Abstract
Since the advent of the postgenomic era, efforts have focused on the development of rapid strategies for annotating plant genes of unknown function. Given its simplicity and rapidity, virus-induced gene silencing (VIGS) has become one of the preeminent approaches for functional analyses. However, several problems remain intrinsic to the use of such a strategy in the study of both metabolic and developmental processes. The most prominent of these is the commonly observed phenomenon of “sectoring” the tissue regions that are not effectively targeted by VIGS. To better discriminate these sectors, an effective marker system displaying minimal secondary effects is a prerequisite. Utilizing a VIGS system based on the tobacco rattle virus vector, we here studied the effect of silencing the endogenous phytoene desaturase gene (pds) and the expression and subsequent silencing of the exogenous green fluorescence protein (gfp) on the metabolism of Arabidopsis (Arabidopsis thaliana) leaves and tomato (Solanum lycopersicum) fruits. In leaves, we observed dramatic effects on primary carbon and pigment metabolism associated with the photobleached phenotype following the silencing of the endogenous pds gene. However, relatively few pleiotropic effects on carbon metabolism were observed in tomato fruits when pds expression was inhibited. VIGS coupled to gfp constitutive expression revealed no significant metabolic alterations after triggering of silencing in Arabidopsis leaves and a mild effect in mature green tomato fruits. By contrast, a wider impact on metabolism was observed in ripe fruits. Silencing experiments with an endogenous target gene of interest clearly demonstrated the feasibility of cosilencing in this system; however, carefully constructed control experiments are a prerequisite to prevent erroneous interpretation.
To date, the manipulation of gene expression for the analysis of gene function in plants has largely been carried out via stable Agrobacterium tumefaciens-mediated transformation protocols. However, the use of mutagenesis approaches, either in isolation or in combination with TILLING, has also provided considerable advances in our understanding of gene function (Xin et al., 2008; Dong et al., 2009; Gady et al., 2009; Perry et al., 2009; Chawade et al., 2010). More recently, the rapid increase in the availability of entire genome sequences has led to the development of faster yet reliable strategies for reverse genetics. Transient virus-induced gene silencing (VIGS) is a powerful tool for this purpose and considerably less time consuming than classical stable transformation approaches (Lu et al., 2003; Burch-Smith et al., 2004). This technique has proven to be useful for gene functional characterization in several plant organs, including leaves (Liu et al., 2002), roots (Ryu et al., 2004), tubers (Faivre-Rampant et al., 2004), flowers (Chen et al., 2004; Liu et al., 2004), and, more recently, tomato (Solanum lycopersicum) fruits (Fu et al., 2005; Orzaez et al., 2006, 2009). Moreover, VIGS simplifies biological assays, since several replicates can be obtained from a single plant instead of the one plant/one biological replicate sampling system required for stable transformants. Furthermore, depending on the gene under study, this transient technique can avoid complexities associated with the silencing of essential genes via stable transformation, especially in the case of genes involved in vital cell processes. The reason for this is that, similar to inducible transgene expression systems (Junker et al., 2003), VIGS manipulation can potentially be triggered at any stage in which the system can be used. However, given that the induced silencing may present an irregular dispersion displaying a patchy or “sectored” distribution, the development of a visible marker associated with silencing is essential for the identification and further quantification of nonvisual phenotypes (Burch-Smith et al., 2006). For an efficient VIGS system, such a visible marker should not dramatically affect the biochemical or the developmental network underlying the process under study. This is especially important in cases where such processes comprise rapid and dramatic changes in metabolism or development, which could be evident in relatively narrow time windows, such as those that occur continually during the development and ripening of tomato fruits (Carrari and Fernie, 2006; Carrari et al., 2006). In addition to its nutritional importance for the human diet, this species has become a model for physiological studies of fleshy fruits, and in recent years, several reports have identified a number of candidate genes associated with metabolic pathways underlying different fruit quality traits (Schauer et al., 2006; Tieman et al., 2006; Stevens et al., 2007; Bermúdez et al., 2008).
To date, relatively few reporter genes, including the encoding phytoene desaturase (pds) enzyme, have been tested in tomato VIGS experiments (Fu et al., 2005; Orzaez et al., 2006). Most recently, Orzaez et al. (2009) proposed a VIGS system for tomato based on purple fruit-producing transgenic plants via the ectopic expression of two transcription factors that induce the anthocyanin biosynthesis pathway during ripening (Butelli et al., 2008). The silencing of these transcription factors, together with the gene of interest (GOI), switches off the anthocyanin pathway by reversing the purple phenotype, rendering the approach a clearly tractable system. Although the utility of this approach is clearly proven, it is appropriate only for studies focused exclusively on later processes of tomato fruit development due to the fact that the driving promoter (E8) of the two transcription factors is active only from the onset of ripening (Deikman and Fischer, 1988). Given the intense interest in early fruit development (Garcia et al., 2009; Wang et al., 2009; Mathieu-Rivet et al., 2010), a system capable of studying this stage of development would clearly represent an important resource for the fleshy fruit research community.
This work evaluates a VIGS system based on the constitutive expression of the GFP and its subsequent silencing as a traceable marker for functional genomics analyses from very early stages of tomato fruit development throughout the ripening process. In addition, the impact of the exogenous gfp and the endogenous pds reporter genes on metabolism was evaluated both in photosynthetic tissues of Arabidopsis (Arabidopsis thaliana) and in tomato fruit pericarp. A comprehensive metabolite profiling analysis revealed that pds silencing, which affects carotenoid, chlorophyll, vitamin E, and gibberellin biosynthesis (Qin et al., 2007), causes a dramatic effect on the primary metabolism of photosynthetic tissues and, to a lesser extent, in tomato pericarp. By contrast, the gfp-based system produced only a negligible impact on the metabolism of the leaves and mature green (MG) fruits, although admittedly a wider perturbation in certain areas of metabolism was observed in red ripe fruits. That said, this system was shown to be efficient in cosilencing experiments with the GOI we chose to use as a case study. However, our results indicate that reporter genes used in VIGS experiments must be carefully selected and empirically evaluated in order to avoid data misinterpretation, especially when pathways involved in primary metabolism are under scrutiny.
RESULTS
Establishment of the Experimental System
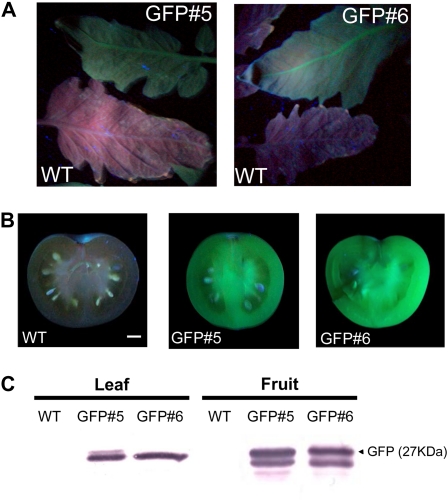
The experiments presented here were designed to provide a rapid and efficient plant gene-silencing method for tomato fruit by means of a tobacco rattle virus (TRV)-based VIGS system. Although this system is broadly used, it results in a heterogeneous and patchy pattern of (nontraceable) silenced tissues. To overcome this limitation, we developed a set of tomato transgenic plants expressing the gfp-encoding gene under the control of the cauliflower mosaic virus 35S promoter and produced gfp-expressing Arabidopsis plants as a model for photosynthetic tissues. Twenty-five tomato plants obtained by Agrobacterium-mediated transformation were transferred to the greenhouse and screened for the level of GFP by both visual fluorescence under UV illumination and western-blot experiments. Following these analyses, two lines (lines 5 and 6), which exhibit nonvisual phenotypic alterations, were selected and vegetatively propagated for further experiments. Fluorescence was shown to be homogeneously distributed in vegetative and fruit tissues (Fig. 1, A and B). Moreover, an approximately 27-kD protein was detected in these tissues by an anti-GFP antibody (Fig. 1C). Also, a slower running band was detected in fruit samples, which could account for posttranslational modifications of GFP occurring specifically in this tissue, as was described previously in tobacco (Nicotiana tabacum) BY-2 cells (Tamura et al., 2004).
Figure 1.
