Abstract
Microarray experiments have yielded massive amounts of expression information measured under various conditions for the model species Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Expression compendia grouping multiple experiments make it possible to define correlated gene expression patterns within one species and to study how expression has evolved between species. We developed a robust framework to measure expression context conservation (ECC) and found, by analyzing 4,630 pairs of orthologous Arabidopsis and rice genes, that 77% showed conserved coexpression. Examples of nonconserved ECC categories suggested a link between regulatory evolution and environmental adaptations and included genes involved in signal transduction, response to different abiotic stresses, and hormone stimuli. To identify genomic features that influence expression evolution, we analyzed the relationship between ECC, tissue specificity, and protein evolution. Tissue-specific genes showed higher expression conservation compared with broadly expressed genes but were fast evolving at the protein level. No significant correlation was found between protein and expression evolution, implying that both modes of gene evolution are not strongly coupled in plants. By integration of cis-regulatory elements, many ECC conserved genes were significantly enriched for shared DNA motifs, hinting at the conservation of ancestral regulatory interactions in both model species. Surprisingly, for several tissue-specific genes, patterns of concerted network evolution were observed, unveiling conserved coexpression in the absence of conservation of tissue specificity. These findings demonstrate that orthologs inferred through sequence similarity in many cases do not share similar biological functions and highlight the importance of incorporating expression information when comparing genes across species.
Comparative sequence analysis provides valuable information about the functional parts encoded by a genome, mainly by exploring the conserved DNA sequences that code for proteins, RNAs, or regulatory elements (Hardison, 2003). Interspecies comparisons have two major applications: conservation between species helps to detect and characterize functional elements, whereas differences can reveal biological adaptations linking genotype with phenotype (Tirosh et al., 2007). Recently, the increase in functional genomics data has transformed comparative approaches from basic sequence analysis to detailed studies of functional attributes such as gene expression or protein-protein interactions (Stuart et al., 2003; Jensen et al., 2006). For instance, microarray experiments have yielded large amounts of genome-wide expression information under various conditions or in different tissues for several model species. Expression compendia grouping multiple microarray experiments make it possible to define correlated expression patterns between genes (Eisen et al., 1998; Lee and Tzou, 2009). Genes within a coexpression cluster are expected to have more similar functionality than those without expression similarity. In other words, functionally related genes are often coexpressed, both in closely related and more distantly related species (Stuart et al., 2003; Bergmann et al., 2004).
To compare expression data between different species, microarrays can be processed in three different manners (Lu et al., 2009). One approach combines multiple microarray experiments to identify differentially expressed genes in each species independently and then compare these genes among different species (Mustroph et al., 2010). This method requires for each species orthology information to link gene expression states across species and can be applied to both closely related or more distantly related species. Although orthologs are defined as homologs derived by a speciation event, it is important to note that sequence-based orthology inference does not necessarily imply functional equivalence (Studer and Robinson-Rechavi, 2009). Another approach hybridizes samples from different but closely related species to the same microarray and requires similar experimental conditions as well as orthology information. Finally, separate arrays can be used to sample similar experimental conditions for different species, and all of them are analyzed together to investigate expression evolution of orthologs, paralogs, or specific functional categories (Lu et al., 2009). The latter approach was used, for example, to investigate species-specific gene duplications in human and mouse (Huminiecki and Wolfe, 2004). In addition to comparison of expression profiles between orthologous genes, some studies also try to combine expression information with sequence evolution and gene function. Bergmann et al. (2004) integrated expression data of six species with their genomic sequence information to identify coexpression conservation and to improve functional gene annotation. Based on graph theory, transcriptional networks were inferred revealing that highly connected genes are often conserved and have essential functional rules. Sets of genes that are conserved at both sequence and expression levels among multiple species are expected to play a key role in biological responses (Stuart et al., 2003; Lu et al., 2009).
Although comparative expression analysis is most straightforward when compatible expression data sets are used that cover equivalent conditions for all species, in this approach only a small fraction of all available data in different species can be utilized (Tirosh et al., 2007). Furthermore, how similar gene expression is modulated in distantly related species is still not understood, even when considering compatible conditions. To overcome these limitations, more advanced transcriptomics studies have shown that comparing coexpression, instead of the raw expression values, provides a valid alternative to identifying regulatory modules and studying their evolution (Stuart et al., 2003). For instance, gene expression between the four distantly related species Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, and Saccharomyces cerevisiae was compared by means of a coexpression meta-analysis (Dutilh et al., 2006). Pairs of species were considered, and for each gene the “expression context,” which is based on the coexpression with all other genes that have unequivocal orthologous counterparts in both genomes, was compared. Significant expression context conservation (ECC) was found for many orthologs; but sequence and coexpression context evolution did not strongly correlate after duplication and speciation (Dutilh et al., 2006).
Rice (Oryza sativa) is one of the most important alimentary crops, with a relatively small genome size compared with many other cereals, and serves as a model for monocotyledons. Although the genome size of Arabidopsis (Arabidopsis thaliana), a model for dicotyledonous plants, is much smaller than that of rice (115 and 420 Mb, respectively), both species share a large number (56%–77%) of homologous genes (Vandepoele and Van de Peer, 2005; Sterck et al., 2007). Analysis of similarities and differences between the Arabidopsis and rice transcriptomes for similar organ types with custom-made oligomer microarrays revealed that similar portions were expressed in their corresponding organ types (Ma et al., 2005). In addition, evidence was found that a large fraction of rice genes lacking Arabidopsis homologs were expressed (Ma et al., 2005). Here, we developed a statistical framework to compare the expression of orthologous genes in rice and Arabidopsis. The importance of gene function and tissue specificity in correlation with the coexpression conservation was analyzed as well as the relationship between coding sequence and expression evolution.
RESULTS
Relationship between Expression Similarity and Gene Function
To compare gene expression between the two model species Arabidopsis and rice, we assembled for both organisms, an expression compendium based on Affymetrix microarray experiments, retrieved from the National Center for Biotechnology Information Gene Expression Omnibus. Starting from a set of 322 Arabidopsis and 203 rice microarray slides, data normalization and averaging of replicates resulted in an initial expression data set of 129 Arabidopsis and 84 rice experiments. Subsequently, the effect of highly similar experiments per data set was reduced by the identification and removal of highly redundant samples (see “Materials and Methods”) because they can introduce functional biases (De Bodt et al., 2010). After collapsing redundant conditions and removing transgenic or mutant experiments, we obtained a final data set covering 76 Arabidopsis and 63 rice experiments (Supplemental Table S1). By means of a custom-made chip description file (CDF) grouping only non-cross-hybridizing probes in probe sets, the expression patterns of 19,937 Arabidopsis genes and 32,004 rice genes were monitored (see “Materials and Methods”).
As both compendia contained experiments covering different tissues, developmental stages, or (a)biotic treatments, we first determined whether biologically relevant information could be retrieved from both expression data sets. Starting from predefined gene sets that grouped genes based on Gene Ontology (GO) annotations, we used the expression coherence (EC) to quantify the level of expression similarity for functionally related genes in one species. EC is a measure for the amount of expression similarity within a set of genes and is high for a set of genes that converges into one cluster or a few tight coexpression clusters (Pilpel et al., 2001). Expression similarities between gene pairs were calculated with the Pearson correlation coefficient (PCC), and an EC value of 100% indicated that all genes were coexpressed with each other (see “Materials and Methods”). For both species, 16% to 25% of all 1,550 GO functional categories (with five or more genes) had high coexpression levels (EC > 10%). As a control experiment, the influence of individual microarray experiments on the globally observed coexpression pattern was determined with jackknifing. The application of this bootstrapping procedure, which iteratively removes a subset of the initial data, showed that the observed EC values are robust for both species (Supplemental Fig. S1).
