Abstract
Rubisco limits photosynthetic CO2 fixation because of its low catalytic turnover rate (kcat) and competing oxygenase reaction. Previous attempts to improve the catalytic efficiency of Rubisco by genetic engineering have gained little progress. Here we demonstrate that the introduction of the small subunit (RbcS) of high kcat Rubisco from the C4 plant sorghum (Sorghum bicolor) significantly enhances kcat of Rubisco in transgenic rice (Oryza sativa). Three independent transgenic lines expressed sorghum RbcS at a high level, accounting for 30%, 44%, and 79% of the total RbcS. Rubisco was likely present as a chimera of sorghum and rice RbcS, and showed 1.32- to 1.50-fold higher kcat than in nontransgenic rice. Rubisco from transgenic lines showed a higher Km for CO2 and slightly lower specificity for CO2 than nontransgenic controls. These results suggest that Rubisco in rice transformed with sorghum RbcS partially acquires the catalytic properties of sorghum Rubisco. Rubisco content in transgenic lines was significantly increased over wild-type levels but Rubisco activation was slightly decreased. The expression of sorghum RbcS did not affect CO2 assimilation rates under a range of CO2 partial pressures. The Jmax/Vcmax ratio was significantly lower in transgenic line compared to the nontransgenic plants. These observations suggest that the capacity of electron transport is not sufficient to support the increased Rubisco capacity in transgenic rice. Although the photosynthetic rate was not enhanced, the strategy presented here opens the way to engineering Rubisco for improvement of photosynthesis and productivity in the future.
Rubisco is the key photosynthetic enzyme responsible for CO2 fixation. Many agriculturally important crops, including cereals such as rice (Oryza sativa) and most woody perennials, are classified as C3 plants. Their photosynthetic rate at present atmospheric CO2 level is limited by the activity of Rubisco because of its extremely low catalytic turnover rate (kcat) and competing oxygenase reaction, which initiates wasteful photorespiration (von Caemmerer and Quick, 2000; Parry et al., 2003). To compensate for these detrimental enzymatic properties, C3 plants invest 15% to 35% of total leaf nitrogen in this single enzyme (Evans, 1989; Makino et al., 1992). These observations lead us to the notion that Rubisco significantly limits agricultural productivity and carbon cycling and sequestration in the global ecosystem.
Many attempts to improve photosynthetic efficiency by engineering Rubisco have gained little progress (Andrews and Whitney, 2003; Whitney et al., 2011). Most of these efforts were focused on altering the CO2/O2 specificity of Rubisco to suppress photorespiration. Rubisco in nongreen algae such as Galdieria and Griffithsia are more specific to CO2 than those of vascular plants (Whitney et al., 2001). However, it is difficult to express these genetically distant enzymes in the chloroplast of vascular plants (Whitney et al., 2001). Considering the recent rapid increase in atmospheric CO2 partial pressure, it is likely that the relative frequency of the oxygenase reaction will be decreased in the near future, though a concomitant increase in global temperature might somewhat negate advantages of elevated CO2. In C4 plants, carrying CO2 concentrating mechanism, high kcat Rubisco suggested to be an important determinant of nitrogen-use efficiency of photosynthesis and growth (Ghannoum et al., 2005). Thus, high kcat Rubisco is expected to have a potential for improvement of photosynthetic efficiency under elevated CO2. There is significant natural variation in the kcat of Rubisco even in vascular plants (Seemann et al., 1984; Sage, 2002). C3 plants from cool habitats and C4 plant Rubiscos have higher kcat than those of C3 plants from warm habitats, such as rice (Sage, 2002). To find an efficient Rubisco for rice photosynthesis, we analyzed the kinetic properties of Rubisco in plants categorized into these functional groups among Poaceae (Ishikawa et al., 2009). The sorghum (Sorghum bicolor) Rubisco showed a significantly higher kcat and relatively high amino acid sequence identity to rice, making it a potential candidate for the improvement of rice photosynthesis under elevated CO2.
