Abstract
The major quantitative trait locus (QTL) Phosphorus uptake1 (Pup1) confers tolerance of phosphorus deficiency in soil and is currently one of the most promising QTLs for the development of tolerant rice (Oryza sativa) varieties. To facilitate targeted introgression of Pup1 into intolerant varieties, the gene models predicted in the Pup1 region in the donor variety Kasalath were used to develop gene-based molecular markers that are evenly distributed over the fine-mapped 278-kb QTL region. To validate the gene models and optimize the markers, gene expression analyses and partial allelic sequencing were conducted. The markers were tested in more than 80 diverse rice accessions revealing three main groups with different Pup1 allele constitution. Accessions with tolerant (group I) and intolerant (group III) Pup1 alleles were distinguished from genotypes with Kasalath alleles at some of the analyzed loci (partial Pup1; group II). A germplasm survey additionally confirmed earlier data showing that Pup1 is largely absent from irrigated rice varieties but conserved in varieties and breeding lines adapted to drought-prone environments. A core set of Pup1 markers has been defined, and sequence polymorphisms suitable for single-nucleotide polymorphism marker development for high-throughput genotyping were identified. Following a marker-assisted backcrossing approach, Pup1 was introgressed into two irrigated rice varieties and three Indonesian upland varieties. First phenotypic evaluations of the introgression lines suggest that Pup1 is effective in different genetic backgrounds and environments and that it has the potential to significantly enhance grain yield under field conditions.
Phosphorus uptake1 (Pup1) is a major quantitative trait locus (QTL) located on rice (Oryza sativa) chromosome 12 that is associated with tolerance of phosphorus (P) deficiency in soil (Wissuwa et al., 1998, 2002). Kasalath, the Pup1 donor variety, was initially identified in a screening of 30 diverse rice genotypes in a P-deficient soil in Japan under rain-fed conditions. Subsequently, phenotypic data derived from Nipponbare contrasting near isogenic lines (NILs) with and without the QTL showed that Pup1 increases P uptake (Wissuwa and Ae, 2001a; Wissuwa et al., 2002) and confers a significant yield advantage (2- to 4-fold higher grain weight per plant) in pot experiments using different P-deficient soil types and environments (Chin et al., 2010).
Since the identification of Pup1 almost a decade ago, considerable efforts have been made to understand the underlying tolerance mechanism (Wissuwa and Ae, 2001a; Wissuwa, 2005). However, detailed analyses of common plant P responses (e.g. organic acid exudation, root hair growth) are difficult in soil, and the P responses could not be detected under the experimental conditions applied (Wissuwa, 2005). Likewise, gene expression analyses (Agilent gene chips) of root samples did not reveal specific up-regulation of P transporters or other P uptake-related genes in Pup1 NILs (Pariasca-Tanaka et al., 2009). In the original QTL mapping as well as in a parallel study, an intermediate-effect QTL was identified on the short arm of chromosome 6 (Ni et al., 1998; Wissuwa et al., 1998). Many P-responsive genes colocalize with this QTL region in the Nipponbare reference genome, including the phosphate starvation-induced transcription factor gene OsPTF1. As was shown by Yi et al. (2005), overexpression of OsPTF1 in Nipponbare increased P uptake and vegetative growth in soil and in P-deficient culture solution. However, no obvious P uptake-related gene is located in the Pup1 region on chromosome 12, although this region had a larger effect on tolerance (Heuer et al., 2009).
To identify the genes associated with Pup1, the genomic region had been sequenced in the donor variety Kasalath, revealing a complex locus with large insertions/deletions (INDELs) and overall low conservation compared with the corresponding genomic regions in the Nipponbare and 93-11 reference genomes. Of the 68 preliminary gene models, 16 genes code for transposable elements, more than 40 show partial sequence similarity to transposons, and other genes cannot currently be annotated with certainty. The remaining candidate genes encode a putative fatty acid oxygenase, a dirigent-like protein, an aspartic proteinase, hypothetical proteins, and a putative protein kinase (Heuer et al., 2009; this paper). In parallel to the identification of the Pup1 genes, molecular markers have been developed to initiate marker-assisted breeding. A first set of gene-based markers was developed based on the preliminary Pup1 gene models and published by Chin et al. (2010). The germplasm survey conducted with these markers revealed that Pup1 is largely absent from modern irrigated rice varieties but that it is highly conserved in drought-tolerant breeding lines and upland varieties. This suggested that breeders are unknowingly selecting for Pup1 in breeding programs that target drought-prone environments (Chin et al., 2010).
The impact of Pup1 and other QTLs that enhance yield in P-deficient soil and/or under drought stress (Bernier et al., 2009; Venuprasad et al., 2009) is potentially very high, since about 50% of the rain-fed rice in Asia is grown on problem soils (Haefele and Hijmans, 2007). Both P deficiency and drought are widespread problems, particularly in soils with acidic pH and high concentrations of cations, for example, aluminum and iron, which complexate phosphate (Kochian et al., 2005; Ismail et al., 2007; Xue et al., 2007). Due to the poor productivity of soils as well as the frequent occurrence of drought and floods in the geographical regions concerned, the local incidence of poverty is overproportionally high. In addition, poor farmers often have no access to or means to purchase fertilizer and other agrochemicals, which further aggravates the situation. In the future, increasing energy costs and limited P resources (Cordell et al., 2009; Van Kauwenbergh, 2010) will lead to rising fertilizer prices, which will increase the costs of food production and consequently food prices and availability. The recent food crisis had severe and long-lasting effects on resource-poor farmers and consumers in Asia and Africa, which often spend a large proportion (25%–40%) of their income on rice alone (Dawe et al., 2010). Efforts have since been intensified to develop rice that is tolerant of major abiotic (and biotic) stresses, and the first rice varieties tolerant of submergence and drought have been released in Asia (Bernier et al., 2009; Singh and Singh, 2010; Manzanilla et al., 2011). In addition, the Global Rice Science Partnership has been put in place to enhance the efficiency of rice breeding and increase the likelihood of impact (International Rice Research Institute, 2010).
The importance of rice varieties with enhanced yield in P-deficient soils and/or enhanced P fertilizer use efficiency has now been recognized and requires large-scale breeding efforts. However, phenotypic screenings in problem soils are often constrained by the occurrence of companion stresses (e.g. aluminum toxicity) that restrict root growth and impair phenotyping for P efficiency. For those soils, varieties with multiple tolerances need to be developed. On the other hand, favorable soils without major stresses are often saturated with P due to continuous P fertilizer application, and endogenous P concentrations of the soil need to be reduced before genotypic differences can be determined. This requires several years of continuous cropping without application of P fertilizer. For some abiotic stresses (e.g. salinity and aluminum toxicity), reliable culture solution-based phenotyping systems are available that can be used for large-scale screenings. However, it has repeatedly been shown that the phenotypic differences between contrasting Pup1 NILs cannot be observed in culture solution; therefore, soil-based screenings are inevitable. The availability of molecular markers that can at least partially replace and/or complement phenotypic evaluations in the field, therefore, is of particular value for the development of P-efficient rice varieties. A first generation of gene-based Pup1 molecular markers for the development of Pup1 introgression lines and an assessment of the Pup1 locus in diverse rice genotypes have recently been published (Chin et al., 2010). However, the germplasm survey conducted showed that individual Kasalath Pup1 alleles are present (“partial Pup1”) in many of the analyzed rice accessions. This is likely due to the large number of transposable elements within the Pup1 region (Heuer et al., 2009) causing genomic rearrangements or to unspecific amplification of markers in certain genotypes. Because of the genetic instability of the Pup1 locus, many of the developed markers were not informative when tested in a wider range of rice accessions, and the most reliable markers were located in a large Kasalath-specific INDEL and therefore were dominant (Chin et al., 2010). Dominant markers are of some limitation in breeding programs, since they do not differentiate homozygous and heterozygous progeny plants and are also not suitable for the development of single-nucleotide polymorphism (SNP) markers, which are increasingly used for genotyping in high-throughput breeding programs (McNally et al., 2009; McCouch et al., 2010; Zhao et al., 2010).
Here, we report on the development of an extended and improved set of Pup1-specific molecular markers based on gene models that have now been verified through gene expression and allelic sequencing data. The markers differentiate between three main groups of genotypes with different Pup1 haplotypes and were applied for the development of two irrigated and three Indonesian upland Pup1 introgression lines by marker-assisted backcrossing (MABC). Initial field-based phenotypic data are presented indicating that Pup1 has the potential to enhance yield in different genetic backgrounds and P-deficient environments.
RESULTS
The Kasalath genomic Pup1 sequence (GenBank accession no. AB458444.1) and the preliminary gene models published by Heuer et al. (2009) were initially used as a basis for the development of an improved and extended set of molecular markers. The Kasalath Pup1 locus is structurally defined by the presence of a large (greater than 90 kb) INDEL that is absent from the Nipponbare reference genome, and an earlier set of Pup1 markers targeting this region was published by Chin et al. (2010). Since all of these markers were clustered in the INDEL and therefore were dominant, additional genes that are at least partially conserved in Nipponbare were targeted in this study to develop codominant markers and to identify SNPs suitable for high-throughput marker applications.