Screening and analysis of transgenic tomato plants expressing GFP. GFP detection is shown in tomato transgenic lines GFP#5 and GFP#6 and wild-type (WT) controls. A and B, Leaves from 6-week-old plants (A) and fruits at MG stage (B) illuminated with a hand-held UV lamp. Bar = 1 cm. C, Western-blot analysis of tomato leaves and fruit pericarp. [See online article for color version of this figure.]
Analyses of the self-pollinated progeny (T1) from one of these lines (GFP#6) suggested a single-copy insertion of the transgene and also revealed a homogeneous distribution of fluorescence both in vegetative and reproductive tissues (data not shown). Arabidopsis transgenic lines carrying the same construct obtained by floral dip transformation were also selected on the basis of the level of the fluorescence they emitted. The selected line, C-GFPpbin#3, was selfed until homozygous status was achieved (T4).
Endogenous pds and gfp Transgene-Silencing Experiments
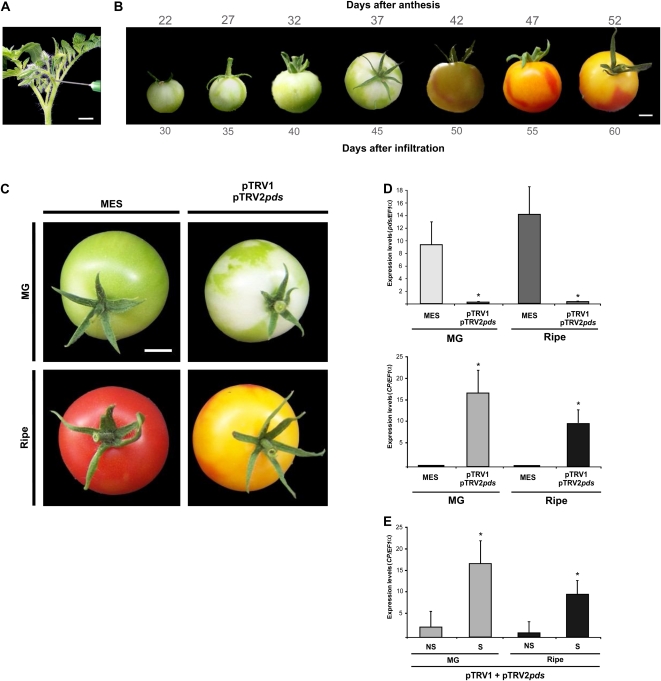
In order to evaluate the time course of silencing across tomato development, inflorescence peduncles at the preanthesis stage (Fig. 2A) were infiltrated with pTRV2-pds and the fruits were visually inspected daily. Silenced areas were clearly detected by following the characteristic bleaching phenotype, which became apparent as early as 30 d after infiltration, and the area continued expanding until ripening (Fig. 2B). More than 90% of the infiltrated inflorescence resulted in at least one fruit per truss exhibiting silenced sectors, and at MG stage, about 60% to 70% of the fruit surfaces was bleached (Fig. 2C). Based on these results, subsequent experiments were sampled at MG and ripe stages. Neither the diameter nor the weight differed between control and silenced fruits (data not shown).
Figure 2.
Silencing of the pds gene in tomato fruits. A, Inoculation of inflorescence peduncles of preanthesis flowers. B, Time course of pds gene silencing in wild-type tomato during fruit development. C, Eight-week-old wild-type plants were infiltrated with MES buffer or pTRV1/pTRV2pds Agrobacterium cultures. Fruits were sampled 45 d (MG) and 60 d (ripe) after infiltration. Bars = 1 cm. D, Expression analysis of the pds and CP genes by qRT-PCR. E, Expression analysis of the CP gene by qRT-PCR in silenced (S) and nonsilenced (NS) regions of pTRV1/pTRV2pds-infiltrated fruits. Data indicate relative expression means from five replicates ± se. Asterisks indicate statistically significant differences (P < 0.05). All qRT-PCRs were performed in triplicate. [See online article for color version of this figure.]
The level of pds mRNA was measured by quantitative reverse transcription (qRT)-PCR in the fruit pericarp of pTRV2-pds and MES buffer-infiltrated plants; reductions of approximately 80% were observed in the bleached areas of both MG and ripe stages. Inversely, levels of TRV capsid protein (CP) mRNA were detected exclusively in the silenced tissues (Fig. 2D). Moreover, levels of CP mRNA were significantly lower in nonsilenced tissues of infected fruits in comparison with those found in silenced tissues (Fig. 2E).
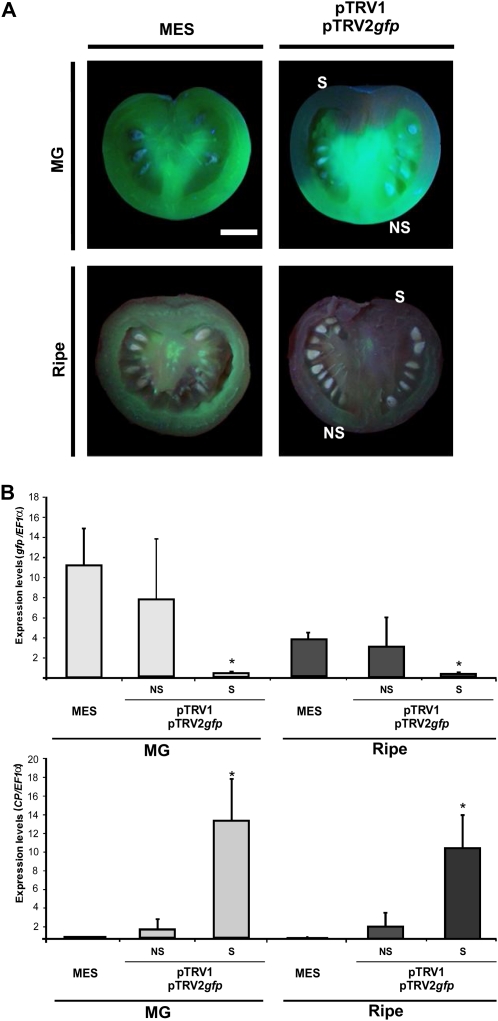
In parallel, GFP#5 and GFP#6 transgenic plants were infiltrated with a pTRV2-gfp vector, harboring a gfp fragment of 695 bp, as described above. Daily inspection with a hand-held UV lamp allowed detection of gfp silencing from very early developmental stages of the tomato fruits (Fig. 3). Moreover, quantitative analyses of gfp mRNA in fluorescent sectors of the same fruits revealed similar results to those found in MES-infiltrated fruits (Fig. 3B). This result indicates that there was no reduction in the gfp mRNA levels on those fluorescent fruit sectors.
Figure 3.
Silencing of the gfp gene in tomato fruits. A, Transverse sections of tomato fruits from GFP#5 transgenic plants, illuminated with a hand-held UV lamp, infiltrated in the inflorescence peduncle with MES buffer or pTRV1/pTRV2gfp Agrobacterium cultures. Fruits were sampled 45 d (MG) and 60 d (ripe) after infiltration. NS indicates nonsilenced fruit regions, while S indicates silenced regions. Bar = 1 cm. B, Expression analysis of the gfp and CP genes by qRT-PCR in MG and ripe tomatoes. Data indicate relative expression means from five replicates ± se. Asterisks indicate statistically significant differences (P < 0.05). All qRT-PCRs were performed in triplicate. [See online article for color version of this figure.]
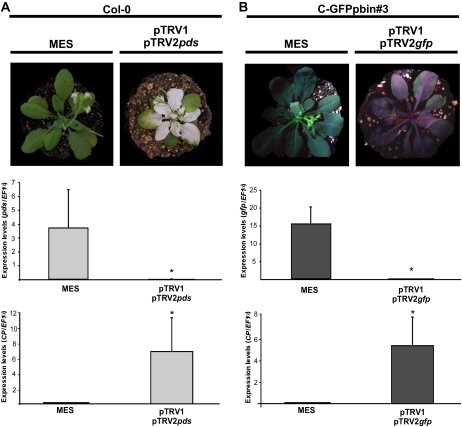
The same two reporter genes analyzed in tomato fruits were also used in Arabidopsis as a model system for photosynthetic tissues (Burch-Smith et al., 2006). Figure 4 presents the phenotypes of control and silenced plants at bolting stage as well as the quantification of the endogenous pds, the gfp transgene, and the CP mRNA levels in rosette leaves. Notably, there was a reduction in the rosette size of the pds-silenced plants in comparison with all the other treated plants. This observation correlates with the sharp reduction of the pds mRNA level (Fig. 4A). Although the target gene expression was also dramatically reduced in the gfp-based system, the size of silenced plants was not altered (Fig. 4B). These results reveal that the silencing signal spreads and replicates efficiently in both tomato fruits and Arabidopsis leaves for the gfp transgene. The same is true for the endogenous pds gene, as has been demonstrated previously by Burch-Smith et al. (2006) in Arabidopsis and by Orzaez et al. (2006) in tomato. Moreover, they are indicative of impairment of the normal growth of Arabidopsis plants when the endogenous pds gene is silenced.