In vascular plants, Rubisco is composed of eight large subunits (encoded by the chloroplast Rubisco large subunit [RbcL]) and eight small subunits (encoded by a family of nuclear Rubisco small subunit [RbcS]) to form the functional holoenzyme. RbcL contains most of the amino acids important for catalysis (Andersson and Backlund, 2008). By contrast, the function of RbcS is uncertain and it has been suggested that RbcS could have a role in determining the carboxylation efficiency and CO2 specificity (Spreitzer, 2003). In this study, transgenic rice plants expressing the sorghum RbcS were produced and the effects on the kinetic properties of Rubisco and photosynthesis were analyzed. Our results clearly show that the introduction of sorghum RbcS increases the kcat of Rubisco in rice, suggesting that RbcS could be an important determinant of the kinetic properties of Rubisco and a useful target for genetic engineering to improve photosynthesis.
RESULTS AND DISCUSSION
Expression of Sorghum RbcS in Transgenic Rice
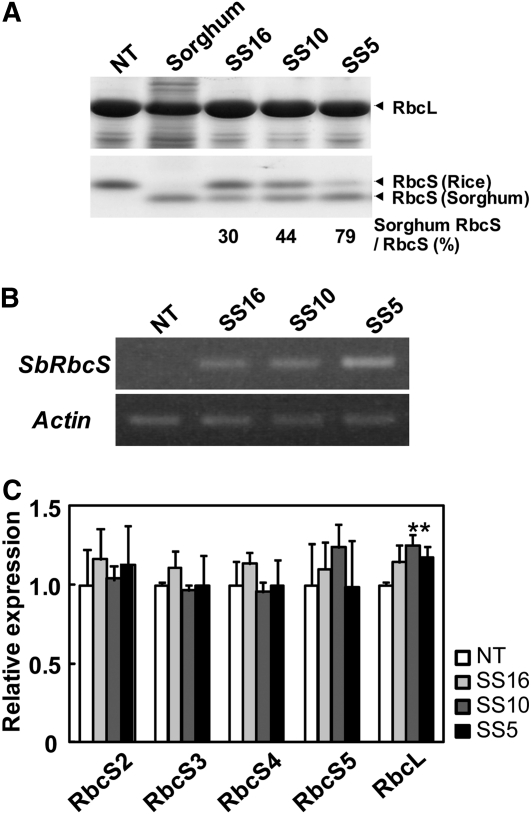
The full-length cDNA encoding the major RbcS isoform expressed in sorghum leaf was amplified by reverse transcription (RT)-PCR and fused to the rice chlorophyll a/b-binding protein promoter, which directs mesophyll-specific and light-regulated expression in the leaves of C3 plants (Tada et al., 1991). This construct was introduced into rice and antibiotic-resistant transgenic rice plants were regenerated. The apparent molecular mass of sorghum RbcS expressed in rice was analyzed by SDS-PAGE and found to be slightly smaller than that of rice RbcS (Fig. 1A). Considering this difference, the relative expression level of sorghum RbcS in transgenic rice was estimated by SDS-PAGE with Coomassie Blue staining. The expression levels of sorghum RbcS were easily detected and the band was present in most primary transgenic rice plants. Among these progeny, homozygous lines were selected from the segregation pattern of sorghum RbcS, as judged by Coomassie-stained SDS-PAGE gels. The expression levels of sorghum RbcS in three homozygous lines designated as SS16, SS10, and SS5 were 30%, 44%, and 79% of total RbcS, respectively (Fig. 1A). The relative expression levels of rice RbcS were reduced by overexpression of sorghum RbcS, suggesting that expression of a foreign RbcS down-regulates the native protein, keeping the total RbcS population relatively constant. The expression of sorghum RbcS transcript was analyzed by semiquantitative RT-PCR (Fig. 1B). The expression of sorghum RbcS was only detected in transgenic rice and the expression levels were largely correlated with the expression of sorghum RbcS protein. These results suggest that the expression level of sorghum RbcS protein would be determined by the transcript level of sorghum RbcS. To study the effects of overexpression of sorghum RbcS on the expression of RbcS gene family and RbcL in rice, quantitative RT-PCR was carried out (Fig. 1C). Rice contains five RbcS genes (OsRbcS1-5) in nuclear genome and one RbcL gene in chloroplast genome (Suzuki et al., 2007). The expression of OsRbcS1 is quite low in leaves compared with other OsRbcSs (Suzuki et al., 2009b). Thus, OsRbcS1 was omitted from our experiment. The expression of sorghum RbcS did not significantly affect the expression of OsRbcS2-5. However, the expression of OsRbcL was slightly increased in transgenic rice. These results suggest that the overexpression of sorghum RbcS would not down-regulate the expression of RbcS gene family and possibly stimulate the expression of RbcL in rice.