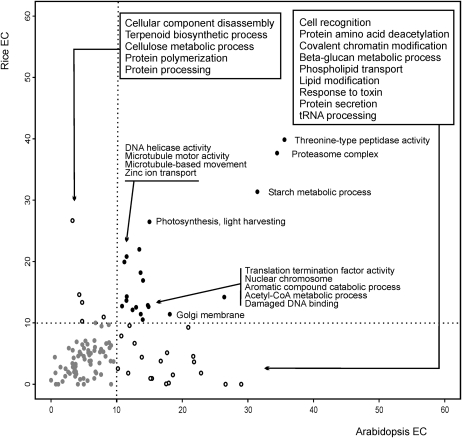
Many functional GO terms had significant EC in both species (Fig. 1, black dots), and examples included general housekeeping functions related to DNA and RNA metabolism, ribosome biogenesis, translation, photosynthesis, tricarboxylic acid cycle, starch metabolic processes, and cell cycle (Supplemental Table S2). Some GO terms only showed high EC values in one organism, and examples covered protein polymerization, defense response to fungus, and response to brassinosteroid stimulus in rice and cell recognition, phospholipid transport, and 1,3-β-glucan metabolism in Arabidopsis.
Figure 1.
EC values for different GO categories. EC is shown for 114 nonredundant GO biological process terms in rice and Arabidopsis. GO terms with elevated coexpression in both organisms are shown as black dots, whereas those significantly coexpressed in only one species are shown as open dots. Only GO terms covering between 10 and 80 genes are shown, and redundant GO terms are omitted (for full list, see Supplemental Table S2).
Measuring ECC between Arabidopsis and Rice Orthologs
Whereas EC values indicated that for some predefined functional categories differences in global coexpression levels existed, this measure did not report whether orthologous genes had similar expression patterns in different species or whether, for specific gene functions, the underlying coexpression network had diverged during evolution. To determine whether the expression profiles were conserved between two species, we developed an ECC score. According to Dutilh et al. (2006), the expression context is based on the expression correlations between a query gene and all other genes in that species (or gene-centric coexpression cluster). The ECC was obtained by starting from a 1:1 orthologous seed gene pair, retrieving all coexpressed genes per species, and calculating how many orthologous genes were coexpressed in both species (Fig. 2; see “Materials and Methods”). High ECC values indicated that in both species, the same genes were coexpressed, potentially reflecting the conservation of an ancestral coexpression module, whereas low ECC values suggested that since the divergence of both species, a substantial number of coexpression partners had been gained or lost. The OrthoMCL algorithm was applied to identify orthologous genes from the full set of Arabidopsis and rice proteins. In total, 7,911 orthologous gene families were found that covered approximately 12,000 genes for both species (see “Materials and Methods”). Based on the functional annotations for both species, all orthologous families were annotated with GO and Reactome (see “Materials and Methods”).
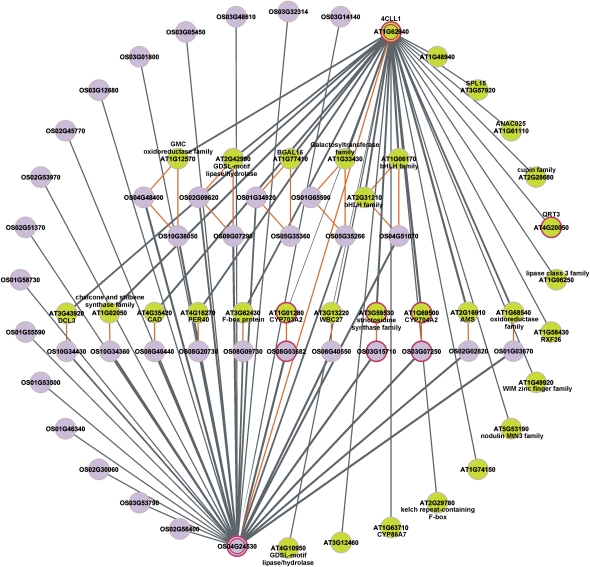
Figure 2.
Calculation of ECC scores. Starting from an orthologous gene pair (rice gene OS04G24530 and Arabidopsis gene AT1G62940, marked with double circles), all coexpressed genes per species were retrieved (solid gray lines). The thickness of the line indicates the expression similarity measured with the PCC. All orthologous relationships are linked with orange lines and are used to determine the number of shared families between both coexpression clusters. Red circles represent GO functional annotations enriched in both clusters (GO:0009555 pollen development). The Jaccard Index of the depicted ECC conserved gene pair is 0.088 (16 shared families over 182 families in total). Note that for clarity, not all coexpressed genes and GO terms are depicted.
The biological relevance of ECC was evaluated by comparing the obtained ECC score for an orthologous gene pair against a null model in which neutral expression evolution was assumed. Application of an iterative sampling procedure generating 1,000 random pairs of gene-centric clusters (per species and per orthologous gene pair) and comparison with the expected ECC scores made it possible to classify the orthologous expression contexts as significantly conserved, diverged, or not significant (called ECC category in Supplemental Fig. S2). Whereas ECC scores nonsignificantly differing from the expected background distribution were simply considered as nonsignificant, significantly diverged ECC scores referred to orthologous gene pairs with fewer shared coexpression partners between both species than expected by chance, possibly reflecting positive selection. As the false discovery rate for ECC diverged genes was much higher than that of ECC conserved genes (25% and 0.82%, respectively), it should be treated with caution. Application of two different null models to estimate significance levels, one controlling tissue-specific expression and one correcting the degree of distribution in the network (or connectivity), yielded highly similar results (see “Materials and Methods”). Results from the connectivity model are reported throughout, whereas genes with nonconserved ECC refer to the categories diverged and nonsignificant.
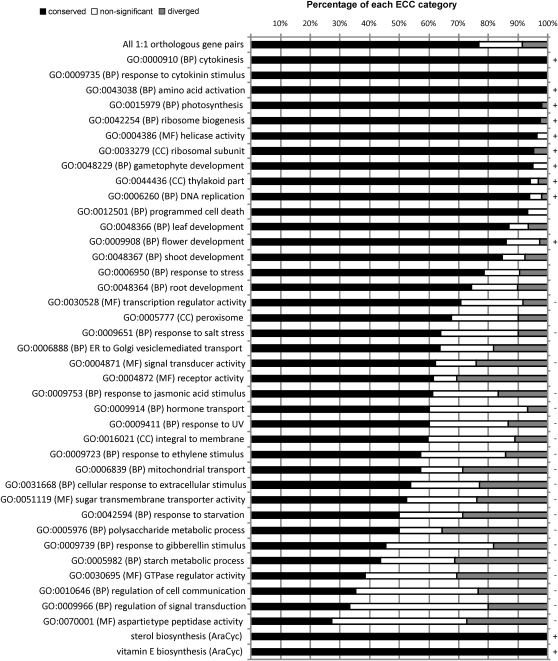
ECC scores for all 4,630 1:1 orthologous rice-Arabidopsis gene pairs revealed that 77% had a conserved expression context, whereas 8.5% and 14.5% had a diverged and nonsignificant ECC, respectively (Fig. 3). As expected, low ECC scores primarily included diverged and nonsignificant genes, whereas high values were mostly classified as significantly conserved (Supplemental Table S3). Functional biases determined with GO and MAPMAN enrichment analysis indicated that several biological processes linked with general housekeeping functions had highly conserved expression contexts (e.g. photosynthesis, plastid organization, DNA replication, RNA processing, cell division, and reproductive structure development). Interestingly, several functional categories were significantly underrepresented in ECC conserved genes, and examples include transcription factor (TF) activity, cell communication, ubiquitin-protein ligase activity, response to salt stress, and hormone stimulus. MAPMAN annotations indicated that different specific TF families (GRAS, Homeobox, WRKY, bZIP, and JUMONJI) were enriched in the set of nonconserved ECC genes.
Figure 3.
Comparison of ECC scores for different functional categories between Arabidopsis and rice. The fractions of genes with conserved, diverged, and nonsignificant ECC scores for different gene sets are shown. The first line reports the results for all 4,630 1:1 orthologous gene pairs, while the other lines refer to different functional sets delineated with GO and AraCyc (Reactome). The plus or minus sign indicates that the fraction of ECC conserved genes is significantly higher or lower compared with the overall ECC conservation level, respectively.
ECC Varies for Different Functional Categories
The distributions of conserved, diverged, and nonsignificant ECC categories differed for various functional classification systems (Supplemental Table S4). Average ECC scores indicated that genes assigned to KEGG/AraCyc pathways had a more conserved coexpression than those of the GO Biological Process or Molecular Function categories (average fractions of conserved ECC genes were 86%, 79%, and 76%, respectively). Categories with 50% or fewer conserved genes included regulation of signal transduction and cell communication, GTPase regulatory activity, starch metabolism, response to ethylene and GA stimulus, and response to starvation (Fig. 3; Supplemental Table S4). A high fraction of ECC diverged genes were found for receptor activity (31%), polysaccharide metabolism (36%), and starch metabolism (31%).