Figure 4.
Silencing of pds and gfp genes in Arabidopsis. A, Wild-type (Col-0) Arabidopsis plants with two to three leaves infiltrated with MES buffer or pTRV1/pTRV2pds Agrobacterium cultures. Expression analysis of pds and CP genes was by quantitative RT-PCR. B, C-GFPpbin#3 Arabidopsis plants with two to four leaves infiltrated with MES buffer or pTRV1/pTRV2gfp Agrobacterium cultures illuminated with a hand-held UV lamp. Expression analysis of gfp and CP genes was by qRT-PCR. Data indicate relative expression means from five replicates ± se. Asterisks indicate statistically significant differences (P < 0.05) in comparison with MES-infiltrated plants. All qRT-PCRs were performed in triplicate. [See online article for color version of this figure.]
Assessment of Metabolic Changes Associated with the gfp-Based VIGS System
Having demonstrated that the VIGS system was equally efficient with both reporter genes in terms of the silencing percentage, we next turned our attention to assessing the impact of GFP constitutive expression, and its subsequent suppression by VIGS, on the metabolism of the treated tissues. The metabolite composition of both the tomato fruit pericarp in two developmental stages (MG and ripe) and expanded Arabidopsis leaves was analyzed by applying a combination of HPLC and gas chromatography-mass spectrometry (GC-MS) techniques (Lisec et al., 2006). Analyses of targeted metabolite profiles resulted in the detection and relative quantification of 52, 56, and 59 different compounds of defined chemical structures in MG and ripe tomato pericarp and Arabidopsis leaves, respectively. These metabolites correspond to amino acids and nitrogen compounds, organic acids, sugars and alcohols, lipids, and a few phosphorylated intermediates and terpenoids, thus giving broad coverage of metabolism.
First, we compared the relative amounts of every metabolite between the two tomato lines (GFP#5 and GFP#6) and detected no significant differences (data not shown). Thus, for subsequent analyses, metabolite profile data from four to six plants from these two lines were pooled to increase the power of the tests applied. Variations detected in each metabolite were analyzed from the following: (1) GFP-expressing plants versus wild-type plants, both infiltrated with MES buffer (GFP MES/MM MES); (2) GFP-expressing plants infiltrated with pTRV2-empty vector versus gfp-expressing plants infiltrated with MES (GFP TRV-/GFP MES); (3) GFP-expressing plants infiltrated with pTRV2-gfp versus gfp-expressing plants infiltrated with MES buffer (GFP TRV-gfp/GFP MES); (4) GFP-expressing plants infiltrated with pTRV2-gfp versus the same transgenics infiltrated with pTRV2-empty vector (GFP TRV-gfp/GFP TRV-); and (5) GFP-expressing plants infiltrated with pTRV2-gfp versus wild-type plants infiltrated with buffer (GFP TRV-gfp/MM MES; Supplemental Tables S1–S3). The number of metabolites showing significant changes in the first comparison revealed no apparent effects of the gfp transgene expression on the metabolism of the Arabidopsis leaves and significant reductions in the levels of two metabolites (glycerol and malonate) in MG fruits. However, a wider impact on amino acid and organic acid levels was observed in ripe tomatoes. The second comparison again showed no effects of the viral vector on the metabolism of Arabidopsis leaves but significant increments in the levels of one (putrescine) and three (γ-aminobutyrate, arachidic acid, and Suc) metabolites in MG and ripe tomatoes, respectively. When the metabolism of silenced tissues was compared with tissues infiltrated with buffer or pTRV2-empty vector, no changes were detected in Arabidopsis leaves and only one metabolite (maleate) was significantly altered in MG fruits. In ripe tomatoes, however, GFP silencing affected the levels of malate consistently either in comparison with GFP-expressing fruits infiltrated with buffer or with the pTRV-empty vector and the levels of putrescine in the comparison with GFP-expressing fruits infiltrated only with buffer.
Interestingly, when the transgene was silenced, no significant changes were observed on any of the metabolites measured in comparison with the leaves of Arabidopsis wild-type plants, and only the levels of glycerol and α-carotene showed significant variations in MG fruits. However, in ripe tomatoes, GFP silencing only partially reverted the chemotype of the wild-type fruits.
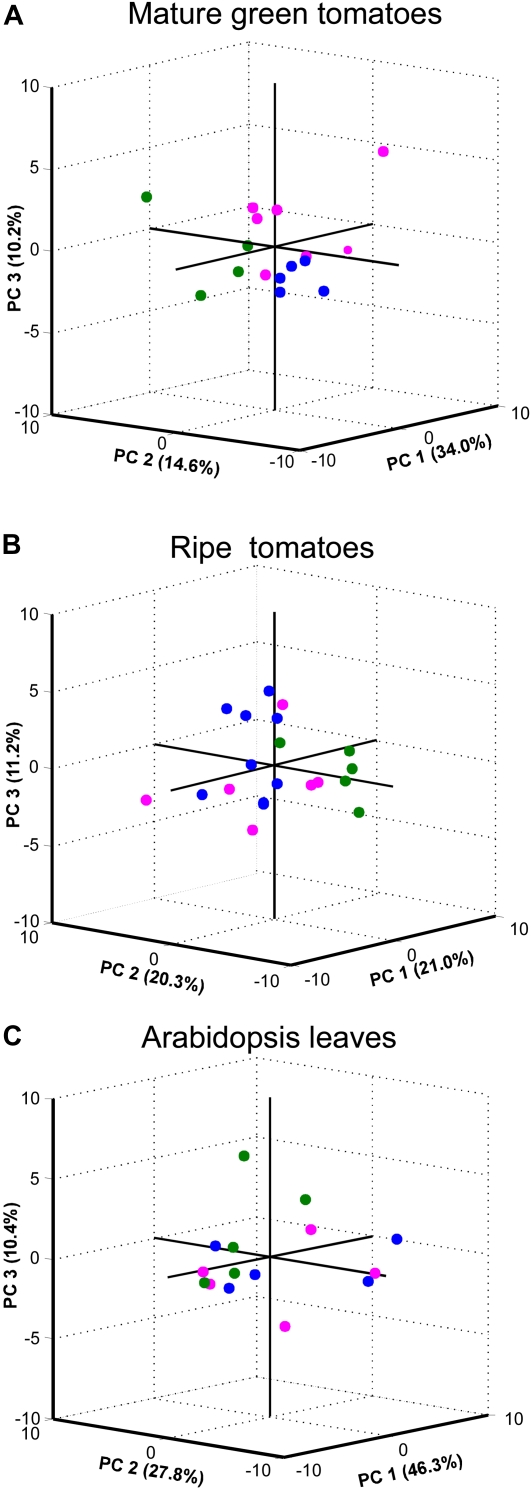
In order to better interpret the differences found in metabolism associated with the gfp-based VIGS system, nonparametric analyses were applied to the full data set. Models of principal component analysis (PCA) explaining 58.8%, 52.5%, and 84.5% of the data variance for MG and ripe tomato pericarps and Arabidopsis leaves, respectively, are shown in Figure 5. PCA results showed that MG fruits sampled from wild-type plants separate from both those expressing GFP and those infiltrated with pTRV2-gfp (Fig. 5A). In the case of the samples harvested from ripe fruits, none of the three components was able to discriminate either between gfp-overexpressing plants and silenced plants or even wild-type plants (Fig. 5B). Similar results were obtained with the PCA model applied to the metabolite profile data from Arabidopsis leaves, with samples harvested from the three treatments being indistinguishable from one another (Fig. 5C).
Figure 5.