Figure 1.
Expression of sorghum RbcS in transgenic rice. A, Expression levels of sorghum RbcS analyzed by SDS-PAGE. Total soluble proteins (3.2 μg for rice and 6.4 μg for sorghum) were separated by SDS-PAGE and stained with Coomassie Blue. Intensities of RbcS bands were quantified using the National Institutes of Health image program. The average ratios of sorghum RbcS to total RbcS of five biological replicates (average ± sd) are 30.1 ± 1.14, 43.8 ± 0.39, and 79.1 ± 0.11 for SS16, SS10, and SS5, respectively. These results are significantly different from each other (ANOVA, P < 0.01). B, Expression analysis of sorghum RbcS by RT-PCR. Expression of the actin gene was examined as an internal control. C, Expression of RbcS gene family and RbcL analyzed by quantitative RT-RCR. The expression data of RbcS2-5 and RbcL of rice are normalized by the expression levels of the actin gene and the ratios relative to each gene expression in nontransgenic rice (NT) are shown as relative expression levels. The data are presented as mean ± sd of three biological replicates. One-way ANOVA: *P < 0.05. SS5, SS10, SS16, Transgenic rice that express sorghum RbcS.
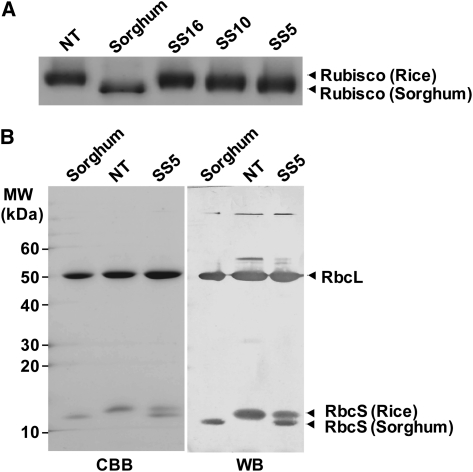
Rubisco in higher plants is present as a multisubunit complex consisting of eight RbcL and eight RbcS: L8S8 structure (Andersson and Backlund, 2008). As both the native rice and sorghum RbcS were expressed in transgenic rice, the possibility of heteropolymers of sorghum and rice Rubisco was investigated. We studied the subunit structure of Rubisco by Blue Native (BN)-PAGE and subsequent SDS-PAGE. In BN-PAGE, sorghum Rubisco showed a slightly lower apparent molecular mass than rice Rubisco (Fig. 2A). This might be due to the smaller molecular mass of sorghum RbcS compared to rice RbcS. In transgenic rice, the expression of sorghum RbcS caused a shift of apparent molecular mass of Rubisco holoenzyme from that of rice to that of sorghum Rubisco, depending on the expression levels of sorghum RbcS. Considering the molecular mass of Rubisco in transgenic rice, sorghum RbcS should be correctly assembled into the L8S8 structure holoenzyme. After BN-PAGE, Rubisco bands were excised from the gel and further separated by SDS-PAGE. Detection of Rubisco bands by Coomassie Blue staining and immunoblotting, confirmed that Rubisco of transgenic rice contained both rice and sorghum RbcS (Fig. 2B). From these results, it seems likely that Rubisco in these transgenic rice plants is present as a chimera of rice and sorghum RbcS.
Figure 2.
Subunit structure of Rubisco. A, Analysis of Rubisco by BN-PAGE. Total soluble proteins (1.0 μg for rice and 2.0 μg for sorghum) were separated on a 3% to 12% gradient gel and stained with Coomassie Blue. B, SDS-PAGE and immunoblotting of Rubisco separated by BN-PAGE. Rubisco bands from BN-PAGE were excised from the gel and separated by SDS-PAGE. Polypeptide bands were detected by Coomassie Blue staining (left section) and immunoblotting with an antiserum raised against the rice Rubisco (right section). NT, Nontransgenic rice; SS5, SS10, SS16, transgenic rice that express sorghum RbcS.