As a substantial number of genes involved in gene regulation, hormone response, and starch metabolism showed nonconserved coexpression contexts, we analyzed these categories in more detail. Focus on TFs revealed that developmental regulators were overall well conserved at the transcriptional level (e.g. ECC conserved TFs: 11 of 14 tissue, four of five leaf, seven of nine shoot, and nine of 12 flower development). Examples of highly diverged hormone-related TFs included HY5 (bZIP involved in light-regulated transcriptional activation), the auxin-responsive NAC domain-containing protein 9 (AT1G26870), MYB26 (AT3G13890; GA responsive), and the ethylene-response factor AT5G25190 (subfamily B-6 of the ERF/AP2 family). In contrast, TFs with highly conserved coexpression patterns covered OCP3, BLH1, and ATCDC5 (involved in response to fungus) as well as CLF, FMA, AGL16, LAS, and ATMYB5 (role in development). For all Arabidopsis genes discussed throughout this paper, a list with orthologous gene identifiers is provided (see “Materials and Methods”).
Several diverged stress-related genes encoded for DNA photolyases (CRY3 and PHR1), were responsive to DNA damage stimulus, or were involved in DNA repair (ATRAD51B/RAD51B and AT1G49980). GO categories such as “response to stimulus” or “response to stress” shared 78% conserved ECC genes, suggesting that general stress-related signaling was largely conserved between both species. Interestingly, the ECC conservation levels varied largely for some specific response categories: all responses to cytokinin stimulus genes were ECC conserved (e.g. GCR1, ARR11/22, TSD2, ADA2B, MCB1, PAS1, AHK5, and CKI1), 62% of the responses to UV genes, but only 50% of the genes responsive to GA or Suc stimulus (Supplemental Table S4).
Although starch metabolic genes had elevated levels of coexpression per species (Fig. 1), many genes had nonconserved coexpression patterns. Starch metabolism is tightly coupled with photosynthesis, resulting in biosynthesis in transient starch granules during the day and nocturnal breakdown. Consequently, both starch synthases and different degrading enzymes, phosphatases, and transporters are required to maintain correct sugar levels in different plant tissues (Orzechowski, 2008). Starch, stored in tuberous tissues or seeds, plays a central role as an energy source during germination. Whereas in most plants, ADPglucose, a substrate for starch synthases, is produced in the plastid through a ADPglucose pyrophosphorylase, cereals possess a cytosolic ADPglucose pyrophosphorylase-synthesizing ADPglucose in the developing endosperm that is then imported into the plastid for starch synthesis. Nonconserved ECC genes involved in starch metabolism included both genes involved in starch synthesis (ATSS3) and degradation (isoamylases ISA2 and ISA3, α-amylase ATPU1, and β-amylase CT-BMY). Whereas most Arabidopsis starch-related genes were expressed in several tissues, including leaves and seeds, the corresponding rice genes were expressed in fewer tissues, with ECC diverged starch genes being primarily expressed in seed endosperm and embryos. These results suggest that, at the transcriptional level, the integration of light perception coupled with the complex regulation of starch metabolism in Arabidopsis and rice has diverged substantially since the divergence of dicots and monocots (Fu and Xue, 2010).
ECC Patterns for Evolutionarily Conserved Plant Genes
In addition to the study of expression conservation for different functional categories, we also delineated specific gene sets focusing on genome organization and phyletic distribution. Closely related species have extensive regions of shared gene content and order (or colinearity), but as the evolutionary distance between two species increases, colinear segments erode due to gene loss and rearrangements (Liu et al., 2001; Vandepoele et al., 2002). Starting from a set of 76 regions with conserved gene content and order between Arabidopsis and rice, 116 1:1 orthologous colinear gene pairs were extracted. Comparison of the degree of coexpression of neighboring genes in colinear regions revealed that only 5.3% and 5.9% of these genes had coordinated expression profiles in Arabidopsis and rice, respectively. Compared with the expected degree of coexpression (i.e. maximum 10% with the 90th percentile PCC threshold), these genes showed no strong evidence for large-scale coregulation (Ren et al., 2005). Of all orthologous colinear gene pairs, 69% had a conserved ECC.
Genes with homologs in all other plants are known as “core” genes and include essential genes covering the basic genetic toolbox in plants. The study of core plant genes combined with expression states provides a simple means to enlarge our understanding of the evolution of gene function versus regulation. From the 8,478 conserved Arabidopsis core genes (with homologs in all nine plant species present in the PLAZA comparative genomics platform; Proost et al., 2009), 1,787 genes were present in 1:1 orthologous families. Of these core genes, 80% (1,426 of 1,787) had conserved ECC patterns, revealing a 3% increase in conserved coexpression state compared with the average level in the complete data set (P < 8.5e-05; hypergeometric distribution).
Organization and Conservation of cis-Regulatory Elements in ECC Conserved Genes
To characterize the underlying mechanism of conserved coexpression patterns in Arabidopsis and rice, we analyzed the promoter regions of all genes with known cis-regulatory elements. First, known plant DNA motifs from AGRIS (Palaniswamy et al., 2006) and PLACE (Higo et al., 1999) were mapped onto the 1-kb upstream promoters followed by motif enrichment analysis per gene-centric cluster (Vandepoele et al., 2009). To reduce the inclusion of false-positive motif instances, for a given gene-centric cluster, only motifs present in the seed gene promoter and significantly enriched in the corresponding coexpression cluster were retained for further analysis. A total of 308 and 276 DNA motifs were found to be enriched in Arabidopsis and rice, respectively, with an average number of 15 and 21 enriched motifs per promoter (covering 3,242 Arabidopsis and 3,267 rice genes).
For the 3,270 ECC conserved orthologous genes, all promoter motifs conserved between Arabidopsis and rice were determined to study whether conserved coexpression patterns correlated with shared cis-regulatory elements. As a control, we shuffled all enriched gene-motif annotations and compared the real motif conservation rates with the expected values. In total, 3,003 (84%) Arabidopsis genes were found with one or more conserved motif, whereas 161 DNA motifs were conserved in orthologous genes from both species. In contrast, the motif conservation rate in the nonconserved ECC genes and the control set was 75% and 73%, respectively. When genes with at least five or 15 conserved motifs were considered, the ECC conserved category contained 1,240 and 608 genes (i.e. 2- and 3.7-fold more than in the control data set, respectively).
At least 14 different regulatory elements were much more conserved between orthologous genes than expected by chance (P < 0.05), and several of them were related to (or showed GO enrichment for) conserved processes, such as photosynthesis (IBOXCORE, MYBST1, SORLIP1AT, and ABRELATERD1), ribosome biogenesis (SITEIIATCYTC), or basic transcriptional control (INRNTPSADB). Motifs enriched in more than 50 Arabidopsis genes, but not conserved in the orthologous rice genes, covered CBFHV (C-repeat-binding factors; dehydration-responsive element), ARR1AT (ARR1-binding element; response regulator), MYBGAHV (central element of the GA response complex), AP3SV40 (AP-3-binding site consensus sequence), and SV40COREENHAN (SV40 core enhancer).
Influence of Tissue Specificity, Protein Evolution, and Connectivity on ECC
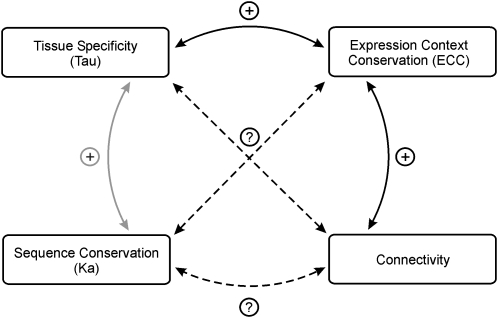
Tissue-specific gene expression plays a fundamental role in multicellular systems, underlying development, function, and maintenance of diverse cell types within an organism. Here, we used τ (Tau in the figures) to quantify the level of differential expression across conditions (accounting for the quantitative variations in transcript levels) and to identify tissue specificity (Liao and Zhang, 2006). τ ranges from 0 to 1, and high values correspond with low expression breadth (i.e. expressed in a few tissues). First, we investigated how tissue-specific or constitutive expression was linked with ECC and protein evolution (Fig. 4). For orthologs with different expression patterns, the fraction of genes with conserved ECC rose with increasing τ values (Supplemental Fig. S3). Both for Arabidopsis and rice, the median τ values were significantly higher for ECC conserved than ECC nonconserved genes (Arabidopsis τ = 0.184 and 0.169, respectively [P < 2.6e-05]; rice τ = 0.200 and 0.175, respectively [P < 2.2e-10]). For all orthologous gene pairs, the rate of nonsynonymous substitutions (Ka) was used to measure evolution at the protein-coding level. For both species, comparison of τ as a function of Ka showed that tissue specificity correlated with increasing rates of protein evolution (Supplemental Fig. S4).