PCA of metabolic variations in tomato fruits and Arabidopsis leaves transiently silenced for the exogenous gfp gene. GC-MS and HPLC metabolite profile data are from MG (A) and ripe (B) tomatoes and from Arabidopsis leaves (C) subjected to PCA models. Raw data were collected from wild-type tomatoes or Arabidopsis plants infiltrated with MES buffer (green circles), GFP#5, GFP#6, or C-GFPpbin#3 Arabidopsis plants infiltrated with MES buffer (blue circles), and GFP#5, GFP#6, or C-GFPpbin#3 plants infiltrated with pTRV1/pTRV2gfp (magenta circles). [See online article for color version of this figure.]
Metabolic Changes Associated with the Silencing of the Endogenous pds Gene
To evaluate the impact of the pds gene silencing on metabolism, the variations detected in each metabolite were analyzed from wild-type plants infiltrated with pTRV2-pds versus wild-type plants infiltrated with MES buffer. As may be expected, the silencing of the endogenous pds gene altered terpenoid levels in tomato fruits as well as in Arabidopsis leaves. Moreover, Arabidopsis photosynthetic tissues displayed a completely altered metabolite profile, whereby a decrease in the level of all pigments measured correlated well with the dramatic depletion in almost all sugars and organic acids. Of particular note is the large excess of Asn, which is considered a marker molecule for carbon starvation (Lam et al., 1996; Supplemental Table S3). However, with regard to all other metabolites measured, pds silencing displayed only a mild effect on tomato fruit metabolism in comparison with that observed in photosynthetic Arabidopsis tissues. Putrescine and citrate contents decreased dramatically in MG tomatoes, while chlorogenate and galactinol displayed significant increases. In ripe fruits, glucuronate content dropped significantly while maltose and glycerol-3-phosphate displayed significant increases (Supplemental Tables S1–S3). As might be expected, in both ripening stages, the pigment contents and also the levels of γ-tocopherol in the case of the ripe fruits were significantly altered.
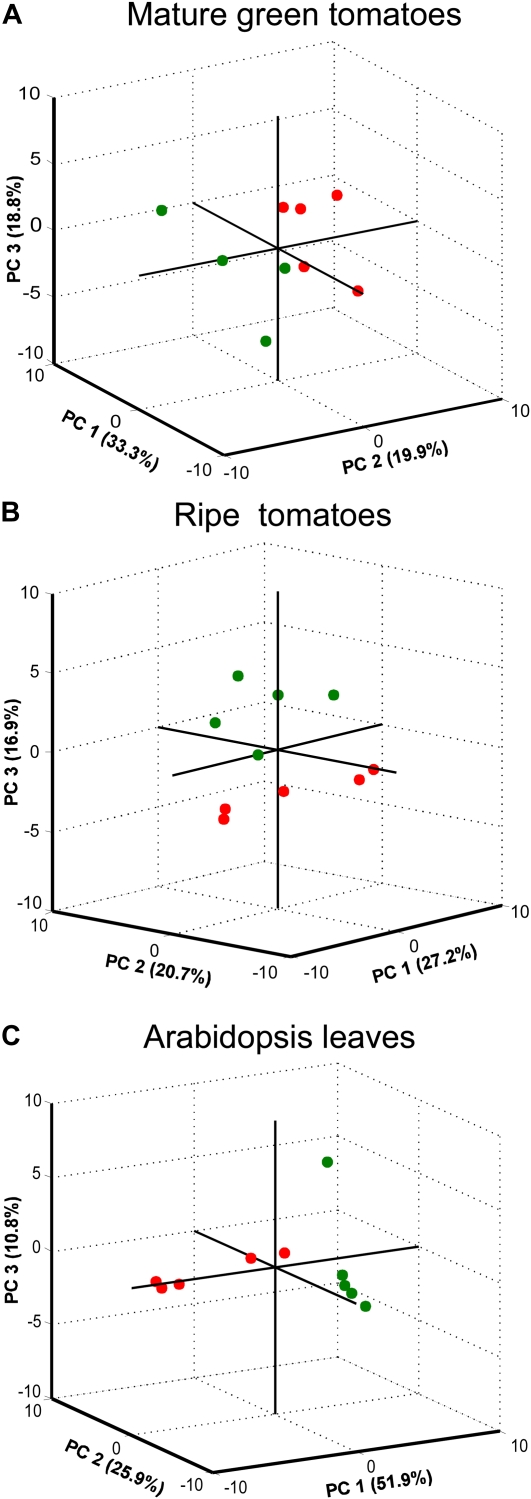
Sample separation by PCA allowed the discrimination of fruits harvested from wild-type plants from those harvested from pds-silenced plants. This was true for both MG and ripe tomatoes, where the first three dimensions of the analysis explain 72% and 64.8% of the data variance, respectively (Fig. 6, A and B). When PCA was applied to metabolite profiling data from Arabidopsis leaves, it clearly revealed that pds-silenced plants were different from wild-type plants (Fig. 6C).
Figure 6.
PCA of metabolic variations in tomato fruits and Arabidopsis leaves transiently silenced for the endogenous pds gene. GC-MS and HPLC metabolite profile data are from MG (A) and ripe (B) tomatoes and from Arabidopsis leaves (C) subjected to PCA models. Raw data were collected from wild-type tomatoes or Arabidopsis plants infiltrated with MES buffer (green circles) and from wild-type tomatoes or Arabidopsis plants infiltrated with pTRV1/pTRV2pds (red circles). [See online article for color version of this figure.]
gfp and GOI Cosilencing Experiments
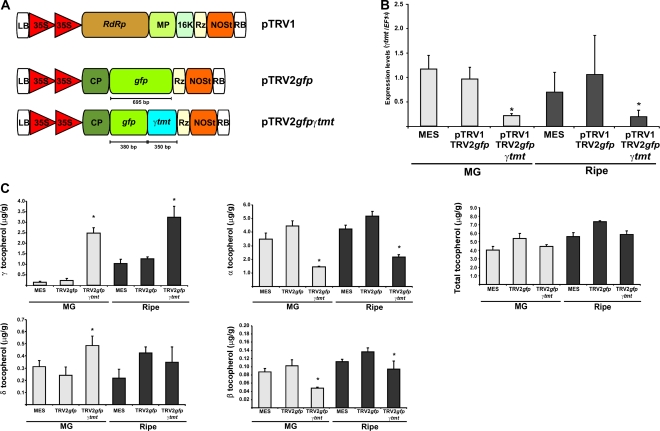
In order to prove the utility of the gfp-based VIGS system described above in reverse genetic studies, focused on genes operating in metabolic processes in tomato fruits, we next performed an experiment wherein a 380-bp fragment of the gfp coding region was fused to a 351-bp fragment of the tomato gene encoding a plastidial γ-tocopherol C-methyl transferase (γ-TMT; EC 2.1.1.95; Fig. 7A). This enzyme catalyzes the last step of the tocopherol biosynthetic pathway in higher plants (for review, see DellaPenna and Pogson, 2006) by ring methylation of δ- and γ-tocopherol, yielding β- and α-tocopherol. Following the infiltration of inflorescence peduncles as described above, gfp-silenced regions were monitored in planta across fruit development and MG and ripe tomatoes were harvested 42 and 60 d after infiltration. More than 85% of pTRV2-gfp-380-γ-tmt-infiltrated inflorescences resulted in at least one fruit showing silenced sectors. Sectors lacking fluorescence were dissected from MG and ripe fruits, and the levels of γ-tmt mRNA were measured by qRT-PCR. Reductions of approximately 75% were observed in the gfp-silenced pericarp sectors with respect to fruits set from plants infiltrated with MES buffer and pTRV2-gfp alone (Fig. 7B).
Figure 7.
Marker (gfp) and GOI (γ-tmt) cosilencing. A, Schematic view of the pTRV1, pTRV2-gfp, and pTRV2-gfp-γ-tmt constructs used in the VIGS experiments. B, Expression analysis of γ-tmt by qRT-PCR in MG and ripe tomatoes. C, Tocopherol profiles in silenced MG and ripe tomatoes. Data indicate relative expression means from six replicates ± se. Asterisks indicate statistically significant differences (P < 0.05) in comparison with both MES- and pTRV2gfp-infiltrated plants. qRT-PCRs were performed in triplicate. [See online article for color version of this figure.]