In our transgenic plants, the expression of foreign RbcS was significant compared to that of the native protein (Fig. 1A). A previous report of chimeric Rubisco in transgenic plants indicated that the expression of pea (Pisum sativum) RbcS in transgenic Arabidopsis (Arabidopsis thaliana) was at most 15% to 18% of total RbcS (Getzoff et al., 1998). In addition, Rubisco carbamylation was reduced in those plants. In another attempt to engineer a chimeric Rubisco, the tobacco (Nicotiana tabacum) RbcL was replaced with sunflower (Helianthus annuus) RbcL by chloroplast transformation. However, in these experiments the accumulation of the resultant hybrid enzyme was very low (Sharwood et al., 2008). Recently, a functional hybrid Rubisco consisting of algal RbcL and vascular plant RbcS was successfully expressed in Chlamydomonas, although the transformants exhibited growth retardation in the absence of supplemental CO2 (Genkov et al., 2010). These findings suggest that certain species-specific interactions of RbcS and RbcL, Rubisco activase, and chaperons are crucial for the assembly, accumulation, and activation of Rubisco. In this study, we used Rubisco from sorghum that is a related species of rice classified into the same Poaceae and its Rubisco shares relatively high identity of amino acid sequence to rice Rubisco (Ishikawa et al., 2009). These high sequence identities may reduce or eliminate the potential for incompatibility between sorghum RbcS and various interacting proteins, making possible the accumulation of high levels of foreign RbcS in rice.
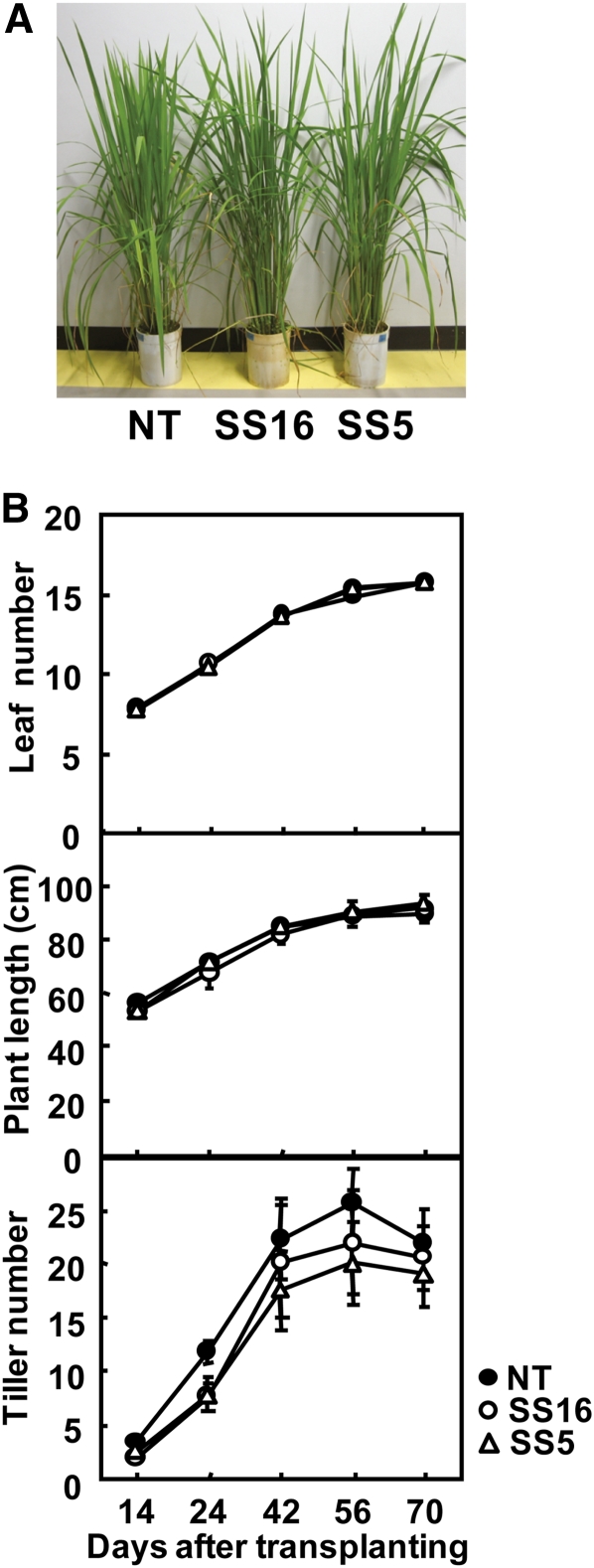
Despite high levels of expression of sorghum RbcS in the transgenic rice plants reported here, they exhibited a relatively normal phenotype with similar leaf expansion rate and plant length, whereas tiller number was slightly decreased in transgenic rice during the active tillering stage (Fig. 3). After flowering (70 d after transplanting), the difference in tiller number became marginal between the transgenic and the control lines, indicating that the number of nonproductive tillers was reduced in transgenic rice compared with nontransgenic rice.