Figure 4.
Summary of the correlations between expression and sequence evolution, connectivity, and tissue specificity. Plus signs denote positive correlations, whereas question marks and dotted lines indicate that no positive or negative correlation is found. Correlations deemed significant with the Mann-Whitney U test are highlighted as solid black arrows.
Ka distributions for genes with different ECC patterns did not differ significantly in protein evolution for genes with conserved and nonconserved coexpression contexts (median Ka values of 0.323 and 0.329, respectively [P = 0.28]). Plotting the distribution of ECC categories for genes with increasing rates of protein evolution confirmed the absence of a strong association between expression and protein evolution (Supplemental Fig. S5). The ratio of the number of nonsynonymous substitutions to the number of synonymous substitutions (Ka/Ks) was used to evaluate whether the selective pressure that acted on a protein-coding gene varied per ECC category. Although significant, the difference in the level of purifying selection between different ECC types was small (median Ka/Ks values for conserved and nonconserved genes were 0.087 and 0.084, respectively [P = 0.023]).
Finally, we evaluated whether the connectivity of a gene, defined as the number of coexpressing partners, had an influence on the evolution of orthologs or tissue specificity. As shown in Supplemental Figure S6, both in Arabidopsis and rice highly connected genes were enriched for ECC conserved genes, while no clear trend was observed for τ or Ka (data not shown). Control for rates of protein evolution (by binning genes based on Ka) confirmed that, overall, ECC conserved genes were more tissue specific than their nonconserved counterparts (Supplemental Fig. S7). The presence of a TATA promoter motif did not correlate with the level of coexpression conservation.
Concerted Network Evolution for Genes without Conserved Tissue Specificity
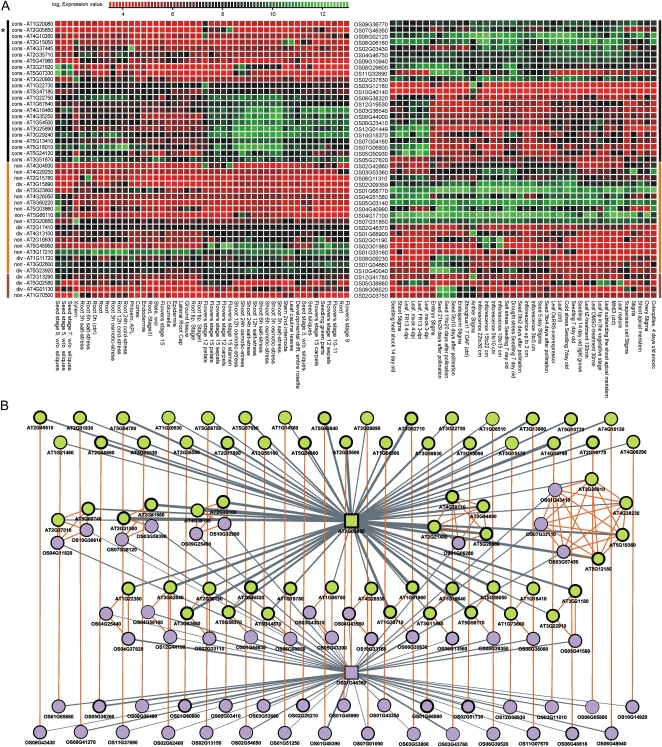
As in vertebrates (Chan et al., 2009), in Arabidopsis and rice many genes show tissue specificity as well as expression conservation (e.g. AT3G04700, AT3G17060, GATL4, AMS, and AG in flower; AT4G31830, AT2G28420, AT1G05510, AT3G12960, and ATOEP16-2 in seed; AT1G30870 in root). However, based on a set of tissue-specific Arabidopsis genes (Schmid et al., 2005), several orthologs (five of 14) were broadly expressed in rice. By including additional Arabidopsis and rice genes with high τ values in our expression compendia, we found several orthologs without conserved tissue specificity (malate synthase [AT5G03860], seed; C3HC4-type RING finger [AT2G20650], root xylem; selenium-binding protein [AT3G23800], root cortex; TET4, stage 8 seeds without siliques; Fig. 5A). Whereas for a large number of genes the loss of tissue specificity in one species coincided with nonconserved coexpression patterns (Table I; type II), more than 20 genes were identified with tissue-specific expression in only one species but also with ECC (type I). In this case, the tissue-specific gene under investigation as well as the genes showing a conserved expression adopted, in a concerted manner, a different expression pattern in the other species. Figure 5B depicts a flower-specific (stage 15 stamen pollen) Arabidopsis Ser carboxypeptidase (SCPL38) with functional enrichment for reproduction and flower development. Although the orthologous gene had a broad expression pattern in rice (thin borders of rice genes in Fig. 5B), the coexpression with several known flower development genes such as SEP1-3, AG, and AP3 was conserved in both species. Another example is the concerted network for Arabidopsis IQD10, a gene involved in phloem or xylem histogenesis but with ubiquitous expression in rice (Supplemental Fig. S8). Finally, a small set of genes was specifically expressed in both species, but for different tissues (type III). Although all these genes have a nonconserved ECC, in at least one case (AT1G70500, pectin lyase) the coexpression contexts in both species had a similar GO enrichment (carbohydrate metabolism), suggesting a similar molecular function.
Figure 5.
Expression evolution of tissue-specific genes. A, Expression heat map of orthologous Arabidopsis genes (left) and rice genes (right) lacking conservation of tissue specificity. Green values indicate expression above background, whereas the prefixes “cons,” “div,” and “non” indicate the ECC category. Black, orange, and brown bars correspond to type I, II, and III genes, respectively (for details, see Table I). B, Expression network of concerted expression divergence for the flower-specific Arabidopsis gene SCPL38. Gray and orange lines show coexpression and orthology relationships, respectively, whereas the thickness of the gray lines indicates the expression similarity. Green and purple nodes denote Arabidopsis and rice genes, respectively, whereas the orthologous seed genes are drawn as boxes (and indicated by the asterisk in the heat map shown in A). Node border thickness gives the tissue-specific expression measured with τ.
Table I. Genes showing nonconserved tissue-specific expression in Arabidopsis and rice.