Pup1 Gene Models
The annotation of Pup1 genes in the Kasalath genomic region is challenged by the presence of many transposable elements and truncated genes (Heuer et al., 2009). Based on in silico analyses, a set of eight priority candidate genes that are non-transposon-related protein-coding genes has now been selected for in-depth analyses (Table I). For the development of molecular markers, these genes were prioritized, since coding genes are more likely to be conserved and stable than transposable elements across diverse rice accessions. Detailed marker information on target genes can additionally provide supportive information on the Pup1 major gene and therefore is complementary to the ongoing cloning efforts (International Rice Research Institute, unpublished data). An overview of the relative position of the genes and current gene models is given in Figure 1.
Table I. Pup1 candidate genes and RT-PCR primers.
| Gene Name | Annotation | TIGR6 BLASTp | E Value | RT-PCR Primers (5′–3′)a | Gene Expressed |
| OsPupK01-1 | Hypothetical protein | No significant similarity; located in intergenic region between LOC_Os12g26260 and LOC_Os12g26270 | 4.10E-07 | F: ACACGTACAATGTCACCGGG R: ATGTCAACGCCACGTTCACG | No |
| OsPupK04-1 | Highly similar to α-DOX2 (LOC_Os12g26290) | α-DOX2, putative, expressed | 0 | F: GGGATATCAAGCTTGTGGTGR: GAATGCTGTTTCGCTTATGG | Yes |
| OsPupK05-1 | Located within OsPupK04, expressed | Transposon protein, putative, CACTA, En/Spm subclass | 0.97 | F: AGTACAGTCCGGCGTCATACR: CCGAGATCTGGTCCTCAATA | Yes |
| OsPupK20-2 | Highly similar to dirigent protein, putative, expressed (LOC_Os12g26380) | Dirigent, putative, expressed | 3.50E-100 | F: CTGGACTTGACCCCAATGTAR: TCTGATGGAGTGTTCGGAGT | Yes |
| OsPupK29-1 | Similar to expressed proteins LOC_Os12g26390 and LOC_Os12g26410 | LOC_Os12g26390, expressed, LOC_Os12g26410, expressed | 0 6.2E-101 | F: CCAATGCATCCAATTCTTGTR: ATGAGCCCAGATTACGAATG | Yes |
| OsPupK46-2 | Most similar to protein kinase genes (LOC_Os01g49580, LOC_Os01g49614, LOC_Os08g24630) | LOC_Os01g49580, expressed, LOC_Os01g49614, expressed, LOC_Os08g24630, expressed | 1E-93 1.3E-91 1.1E-87 | F: AGGAAGATGGTTGTCGTTGGR: TTCACACCAAACAGTGTTGTC | Yes |
| OsPupK53-1 | Insignificant similarity to zinc finger CCCH-type family protein, expressed (LOC_Os05g10670) | Zinc finger CCCH-type family protein, putative, expressed | 6.40E-37 | F: TTCAGCTTCTTCGCCGACACR: TAGACGAGCACGTCGTTGAC | No |
| OsPupK67-1 | Highly similar to aspartic proteinase precursor nepethesin-1 precursor (LOC_Os12g262470) | Aspartic proteinase nepenthesin-1 precursor, putative, expressed | 9.50E-211 | F: CACGAGCTATTTCGTGTGGGR: GGTACGAGAACTTGCCGAAC | No |
F, Forward primer; R, reverse primer.
Figure 1.
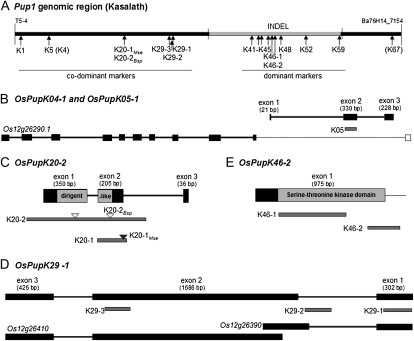
Pup1 gene models and positions of markers. A, The genomic Pup1 region of the tolerant donor Kasalath is defined by the markers T5-4 and Ba76H14_7154 (Heuer et al., 2009; accession no. AB458444.1) and characterized by a large INDEL (gray line) with adjacent regions partially conserved in Nipponbare (black lines). The positions and names of the developed codominant and dominant markers are indicated. Markers K4 and K67 were not polymorphic. B to E, The targeted gene models and marker positions within the models. B, The putative gene OsPupK05-1 is located within an intron of gene OsPupK04-1, which is highly similar to the shown Nipponbare gene model Os12g26290. C, For the dirigent-like gene model OsPupK20-2, two markers were developed, and PCR products can be digested with Bsp1286I (white triangles) and MseI (gray triangle). D, The hypothetical protein gene OsPupK29-1 is partially similar to two genes in Nipponbare, and the markers target exon 1, exon 2, and an intron. E, For the protein kinase OsPupK46-2, the markers amplify a region flanking the protein kinase domain (K46-1) and the 3′ region (K46-2). Black boxes in the gene models indicate exons, and lines indicate introns and untranslated regions. Conserved protein domains are highlighted in gray.
Based on the below-described gene expression analyses in conjunction with detailed in silico sequence analyses, two Pup1 gene models have been revised. The new model for the dirigent-like gene (OsPupK20-2) has a different intron-exon structure, which changes the N terminus of the predicted protein sequence (Fig. 1C), and is highly similar to Os12g26380 (3.5E-100). The new model for the Pup1 protein kinase gene (OsPupK46-2) has a revised coding region and predicts an intronless gene (Fig. 1E) most similar to Os01g49580 (1E-93). Both revised models are supported by cDNA sequence data (data not shown). All Pup1 gene models targeted for marker design in this study are published under GenBank accession number AB458444.1 (Heuer et al., 2009).
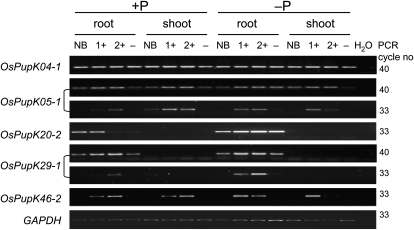
To assess whether the short-listed Pup1 candidate genes are expressed and responsive to P, reverse transcription (RT)-PCR analyses were conducted using root and shoot samples derived from Nipponbare and contrasting Pup1 NILs grown in P-deficient soil with and without application of P fertilizer (Fig. 2). The data show that OsPupK04 and OsPupK05 are both ubiquitously expressed. However, higher expression of OsPupK05 in the Pup1 NILs was detected at a lower PCR cycle number (Fig. 2, panels 1–3). The gene model of OsPupK05 is a hypothetical protein located in the reverse direction within a putative intron of OsPupK04, a putative fatty acid oxygenase (Fig. 1B). Expression of the dirigent-like gene OsPupK20 (Fig. 1C) is restricted to roots with an apparent higher transcript abundance under P-deficient conditions (Fig. 2, panel 4). Likewise, the hypothetical protein OsPupK29 (Fig. 1D; for details, see Heuer et al., 2009) is specifically expressed in roots with a higher transcript abundance under P-deficient conditions, especially in the Pup1 NILs (Fig. 2, panels 5 and 6). The Pup1 protein kinase gene OsPupK46-2 is expressed under −P and +P conditions in roots and in shoots (Fig. 2, panel 7). Since this gene is absent from the Nipponbare genome, no expression was detected in Nipponbare and sister NILs without the tolerant Pup1 locus.
Figure 2.
RT-PCR gene expression analysis. The Pup1 genes indicated were analyzed by RT-PCR using RNA samples derived from roots and shoots of plants grown in P-deficient soil with (+P) and without (−P) P fertilizer application. Two different tolerant Nipponbare-Pup1 NILs with the Kasalath Pup1 locus were used (NIL24-4 = 1+; NIL14-4 = 2+). Nipponbare (NB) and a NIL14-4 sister line without the tolerant Pup1 locus (NIL14-6 = −) were used as intolerant controls. cDNA samples were analyzed by PCR using the cycle number indicated on the right. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included as a control. H2O, Water control.
For the genes OsPupK01, OsPupK53, and OsPupK67, several primer pairs were designed targeting different regions of the predicted gene models, but expression was never observed in any of the root and shoot RNA samples analyzed (data not shown).
Gene-Based Molecular Markers
As mentioned above, the hypothetical gene OsPupK05 is located within a predicted intron of OsPupK04 (Fig. 1B), based on a sequence comparison with the most similar Nipponbare gene coding for a fatty acid α-oxygenase (α-DOX2; Os12g26290). The genomic sequence of the Nipponbare gene is more than 10 kb, and several alternative splice products have been identified (Rice Genome Browser version 6.1 at the Rice Genome Annotation Project). More detailed analyses are needed to clarify whether OsPupK05 is an independent gene or if it represents a splice variant of OsPupK04. Several markers specifically targeting OsPupK04 were designed but failed to reveal reliable data (data not shown). Therefore, a marker (K5) located within OsPupK05 was selected to target this locus and accordingly represents both genes (Fig. 1, A and B; Table II).