Figure 3.
Growth of transgenic rice plants. A, Photograph of rice plants grown for 60 d after transplanting. B, Leaf number of the main culms, plant length, and tiller number. The growth parameters were measured after transplanting of young seedlings (leaf number of 4.5). NT, Nontransgenic rice; SS5 and SS16, transgenic rice that express sorghum RbcS. The data were expressed as mean ± sd obtained from five biological replicates. [See online article for color version of this figure.]
Kinetic Properties of Rubisco
As reported previously (Ishikawa et al., 2009), sorghum Rubisco has a higher kcat and Km for CO2 (Kc) than rice Rubisco (Table I). Rubisco extracted from transgenic rice also had a significantly higher kcat than that in nontransgenic rice. The values ranged from 1.32- to 1.50-fold relative to nontransgenic rice controls and largely correlated with the expression level of sorghum RbcS. To the best of our knowledge, such a significant enhancement of the kcat of Rubisco by genetic engineering has not been achieved previously. The Kc of Rubisco from transgenic rice was also higher than that of the nontransgenic controls, suggesting that the affinity for CO2 was reduced. The specificity of Rubisco (Sc/o) was slightly reduced in transgenic rice compared with nontransgenic rice. It has been reported that Sc/o in C4 plants is slightly lower than that in C3 plants (Kubien et al., 2008). Thus, it is likely that the kinetic properties of Rubisco in transgenic rice tend toward those of sorghum Rubisco by incorporation of sorghum RbcS.
Table I. Kinetic properties of Rubisco.
kcat and Kc values are the means ± se of three to four biological replicates. Sc/o values are the means ± se of four to five biological replicates. NT, Nontransgenic rice; SS5, SS10, SS16, transgenic rice that express sorghum RbcS. One-way ANOVA: *P < 0.05; **P < 0.01.
| Plant | kcat | Kc | Sc/o |
| mol CO2 mol sites−1 s−1 | μm | mol mol−1 | |
| NT | 1.68 ± 0.06 | 17.3 ± 0.28 | 117 ± 9.2 |
| Sorghum | 4.09 ± 0.10** | 41.5 ± 0.71** | |
| SS16 | 2.22 ± 0.06** | 22.4 ± 2.91* | 108 ± 6.1 |
| SS10 | 2.52 ± 0.07** | 21.7 ± 0.83** | |
| SS5 | 2.48 ± 0.10** | 24.0 ± 2.88* | 103 ± 5.5* |
The chloroplast-encoded RbcL is thought to contain most of the important amino acids determining the kinetic properties of catalysis (Andersson and Backlund, 2008). Consistent with this, the specific activity of Rubisco in intergenetic hybrids shows maternal inheritance (Evans and Austin, 1986). In contrast, the importance of RbcS in determining catalytic performance of Rubisco has been suggested by the genetic engineering of Chlamydomonas Rubisco with RbcS from different species or with chimeric RbcS (Karkehabadi et al., 2005; Genkov et al., 2010). In addition to those reports, the results in this study clearly show that RbcS of high kcat Rubisco greatly enhanced the kcat of Rubisco in rice (Table I). Therefore, it is proposed that RbcS as well as RbcL can make a significant contribution to overall catalytic performance and diversity of Rubisco across species.