| Ath Genea | Osa Genea | Ath E Breadth (τ) | Osa E Breadth (τ) | ECC Categoryb | Ath Tissuec | Osa Tissuec | Typed | Descriptione |
| AT1G20080 | OS09G36770 | 1 (0.492) | 58 (0.130) | cons | Flower | – | I | SYTB |
| AT1G22730 | OS03G12180 | 19 (0.309) | 2 (0.706) | cons | – | Anther-stigma | I | MA3 domain-containing protein |
| AT1G22750 | OS06G38320 | 63 (0.136) | 2 (0.407) | cons | – | Leaf | I | Cofactor metabolism* |
| AT1G54500 | OS08G23410 | 55 (0.191) | 6 (0.425) | cons | – | Leaf, seedling | I | Rubredoxin family protein |
| AT1G67840 | OS12G19530 | 50 (0.156) | 4 (0.468) | cons | – | Leaf | I | Chloroplast sensor kinase (CSK) |
| AT2G05850 | OS07G46350 | 3 (0.593) | 36 (0.232) | cons | Flower | – | I | Ser carboxypeptidase-like 38 (SCPL38) |
| AT2G21820 | OS08G29600 | 8 (0.601) | 21 (0.328) | cons | Seed, shoot osmotic stress | – | I | Reproductive structure development* |
| AT2G35710 | OS04G46750 | 6 (0.405) | 61 (0.137) | cons | Root | – | I | Glycogenin glucosyltransferase (glycogenin)-related |
| AT3G15050 | OS06G06160 | 4 (0.558) | 59 (0.152) | cons | Leaf, xylem, stem | – | I | IQ-domain 10 (IQD10) |
| AT3G20860 | OS02G37830 | 3 (0.451) | 61 (0.122) | cons | Xylem, phloem, root salt stress | – | I | Member of the NIMA-related Ser/Thr kinases (ATNEK5) |
| AT3G25690 | OS12G01449 | 52 (0.247) | 9 (0.626) | cons | – | Leaf, shoot-stigma, seedling | I | Actin-binding protein required for normal chloroplast positioning (CHUP1) |
| AT3G29240 | OS10G18370 | 67 (0.156) | 9 (0.504) | cons | – | Leaf, shoot-stigma, seedling | I | Chlorophyll metabolic process* |
| AT3G51670 | OS05G27820 | 69 (0.172) | 1 (0.402) | cons | – | Shoot | I | SEC14 cytosolic factor family protein/phosphoglyceride transfer family protein |
| AT4G10260 | OS08G02120 | 3 (0.460) | 61 (0.131) | cons | Flower | – | I | pfkB-type carbohydrate kinase family protein |
| AT4G18480 | OS03G36540 | 55 (0.204) | 2 (0.521) | cons | – | Leaf | I | CHLI subunit of magnesium chelatase, which is required for chlorophyll biosynthesis (CHLI1) |
| AT4G35250 | OS08G44000 | 51 (0.250) | 3 (0.413) | cons | – | Leaf | I | Vestitone reductase-related |
| AT4G37445 | OS02G03430 | 1 (0.547) | 25 (0.306) | cons | Phloem, APL | – | I | Disaccharide biosynthesis* |
| AT5G07330 | OS11G32890 | 6 (0.618) | 13 (0.519) | cons | Seed, shoot osmotic stress | – | I | Response to abscisic acid stimulus* |
| AT5G13410 | OS07G04160 | 50 (0.169) | 6 (0.429) | cons | – | Leaf, shoot-stigma, seedling | I | Immunophilin/FKBP-type peptidyl-prolyl cis-trans-isomerase family protein |
| AT5G16010 | OS07G06800 | 64 (0.230) | 6 (0.584) | cons | – | Leaf, shoot-stigma, seedling | I | 3-Oxo-5-α-steroid 4-dehydrogenase family protein/steroid 5-α-reductase family protein |
| AT5G24120 | OS05G50930 | 35 (0.323) | 6 (0.574) | cons | – | Leaf, shoot-stigma, seedling | I | σ-factor E (SIGE) |
| AT5G47180 | OS10G40140 | 13 (0.261) | 1 (0.676) | cons | – | Anther-stigma | I | Vesicle-associated membrane family protein/VAMP family protein |
| AT5G47960 | OS09G10940 | 3 (0.415) | 59 (0.089) | cons | Root | – | I | Encodes a small-Mr G-protein (ATRABA4C) |
| AT1G11720 | OS08G09230 | 34 (0.224) | 7 (0.548) | div | – | Seed | II | ATSS3 (starch synthase 3) |
| AT2G15780 | OS06G11310 | 2 (0.604) | 31 (0.291) | non | Root | – | II | Gly-rich protein |
| AT2G16630 | OS02G01190 | 33 (0.301) | 6 (0.620) | non | – | Inflorescence | II | Pro-rich family protein |
| AT2G17410 | OS02G48370 | 22 (0.113) | 1 (0.514) | div | – | Anther-stigma | II | ARID/BRIGHT DNA-binding domain-containing protein |
| AT2G20650 | OS07G31850 | 6 (0.471) | 63 (0.059) | non | Xylem, cortex, root salt stress | – | II | Zinc finger (C3HC4-type RING finger) family protein |
| AT3G02600 | OS01G04660 | 73 (0.172) | 4 (0.492) | non | – | Seed, embryo-stigma | II | Phosphatidic acid phosphatase (LPP3) |
| AT3G15890 | OS02G09359 | 1 (0.416) | 59 (0.132) | div | Root | – | II | Protein kinase family protein |
| AT3G17210 | OS01G33160 | 76 (0.123) | 3 (0.537) | non | – | Salt/drought/cold stress seedling | II | HS1 (heat-stable protein 1) |
| AT3G23800 | OS01G68770 | 7 (0.437) | 63 (0.164) | div | Root, xylem, phloem, cortex | – | II | Selenium-binding protein 3 (SBP3) |
| AT4G04930 | OS02G42660 | 4 (0.591) | 21 (0.350) | non | Flower | – | II | Sphingolipid Δ4-desaturase, involved in sphingolipid biosynthesis (DES-1-LIKE) |
| AT4G13100 | OS01G68900 | 16 (0.200) | 2 (0.519) | div | – | Anther-stigma | II | Zinc finger (C3HC4-type RING finger) family protein |
| AT4G26050 | OS04G51580 | 1 (0.416) | 44 (0.229) | non | Seed | – | II | Leu-rich repeat family protein |
| AT4G29250 | OS03G53360 | 1 (0.402) | 21 (0.363) | non | Flower | – | II | Transferase family protein |
| AT5G03860 | OS04G40990 | 5 (0.566) | 30 (0.311) | non | Seed, cortex | – | II | Protein with malate synthase activity (MLS) |
| AT5G23920 | OS10G40040 | 60 (0.174) | 9 (0.516) | div | – | Seed, embryo-stigma | II | Unknown |
| AT5G45950 | OS02G01980 | 44 (0.390) | 3 (0.676) | non | – | Inflorescence | II | GDSL-motif lipase/hydrolase family protein |
| AT5G60220 | OS05G03140 | 1 (0.538) | 63 (0.080) | non | Seed | – | II | Member of TETRASPANIN family (TET4) |
| AT5G66110 | OS04G17100 | 3 (0.537) | 63 (0.129) | non | Seed, shoot osmotic stress | – | II | Metal ion binding |
| AT1G70500 | OS02G03750 | 2 (0.630) | 4 (0.528) | non | Xylem, root | Seed | III | Polygalacturonase, putative/pectinase, putative |
| AT2G13290 | OS12G41780 | 5 (0.283) | 1 (0.565) | div | Seed | Anther-stigma | III | Glycosyl transferase family 17 protein |
| AT4G01130 | OS06G06520 | 5 (0.344) | 5 (0.640) | div | Shoot | Seed | III | Acetylesterase, putative |
| AT5G02580 | OS05G38680 | 7 (0.479) | 9 (0.557) | div | Flower | Leaf | III | Unknown |
Ath, Arabidopsis; Osa, rice.
The categories “cons,” “div,” and “non” indicate conserved, diverged, and not significant, respectively.
Dashes indicate expression in a large number of tissues.
Type I, ECC conserved, tissue-specific expression in only one species; type II, ECC nonconserved, tissue-specific expression in only one species; type III, ECC nonconserved, tissue-specific expression in both species but different tissue.
Descriptions indicated by asterisks denote GO enrichments based on the coexpression context of the tissue-specific gene with unknown function.
DISCUSSION
Whereas comparative sequence analysis is a powerful tool to study genome evolution, to discover conserved orthologous genes, and to characterize species-specific gene families, the integration of functional genomics data provides an additional layer of information to study gene function and regulation. Recently, expression data, functional gene annotations, protein-protein interaction data, knockout phenotype information, and cis-regulatory elements have been combined to delineate coexpressed modules, to predict new gene functions, and to identify transcriptional regulatory interactions (Aoki et al., 2007; Lee et al., 2009; Vandepoele et al., 2009; De Bodt et al., 2010; Mutwil et al., 2010; Obayashi and Kinoshita, 2010). Although most coexpression approaches have been used to predict different types of gene-gene interactions in model species such as Arabidopsis and rice, interspecies comparisons have identified examples of conserved coexpression modules in plants (Ma et al., 2005; Street et al., 2008).