We next assayed the effect of gfp and γ-tmt cosilencing on fruit tocopherol metabolism by measuring the four isoforms of this vitamin in fruits. As would be expected, γ- and δ-tocopherols were significantly increased in MG fruits with respect to the levels observed in control treatments (MES- and pTRV2-gfp-infiltrated fruits). These increments were accompanied by significant reductions in the levels of α- and β-tocopherols in the same fruits. With the exception of δ-tocopherol, which did not show a significant variation, ripe tomatoes displayed the same tocopherol profile as that observed for MG fruits (Fig. 7C). However, when the total pool of tocopherol was considered, no significant changes were observed in γ-tmt-silenced fruits.
DISCUSSION
Although the use of the VIGS system approach has facilitated the elucidation of gene function in a handful of plant species (Page et al., 2004; Manning et al., 2006; Lin et al., 2007; Oikawa et al., 2007; Ye et al., 2009; Ballester et al., 2010), one of the main constraints to its broader application is the irregular distribution of silencing across target tissues. This phenomenon thus requires the use of easily traceable marker genes that have no, or at least only a controlled, impact on the process under study, especially in cases where metabolism is the major focus of interest. The motivation of this report was to identify a marker gene that would ultimately, once silenced, have only minor consequences on the metabolism of the target tissue. That said, in choosing a GFP-based reporter system, this approach will be equally applicable in studies of plant development. In this report, however, we concentrated on testing the hypothesis regarding metabolism. In order to do so, we established a VIGS system based on the expression of the exogenous gfp gene and evaluated its impact on metabolism by comparison with the silencing of the pds gene in both photosynthetic and sink organs. Transgenic plants expressing the exogenous GFP protein constitutively and a very early delivery of silencing vectors in inflorescence peduncles by agroinjection allowed the establishment of a VIGS system for the study of genes involved in tomato fruit metabolism. Moreover, we proved the feasibility of the system by cosilencing the gfp marker gene along with one of the key genes that determines the types of tocochromanols that considerably accumulate in tomato pericarp and represent a key target for metabolic engineering within plants (Collakova and DellaPenna, 2003a, 2003b).
The gfp-based system assayed here showed comparable efficiency levels to other systems using endogenous genes as silencing markers in tomato fruits (Fu et al., 2005; Orzaez et al., 2006). Moreover, silenced sectors were readily identified with a hand-held UV lamp. This was also valid for Arabidopsis leaves, as demonstrated previously by Burch-Smith et al. (2006).
A number of genes with significant effects on fruit characteristics are expressed early in tomato fruit development (Fridman et al., 2004; Xiao et al., 2008; Mounet et al., 2009; Wang et al., 2009). Moreover, a compendium of candidate genes related to metabolic pathways important for nutritional traits has been recently reported (Bermúdez et al., 2008), and several papers reporting quantitative trait loci for volatile organic compounds of known relevance to taste have been published (Schijlen et al., 2006; Tieman et al., 2006; Zanor et al., 2009). Thus, the VIGS system presented here was established in order to trigger silencing as early in fruit development as possible by infiltrating the peduncles of inflorescence at the preanthesis stage. The results demonstrated that the silencing phenotype can be visually detected from 22 d post anthesis until fruit ripening (Fig. 2B), thus affording a highly versatile system that is capable of evaluating the role of a gene across the entire lifespan of the fruit. Such a long-term evaluation has not been possible utilizing previously reported strategies. Direct infiltration into young fruits was also tested, and although silencing was successfully triggered as reported previously (Fu et al., 2005; Orzaez et al., 2006), phenotypes associated with wounding and pathogen defense were apparent (data not shown). Thus, we decided to discard this strategy, since we believed it could produce undesirable pleiotropic effects on metabolism (and/or development) and unnecessarily complicate experimental interpretation (Hammond-Kosack and Jones, 1996).
Recently, Orzaez et al. (2009) reported the establishment of an elegant reporter system for VIGS in tomato fruits based on anthocyanin accumulation induced by endogenous ethylene. The ectopic expression of two transcription factors, Rosea1 and Delila, under the ripening-induced E8 promoter resulted in up-regulation of the anthocyanin biosynthesis enzyme pathway, leading to a gradual accumulation of pigments after the MG stage, finally resulting in a purple phenotype. Cosilencing the transcription factors, together with the GOI, reverses the purple phenotype, thus allowing the identification of the silenced region. This system is attractive for testing genes exclusively active after the triggering of fruit ripening. However, it is quite conceivable that the activation of anthocyanin metabolism may alter either or both primary and secondary metabolism in adjacent silenced fruit areas, thus potentially rendering the constructs inappropriate for testing certain GOIs.
In this sense, the PCA results, which did not reveal the number and extent of significant changes but only the major variances, clearly showed that the silencing of the pds gene has an impact on the metabolism of the evaluated tissues. In contrast, except for the case of MG fruits, for which the wild-type genotype separates from GFP-expressing and -silenced samples, PCA was unable to separate metabolic profiles from Arabidopsis and ripe tomatoes either from gfp-expressing or -silenced tissues. Thus, it is worth considering here that the failure in total reversion observed in the point-by-point analyses could be more related to a transgenesis effect than to the presence of GFP itself. This said, it is clearly important to consider GFP-expressing plants (infiltrated with MES buffer or with a pTRV2-empty vector) as a control in VIGS experiments when using the system proposed here. Using this approach, by comparing metabolite changes in silenced versus nonsilenced tissues, only relatively few significant alterations were detected in both MG and ripe fruits (Supplemental Tables S1 and S2).
Phytoene desaturation is the second step in carotenoid biosynthesis, converting phytoene into ζ-carotene (Chamovitz et al., 1993). Two Arabidopsis albino mutants (pds1 and pds2) defective for phytoene desaturation, which affect 4-hydroxyphenylpyruvate dioxygenase and prenylphytyltransferase, respectively, thus suggesting that plastoquinone is an indispensable electron carrier for the phytoene desaturation, have been characterized. Phytoene desaturase has itself been characterized lately by a different mutant, pds3. This mutant also presents albino and dwarf phenotypes, because the accumulation of phytoene down-regulates the geranylgeranyl pyrophosphate synthase, limiting the level of the common precursor for the synthesis of both chlorophyll and gibberellins (Qin et al., 2007).
Results from the metabolite profiling in Arabidopsis leaves presented here reveal a dramatic decrease in sugars, deregulation of the tricarboxylic acid cycle, and an accumulation of most of the amino acids detected when the pds gene is silenced. This metabolic pattern is quite similar to that obtained in plants grown in the dark or when photosynthesis is impaired. In these cases, nitrogen is stored in molecules as Asn (2N/4C) instead of Gln (2N/5C), due to its higher nitrogen-carbon ratio (Lam et al., 1996). As expected, Arabidopsis silenced leaves displayed dramatic increases in Asn and Arg. These results are largely consistent with those reported by Rodríguez-Villalón et al. (2009), who demonstrated that the first committed enzyme of carotenogenesis, phytoene synthase, controls metabolic fluxes to this pathway in etiolated seedlings.
Despite the fact that the characteristic photobleached phenotype was observed in pds-silenced tomatoes very early during fruit development, only a few metabolic alterations were detected. These results may merely indicate the nature of fruit organs as assimilate sinks, where light reactions may have no major influence on carbon partitioning. Nevertheless, the fact that a small number of metabolites did show significant alterations in pds-silenced tomatoes counters this argument. Of particular note are the increases detected in α-carotene and lutein at the MG stage and γ-tocopherol at the ripe stage, which suggest that the impairment in photosynthetic pigments could induce the accumulation of other pigments in an attempt to ameliorate photooxidation. In the context of a system for functional genomics studies, this is very undesirable when research is focused on genes involved in antioxidant compounds, which are of particularly high importance for tomato production. When fruit ripening proceeded, five metabolites displayed significant changes. Diminished levels of glucuronate at this stage of fruit development may be indicative of impairments in ascorbate and noncellulosic cell wall polysaccharide biosynthetic pathways (Gilbert et al., 2009). Maltose, the other metabolite that showed significant changes in pds-silenced tomatoes, is the main form of carbon exported from the chloroplast at night (Lu and Sharkey, 2004; Niittylä et al., 2004). It could thus be speculated that its accumulation feeds back to limit starch degradation reactions in chromoplasts. The dramatic decrease in lycopene content is entirely expected, since this metabolite is two steps downstream from PDS activity in the carotenoid biosynthesis pathway. This suggests that, in contrast to the effects observed in the MG stage, at the end of ripening, sink strength decreases and the fruit only reallocates assimilates depending on its own metabolism for lycopene production.