Table II. Primer sequences of gene-specific Pup1 markers.
| Marker Name | Pup1 Gene Model | Marker Type | Physical Location |
Amplicon Size K/N | Primer Sequence (5′−3′)a | |
| Kasalath AB458444.1 | Nipponbare Chromosome 12 | |||||
| bp | bp | |||||
| K1 | OsPupK01-1 | Codominant | 96,175−96,299 | 15,315,156−15,315,277 | 125/122 | F: AGTCTGGATGGACAACTCTGCCTG |
| R: TGCTAGCTCATTGCCGTTACGTCG | ||||||
| K5 | OsPupK05-1 | Codominant | 116,430−116,701 | 15,336,063−15,336,342 | 272/280 | F: ATTCAGACATCGACGGCGAC |
| R: TCCTCGTAAACATGGCTTGC | ||||||
| K20-1 | OsPupK20-2 | Codominant | 169,881−170,120 | 15,410,254−15,410,496 | 240/243 | F: TCAGGTGATGGGAATCATTG |
| R: TGTTCCAACCAAACAACCTG | ||||||
| K20-1Mse | OsPupK20-2 | CAPS (Mse1) | 201/243 | |||
| K20-2 | OsPupK20-2 | Codominant | 169,290−170,607 | 15,409,652−15,410,981 | 982/995 | F: TCAAAAATTTCTTCAGGTATGTACTCC |
| R: TTGGGTGATCAGCTTTCAGA | ||||||
| K20-2Bsp | OsPupK20-2 | CAPS (Bsp1286I) | K, 231 + 349 + 402; | |||
| N, 413 + 582 | ||||||
| K29-1 | OsPupK29-1 | Codominant | 205,067−205,287 | 15,431,572−15,431,786 | 212/206 | F: ATGGCCAACGGGGTAGAG |
| R: GTCCAGGTAACCACGAGGAA | ||||||
| K29-2 | OsPupK29-1 | Codominant | 204,398−204,616 | 15,430,672−15,430,883 | 291/212 | F: CCCGTCTGCGTTCTACCTTA |
| R: CTCCCGTCAAGCACAAATCT | ||||||
| K29-3 | OsPupK29-1 | Codominant | 202,698−202,933 | 15,419,578−15,419,825 | 236/248 | F: TTCGTCCAGATGCTGCTATG |
| R: TCTTCGGTGTAATTGGCACA | ||||||
| K41 | OsPupK41-1 | Dominant | 262,050−262,431 | Absent | 382/− | F: TGATGAATCCATAGGACAGCGT |
| R: TCAGGTGGTGCTTCGTTGGTA | ||||||
| K42 | OsPupK42-1 | Dominant | 267,154−268,071 | Absent | 918/− | F: CCCGAGAGTTCATCAGAAGGA |
| R: AGTGAGTGGCGTTTGCGAT | ||||||
| K43 | OsPupK43-1 | Dominant | 268,590−269,501 | Absent | 912/− | F: AGGAGGATGAGCCTGAAGAGA |
| R: TCGCACTAACAGCAGCAGATT | ||||||
| K45 | OsPupK45-1 | Dominant | 274,072−274,344 | Absent | 276/− | F: GCGGAAGAAGAGGATAACGA |
| R: TCTAGGCTTCGTTTGGCAAG | ||||||
| K46-1 | OsPupK46-2 | Dominant | 275,710−276,232 | Absent | 523/− | F: TGAGATAGCCGTCAAGATGCT |
| R: AAGGACCACCATTCCATAGC | ||||||
| K46-2 | OsPupK46-2 | Dominant | 276,371−276,597 | Absent | 227/− | F: AGGAAGATGGTTGTCGTTGG |
| R: TTCACACCAAACAGTGTTGTC | ||||||
| K48 | OsPupK48-1 | Dominant | 282,795−283,640 | Absent | 847/− | F: CAGCATTCAGCAAGACAACAG |
| R: ATCCGTGTGGAGCAACTCATC | ||||||
| K52 | OsPupK52-1 | Dominant | 300,870−301,374 | Absent | 505/− | F: ACCGTTCCCAACAGATTCCAT |
| R: CCCGTAATAGCAACAACCCAA | ||||||
| K59 | OsPupK59-1 | Dominant | 324,843−325,392 | Absent | 550/− | F: GGACACGGATTCAAGGAGGA |
| R: TGCTTTCCATTTGCGGCTC | ||||||
F, Forward primer; R, reverse primer.
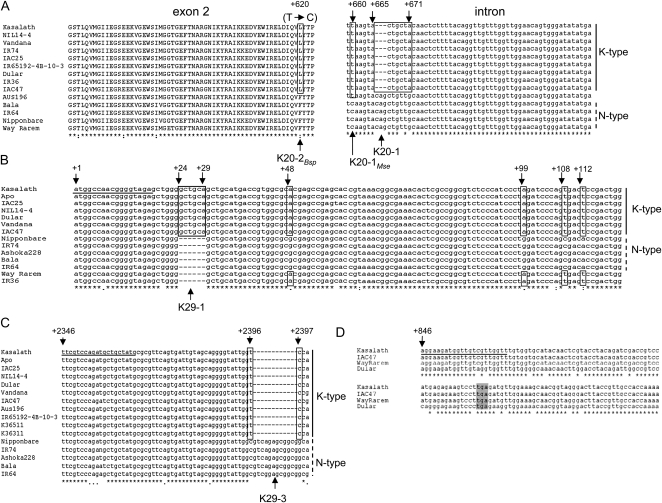
For the dirigent-like gene, two markers (K20-1 and K20-2) were designed that target different regions of the gene (Figs. 1, A and C, 3, and 4; Table II). To validate sequence polymorphisms between the Kasalath gene and the most similar Nipponbare gene (Os12g26380), K20-2 PCR amplicons of 14 rice accessions were sequenced. The data revealed four SNPs and a Kasalath-specific 3-bp deletion (Fig. 3A; only the polymorphic region is shown). The detected polymorphisms are highly conserved, and only three different alleles can be distinguished. Based on this information, the marker K20-1 was designed targeting the INDEL mutation. The resolution of this marker can be enhanced by subsequent digestion of the PCR product with MseI (K20-1Mse), which recognized the TTAA motif located 2 bp upstream of the INDEL in the Kasalath allele (Figs. 3A and 4). A nonsynonymous SNP (at 620 bp) creates a mutation in a Bsp1286I restriction site (GDGCHC; D = A/G/T, H = A/C/T) and causes a change in the amino acid sequence (Leu-177 in Kasalath, Phe-177 in Nipponbare). Digestion of PCR amplicons with Bsp1286I (K20-2Bsp) generates three DNA fragments specific for the Kasalath allele and two DNA fragments in non-Kasalath alleles (Figs. 3A and 4).
Figure 3.
Sequence comparisons of marker amplicons. A, PCR amplicons of five markers were sequenced in the rice varieties indicated to determine the specificity of the primers and validate sequence polymorphisms. For OsPupK20-2, a nonsynonymous SNP at +620 bp was identified that changed the amino acid sequence in exon 2 (T in Nipponbare, C in Kasalath; left) and created a Bsp1286I site in Kasalath-type alleles but not in non-Kasalath (N-type) alleles. In the adjacent intron, one SNP (+660 bp) and one INDEL (+665–671 bp) were identified. The SNP creates an MseI restriction site (right). B, For OsPupK29-1, a polymorphic region with one INDEL and four SNPs is amplified by the marker K29-1. C, Another INDEL in OsPupK29-1 is targeted by the marker K29-3. D, PCR products of the dominant marker K46-2 (stop codon is highlighted in gray) were sequenced in three varieties that possess the OsPupK46 gene. Only polymorphic regions of the sequenced DNA fragments are shown.
Figure 4.
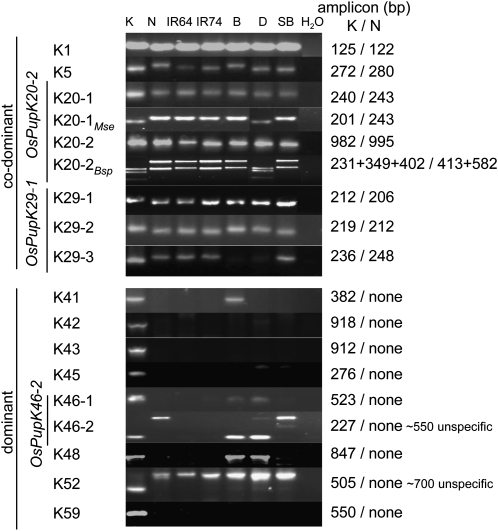
PCR amplicons of Pup1 gene-specific markers. Seven representative rice varieties were genotyped with nine codominant (top panels) and nine dominant (bottom panels) Pup1-specific markers. The markers target a total of 12 Kasalath Pup1 gene models. PCR products K20-1 and K20-2 were digested with MseI (K20-1Mse) and Bsp1286I (K20-2Bsp), respectively, to enhance resolution. K, Kasalath; N, Nipponbare; B, Batur; D, Dodokan; SB, Situ Bagendit; H2O, water control. The sizes of the DNA fragments in Kasalath and Nipponbare (K/N) and unspecific amplicons that sometimes occur are indicated on the right. The absence of PCR amplicons with dominant markers is indicated as “none.”