X-ray crystallographic analysis of the quaternary complex of Rubisco determined the interface between RbcS and RbcL (Knight et al., 1990). Mutations of some amino acids at the interface altered the kinetic properties of Rubisco (Spreitzer, 2003; Karkehabadi et al., 2005). Relatively large differences in amino acid sequence between rice and sorghum RbcS were found in the βA-βB loop and C terminus (Supplemental Fig. S1). In the βA-βB loop, four amino acids differ between rice and sorghum RbcS and two of the four are located at the interface between RbcS and RbcL. These are His-56 and Gly-60 (numbering based on spinach [Spinacia oleracea] RbcS) in rice RbcS and Ser and Cys in sorghum RbcS, respectively. Although the amino acids responsible for catalytic efficiency have not been identified, the substitution of land-plant or bacterial Chlamydomonas RbcS in this region alters the carboxylation efficiency and CO2/O2 specificity (Karkehabadi et al., 2005), suggesting that amino acid sequence in the βA-βB loop could be an important determinant of the CO2 specificity and catalytic efficiency of Rubisco. Thus, it is likely that the interaction of amino acid residues between rice RbcL and sorghum RbcS at this interface could contribute to the enhancement of Rubisco kcat in the transgenic rice lines we have studied. In contrast, the C terminus of rice RbcS is also significantly different and five amino acids longer than that of sorghum RbcS. Notable differences in the amino acid sequences of RbcS among photosynthetic organisms have also been found in the C terminus (Spreitzer, 2003). Nongreen algae and prokaryotes have a shorter βA-βB loop than higher plants and green algal Rubisco, whereas they have a long C terminus that forms an extra βE-βF loop. This βE-βF loop resides in the spaces that would normally be occupied by the βA-βB loop in vascular plants (Spreitzer, 2003) and could affect the kinetic properties of Rubisco. Using the SWISS-MODEL internet server described by Arnold et al. (2006), the structural predictions of sorghum RbcS was carried out based on the structure of rice RbcS (Supplemental Fig. S2). It was anticipated that their C termini should be located on the surface of Rubisco, suggesting that this C-terminal region may not be essential to the enhancement of catalytic efficiency of Rubisco in transgenic rice.
Content and Activation of Rubisco in Transgenic Rice
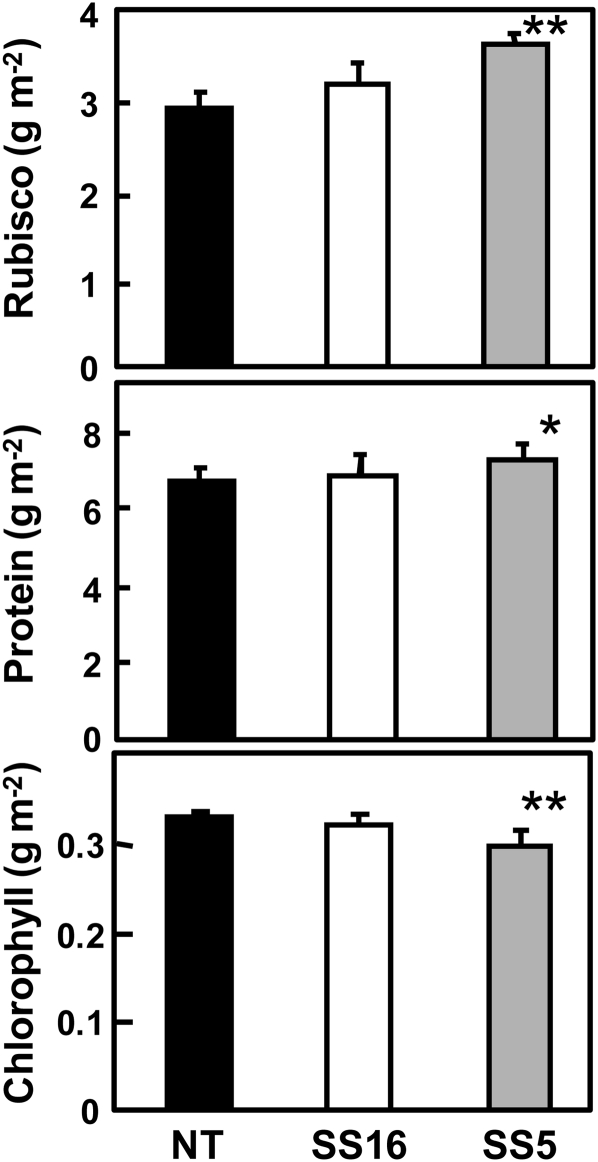
Rubisco content was increased in transgenic rice and its increase was dependent on the expression levels of sorghum RbcS (Fig. 4). In particular, the Rubisco content in SS5, a high expressing line, was 24% higher than that in nontransgenic rice. Rubisco content is not always regulated by the transcript abundance of RbcS when part of RbcL was truncated or Rubisco-specific chaperone was knocked down in tobacco (Wostrikoff and Stern, 2007). In this report, RNAi knockdown of RbcS did not influence the expression of RbcL. However, in rice, the expression levels of RbcS, RbcL, and Rubisco were apparently correlated with each other among nontransgenic, RbcS-antisense and RbcS-sense lines (Suzuki et al., 2009a). Consistent with this, the overexpression of sorghum RbcS led to increase in the expression of RbcL and the content of Rubisco in this study (Figs. 1 and 4). Thus, it is likely that the expression of RbcS is more tightly coordinated with the expression of RbcL in rice, and then the overexpression of foreign RbcS from closely related species in the presence of endogenous RbcS increases the expression of Rubisco protein in rice.
