance to environmental extremes, including Pi starvation. The continued identification, and biochemical and functional characterization of intracellular and secreted Pi scavenging and recycling enzymes up-regulated by −Pi plants will also contribute to the development of rational strategies for engineering Pi-efficient crops. These strategies may include the overexpression of PSI PAPs, high-affinity Pi transporters, and enzymes and membrane transporters that result in enhanced organic acid excretion by roots during nutritional Pi deficiency. Pi-efficient crops are urgently needed to optimize inputs of unsustainable and nonrenewable Pi fertilizers for maximum agronomic benefit.
Acknowledgments
W.C.P. thanks past and present members of his laboratory who have examined various aspects of the metabolic adaptations of plants to nutritional Pi deprivation. W.C.P. is also grateful to collaborators who have contributed to our studies of −Pi plants, particularly Drs. Daniel Lefebvre, David Turpin, Bruce Grant, Julie Niere, Susan Wood, Wayne Snedden, Yi-min She, Alexander Valentine, Robert Mullen, Eduardo Blumwald, Daowen Wang, and Kashchandra Raghothama.
References
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett-Lennard EG, Dracup M, Greenway H. (1993) Role of extracellular phosphatases in the phosphorus-nutrition of cover. J Exp Bot 44: 1595–1600 [Google Scholar]
- Bozzo GG, Dunn EL, Plaxton WC. (2006) Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ 29: 303–313 [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 [DOI] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. (2004a) Structural and kinetic properties of a novel purple acid phosphatase from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Biochem J 377: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Singh VK, Plaxton WC. (2004b) Phosphate or phosphite addition promotes the proteolytic turnover of phosphate-starvation inducible tomato purple acid phosphatase isozymes. FEBS Lett 573: 51–54 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Nimmo GA, Jenkins GI, Nimmo HG. (2007) BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem J 405: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilizing/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Diatloff E, Roberts M, Sanders D, Roberts SK. (2004) Characterization of anion channels in the plasma membrane of Arabidopsis epidermal root cells and the identification of a citrate-permeable channel induced by phosphate starvation. Plant Physiol 136: 4136–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GB, Lefebvre DD, Plaxton WC. (1989) Phosphate starvation inducible “bypasses” of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol 90: 1275–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZY, Shao C, Meng YJ, Wu P, Chen M. (2009) Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci 176: 170–180 [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ. (2002) Activation of pyrophosphate:fructose-6-phosphate 1-phosphotransferase by fructose 2,6-bisphosphate stimulates conversion of hexose phosphates to triose phosphates but does not influence accumulation of carbohydrates in phosphate-deficient tobacco cells. Physiol Plant 114: 172–181 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Saito A, Wasaki J, Shinano T, Osaki M. (2007) Metabolic alterations proposed by proteome in rice roots grown under low P and high Al concentration under low pH. Plant Sci 172: 1157–1165 [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dormann P. (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Gregory AL, Hurley BA, Tran HT, Valentine AJ, She YM, Knowles VL, Plaxton WC. (2009) In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem J 420: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi? Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Hurley BA, Tran HT, Marty NJ, Park J, Snedden WA, Mullen RT, Plaxton WC. (2010) The purple acid phosphatase AtPAP26 is essential for efficient acclimation of Arabidopsis thaliana to nutritional phosphate deprivation. Plant Physiol 153: 1112–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida R, Satoh Y, Bulone V, Yamada Y, Kaku T, Hayashi T, Kaneko TS. (2009) Activation of beta-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol 150: 1822–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xu C, Li Z, Zhang K, Yang A, Zhang J. (2008a) Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-P-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J 55: 927–939 [DOI] [PubMed] [Google Scholar]
- Li M, Qin C, Welti R, Wang X. (2006) Double knockouts of phospholipases D1 and D2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Physiol Plant 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Shao G, Lam HM. (2008b) Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol 178: 80–91 [DOI] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. (2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lin SI, Chiou TJ. (2009) Molecular regulators of phosphate homeostasis in plants. J Exp Bot 60: 1427–1438 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Albrecht G, Hajirezaei M, Grimm B, Biemelt S. (2005) Low levels of pyrophosphate in transgenic potato plants expressing E. coli pyrophosphatase lead to decreased vitality under oxygen deficiency. Ann Bot 96: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Hachiya A, Ashihara H. (1994) Phosphate starvation and a glycolytic bypass catalyzed by phosphoenolpyruvate carboxylase in suspension-cultured Catharanthus roseus cells. Z Naturforsch C 49: 742–750 [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Palma DA, Blumwald E, Plaxton WC. (2000) Upregulation of vacuolar H(+)-translocating pyrophosphatase by phosphate starvation of Brassica napus (rapeseed) suspension cell cultures. FEBS Lett 486: 155–158 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Podestá FE. (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Ricard B, Van Toai T, Chourey P, Saglio P. (1998) Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol 116: 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE. (2009) Regulating the phosphorus nutrition of plants: molecular biology meeting agronomic needs. Plant Soil 322: 17–24 [Google Scholar]
- Rychter AE, Mikulska M. (1990) The relationship between phosphate status and cyanide-resistant respiration in bean roots. Physiol Plant 79: 663–667 [DOI] [PubMed] [Google Scholar]
- Sieger SM, Kristensen BK, Robson CA, Amirsadeghi S, Eng EWY, Abdel-Mesih A, Møller IM, Vanlerberghe GC. (2005) The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J Exp Bot 56: 1499–1515 [DOI] [PubMed] [Google Scholar]
- Stark DM, Timmerman KP, Barry GF, Preiss J, Kishore GM. (1992) Regulation of the amount of starch in plant-tissues by ADP-glucose pyrophosphorylase. Science 258: 287–292 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Tran HT, Hurley BA, Plaxton WC. (2010a) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179: 14–27 [Google Scholar]
- Tran HT, Qian W, Hurley BA, She YM, Wang D, Plaxton WC. (2010b) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33: 1789–1803 [DOI] [PubMed] [Google Scholar]
- Tran HT, Plaxton WC. (2008) Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 8: 4317–4326 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 427–447 [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles VL, Snedden WA, Plaxton WC. (2006) Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol 142: 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H. (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasaki J, Maruyama H, Tanaka M, Yamamura T, Dateki H, Shinano T, Ito S, Osaki M. (2009) Overexpression of the LASAP2 gene for secretory acid phosphatase in white lupin improves the phosphorus uptake and growth of tobacco plants. Soil Sci Plant Nutr 55: 107–113 [Google Scholar]
- Wasaki J, Omura M, Ando M, Dateki H, Shinano T, Osaki M, Ito H, Matsui H, Tadano T. (2000) Molecular cloning and root specific expression of secretory acid phosphatase from phosphate deficient lupin (Lupinus albus L.). Soil Sci Plant Nutr 46: 427–437 [Google Scholar]
- Yan F, Zhu YY, Müller C, Zörb C, Schubert S. (2002) Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol 129: 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gaxiola RA. (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol J 5: 735–745 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu CC, Benning C. (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng XQ, Chow WS, Su LJ, Peng XX, Peng CL. (2010) Protective effect of supplemental anthocyanins on Arabidopsis leaves under high light. Physiol Plant 138: 215–225 [DOI] [PubMed] [Google Scholar]