The Pup1 gene OsPupK29 codes for a hypothetical protein with high but partial sequence similarity to two expressed protein genes (Os12g26390 and Os12g26410) in the Nipponbare reference genome (Fig. 1D; Heuer et al., 2009). Based on a comparative sequence analysis of the three genes, three markers were developed that target INDELs located in the first exon (K29-1), first intron (K29-2), and second exon (K29-3; Figs. 1, A and D, and 4; Table II). The markers K29-1 and K29-3 were the most reliable (see below), and PCR amplicons were sequenced in representative rice varieties revealing high conservation of the targeted polymorphism (Fig. 3, B and C; only polymorphic regions are shown).
For the Pup1 protein kinase gene, two markers were designed amplifying a region including the conserved kinase domain (marker K46-1) and the 3′ untranslated region (marker K46-2; Fig. 1E). Both markers are dominant, since this gene is located in the large approximately 90-kb Kasalath-specific INDEL region (Fig. 1A), and therefore are absent from the Nipponbare reference genome and other intolerant rice accessions (Fig. 4; see below). The Pup1 gene OsPupK67 is nearly identical to an aspartic proteinase gene (Os12g26470), and only four SNPs are present that distinguish the two sequences (data not shown). Since we were unable to detect expression of this gene (see above), this gene was excluded from further analyses in this study.
The gene model OsPupK01 is located in the Pup1 region partially conserved in Nipponbare and is highly similar (96.0%) to an intergenic region between the Nipponbare genes Os12g26260 and Os12g26270 annotated as Gypsy-type transposon and hypothetical protein, respectively. RT-PCR analyses of roots and shoots showed that this putative gene is not expressed (data not shown); therefore, it is likely that OsPupK01 is a pseudogene or incorrectly predicted. However, a molecular marker (K1) for this gene was included in the study because of its physical location at the 5′ border of the Pup1 locus (Fig. 1A). This marker amplifies a DNA fragment flanking a 3-bp insertion (GTC) specific to the Kasalath gene model (Fig. 4; Table II). The remaining markers target genes coding for putative transposable elements (OsPupK41–OsPupK44), hypothetical proteins (OsPupK45 and OsPupK48), and an unknown gene (OsPupK52) located in the Kasalath-specific INDEL region (Chin et al., 2010; Figs. 1a and 4; Table II).
Determination of Pup1 Haplotypes in Breeding Materials
To standardize PCR conditions and evaluate the markers, genomic DNA from Kasalath, Nipponbare, and five rice varieties that had been selected for Pup1 breeding was analyzed. The data show that the developed markers clearly differentiate between Kasalath and Nipponbare (non-Kasalath) alleles (Fig. 4). With the dominant markers K46-2 and K52, unspecific PCR amplicons are sometimes observed in Nipponbare and other genotypes that can be distinguished from the specific amplicons due to their larger size.
The marker data revealed that IR64 and IR74, the two irrigated varieties selected as Pup1-recipient parents, did not possess Kasalath alleles at any of the analyzed loci, with the exception of marker K5. As mentioned above, the gene models targeted by this marker are currently under validation. The data obtained for the three Indonesian varieties selected as Pup1-recipient parents suggest that they possess different haplotypes with at least the partial presence of Kasalath Pup1 alleles. Although Situ Bagendit is most similar to IR64 and IR74 (only the K5 Kasalath allele), the other two Indonesian varieties possess additional Kasalath alleles, including the Pup1 protein kinase (Fig. 4). In both varieties, the markers targeting OsPupK29 revealed inconsistent results, suggesting that Batur and Dodokan may have a different allele of this gene (Fig. 4).
Germplasm Survey with Pup1 Markers
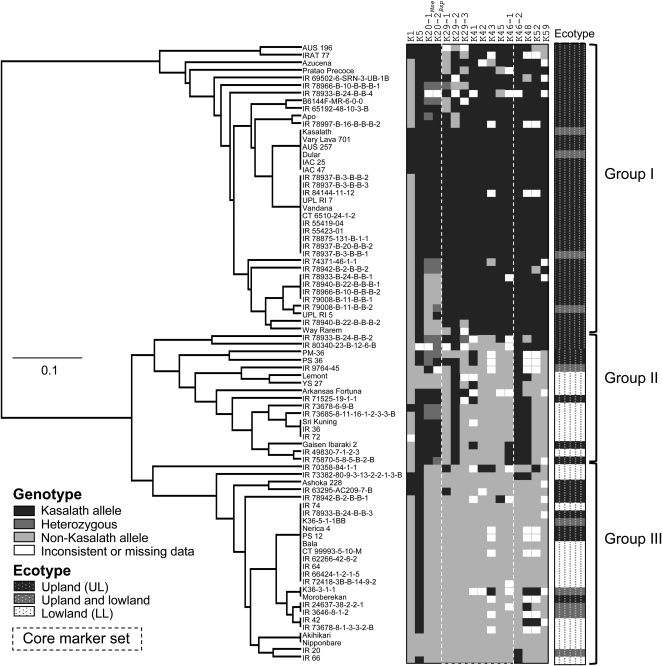
In order to determine the Pup1 haplotype in a wider range of genotypes, 81 rice varieties and breeding lines were genotyped. The number of accessions that possess Kasalath alleles at all analyzed loci was small and was, apart from Kasalath, observed in only five genotypes. Among these were three varieties (Dular, IAC25, and IAC47) that are known to be highly tolerant of P deficiency (Wissuwa and Ae, 2001b). Likewise, only a few varieties (Akihikari and Nipponbare from Japan) lacked Kasalath alleles at all loci.
More detailed analyses showed that several markers are not associated and are not suitable to differentiate between Pup1 haplotypes within the analyzed genotypes. This was especially the case for the markers K1 (14.8% Kasalath allele frequency) and K5 (90.3% Kasalath allele frequency; Fig. 5), although K1 showed some specificity for the analyzed aus-type varieties (Kasalath, Dular, AUS257, and AUS196) and traditional upland-adapted genotypes (e.g. IAC47, IAC25, Vary Lava 701, and Ashoka 228). A subsequent analysis of all Pup1 markers with GGT 2.0 (Van Berloo, 2008) revealed very low r2 values for K1 and K5, suggesting that they are not associated with the other Pup1 markers (Fig. 6).
Figure 5.
Pup1 haplotype in diverse rice genotypes. Eighty-one diverse rice varieties and breeding lines were genotyped using the Pup1-specific markers indicated. Based on the Pup1 core marker set (highlighted with dashed lines; see text for details), an UPGMA cluster analysis was conducted using Powermarker version 3.25 (Sokal and Michener, 1958; Liu and Muse, 2005). Three main groups (I, II, III) with different allelic composition at the Pup1 locus were identified. The different allele calls (Kasalath, non-Kasalath, and heterozygous) are color coded as indicated at bottom left. The rice genotypes analyzed are adapted to different rice agroecosystems [upland (UL): aerobic, unfavorable, and drought prone; lowland/irrigated (LL): anaerobic, generally favorable conditions] as shown at right. For details on the genotypes analyzed, see text and Supplemental Table S1.
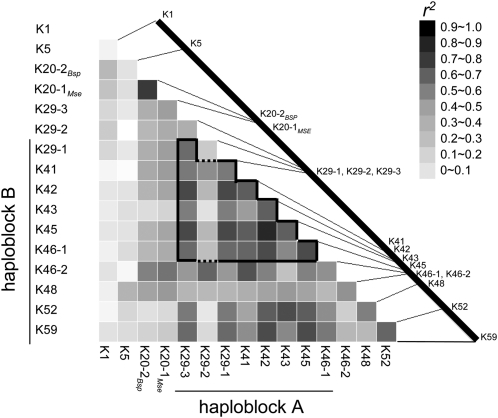
Figure 6.
Pup1 core markers. A GGT 2.0 analysis (Van Berloo 2008) was conducted with the marker data of the 81 genotypes analyzed with the indicated Pup1-specific markers. The r2 values of the individual markers are represented as a heat plot (black indicates r2 = 1). The approximate physical locations of the markers are indicated on the black line, which represents the Kasalath Pup1 locus. Two haploblocks can be identified that define the core marker set (thick lines). The marker K29-2 is not associated with the other two markers for the OsPupK29-1 gene (dashed lines; see text for details).
The highest r2 values were identified for markers targeting the hypothetical protein OsPupK29 (markers K29-1 and K29-3), two transposable elements (markers K42 and K45), and the Pup1 protein kinase gene (marker K46-1; Fig. 6). With the exception of K29-1 and K29-3, all markers are located within the Kasalath-specific INDEL region (Fig. 1A; Chin et al., 2010).
It is noteworthy that the intron-specific marker K29-2, in contrast to markers K29-1 and K29-3 that target exons of the same gene, is not associated with the other Pup1 markers (r2 = 0.34; Fig. 6). As mentioned above, this Kasalath gene corresponds to two genes in Nipponbare that might have evolved by a duplication event and several transposon integrations (Fig. 1D; Heuer et al., 2009). The low r2 value of marker K29-2, therefore, suggests that some regions of the OsPupK29 gene are unstable and are still evolving in the genotypes analyzed (Fig. 5; Supplemental Table S1).