To determine which factors influence expression evolution in plants, we performed a comparative transcriptomics analysis with large-scale expression data from Arabidopsis and rice. ECC represents an unsupervised approach to systematically compare gene expression networks between two species, including a statistical framework to quantify coexpression conservation. Although this approach does not directly compare expression profiles from identical experiments between species (Tirosh et al., 2007), comparison of coexpression clusters provides a valuable alternative to studying expression between less and more distantly related species lacking perfectly matched data sets. In contrast to two recent methods that also compare conserved expression patterns to identify functional analogy among homologous genes (Chikina and Troyanskaya, 2011; Mutwil et al., 2011), the presented ECC method includes different null models to reliably estimate the significance levels of conserved coexpression that control network properties, such as connectivity or tissue specificity. Overall, conserved coordinated expression occurred in 77% of the analyzed genes of Arabidopsis and rice. This degree of expression similarity is higher than the 60% reported for Arabidopsis and Populus orthologs that have broadly similar expression patterns during leaf development (Street et al., 2008). Whereas the set of genes with expression conservation delineated here is robust when alternative models are used to estimate significance levels (77% and 75% ECC conservation with the connectivity and tissue-specific null models, respectively), the study of Street et al. (2008) monitored conservation of leaf expression in the absence of a statistical framework. Although only genes with orthologs in both species have been retained, almost 25% of all analyzed genes had no significant coexpression conservation, indicating that a substantial fraction of coexpression links have been rewired during plant evolution.
The observed conservation of many developmental regulators generates valuable information for the transfer of biological knowledge between different species in plant biotechnology. In contrast, many genes that respond to a specific stress stimulus or are involved in signal transduction had low conservation levels, suggesting a link between regulatory evolution and adaptation to lifestyle or environment. As the degree of coexpression conservation varied for different functional categories, the relationship between protein evolution (Ka), tissue specificity (τ), and ECC was analyzed in more detail considering different GO categories (Supplemental Table S5). Notably, several GO terms deviated significantly from the general trends. Genes evolving rapidly at the expression level but slowly at the protein level (low fraction of ECC conserved genes and low Ka) include categories such as cellular response to extracellular stimulus, oxygen and reactive oxygen species metabolism, response to salt stress, and mitochondrial and carbohydrate transmembrane transport. Expression divergence of duplicate genes under environmental stress has also been found to be significantly greater than that under developmental stress (Ha et al., 2007). In addition, the levels of expression divergence between gene duplicates were the highest in extracellular transport, signal transduction, stress response, and transcription and were the lowest in the cellular and developmental processes, such as energy pathway, protein metabolism, intracellular transport, DNA and RNA metabolism, and cell organization and biogenesis. These functional categories are highly congruent with the ECC results (Fig. 3) and indicate that expression divergence in response to external processes not only acts on recent duplicates but also on orthologous genes that have been present for hundreds of millions of years within the genomes of flowering plants. Moreover, both the diverged and conserved gene functions highlight the robustness and stochasticity of gene regulatory networks in the control of gene expression. Ninety-three percent of a set of 147 essential Arabidopsis embryo-defective genes (Meinke et al., 2008) were ECC conserved, confirming the robustness of regulatory developmental programs across species (Macneil and Walhout, 2011).
Although it is plausible to assume that conserved expression contexts are the output of ancestral regulatory interactions that have been conserved in extant species (Humphry et al., 2010), the absence of large-scale TF-target data makes it difficult to construct genome-wide regulatory networks in plants and to directly study their evolution. Therefore, information about known cis-regulatory elements was integrated to annotate coexpression contexts. Tight regulatory promoter conservation (i.e. five or more conserved motifs) explains the observed coexpression conservation for 41% of all analyzed genes, revealing a 2-fold enrichment compared with a control data set. Corroborating the relationship between expression evolution and adaptation, several motifs enriched only in the context of Arabidopsis have been annotated as responsive elements, suggesting that different cis-regulatory elements and/or other TFs regulate these genes in each organism. The contribution of cis-regulatory elements to conserved transcriptional control should be interpreted cautiously, because the knowledge of plant promoter elements is far from complete.
Although “ancient” core plant genes with a broad phyletic distribution displayed a small, but significant, increase in the fraction of ECC conserved genes (80% versus 77% considering all genes), we found no strong evidence for the hypothesis that genes with conserved gene organization are highly conserved in expression (69% ECC conservation). These results imply that colinear genes between Arabidopsis and rice are, at the regulatory level, not more conserved than genes that have been rearranged since the divergence of both species (Ren et al., 2007). Despite the coexpression of neighboring genes in rice (Ma et al., 2005), currently there is little evidence that general coregulatory mechanisms, complementary to, for example, bidirectional promoters, act on a global scale in plant genomes. Altogether, our findings indicate that orthologs inferred through sequence similarity in many cases do not resemble similar biological gene functions and highlight the importance of incorporating expression information when homologous genes between different species are analyzed.
The combined information about tissue-specific gene expression (τ) and protein sequence evolution (Ka) indicates that high tissue specificity is linked with conserved ECC and high rates of protein evolution (Fig. 4). Reversely, this pattern suggests that genes expressed in many tissues or conditions, potentially with a pleiotropic function, are strongly constrained at the sequence level. Network connectivity analysis confirmed that genes expressed coordinately with many other partners have more conserved patterns of expression evolution. A large-scale comparative expression study in mammals confirmed that highly tissue-specific genes tend to evolve rapidly at the sequence level but slowly at the expression level (Liao and Zhang, 2006). The high divergence at the protein level seems to coincide with the assumption that tissue-specific genes have less pleiotropic effects (Hastings, 1996; Duret and Mouchiroud, 2000), whereas carefully tuned expression of, for example, tissue-specific TFs is essential for cell differentiation and the proper execution of developmental programs. The functional importance hypothesis (Rocha and Danchin, 2004), in which highly expressed genes are functionally more important and therefore more conserved in their coding sequences, corresponds with the high levels of sequence conservation for genes with low τ values. Also in yeast, genes with many protein-protein interactions or functional protein sites negatively correlate with coding-sequence divergence, confirming the strong constraint to maintain physical interactions (Tirosh and Barkai, 2008). We observed no significant correlation between protein and expression evolution, indicating that both modes of gene evolution are not coupled in plants. Similarly, after comparison of expression similarity with coding sequences for more than 10,000 human-mouse orthologs, no strong correlation between expression and sequence evolution was found in mammals (Liao and Zhang, 2006). Although contradictory conclusions have been drawn in different species (Makova and Li, 2003; Jordan et al., 2005; Lemos et al., 2005; Liao and Zhang, 2006; Sartor et al., 2006), also in yeast no evidence has been found for a strong correlation between these two modes of gene evolution (Tirosh and Barkai, 2008).
Changes in the expression pattern of a tissue-specific gene will have important consequences for the interacting genes and the biological function of the underlying network. Complementary with a report of concerted expression divergence after large-scale duplications in Arabidopsis (Blanc and Wolfe, 2004), we have identified more than 20 genes in which the expression contexts are conserved and tissue specificity is not. Although in the case of gene duplication, evolution might work on the initially redundant version of a specific pathway or a set of genetically interacting genes, the pattern of concerted expression evolution for different single-copy tissue-specific markers in Arabidopsis and rice is intriguing. In yeast, cross-species promoter analysis has shown how the regulation of ribosomal proteins evolved via intermediate redundant programs in which the concurrent emergence of cis-regulatory elements was followed by the loss of more ancient elements (Tanay et al., 2005). A similar mechanism might provide a mechanistic explanation for the concerted tissue-specific divergence (Table I), but the knowledge about regulatory control in plants is currently insufficient to determine the role of cis- versus trans-regulatory changes in this network rewiring. The developed ECC framework provides a practical approach to compare expression patterns and molecular phenotypes between species. Nevertheless, the detailed characterization of target genes for orthologous regulators in different species will be required to obtain a more detailed view of the regulatory evolution of signaling networks and the rate of expression evolution in different plant families.
MATERIALS AND METHODS
Expression Data
We obtained 203 and 322 Affymetrix CEL files for rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), respectively, from the National Center for Biotechnology Information Gene Expression Omnibus database monitoring the transcriptional activity in different tissues and developmental stages (Schmid et al., 2005; Jain et al., 2007; Li et al., 2007), root cell types (Birnbaum et al., 2003; Norton et al., 2008), and under different stress conditions (Walia et al., 2005; Kilian et al., 2007; Swarbrick et al., 2008). The ATH1 microarray slides were processed with a custom-made CDF measuring 19,937 genes as described before (Vandepoele et al., 2009). Briefly, all raw data were processed with the robust multichip analysis algorithm implementation (Irizarry et al., 2003) in Bioconductor and with custom-made CDFs (background adjustment, quantile normalization, and finally summarization). A remapping of all 631,066 rice probes to the rice gene models (The Institute for Genome Research 5) showed that 43% of the original probe set (as defined by Affymetrix CDF ricecdf) contained one or more cross-hybridizing probes (S. Movahedi and K. Vandepoele, unpublished data). These cross-hybridization effects could lead to significant errors, especially under conditions where changes in expression are not dramatic. For all microarray analyses, it is important to ascertain that a probe really measures the intended gene. Therefore, this inaccuracy in the original rice Affymetrix probe set urged us to design a new set of probe set (or CDF)-containing probes unique at the gene level, as described for Arabidopsis (Casneuf et al., 2007). The new rice gene CDF allowed us to reliably measure expression levels for 32,004 rice gene models and is available upon request. The mean intensity values were calculated for the replicated slides. To identify and remove redundant experiments, we first clustered all experiments with hierarchical clustering (considering all genes and the PCC as a measure) and retained only one experiment as representative for a set of similar experiments (De Bodt et al., 2010).