As a proof of concept of the system presented here, a tandem construct of gfp and a GOI was used for peduncle infiltration. Taking into account that the size of the pTRV2 insert may be an important factor in the stability of the virus (Chung et al., 2007), we decided to construct a fused insert with a similar length to the pTRV2-gfp. We selected a gene encoding one of the last key steps in determining the type of tocopherol that accumulates in photosynthetic organs. Despite the fact that this pathway has been fully elucidated in Synechocystis sp. PCC6803 (for review, see Grusak and DellaPenna, 1999) and Arabidopsis (Shintani and DellaPenna, 1998), relatively little information exists for tomato. This is somewhat surprising given that tomato represents one of the main sources of this compound in the human diet. We next decided to use the tomato γ-tmt-encoding gene, recently cloned in our laboratories, as a test case to assess the viability of manipulating fruit metabolism by applying the established VIGS system. γ-TMT is encoded by a single gene in tomato (Almeida et al., 2011) and is expressed from very early developmental stages until harvest maturity in tomato fruits (data not shown). qRT-PCR results demonstrated that the lack of fluorescence colocates with a reduction in the γ-tmt mRNA levels. Moreover, HPLC measurements demonstrated that this reduction correlates with substantial changes in the tocopherol profiles of the fruits. Although more research is needed to elucidate the regulatory mechanism of vitamin E biosynthesis in tomato, the results presented here constitute, to our knowledge, the first step toward manipulating the amount of these compounds in tomato fruits following the application of a fruit-specific silencing system.
CONCLUSION
The results presented in this work show that the gfp-based VIGS system functions properly both in tomato fruits and Arabidopsis leaves. Moreover, agroinjection in tomato inflorescence peduncles allowed the recovery of silenced fruits from very early stages of development. A comprehensive metabolite profiling of the silenced tissues allowed us to propose that choosing a silencing system should consider not only its efficiency but also possible side effects on the metabolism and development of the targeted tissues. The gfp-based system has no effects on the primary carbon metabolism of photosynthetic tissues, in contrast to the widely used pds marker gene. In fruit, it is particularly important to note that the absence of changes in pigmentation in the control lines would greatly simplify the interpretation of studies focused on understanding the essential role of pigment biosynthesis during ripening as opposed to that possible using previously reported approaches (Fernandez et al., 2009; Hueso Estornell et al., 2009). However, it must be noted that this system can be proposed as a good alternative for some tissues, but caution should be taken in using a set of proper controls (i.e. a GFP-expressing plant infiltrated with MES buffer or with a pTRV2-empty vector) when analyzing primary metabolism in tomatoes harvested at a more mature developmental stage. Here, we demonstrate the utility of this approach both by down-regulating the pds gene in Arabidopsis and tomato and by silencing experiments targeting the tomato γ-tmt gene, which clearly demonstrated the feasibility of this system in metabolic studies in tomato fruits. Taken together with previous demonstrations of the efficacy of the coincidence of expression of gfp and gene silencing via VIGS (Burch-Smith et al., 2006; Orzaez et al., 2009), our results demonstrate the power of such approaches as a tool for addressing the roles of various pathways of primary metabolism and carotenoid biosynthesis. Moreover, the fact that the agroinjection of tomato inflorescence peduncles allows the recovery of silenced tissues from very early during fruit development ensures that the pathway(s) under analysis can be followed under strict spatiotemporal regulation. The application of this system to test candidate regulatory genes would likely be exceptionally useful in the confirmation of candidate genes identified in forward genetic screens of this important crop species (Schauer et al., 2006, 2008; Do et al., 2010; Maloney et al., 2010).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tomato seeds (Solanum lycopersicum ‘Moneymaker’ [MM]) were obtained from Meyer Beck. Tomato plants were grown in 20-L pots under greenhouse conditions: 16-h/8-h photoperiod, 24°C ± 3°C, 60% humidity, and 140 ± 40 μmol m–2 s–1 incident photoirradiance. Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) seeds were obtained from the Arabidopsis Biological Resource Center. All Arabidopsis plants were grown in pots at 23°C ± 1°C in a growth chamber (Percival AR-95L) under a 16-h/8-h photoperiod with 60% humidity and 100 μmol m–2 s–1 incident photoirradiance.
Generation of Transgenic Plants
The pBIN-35S-gfp construct (Haseloff et al., 1997) was used to transform, via Agrobacterium tumefaciens-mediated transformation, Arabidopsis plants by the floral dip method (Clough and Bent, 1998) and tomato as described previously by Nunes-Nesi et al. (2005). After selection on culture medium, plant screening was carried out on the basis of visual fluorescence in leaves and fruits using a 100-W hand-held long-wave ultraviolet lamp (UV Products; Black Ray model B 100AP). The presence of the GFP protein was tested by western-blot analysis with a monoclonal rabbit antibody (Invitrogen). This screening allowed the identification of two tomato lines (GFP#5 and GFP#6) that were in vitro propagated and used for subsequent VIGS experiments and an Arabidopsis line (C-GFPpbin#3) that was self-crossed until homozygous status was achieved (T4). Seeds from the T1 generation of the GFP#6 tomato line are available upon request.
Cloning Procedures and Vector Construction
gfp fragments of 695 and 380 bp were amplified by PCR from the pBIN-35S-gfp vector with the following primers: GFP-For (5′-GGGTAAAGGAGAAGAACTT-3′) and GFP695-Rev (5′-TTATTTGTATAGTTCATCCATGC-3′) or GFP380-Rev (5′-TCTAGACGATCCTGTTGACGAGGGTG-3′). PCR products were cloned into pCR8/GW/TOPO vector (Invitrogen) and pTOPO 2.1 vector (Invitrogen). In parallel, a 351-bp fragment of the tomato γ-tmt-encoding gene (SGN-U584511; Almeida et al., 2011) was amplified from tomato leaf cDNA by PCR (primers TMT-For [5′-GAGTGGAGAACACATGCCGA-3′] and TMT-Rev [5′-ATTGTCTTCCATCCACTGCGT-3′]) and cloned into the pTOPO 2.1 vector (Invitrogen). pTOPO-gfp380 and pTOPO-γ-tmt vectors were digested with EcoRI restriction enzyme and ligated at room temperature for 2 h. Ligation products were subsequently amplified by PCR (primers GFP-For and TMT-Rev), and a 731-bp PCR product was inserted into the pCR8/GW/TOPO vector (Invitrogen). Both resulting vectors (pCR8/GW/gfp-γ-tmt and pCR8/GW/gfp) were recombined into the pTRV2-GW vector (Liu et al., 2002) resulting in pTRV2-gfp380-γ-tmt and pTRV2-gfp vectors and subsequently checked by sequencing with the following primers: TRV1536-For (5′-ATGTTCAGGCGGTTGTGTG-3′) and TRV1793-Rev (5′-ATGTCAATCTCGTAGGTTTA-3′).
Agroinfiltration and Plant Treatments
For VIGS experiments, pTRV1 and pTRV2-empty, pTRV2-pds (Liu et al., 2002), pTRV2-gfp, or pTRV2-gfp380-γ-tmt were introduced into Agrobacterium strain GV3101. A 5-mL culture was grown overnight at 28°C in 50 mg L−1 gentamycin and 50 mg L−1 kanamycin in Luria-Bertani medium and used to inoculate 50 mL of Luria-Bertani medium containing the same antibiotics. After an overnight culture at 28°C, the cells were harvested by centrifugation and resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, and 200 mm acetosyringone), adjusted to an optical density at 600 nm of 1.5 for Arabidopsis and 1.0 for tomato, and left at room temperature for 3 to 4 h. Equivalent aliquots of GV3101-pTRV1 and -pTRV2 were mixed immediately before inoculation (Liu et al., 2002). Arabidopsis plants with two to four leaves were agroinfiltrated as described previously by Burch-Smith et al. (2006). One week preanthesis, inflorescence peduncles of 8-week-old tomato plants were wounded with a needle (size: 27 G 1/2 inches), and approximately 40 μL of the Agrobacterium suspension was delivered (Ryu et al., 2004).