Likewise, data derived with the two markers targeting the Pup1 protein kinase gene are not always consistent. Whereas marker K46-2 indicates the presence of the gene in 57 genotyes, the data derived with marker K46-1 suggest that the gene is present in only 38 genotypes (Figs. 5 and 6; Supplemental Table S1). Sequencing of the K46-2 PCR amplicon subsequently confirmed the specificity of this marker and showed a high conservation between the four analyzed sequences (Fig. 3D). However, a low r2 value (r2 = 0.36; Fig. 6) indicates that marker K46-2 is less informative than K46-1; therefore, the latter is recommended for breeding applications and allelic surveys.
Based on this assessment, a core set of the six most informative Pup1 markers was defined (K29-1, K29-3, K41, K43, K45, and K46-1; Fig. 6). Using these core markers, an Unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis was conducted, revealing three main groups of genotypes with different Pup1 allele constitutions (Fig. 5; Supplemental Table S1). Group I includes genotypes with all or most Kasalath Pup1 alleles. In contrast, Kasalath alleles for the core markers are absent in genotypes within groups II and III. Interestingly, group II is mainly defined by the presence of Kasalath alleles for the markers K29-2 and K46-2 (see above), whereas genotypes in group III do not possess any Kasalath allele, with the exception of K5.
In agreement with earlier data (Chin et al., 2010), all genotypes within group I are adapted to rain-fed drought-prone environments, whereas the majority (63%) of genotypes within group II and group III are adapted to lowland/irrigated conditions (Fig. 5; Supplemental Table S1). The japonica-type accessions that were included in this study are represented in all three groups, suggesting that Pup1 is not an indica/aus type-specific locus.
Development of Pup1 Varieties by MABC
The Pup1 marker survey described above has revealed the presence or partial presence of the Kasalath Pup1 locus in the majority of the varieties and breeding lines analyzed. For marker-assisted breeding applications, therefore, it is critically important to first determine the Pup1 haplotype in the parental lines. The core marker data indeed indicate that two of the three Indonesian varieties selected for breeding possess some (Batur) or all (Dodokan) Kasalath alleles (Fig. 4). In contrast, Kasalath alleles were not detected with the core markers in IR64 and IR74, and both belong to group III (Figs. 4 and 5).
For the development of Indonesian Pup1 varieties, Kasalath and the Pup1 NILC443 (Wissuwa et al., 2002) were used as donor parents. Starting at the F1 generation, progeny plants were genotyped with a set of best suitable Pup1 foreground markers and backcrossed to the recipient parent. After a repeated genotyping, selected BC1F2 plants were backcrossed again to the respective recipient parent to restore all desirable traits of the upland varieties. A background screening of the three different BC2F2 breeding populations was conducted with, depending on the population, 47 to 61 polymorphic simple sequence repeat markers distributed across the 12 chromosomes. The genotype of the populations is shown in Supplemental Figure S1. In the generation analyzed, 16% to 19% of remaining donor alleles were detected with 1% to 9% heterozygous loci. Analyses of advanced generations are now ongoing.
For the development of irrigated Pup1 varieties, the NIL14-4 (Chin et al., 2010) was used as the Pup1 donor. Progeny plants were backcrossed once each to IR64 and IR74, respectively, and genotyped with at least three Pup1 foreground markers (K20-1Mse, K46-1, and K29-1; data not shown). In the BC2F1 generation, 29 out of 113 plants with the Pup1 introgression were selected for background genotyping using the Illumina Beadexpress SNP chip set that contained 326 markers (M. Thomson, unpublished data). The SNP data revealed that, on average, 88.3% of the recipient genome was recovered. Ten plants with the least number of background introgressions were selected, and the presence of the tolerant Pup1 locus was reconfirmed in the BC2F3 generation (data not shown). From these plants, contrasting sister lines with and without the tolerant Pup1 locus (Pup1+/Pup1−) were selected and again genotyped. The data showed that background introgressions were further reduced and that 89% to 93% of the recipient genome was restored (IR74-Pup1+, 93%; IR74-Pup1−, 93.4%; IR64-Pup1+, 92.6%; IR64-Pup1−, 89.4%).
Phenotyping of Pup1 Breeding Lines under Field Conditions
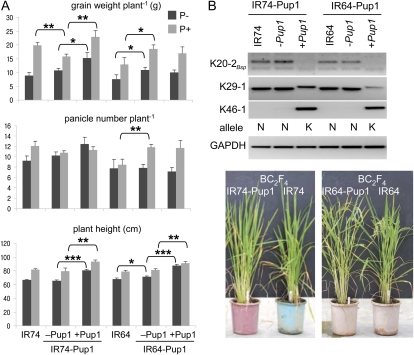
The IR64-Pup1 and IR74-Pup1 breeding lines have been subjected to a very stringent genotypic selection (see above), and only nearly identical and nonsegregating BC2F3 sister lines (Pup1+/Pup1–) from each cross were selected for an initial field evaluation. Plants were grown at the International Rice Research Institute experiment station in the Philippines in 2010 in a P-deficient irrigated plot and a control plot under P-fertilized conditions (equivalent to 60 kg P2O5 ha−1). In the unfertilized P-deficient field plot, IR74-Pup1+ had significantly higher grain yield (+73.1%) and was taller (20.5%) than the IR74-Pup1− sister line and the IR74 parent (Fig. 7A). Some degree of superiority remained under P-fertilized conditions, in which IR74-Pup1+ developed a 15.1% higher grain weight per plant and was 14.2% taller than the controls. The number of panicles was increased in IR74-Pup1+ under both P conditions, but the data were not statistically significant. A higher grain yield (33.0%) was also observed in IR64-Pup1+. However, this cannot be attributed to Pup1, since a significantly higher (44.7%) grain yield was also observed in the sister line without Pup1 (IR64-Pup1–). This is likely due to the presence of remaining background introgressions, and analyses are now ongoing. However, IR64-Pup1+ plants were significantly taller than both checks (IR64 and IR64-Pup1–) under +P and –P conditions (Fig. 7A). Sister lines (BC2F4) of both crosses have now been selected based on genotypic and phenotypic data (Fig. 7B), and seeds are being multiplied for further field evaluations in collaboration with rice breeders in Asia and Africa. In parallel, an additional backcross has been conducted to further remove the remaining background introgressions and to provide additional Pup1 breeding lines.
Figure 7.
Field evaluation and selection of IR64-Pup1 and IR74-Pup1 breeding lines. A, Sister lines with (+Pup1) and without (−Pup1) the tolerant Pup1 locus and control plants were grown under irrigated field conditions in P-deficient soil (P−) and in a P-fertilized parallel control plot (P+) in three replicates. Phenotypic data from the BC2F3 populations were collected in 2010. Data were analyzed by paired t test (95%). Significance levels are as follows: * 0.05 > P ≥ 0.01, ** 0.01 > P ≥ 0.001, *** 0.001 > P. No asterisk indicates not significant. B, For the final selection of IR64-Pup1 and IR74-Pup1 plants, contrasting BC2F4 sister lines were genotyped with selected Pup1 core markers (top). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included as a control. K, Kasalath; N, Nipponbare. Representative plants are shown at bottom.
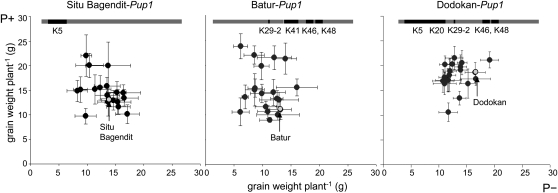
The Indonesian Pup1 breeding lines (BC2F4) were assessed under field conditions in Sukabumi (Indonesia, West Java) in 2010 under rain-fed conditions in a field with an intermediate high endogenous P content (8.3 mg L−1 available P) and in a P-fertilized control plot (39.3 kg P ha−1). The phenotypic data of the BC2F4 generation analyzed showed that most Situ Bagendit-Pup1 lines outperformed the original parent under both P-fertilized and −P conditions (Fig. 8A, left). On average, the best lines developed a 22% (+P) and 14% (−P) higher grain yield than Situ Bangendit. In the Batur-Pup1 population, the largest effect was observed under P-fertilized conditions, and the best lines developed a 2-fold higher grain yield than Batur (Fig. 8, middle). In contrast, only one Dodokan-Pup1 line showed a better performance compared with Dodokan (Fig. 8, right). This finding is in agreement with the genotypic data (Fig. 4) showing that the tolerant Pup1 locus is naturally present in Dodokan, whereas it is absent in Situ-Bangendit, in which we have observed the largest improvement in grain yield. In Batur, some Kasalath Pup1 alleles were detected, including the protein kinase gene (Fig. 4). Interestingly, improved performance of the Batur breeding lines was mainly detected under +P conditions (Fig. 8), suggesting that Batur naturally possesses some tolerance of P deficiency, possibly related to the presence of the Pup1 protein kinase.
Figure 8.
Field evaluation of three Indonesian Pup1 breeding lines. Advanced generations (BC2F4) of Pup1 breeding populations of three Indonesian upland varieties were grown in a field in Sukabumi (West Java, Indonesia) under rain-fed conditions in 2010. The data show average grain yield per plant obtained with (P+; y axis) and without (P−; x axis) P fertilizer application. Grain yield of the Pup1 recipient parents is highlighted in the graphs as light gray marker. Horizontal and vertical lines indicate se. The gray bar at the top of each graph signifies the Pup1 locus of the recipient parent before introgression of the tolerant Pup1 locus. Kasalath-type alleles naturally present in the recipient parent detected with the indicated Pup1 markers are shown as black lines.