Clustering of Expression Data
Expression similarities between gene pairs (per species) were calculated with the PCC. To identify coexpressed genes, a similarity threshold was determined for both expression compendia. Based on the similarity between expression profiles for 1,000 random genes (approximately 1,000 × 999 × 0.5 gene pairs), a PCC threshold of 0.41 and 0.48, corresponding with the 90th percentile of this distribution, was set for rice and Arabidopsis, respectively. Although the absolute PCC threshold differed for both species (because of differences in size and composition of the expression data set), these percentile-based thresholds returned a similar relative cutoff for both species. Application of different percentile thresholds to predict functional enrichments with gene-centric coexpression clusters for genes with known annotations (excluding electronic GO functional assignments) showed that the 90th percentile threshold recovered the largest number of correct gene functions (results for Arabidopsis/rice with a random set of 100 reference genes are as follows: 85th percentile, 32%/22% correct predictions; 90th percentile, 33%/24% correct predictions; 95th percentile, 33%/23% correct predictions).
The EC for a set of N genes was calculated as the fraction of all possible N × (N − 1) × 0.5 gene pairs with a PCC higher than the threshold value defined for that species. To determine the stability of the coexpression on the removal of subsets of experiments from the original expression compendium, a jackknife procedure was applied. Based on 100 iterations, randomly 25% and 50% of the original experiments were removed, percentile-based PCC thresholds were defined for the retained experiments, and the EC per GO category was calculated.
To create gene-centric coexpression clusters, each gene was considered as a seed gene and all genes with a PCC value greater than the determined PCC threshold were assigned to the cluster. Therefore, the number of clusters is equal to the number of genes available in the expression data set, and clusters are potentially overlapping on a genome-wide scale. Data files containing the Arabidopsis and rice gene coexpression clusters as well as the orthologous gene families are available at http://bioinformatics.psb.ugent.be/supplementary_data/Movahedi_ECC.
Identification of Orthologous Gene Families
To identify orthologous genes, we clustered similar protein sequences from rice and Arabidopsis by means of the OrthoMCL algorithm starting from an all-against-all sequence similarity search with BLASTP (E < 1e-05, with a default inflation factor of 1.5). Arabidopsis and rice protein sequences were downloaded from The Arabidopsis Information Resource 7 and The Institute for Genome Research 5, respectively. Among 33,195 families, 7,911 contained at least one gene from each species also present in the expression data. These families covered 12,019 rice genes and 12,419 Arabidopsis genes and could be subdivided into three subcategories: 1:1 orthologous groups (both species contained one gene; 4,630 families), 1:n groups (one of the species contained a single gene and the other contained multiple genes; 2,194 families with 3,748 and 4,031 rice and Arabidopsis genes, respectively), and n:m groups (n,m > 1; 1,087 families with 3,641 and 3,758 rice and Arabidopsis genes, respectively). A list of Arabidopsis gene symbols, Arabidopsis Genome Initiative locus identifiers, and orthologous rice genes is available in Supplemental Table S6. Inparalogs are gene duplicates postdating speciation. Recent benchmark studies have shown that the OrthoMCL gene-clustering method performs well in modeling recent and ancient duplication events compared with more advanced methods based on phylogenetic inference (Chen et al., 2007; Proost et al., 2009). For every functional category (see below), all genes with one or more orthologous gene in the other species were retained for the EC analysis. Genes from 1:1 orthologous families were used as seeds to study the ECC (retaining all genes with orthologs in both plants to calculate the gene-centric coexpression clusters). Because for both species not all genes are present on the Affymetrix microarrays, the reported ECC scores are not exact but represent estimates based on the orthologs available in the CDFs.
Functional Annotation
Genes and orthologous gene families were functionally annotated with GO and Reactome terms. Whereas GO labels were retrieved from the GO Web site (versions of July 21, 2009, for Arabidopsis and April 18, 2009, for rice), Reactome data were downloaded from http://arabidopsisreactome.org/ (Tsesmetzis et al., 2008). The gene-GO annotations were extended to include parental GO terms (i.e. a gene assigned to a given GO category was automatically assigned to all the parent categories as well) by propagating all GO annotations up to all possible edges of the GO graph (with the Perl GO::Parser and GO::Node modules). GO annotations were assigned to all 1:1 orthologous gene pairs starting from the original gene-GO annotation files. ECC scores for different functional categories excluding electronic evidence tags (IEA, ISS, NAS, NR, and ND) are reported in Supplemental Table S7. To calculate the EC for different GO categories (Fig. 1; Supplemental Table S2), the original gene annotations were used.
The statistical significance of the functional enrichment for GO and MAPMAN (Thimm et al., 2004) annotations was evaluated with the hypergeometric distribution adjusted by the false discovery rate correction for multiple hypotheses testing by means of the p.adjust stats package in R (Benjamini and Hochberg, 1995). Corrected values of P < 0.05 were considered significant. MAPMAN results are reported in Supplemental Table S8. Colinear regions between Arabidopsis and rice were computed with i-ADHoRe and retrieved from PLAZA, a comparative genomics resource (Proost et al., 2009).
Calculation ECC Scores
In a first step, for all genes a set of coexpressed genes was defined (gene-centric cluster) by retrieving the neighboring genes in the coexpression network. Next, for gene-centric clusters from 1:1 orthologs, the number of orthologs was determined with coexpression in both species (Fig. 1). The overlap of conserved shared orthologous families in both coexpression clusters was quantified with the Jaccard Index, which measures the ratio of the number of shared families over the total number of families found in the two coexpression clusters. A permutation test was applied to determine whether the observed ECC score for that 1:1 orthologous gene pair could be considered conserved, diverged, or not significant (i.e. significantly larger, smaller, or not different compared with a background distribution of ECC values [P < 0.05]). For each 1:1 orthologous gene pair A and R, 1,000 pairs of gene-centric clusters were randomly sampled, maintaining the gene-centric cluster sizes (A) and (R), respectively. The expected ECC scores, reflecting a neutral null model of expression evolution, were used to estimate a P value for coexpression conservation or divergence. Two null models were used, one controlling tissue-specific expression and one correcting the degree distribution in the network (or connectivity). One model controlled the expression breadth (with τ) in determining the ECC significance, and the other controlled the degree distribution (or connectivity) of the different genes in the network. Briefly, during the permutation-based significance estimation using these models, we randomly sampled genes with properties similar to those of the genes present in the real network (i.e. expression breadth or connectivity). First, all genes were grouped in 50 bins based on τ (or connectivity); subsequently, we sampled from a specific bin to control expression breadth (or the number of coexpressed genes). Orthologous pairs with higher (or lower) Jaccard Index than that of the random values were considered as conserved (or diverged), and genes not belonging to any of these categories were classified as nonsignificant (Supplemental Fig. S2). To correct for multiple comparisons, again the false discovery rate method was applied with p.adjust. Classification of the Jaccard Index with other percentile-based PCC thresholds for expression similarity yielded highly similar results.
Calculation of Ka and Ks
The coding sequences for the gene pairs were aligned with ClustalW version 1.83 (Thompson et al., 2002) with the protein sequences as alignment guides. From this alignment, unambiguously aligned positions were retained for further analysis (Proost et al., 2009). Synonymous and nonsynonymous substitutions (Ks and Ka) were estimated with codeml (part of the PAML package; Yang, 1997) with the following parameter settings: verbose 0, noisy 0, runmode −2, seqtype 1, model 0, NSsites 0, icode 0, fix_alpha 0, fix_kappa 0, and RateAncestor 0. Note that values of Ks > 5 should be considered as unreliable.