Arabidopsis leaves were harvested 25 d after infiltration. Pericarp tissue from MG and ripe fruits were harvested 45 and 60 d after infiltration, respectively. All samples were immediately frozen in liquid N2 and stored at –80°C until use. Ten plants from each genotype (MM, GFP#5, GFP #6, Col-0, and C-GFPpbin#3) were used in all VIGS experiments.
RNA Isolation and Real-Time RT-PCR Analyses
Total RNA from both 200 mg of tomato fruit pericarps and 50 mg of Arabidopsis leaves was extracted with TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. DNA was removed with 1 μL of amplification-grade DNase (Invitrogen) following the recommended protocol. cDNA was synthesized from 1 μg of RNA using oligo(dT) as primer and the SuperScript III kit (Invitrogen). mRNA levels of CP, pds, gfp, γ-tmt, and elongation factor 1-α (EF1-α) were quantified by quantitative real-time PCR using a 7500 real-time PCR system (Applied Biosystem), Taq Platinum (Invitrogen), Mg2+ at a final concentration of 3 mm, SYBR Green I (Sigma-Aldrich) 1:30,000 in dimethyl sulfoxide, and specific primers at final concentrations of 200 nm. gfp primer sequences were as follows: gfp-For (5′-TCATATGAAGCGGCACGACTT-3′) and gfp-Rev (5′-GATGGTCCTCTCCTGCACGTA-3′). EF1-α and pds primer sequences for Arabidopsis were as follows: EF1-αat-For (5′-ATTGGTAACGGTTACGCCCC-3′) and EF1-αat-Rev (5′-TCTCCTTACCAGAACGCCTGTC-3′), and pdsat-For (5′-ACCATCCCAAACTGTGAACCA-3′) and pdsat-Rev (5′-TGAGAGCAGAATTTGCCAGAGA-3′). γ-tmt primer sequences were as follows: TMT-RT-For (5′-GACGCCTTGAATCAGCCTTT-3′) and TMT-RT-Rev (5′-ATGTGTTCTCCACTCTCCATGGA-3′). EF1-α and pds primer sequences for tomato, as well as CP primer sequences, were extracted from Rotenberg et al. (2006). PCR cycles were 95°C for 15s, 60°C for 20s, and 72°C for 45s. All real-time PCRs were performed in triplicate.
Soluble Sugar Determination
Samples of tomato fruit pericarps and Arabidopsis leaves from VIGS experiments were harvested under daylight conditions, and tissue sectors were dissected on the basis of their photobleached phenotypes or under illumination with a hand-held UV lamp. Soluble sugars from these samples were extracted and determined using established protocols (Fernie et al., 2001). Enzymatic reactions were performed in five replicates.
GC-MS
Arabidopsis frozen tissues from leaves (approximately 100 mg) were extracted in 1,400 μL of methanol as described by Roessner et al. (2000) and Roessner-Tunali et al. (2003) with the modifications proposed by Lisec et al. (2006); 60 μL of internal standard (0.2 mg mL−1 water of ribitol) was added for quantification. In the case of tomato fruits (approximately 250 mg), the reagent volumes were adjusted using 2,000 μL of methanol and 120 μL of ribitol. In both cases, the mixture was extracted for 15 min at 70°C, mixed vigorously with 1 volume of water, centrifuged at 2,200g, and subsequently reduced to dryness under vacuum. The residue was redissolved and derivatized for 120 min at 37°C (in 60 μL of 30 mg mL−1 methoxyamine hydrochloride in pyridine) followed by a 30-min treatment at 37°C with 120 μL of N-methyl-N-[trimethylsilyl] trifluoroacetamide. Sample volumes of 1 μL were then injected in splitless and split modes using a hot needle technique. The GC-time of flight-MS system was composed of an AS 2000 autosampler, a GC 6890N gas chromatograph (Agilent Technologies), and a Pegasus III time-of-flight mass spectrometer (LECO Instruments). Mass spectra were recorded at 20 scans s−1 with a scanning range of 70 to 600 mass-to-charge ratio. Both chromatograms and mass spectra were evaluated using ChromaTOF chromatography processing and mass spectral deconvolution software, version 3.00 (LECO Instruments). Identification and quantification of the compounds were performed with TagFinder 4.0 software (Luedemann et al., 2008), and the mass spectra were cross referenced with those in the Golm Metabolome Database (Kopka et al., 2005; Schauer et al., 2005). Five to 10 biological replicates were used for this analysis.
Pigment Quantification
Tomato pericarp (approximately 100 mg fresh weight) was fixed in 50% ethanol and 1% ascorbic acid and subsequently extracted twice with 5 volumes of n-hexane (Mallinckrodt Baker). After evaporation, pellets were resuspended in 100% ethanol (Merck; HPLC grade) and filtered through a 0.45-μm-pore nylon membrane before injection (Buttriss and Diplock, 1984). The HPLC system used consisted of a quaternary pump (model P4000) with a membrane vacuum degasser connected to an autosampler (model AS4000; with a 10- to 100-μL loop injector) and two detectors, UV-diode array (model UV6000) and fluorescence (model FL3000; Thermo Separation Products). Then, 50-μl samples were separated in an Alltima C18 column (250 mm × 4.6 mm, 5-μm particle size; Alltech Associates). The mobile phase was isocratic with ethanol:methanol (60:40; Baker; HPLC grade). The UV-diode array detector was set at 455 nm for the detection of lutein, α- carotene, and β-carotene and at 470 nm for lycopene. For the detection of tocopherols, the fluorescence detector was set at 296 and 330 nm for excitation and emission wavelengths, respectively. Retention times for different compounds were determined in comparison with authentic standard solutions of dl-α-tocopherol, γ-tocopherol, γ- and β-carotene mixed standard from carrot (Daucus carota), lycopene, all-trans-retinol, and lutein, all purchased from Sigma. Chlorophylls a and b and the carotenoid bulk from Arabidopsis leaves were measured in 80% (w/v) acetone extracts as described by Lichtenthaler (1987).
Tocopherol Quantification by HPLC
Tocopherol extraction was performed as described by Fraser et al. (2000) with the following modifications: tomato fruit were ground to a fine powder in liquid nitrogen, 500 mg of material was extracted with 1.5 mL of methanol, and after vortex mixing, 1 mL of chloroform was added. Following 5 min of sonication, 1 mL of Tris buffer (50 mm Tris, pH 7.5, and 1 m NaCl) was added. The chloroform phase was recovered, and the methanol phase (remaining pellet) was reextracted with chloroform (2 mL). Chloroform extracts were pooled and adjusted to a final volume of 4 mL. Two milliliters were dried under nitrogen gas and resuspended in 0.2 mL of 99.5:0.5 hexane:isopropanol. The tocopherol content was determined using a Hewlett-Packard series 1100 HPLC system coupled with a fluorescence detector (Agilent Technologies series 1200). Separation was carried out on a Metasil Si normal-phase column (250 mm × 4.6 mm, 5 μm; Varian, Metachem) maintained at room temperature using an isocratic solvent system (mobile phase) consisting of 99.5:0.5 hexane:isopropanol with a flow rate at 1 mL min−1. Eluting compounds were detected and quantified by fluorescence with excitation at 296 nm and emission at 340 nm. Identification and quantification of tocopherol compounds were achieved by comparison with the retention times and peak areas of standards purchased from Merck (tocopherol set 613424; Calbiochem). A daily calibration curve was carried out using a tocopherol solution with a concentration range between 0.31 and 5 μg mL−1 for each isoform.
Statistical Analysis
Results from real-time PCR experiments were analyzed by the Kruskal-Wallis test using algorithms incorporated to the InfoStat software (www.infostat.com; Grupo InfoStat, Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba). Metabolite content variations were assessed by t test analyses using the algorithm incorporated into Microsoft Excel considering P < 0.05 as a significant outcome. Since no statistical differences were observed between GFP#5 and GFP#6 transgenic lines, metabolite profile data were pooled to increase the power of the tests applied. PCA was carried out exactly as detailed by Roessner et al. (2000) using the InfoStat software, and three-dimensional graphs were drawn with MATLAB version 7.4.0.287 (R2007a; The MathWorks). Multivariate outliers were detected with Mahalanobis distance and removed from the PCA analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Relative metabolite contents in MG tomatoes infiltrated with MES, pTRV2-empty, pTRV2gfp, and pTRV2pds.