DISCUSSION
Pup1 Core Markers
The main objective of this study was to develop an ideal set of molecular markers that facilitates targeted introgression of the Pup1 major QTL into diverse intolerant genetic backgrounds and for validation under field conditions. For breeders, only QTLs that express their beneficial effect in different genetic backgrounds and environments are of interest and can be widely applied for the development of tolerant rice varieties.
The gene-based Pup1 markers that are now available (Chin et al., 2010; this paper) have been extensively tested and provide sufficient details on the different Pup1 haplotypes present in the diverse rice genotypes analyzed. Among the tested markers, a core set of six markers is best associated with Pup1. The finding that this core region is flanked by two markers located within targeted genes (OsPupK29 and OsPupK46) is interesting and requires more detailed analyses. As outlined above, OsPupK29 appears to be an unstable gene, and its location at the 5′ border of the Pup1-specific INDEL region suggests that it might have contributed to the evolution of Pup1. The 3′ border of the core marker set is located within the INDEL region and is defined by K46-2, one of the two markers targeting the protein-kinase gene. The K46-2 reverse primer is located within the predicted 3′ untranslated region, and since untranslated regions are generally less conserved, it was expected that this marker is more specific than K46-1, which is located within the coding region. The finding that, in most genotypes within group II, an amplicon was derived only with marker K46-2 but not K46-1 suggests that the coding region is mutated or truncated in some genotypes and that the K46-1 primers do not bind to the mutated gene. These data demonstrate the complexity of functional marker design and suggest that multiple markers should be used to assess major genes in breeding programs.
Recently, rapid progress has been made with the development of SNP markers that facilitate high-throughput genotyping of breeding populations (Kim et al., 2009; McCouch et al., 2010). This technology is a very useful tool for background genotyping of mapping and breeding populations and is already being applied for whole-genome association studies (McNally et al., 2009). The development of SNP chips with a collection of high-value genes for foreground selection is under way (International Rice Research Institute, unpublished data) and will enable breeders to routinely screen their populations for important tolerance genes and other agronomically useful genes. In order to include Pup1 in this panel, it will be necessary to develop SNP markers for the Pup1 core genes. Based on allelic sequencing data, SNPs have been identified for three genes (OsPupK20, OsPupK29, and OsPupK67) that can be targeted for this purpose. However, the allelic sequence data suggest that the INDEL mutations targeted for marker design in this study might be more informative than the SNPs across genotypes (Fig. 3, A and B). Likewise, the genes located in the Pup1-specific INDEL cannot be analyzed by SNP markers, since SNPs are based on sequence polymorphisms between genes present in both targeted genotypes. Therefore, a dominant marker system for the Kasalath-specific genes, most importantly for OsPupK46, is inevitable. However, our sequence data show that multiple alleles exist within genotypes that possess this gene, which might be explored for SNP marker development. Interestingly, the highest number of SNPs was detected in Dular, the aus-type variety that showed the highest tolerance in the original screening from which Pup1 was identified (Wissuwa and Ae, 2001a). However, whether these polymorphisms are of any significance with respect to gene function and tolerance of P deficiency remains to be determined.
Pup1 Candidate Genes
Based on the above-described marker data and the RT-PCR expression analysis, the dirigent-like gene OsPupK20, the hypothetical gene OsPupK29, and the protein kinase gene OsPupK46 have been short-listed as priority candidate genes. Since none of these genes code for a known structural P uptake-related gene, the mode of action of Pup1 is still unclear. However, the expression data suggest that OsPupK20 and OsPupK29 have root-specific functions. Dirigent proteins catalyze the polymerization of lignin monomers, a reaction that involves free radicals, and the dirigent gene family has been shown to play a role in plant defense response (Bhuiyan et al., 2009; Wu et al., 2009). Dirigent proteins also catalyze the synthesis of lignans, a diverse group of polyphenolic substances derived from Phe that may play a role in plant pathogen defense and are important anticancer substances in the human diet (Davin and Lewis, 2000, 2005; Yoo et al., 2010). Up-regulation of dirigent genes under P deficiency in roots (At1g64160) and leaves (At2g21100) has been reported in Arabidopsis (Arabidopsis thaliana; Misson et al., 2005), which is in agreement with our data. Furthermore, a dirigent-like gene has also been identified as a candidate gene for heat tolerance from the aus-type variety N22 (Jagadish et al., 2010), suggesting that this gene family may have a more general function in responses to abiotic stresses.
The hypothetical protein OsPupK29 is of interest because of its root-specific expression and putative role in Pup1 evolution (see above). The two Nipponbare genes that show high partial sequence similarity are, according to massively parallel signature sequencing data (http://mpss.udel.edu/rice/mpss_index.php), expressed at a low level in crown meristems (Os12g26410) and in young, drought-stressed roots (Os12g26390). The function of these genes is currently unknown, and it will be interesting to conduct comparative studies between the Kasalath and Nipponbare genes.
The presence of a novel protein kinase gene suggests that Pup1 might confer tolerance via a regulatory pathway. OsPupK46 is predicted as a Ser/Thr kinase and, as such, shows homology to many members of this gene family in rice. In Arabidopsis, an Affymetrix microarray analysis has shown up-regulation of 15 Ser/Thr kinase genes under P-deficient conditions (Misson et al., 2005). Of these, At1g67000, which was specifically up-regulated in roots, was most similar to the Pup1 kinase (8e-93). However, a BLASTp search of The Arabidopsis Information Resource revealed a higher sequence similarity with Suppressor of Npr1-1, Constitutive4 (At1g66980; 9e-99) and Pathogen Related5-like Receptor Kinase (At5g38280; 6e-97), two defense-related receptor kinases (Wang et al., 1996; Bi et al., 2010). All of the above-mentioned genes possess an N-terminal receptor/membrane domain that is absent from the Kasalath gene; therefore, it is unlikely that OsPupK46 is an orthologous gene. In yeast, the phosphatase (PHO) regulon has been described in detail (for review, see Oshima, 1997). The regulon involves the cytokinin-dependent kinase PHO85 that regulates the phosphorylation of the PHO4 transcription factor in a P-dependent manner. Under low-P conditions, PHO4 enters into the nucleus in its underphosphorylated form, where it activates transcription of the acid PHO gene PHO5 (Schneider et al., 1994; O’Neill et al., 1996). In contrast to the other genes mentioned above, PHO85 has no N-terminal transmembrane domain and is located in the cytoplasm. Whether the Pup1 kinase has a function similar to PHO85 remains to be analyzed.
Pup1 Breeding
The marker survey confirmed earlier data (Chin et al., 2010) that Pup1 is conserved in most rice varieties developed for rain-fed stress-prone environments, suggesting that breeders are unknowingly selecting for this QTL. The molecular markers developed now provide breeders with a tool to ensure that Pup1 is present in their breeding materials. Among the genotypes with the tolerant Pup1 locus are several varieties that are known for their superior behavior under P-deficient conditions (Dular, IAC47, and IAC25). Dular had shown a better performance under stress than Kasalath in the original screening (Wissuwa and Ae, 2001b), and it will be important to assess whether allelic differences in the Pup1 kinase gene are related to the higher tolerance and to map any additional QTLs that might be present in Dular to further enhance tolerance.
For the development of the Pup1 breeding lines, we have followed a MABC approach that was first applied for the development of submergence-tolerant rice varieties (e.g. Swarna-Sub1 and IR64-Sub1). These Sub1 varieties are now being widely adopted in submergence-prone regions in Asia and Africa (Xu et al., 2006; Septiningsih et al., 2009; Venuprasad et al., 2009). The same approach is also being applied for the development of drought- and salinity-tolerant varieties (Bernier et al., 2007, 2009; Ismail et al., 2007; Kim et al., 2009; Thomson et al., 2010). The principal idea of MABC is to use widely grown and locally well-adapted rice varieties from target countries as recipient parents for the introgression of tolerance QTLs. The desirable plant type of the local variety is subsequently restored by repeated backcrossing and selfing in conjunction with genotypic selection using foreground, background, and QTL-flanking (recombinant) markers (Collard and Mackill, 2008). Tolerant varieties developed by this approach are more likely to be adopted by farmers, since eating quality and other traits remain unchanged.
The development of the Pup1 varieties is still a work in progress, and additional backcrosses and marker selection are ongoing to further remove remaining background introgressions. However, the initial field data of the five breeding populations suggest that Pup1 has the potential to significantly improve plant performance. In Indonesia, the largest beneficial effect of Pup1 on grain yield (up to a 2-fold increase) was obtained in the genetic background of Situ Bagendit and Batur. In Batur, this was mainly observed under P-fertilized conditions. It will be interesting to analyze whether the lack of improvement under P-deficient conditions is related to the Pup1 protein kinase, which is naturally present in Batur, implying that this variety might already be tolerant of P deficiency. In agreement with this, Dodokan-Pup1 breeding lines did not show improved performance in the field experiment, since the tolerant Pup1 locus is already present in Dodokan.