Statistical Analysis
Differences between ECC conserved and nonconserved gene sets were tested with the nonparametric Mann-Whitney U test (also known as the Mann-Whitney-Wilcoxon test) with the “wilcox.test” function in the “stats” R package version 2.9.1. To determine whether the fraction of ECC conserved genes was significantly larger for a specific functional category than that of the overall ECC conservation considering all genes, the hypergeometric distribution was used (Supplemental Tables S4 and S6). Differences between Ka (or τ) values between all 1:1 orthologs and genes annotated with a specific functional category were also evaluated with the Mann-Whitney U test, as were differences between Ka (or τ) values for ECC conserved and nonconserved gene sets within a specific functional category (Supplemental Table S5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of removing 25% and 50% random experiments from expression data on EC values with sd and average measures.
Supplemental Figure S2. Examples of ECC scores assigned to different categories with random Jaccard Index scores.
Supplemental Figure S3. ECC as a function of τ.
Supplemental Figure S4. τ as a function of Ka, Ks, and Ka/Ks.
Supplemental Figure S5. ECC as a function of Ka, Ks, and Ka/Ks.
Supplemental Figure S6. ECC as a function of connectivity.
Supplemental Figure S7. τ as a function of Ka and ECC.
Supplemental Figure S8. Concerted expression network evolution for the tissue-specific Arabidopsis gene AT3G15050.
Supplemental Table S1. Overview of microarray experiments included in the expression compendia for Arabidopsis and rice.
Supplemental Table S2. EC for GO categories with five or more genes.
Supplemental Table S3. ECC distributions for all 1:1 orthologous genes.
Supplemental Table S4. Distributions of ECC conserved, diverged, and nonsignificant genes for different GO and Reactome categories.
Supplemental Table S5. ECC conservation, τ, and Ka for Arabidopsis genes in different functional categories.
Supplemental Table S6. Overview of 1:1 orthologous families.
Supplemental Table S7. Distributions of ECC conserved, diverged, and nonsignificant genes for different GO categories, excluding electronic annotations.
Supplemental Table S8. MAPMAN enrichment for different gene sets.
Supplementary Material
Acknowledgments
We thank Martine De Cock for help in preparing the manuscript.
References
- Aoki K, Ogata Y, Shibata D. (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48: 381–390 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bergmann S, Ihmels J, Barkai N. (2004) Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, Van de Peer Y, Huber W. (2007) In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ET, Quon GT, Chua G, Babak T, Trochesset M, Zirngibl RA, Aubin J, Ratcliffe MJ, Wilde A, Brudno M, et al. (2009) Conservation of core gene expression in vertebrate tissues. J Biol 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina MD, Troyanskaya OG. (2011) Accurate quantification of functional analogy among close homologs. PLoS Comput Biol 7: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Carvajal D, Hollunder J, Van den Cruyce J, Movahedi S, Inzé D. (2010) CORNET: a user-friendly tool for data mining and integration. Plant Physiol 152: 1167–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. (2000) Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol 17: 68–74 [DOI] [PubMed] [Google Scholar]
- Dutilh BE, Huynen MA, Snel B. (2006) A global definition of expression context is conserved between orthologs, but does not correlate with sequence conservation. BMC Genomics 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FF, Xue HW. (2010) Co-expression analysis identifies Rice Starch Regulator1 (RSR1), a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ. (2007) External factors accelerate expression divergence between duplicate genes. Trends Genet 23: 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. (2003) Comparative genomics. PLoS Biol 1: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KE. (1996) Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J Mol Evol 42: 631–640 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki L, Wolfe KH. (2004) Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res 14: 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M, Bednarek P, Kemmerling B, Koh S, Stein M, Gobel U, Stuber K, Pislewska-Bednarek M, Loraine A, Schulze-Lefert P, et al. (2010) A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc Natl Acad Sci USA 107: 21896–21901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. (2006) Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443: 594–597 [DOI] [PubMed] [Google Scholar]
- Jordan IK, Mariño-Ramírez L, Koonin EV. (2005) Evolutionary significance of gene expression divergence. Gene 345: 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Lee TH, Kim YK, Pham TT, Song SI, Kim JK, Kang KY, An G, Jung KH, Galbraith DW, Kim M, et al. (2009) RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol 151: 16–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WP, Tzou WS. (2009) Computational methods for discovering gene networks from expression data. Brief Bioinform 10: 408–423 [DOI] [PubMed] [Google Scholar]
- Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. (2005) Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol 22: 1345–1354 [DOI] [PubMed] [Google Scholar]
- Li M, Xu W, Yang W, Kong Z, Xue Y. (2007) Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol 144: 1797–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang JZ. (2006) Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol 23: 530–540 [DOI] [PubMed] [Google Scholar]
- Liu H, Sachidanandam R, Stein L. (2001) Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res 11: 2020–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Huggins P, Bar-Joseph Z. (2009) Cross species analysis of microarray expression data. Bioinformatics 25: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen C, Liu X, Jiao Y, Su N, Li L, Wang X, Cao M, Sun N, Zhang X, et al. (2005) A microarray analysis of the rice transcriptome and its comparison to Arabidopsis. Genome Res 15: 1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil LT, Walhout AJ. (2011) Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res 21: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova KD, Li WH. (2003) Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res 13: 1638–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. (2008) Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, Yaghoubi-Masihi A, Fukao T, Bailey-Serres J. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Usadel B, Schütte M, Loraine A, Ebenhöh O, Persson S. (2010) Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol 152: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton GJ, Aitkenhead MJ, Khowaja FS, Whalley WR, Price AH. (2008) A bioinformatic and transcriptomic approach to identifying positional candidate genes without fine mapping: an example using rice root-growth QTLs. Genomics 92: 344–352 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. (2010) Coexpression landscape in ATTED-II: usage of gene list and gene network for various types of pathways. J Plant Res 123: 311–319 [DOI] [PubMed] [Google Scholar]
- Orzechowski S. (2008) Starch metabolism in leaves. Acta Biochim Pol 55: 435–445 [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Sudarsanam P, Church GM. (2001) Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XY, Fiers MW, Stiekema WJ, Nap JP. (2005) Local coexpression domains of two to four genes in the genome of Arabidopsis. Plant Physiol 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XY, Stiekema WJ, Nap JP. (2007) Local coexpression domains in the genome of rice show no microsynteny with Arabidopsis domains. Plant Mol Biol 65: 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A. (2004) An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol Biol Evol 21: 108–116 [DOI] [PubMed] [Google Scholar]
- Sartor MA, Zorn AM, Schwanekamp JA, Halbleib D, Karyala S, Howell ML, Dean GE, Medvedovic M, Tomlinson CR. (2006) A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res 34: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sterck L, Rombauts S, Vandepoele K, Rouzé P, Van de Peer Y. (2007) How many genes are there in plants (…and why are they there)? Curr Opin Plant Biol 10: 199–203 [DOI] [PubMed] [Google Scholar]
- Street NR, Sjödin A, Bylesjö M, Gustafsson P, Trygg J, Jansson S. (2008) A cross-species transcriptomics approach to identify genes involved in leaf development. BMC Genomics 9: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. (2009) How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25: 210–216 [DOI] [PubMed] [Google Scholar]
- Swarbrick PJ, Huang K, Liu G, Slate J, Press MC, Scholes JD. (2008) Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica. New Phytol 179: 515–529 [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. (2005) Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA 102: 7203–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: unit 2.3 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. (2008) Evolution of gene sequence and gene expression are not correlated in yeast. Trends Genet 24: 109–113 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Bilu Y, Barkai N. (2007) Comparative biology: beyond sequence analysis. Curr Opin Biotechnol 18: 371–377 [DOI] [PubMed] [Google Scholar]
- Tsesmetzis N, Couchman M, Higgins J, Smith A, Doonan JH, Seifert GJ, Schmidt EE, Vastrik I, Birney E, Wu G, et al. (2008) Arabidopsis reactome: a foundation knowledgebase for plant systems biology. Plant Cell 20: 1426–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Saeys Y, Simillion C, Raes J, Van De Peer Y. (2002) The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res 12: 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Van de Peer Y. (2005) Exploring the plant transcriptome through phylogenetic profiling. Plant Physiol 137: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, et al. (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.