Supplemental Table S2. Relative metabolite contents in ripe tomatoes infiltrated with MES, pTRV2-empty, pTRV2gfp, and pTRV2pds.
Supplemental Table S3. Relative metabolite contents in Arabidopsis leaves infiltrated with MES, pTRV2-empty, pTRV2gfp, and pTRV2pds.
Supplementary Material
Acknowledgments
We thank Valeria Peralta, Valeria Beracochea, and Teresa Cabrera for technical assistance with transgenic plants and Agustín Montenegro and Ignacio Tevez for taking care of the tomato plants.
References
- Almeida J, Quadrana L, Asís R, Setta N, de Godoy F, Bermúdez L, Otaiza SN, Corrêa da Silva JV, Fernie AR, Carrari F, et al. (April 28, 2011) Genetic dissection of vitamin E biosynthesis in tomato. J Exp Bot http://dx.doi.org/10.1093/jxb/err055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester AR, Molthoff J, de Vos R, Hekkert BL, Orzaez D, Fernández-Moreno JP, Tripodi P, Grandillo S, Martin C, Heldens J, et al. (2010) Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol 152: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez L, Urias U, Milstein D, Kamenetzky L, Asis R, Fernie AR, Van Sluys MA, Carrari F, Rossi M. (2008) A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. J Exp Bot 59: 2875–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP. (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EG, Hall RD, Bovy AG, Luo J, et al. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Buttriss JL, Diplock AT. (1984) High-performance liquid chromatography methods for vitamin E in tissues. Methods Enzymol 105: 131–138 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Chamovitz D, Sandmann G, Hirschberg J. (1993) Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J Biol Chem 268: 17348–17353 [PubMed] [Google Scholar]
- Chawade A, Sikora P, Bräutigam M, Larsson M, Vivekanand V, Nakash MA, Chen T, Olsson O. (2010) Development and characterization of an oat TILLING-population and identification of mutations in lignin and beta-glucan biosynthesis genes. BMC Plant Biol 10: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Gookin TE, Hunter DA, Clark DG, Reid MS. (2004) Chalcone synthase as a reporter in virus-induced gene silencing studies of flower senescence. Plant Mol Biol 55: 521–530 [DOI] [PubMed] [Google Scholar]
- Chung BN, Canto T, Palukaitis P. (2007) Stability of recombinant plant viruses containing genes of unrelated plant viruses. J Gen Virol 88: 1347–1355 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D. (2003a) Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol 131: 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collakova E, DellaPenna D. (2003b) The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol 133: 930–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Fischer RL. (1988) Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J 7: 3315–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR. (2010) The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol 154: 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Vincent K, Sharp P. (2009) Simultaneous mutation detection of three homoeologous genes in wheat by high resolution melting analysis and mutation surveyor. BMC Plant Biol 9: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell LH, Orzáez D, López-Peña L, Pineda B, Antón MT, Moreno V, Granell A. (2009) A multisite Gateway-based toolkit for targeted gene expression and hairpin RNA silencing in tomato fruits. Plant Biotechnol J 7: 298–309 [DOI] [PubMed] [Google Scholar]
- Faivre-Rampant O, Gilroy EM, Hrubikova K, Hein I, Millam S, Loake GJ, Birch P, Taylor M, Lacomme C. (2004) Potato virus X-induced gene silencing in leaves and tubers of potato. Plant Physiol 134: 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AI, Viron N, Alhagdow M, Karimi M, Jones M, Amsellem Z, Sicard A, Czerednik A, Angenent G, Grierson D, et al. (2009) Flexible tools for gene expression and silencing in tomato. Plant Physiol 151: 1729–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ. (2001) Fructose 2,6-bisphosphate activates pyrophosphate:fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Pinto ME, Holloway DE, Bramley PM. (2000) Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. (2005) Virus-induced gene silencing in tomato fruit. Plant J 43: 299–308 [DOI] [PubMed] [Google Scholar]
- Gady ALF, Hermans FWK, Van de Wal MHBJ, van Loo EN, Visser RGF, Bachem CWB. (2009) Implementation of two high throughput techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Stevens R, Gil L, Gilbert L, Gest N, Petit J, Faurobert M, Maucourt M, Deborde C, Moing A, et al. (2009) An integrative genomics approach for deciphering the complex interactions between ascorbate metabolism and fruit growth and composition in tomato. C R Biol 332: 1007–1021 [DOI] [PubMed] [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, Bouchet B, Faurobert M, Gouble B, Page D, Garcia V, et al. (2009) GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J 60: 499–508 [DOI] [PubMed] [Google Scholar]
- Grusak MA, DellaPenna D. (1999) Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol 50: 133–161 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD. (1996) Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker BH, Chu CC, Sonnewald U, Willmitzer L, Fernie AR. (2003) In plants the alc gene expression system responds more rapidly following induction with acetaldehyde than with ethanol. FEBS Lett 535: 136–140 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. (1996) The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569–593 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Lin Z, Yin K, Wang X, Liu M, Chen Z, Gu H, Qu LJ. (2007) Virus induced gene silencing of AtCDC5 results in accelerated cell death in Arabidopsis leaves. Plant Physiol Biochem 45: 87–94 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP. (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54: 701–711 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. (2003) Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD. (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Maloney GS, Kochevenko A, Tieman DM, Tohge T, Krieger U, Zamir D, Taylor MG, Fernie AR, Klee HJ. (2010) Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol 153: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mathieu-Rivet E, Gévaudant F, Sicard A, Salar S, Do PT, Mouras A, Fernie AR, Gibon Y, Rothan C, Chevalier C, et al. (2010) Functional analysis of the anaphase promoting complex activator CCS52A highlights the crucial role of endo-reduplication for fruit growth in tomato. Plant J 62: 727–741 [DOI] [PubMed] [Google Scholar]
- Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M, et al. (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149: 1505–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR. (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A, Rahman A, Yamashita T, Taira H, Kidou S. (2007) Virus-induced gene silencing of P23k in barley leaf reveals morphological changes involved in secondary wall formation. J Exp Bot 58: 2617–2625 [DOI] [PubMed] [Google Scholar]
- Orzaez D, Medina A, Torre S, Fernández-Moreno JP, Rambla JL, Fernández-Del-Carmen A, Butelli E, Martin C, Granell A. (2009) A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol 150: 1122–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzaez D, Mirabel S, Wieland WH, Granell A. (2006) Agroinjection of tomato fruits: a tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol 140: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page JE, Hause G, Raschke M, Gao W, Schmidt J, Zenk MH, Kutchan TM. (2004) Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol 134: 1401–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Brachmann A, Welham T, Binder A, Charpentier M, Groth M, Haage K, Markmann K, Wang TL, Parniske M. (2009) TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol 151: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ. (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17: 471–482 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60: 424–435 [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. (2000) Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg D, Thompson TS, German TL, Willis DK. (2006) Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J Virol Methods 138: 49–59 [DOI] [PubMed] [Google Scholar]
- Ryu CM, Anand A, Kang L, Mysore KS. (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse solanaceous species. Plant J 40: 322–331 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L, et al. (2005) GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Schijlen E, Ric de Vos CH, Jonker H, van den Broeck H, Molthoff J, van Tunen A, Martens S, Bovy A. (2006) Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol J 4: 433–444 [DOI] [PubMed] [Google Scholar]
- Shintani D, DellaPenna D. (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282: 2098–2100 [DOI] [PubMed] [Google Scholar]
- Stevens R, Buret M, Duffé P, Garchery C, Baldet P, Rothan C, Causse M. (2007) Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol 143: 1943–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Yamada K, Shimada T, Hara-Nishimura I. (2004) Endoplasmic reticulum-resident proteins are constitutively transported to vacuoles for degradation. Plant J 39: 393–402 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ. (2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latché A, Pech JC, Fernie AR, Bouzayen M. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J. (2008) Applying genotyping (TILLING) and phenotyping analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Qu J, Bui HT, Chua NH. (2009) Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnol J 7: 964–976 [DOI] [PubMed] [Google Scholar]
- Zanor MI, Rambla JL, Chaïb J, Steppa A, Medina A, Granell A, Fernie AR, Causse M. (2009) Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J Exp Bot 60: 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.