For the indica breeding lines, a significant, Pup1-specific increase in grain yield was observed in the genetic background of IR74 under irrigated P-deficient and P-fertilized conditions. Based on earlier data derived from pot experiments with Nipponbare-Pup1 NILs that suggested that Pup1 does not have a large effect under flooded conditions (Chin et al., 2010), the observed yield advantage in IR74-Pup1 is very encouraging. A beneficial effect of Pup1 under P-deficient lowland/irrigated conditions in at least some genetic backgrounds would greatly enhance the potential impact of this QTL.
For the identification and evaluation of major QTLs, a robust and reliable phenotyping system is a prerequisite. Since Pup1 cannot be phenotyped in liquid culture solutions, in contrast to salinity and aluminum toxicity (Chen et al., 2006; Chin et al., 2010; Thomson et al., 2010), phenotyping has to be conducted in soil, which is inconvenient and a problem for physiological and molecular analyses. In addition, many P-deficient soils are constrained by other stresses (e.g. aluminum toxicity, salinity, acidity, nematodes; Wissuwa et al., 2002; Ismail et al., 2007; Kumar et al., 2007) that restrict root growth and interfere with Pup1 phenotyping. In the future, therefore, it will be important to combine tolerance of P deficiency with tolerance of other predominant stresses. Major QTLs for tolerance of drought and salinity have already been identified, and efforts are now under way to identify and validate QTLs for tolerance of aluminum toxicity (Wu et al., 2000; Nguyen et al., 2003; Kochian et al., 2005; Bernier et al., 2009; Venuprasad et al., 2009).
Despite the large number of published QTLs, still only a few are actively used by breeders (for review, see Collins et al., 2008; Jena and Mackill, 2008; Xu and Crouch, 2008). This is partly due to the small effects of published QTLs but mainly due to the absence of data showing that a given QTL is effective in different genetic backgrounds and environments. The recent success in the development of submergence-tolerant rice varieties has demonstrated the advantages of MABC and the potential impact of large-effect QTLs on food security for poor farmers. The data presented here, therefore, are important milestones toward the application of another large-effect rice QTL with potentially high impact in farmers’ fields.
MATERIALS AND METHODS
Plant Materials for RT-PCR
NILs of rice (Oryza sativa) with the tolerant Pup1 locus, NIL24-4 and NIL14-4, a NIL sister line without the tolerant Pup1 locus (NIL14-6), and Nipponbare control plants were grown in P-deficient Philippine soil, and the equivalent of 90:60:40 kg ha−1 nitrogen:P2O5:KCl fertilizer was applied to the pots. No P fertilizer was applied for the –P treatment. Plants were grown in a greenhouse with an extended (16-h) light period for about 6 weeks to prevent early flowering of the photosensitive genotypes. Root and shoot samples were collected 49 d after sowing, frozen in liquid nitrogen, and stored at −80°C until extraction of total RNA using Trizol according to the instructions from the manufacturer (Invitrogen). RNA samples were treated with RNase-free DNase (Promega) to remove any contaminating genomic DNA. cDNA synthesis at 55°C for 1 h was performed in a 20-μL reaction containing 5 μg of DNase-treated total RNA, 2.5 μm oligo(dT), 0.5 mm deoxyribonucleotide triphosphate (dNTP) mix, 0.01 m dithiothreitol, 1× first-strand buffer, and 200 units of SuperScript II reverse transcriptase (Invitrogen). PCR was carried out in a total volume of 20 μL containing 0.5 μL of cDNA, gene-specific primers (0.2 μm each; sequences are provided in Table I), 1× PCR buffer, 0.5 mm dNTP mix, and 1 unit of Taq DNA polymerase (Beijing SBS Genetech). The PCR cycle settings were 94°C for 5 min, followed by 33 or 40 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 10 min. The cytosolic glyceraldehyde-3-phosphate dehydrogenase gene was used as a control for successful amplification and absence of genomic DNA (forward, 5′-GCAGGAACCCTGAGGAGATC-3′; reverse, 5′-TTCCCCCTCCAGTCCTTGCT-3′). RT-PCR products were separated by agarose electrophoresis and stained with SYBR Safe (Invitrogen).
Gene-Specific Marker Development and Pup1 Haplotyping
To develop specific molecular markers, Pup1 gene models and nucleotide sequences were adapted from Heuer et al. (2009) except for OsPupK20 and OsPupK46. For these genes, primer design was based on revised gene models published under accession number AB458444.1. For all Kasalath gene models, nucleotide and amino acid sequences were subjected to BLASTn and BLASTp analyses at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=4530) and the Michigan State University Rice Genome Annotation Project version 6.1 (http://rice.plantbiology.msu.edu/analyses_search_blast.shtml) to identify the most similar sequences in Nipponbare and other reference genomes. For Kasalath genes with similarity to Nipponbare genes in the syntenic region on chromosome 12, codominant markers were developed based on sequence polymorphism (INDEL, SNP) between alleles. Primers targeting specific regions in the genes were designed using Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) or manually. For codominant markers with small size differences between alleles, allele-specific restriction sites were identified to develop cleaved amplified polymorphic site (CAPS) markers using NEB Cutter version 2 software (http://tools.neb.com/NEBcutter2). For genes that were specific to Kasalath, dominant markers were designed generally targeting predicted exon regions. The specificity of all primers was reconfirmed by BLASTn at the National Center for Biotechnology Information and GRAMENE (www.gramene.org/Multi/blastview), and representative PCR amplicons were sequenced at Macrogen.
Genomic DNA was extracted from leaf samples as described by Palotta et al. (2000) from plants grown under control conditions. PCR was carried out in a total volume of 20 μL containing 40 ng of genomic DNA, gene-specific primers (0.2 μm each; sequences are provided in Table II), 0.5 mm dNTP mix, and 1.5 units of Taq DNA polymerase with the provided PCR buffer (Beijing SBS Genetech). The PCR cycle settings were 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C or 58°C for 45 s, and 30°C for 90 s, and a final extension at 72°C for 10 min. Size differentiation of marker amplicons was carried out on 3.5% agarose gels or 8% polyacrylamide gels.
In total, 81 diverse rice accessions from the rice germplasm collection and International Rice Research Institute breeding lines were included in the study (Supplemental Table S1). The Pup1 NIL14-4 (Wissuwa, 2005) and the Indonesian upland varieties Dodokan, Situ Bagendit, and Batur were additionally included.
Development of Pup1 Breeding Lines
For the development of Pup1 breeding materials, NILC443 and Kasalath were used as the Pup1 donor for crosses involving Dodokan, Batur, and Situ Bagendit. Pup1 NIL14-4 was used as the donor for the IR64-Pup1 and IR74-Pup1 populations. For backcrossing, recipient varieties were used as recurrent parents. Crosses were conducted according to standard protocols with plants grown under control conditions in a greenhouse.
Phenotyping of Pup1 Breeding Lines under Field Conditions
The IR64-Pup1 and IR74-Pup1 BC2F3 breeding lines and parents were phenotyped at the International Rice Research Institute experiment station (Philippines) in the dry season of 2010 in an irrigated plot with full fertilizer treatment (equivalent of elemental nitrogen, 120 kg ha−1; K2O, 40 kg ha−1; P2O5, 60 kg ha−1). A duplicate setup was grown in parallel in P-deficient soil without application of P fertilizer. The equivalent of 20 kg ha−1 zinc sulfate was applied to each plot. Plants were grown in rows with 12 to 15 plants per line with spacing of 20 cm × 20 cm. Phenotypic data from three plants located in the center of the rows were collected from three replicated plots.
Situ Bagendit, Batur, and Dodokan BC2F4 breeding lines were grown in 2010 in Sukabumi, West Java (Indonesia), under rain-fed conditions in a field with a medium endogenous P level (8.3 μL L−1 available P). Urea (250 kg ha−1) and KCl (100 kg ha−1) were applied to all plots. P fertilizer (SP18, 500 kg ha−1; equivalent to 39.3 kg P ha−1) was applied only to +P control plots. Breeding lines were grown in three replicates, and three plants from each replicate were phenotyped.
Software Used for Sequence and Marker Analyses
Comparative sequence analyses were conducted using MAFFT (http://mafft.cbrc.jp/ alignment/software). Based on the Pup1 marker data, a dendrogram was developed using UPGMA cluster analysis (Sokal and Michener, 1958). The genetic distance between the analyzed rice genotypes was calculated based on the method published by Nei (1973) using Powermarker version 3.25 (Liu and Muse, 2005). To calculate the r2 value for the individual Pup1 markers, the software GGT 2.0 was used (Van Berloo, 2008).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB458444.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphical genotype of Indonesian Pup1 introgression lines.
Supplemental Table S1. Plant materials used for Pup1 genotyping.
Supplementary Material
Acknowledgments
We thank Anthony Cruz and Ranie Ramos at the International Rice Research Institute for technical assistance and devotion to the project. Special thanks go to Stephan Haefele and Serafin Amarante at the International Rice Research Institute for support of the project as well as to Juan Pariasca-Tanaka at the Japan International Research Center for Agricultural Sciences.
References
- Bernier J, Kumar A, Venuprasad R, Spaner D, Atlin G. (2007) A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47: 507–516 [Google Scholar]
- Bernier J, Kumar A, Venuprasad R, Spaner D, Verulkar S, Mandal NP, Sinha PK, Peeraju P, Dongre PR, Mahto RN, et al. (2009) Characterization of the effect of a QTL for drought resistance in rice, qtl12.1, over a range of environments in the Philippines and eastern India. Euphytica 166: 207–217 [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J. (2009) Role of lignification in plant defense. Plant Signal Behav 4: 158–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Cheng YT, Li X, Zhang Y. (2010) Activation of plant immune responses by a gain-of-function mutation in an atypical receptor-like kinase. Plant Physiol 153: 1771–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RF, Shen RF, Gu P, Dong XY, Du CW, Ma JF. (2006) Response of rice (Oryza sativa) with root surface iron plaque under aluminium stress. Ann Bot (Lond) 98: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JH, Lu X, Haefele SM, Gamuyao R, Ismail A, Wissuwa M, Heuer S. (2010) Development and application of gene-based markers for the major rice QTL Phosphorus uptake 1. Theor Appl Genet 120: 1073–1086 [DOI] [PubMed] [Google Scholar]
- Collard BCY, Mackill DJ. (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363: 557–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Tardieu F, Tuberosa R. (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol 147: 469–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell D, Drangert JO, White S. (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19: 292–305 [Google Scholar]
- Davin LB, Lewis NG. (2000) Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol 123: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Lewis NG. (2005) Lignin primary structures and dirigent sites. Curr Opin Biotechnol 16: 407–415 [DOI] [PubMed] [Google Scholar]
- Dawe D, Block S, Gulati A, Huang J, Ito S. (2010) Domestic rice price, trade, and marketing policies. Pandey S, Byerlee D, Dawe D, Dobermann A, Mohanty S, Rozelle S, Hardy B, , Rice in the Global Economy: Strategic Research and Policy Issues for Food Security. International Rice Research Institute, Los Banos, Philippines, pp 379–407 [Google Scholar]
- Haefele SM, Hijmans RJ. (2007) Soil quality in rice-based rainfed lowlands of Asia: characterization and distribution. Aggarwal PK, Ladha JK, Singh RK, Devakumar C, Hardy B, eds, Science, Technology, and Trade for Peace and Prosperity: Proceedings of the 26th International Rice Research Conference, October 9–12, 2006, New Delhi, India International Rice Research Institute, Indian Council of Agricultural Research, and National Academy of Agricultural Sciences, Los Baňos, Philippines, and New Delhi, India, pp 297–308 [Google Scholar]
- Heuer S, Lu X, Chin JH, Tanaka JP, Kanamori H, Matsumoto T, De Leon T, Ulat VJ, Ismail AM, Yano M, et al. (2009) Comparative sequence analyses of the major quantitative trait locus phosphorus uptake 1 (Pup1) reveal a complex genetic structure. Plant Biotechnol J 7: 456–457 [DOI] [PubMed] [Google Scholar]
- International Rice Research Institute (2010) Global Rice Science Partnership. http://irri.org/our-science/global-rice-science-partnership-grisp (March 1, 2011)
- Ismail AM, Heuer S, Thomson MJ, Wissuwa M. (2007) Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol Biol 65: 547–570 [DOI] [PubMed] [Google Scholar]
- Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. (2010) Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J Exp Bot 61: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena KK, Mackill DJ. (2008) Molecular markers and their use in marker-assisted selection in rice. Crop Sci 48: 1266–1276 [Google Scholar]
- Kim SH, Bhat PR, Cui X, Walia H, Xu J, Wanamaker S, Ismail AM, Wilson C, Close TJ. (2009) Detection and validation of single feature polymorphisms using RNA expression data from a rice genome array. BMC Plant Biol 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Pineros MA, Hoekenga OA. (2005) The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274: 175–195 [Google Scholar]
- Kumar RM, Prasad JS, Sankar M, Murthy AGK. (2007) Influence of phosphorus levels and phosphorus solubilising bacteria (PSB) application methods on rice-root nematode population in rice-based cropping system. Indian J Nematol 37: 50–55 [Google Scholar]
- Liu K, Muse SV. (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129 [DOI] [PubMed] [Google Scholar]
- Manzanilla DO, Paris TR, Vergara GV, Ismail AM, Pandey S, Labios RV, Tatlonghari GT, Acda RD, Chi TTN, Duoangsila K, et al. (2011) Submergence risks and farmers’ preferences: implications for breeding Sub1 rice in Southeast Asia. Agric Syst 104: 335–347 [Google Scholar]
- McCouch SRM, Zhao K, Wright M, Tung CW, Ebana K, Thomson MJ, Reynolds A, Wang D, DeClerck G, Ali Md L, et al. (2010) Development of genome-wide SNP assays for rice. Breed Sci 60: 524–535 [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA 106: 12273–12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70: 3321–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN, Nguyen HT. (2003) Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor Appl Genet 106: 583–593 [DOI] [PubMed] [Google Scholar]
- Ni JJ, Wu P, Senadhira D, Huang N. (1998) Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet 97: 1361–1369 [Google Scholar]
- O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. (1996) Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271: 209–212 [DOI] [PubMed] [Google Scholar]
- Oshima Y. (1997) The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst 72: 323–334 [DOI] [PubMed] [Google Scholar]
- Palotta MA, Graham RD, Langridge P, Sparrow DHB, Barker SJ. (2000) RFLP mapping of manganese efficiency in barley. Theor Appl Genet 101: 1100–1108 [Google Scholar]
- Pariasca-Tanaka J, Satoh K, Rose T, Mauleon R, Wissuwa M. (2009) Stress response versus stress tolerance: a transcriptome analysis of two rice lines contrasting in tolerance to phosphorus deficiency. Rice 2: 167–185 [Google Scholar]
- Schneider KR, Smith RL, O’Shea EK. (1994) Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266: 122–126 [DOI] [PubMed] [Google Scholar]
- Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ. (2009) Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot (Lond) 103: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AN, Singh US. (2010) Targeted dissemination of stress tolerant rice varieties: propagating Swarna-Sub1, Sahbhagi Dhan, and CSR36 in Uttar Pradesh, India. STRASA Newsl 3: 1–2 [Google Scholar]
- Sokal R, Michener C. (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38: 1409–1438 [Google Scholar]
- Thomson MJ, De Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, et al. (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3: 148–160 [Google Scholar]
- Van Berloo R. (2008) GGT 2.0: versatile software for visualization and analysis of genetic data. J Hered 99: 232–236 [DOI] [PubMed] [Google Scholar]
- Van Kauwenbergh (2010) World phosphate rock reserves and resources. International Fertilizer Development Center. www.ifdc.org (March 1, 2011)
- Venuprasad R, Dalid CO, Del Valle M, Zhao D, Espiritu M, Sta Cruz MT, Amante M, Kumar A, Atlin GN. (2009) Identification and characterization of large-effect quantitative trait loci for grain yield under lowland drought stress in rice using bulk-segregant analysis. Theor Appl Genet 120: 177–190 [DOI] [PubMed] [Google Scholar]
- Wang X, Zafian P, Choudhary M, Lawton M. (1996) The PR5K receptor protein kinase from Arabidopsis thaliana is structurally related to a family of plant defense proteins. Proc Natl Acad Sci USA 93: 2598–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M. (2005) Combining a modelling with a genetic approach in establishing associations between genetic and physiological effects in relation to phosphorus uptake. Plant Soil 269: 57–68 [Google Scholar]
- Wissuwa M, Ae N. (2001a) Further characterization of two QTLs that increase phosphorus uptake of rice (Oryza sativa L.) under phosphorus deficiency. Plant Soil 237: 275–286 [Google Scholar]
- Wissuwa M, Ae N. (2001b) Genotypic variation for tolerance to phosphorus deficiency in rice and the potential for its exploitation in rice improvement. Plant Breed 120: 43–48 [Google Scholar]
- Wissuwa M, Wegner J, Ae N, Yano M. (2002) Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor Appl Genet 105: 890–897 [DOI] [PubMed] [Google Scholar]
- Wissuwa M, Yano M, Ae N. (1998) Mapping of QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor Appl Genet 97: 777–783 [Google Scholar]
- Wu P, Liao CX, Hu B, Yi KK, Jin WZ, Ni JJ, He C. (2000) QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet 100: 1295–1303 [Google Scholar]
- Wu R, Wang L, Wang Z, Shang H, Liu X, Zhu Y, Qi D, Deng X. (2009) Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog Nat Sci 19: 347–352 [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Xu Y, Crouch JH. (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48: 391–407 [Google Scholar]
- Xue Y, Jiang L, Su N, Wang JK, Deng P, Ma JF, Zhai HQ, Wan JM. (2007) The genetic basic and fine-mapping of a stable quantitative-trait loci for aluminium tolerance in rice. Planta 227: 255–262 [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo JH, Lee HJ, Kang K, Jho EH, Kim CY, Baturen D, Tunsag J, Nho CW. (2010) Lignans inhibit cell growth via regulation of Wnt/beta-catenin signaling. Food Chem Toxicol 48: 2247–2252 [DOI] [PubMed] [Google Scholar]
- Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, Tyagi W, Ali ML, Tung CW, Reynolds A, et al. (2010) Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE 5: e10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.