Figure 4.
Rubisco, total soluble protein, and chlorophyll contents. Uppermost fully expanded leaves at vegetative growth stage (40–42 d after transplanting) were harvested after the measurement of photosynthesis rate and used for the determination of Rubisco, total soluble protein, and chlorophyll. Data represent means ± sd of five biological replicates. One-way ANOVA: *P < 0.05; **P < 0.01.
The increase in Rubisco content achieved here affected the contents of other components in the leaves (Fig. 4). The soluble protein content was slightly increased and the chlorophyll content was decreased in transgenic rice. In other experiments designed to increase photosynthesis in rice, overexpressed pyruvate orthophosphate dikinase accounted for 6% to 35% of total soluble protein, increased the total nitrogen content, and decreased the chlorophyll content in transgenic plants (Fukayama et al., 2001). Increases in the level of Rubisco in this study are comparable with that of pyruvate orthophosphate dikinase, in terms of leaf protein content, as reported previously. These observations suggest that high levels of some foreign protein expression in the chloroplast can lead to an increase in total soluble protein content and a decrease in other photosynthesis-related components such as chlorophyll.
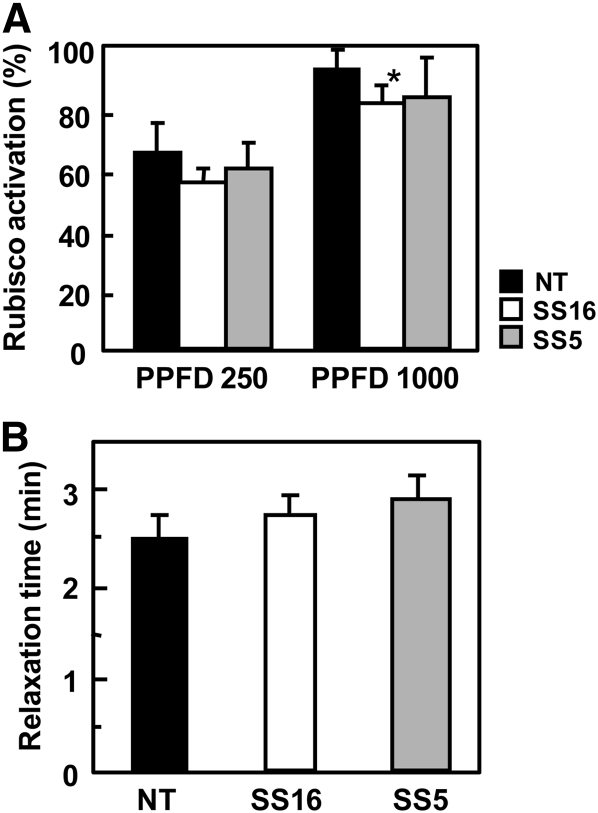
The molecular interaction of Rubisco and Rubisco activase is species specific in some cases (Portis et al., 2008). To ascertain whether rice Rubisco activase could correctly regulate Rubisco-containing sorghum RbcS, Rubisco activation was analyzed under two different light intensities (Fig. 5A). Rubisco activation states were slightly lower in transgenic lines than in nontransgenic rice controls irrespective of light intensity. However, Rubisco activation was higher in high light (photosynthetic photon flux density [PPFD] of 1,000 μmol m−2s−1) than that in low light (PPFD of 200 μmol m−2 s−1) in all plants, suggesting that Rubisco activase in rice can control the activity of Rubisco-containing sorghum RbcS in a manner dependent on light intensity. Function of Rubisco activase can also be analyzed by measuring the gas-exchange rate after a sudden transition from low to high light intensity (Woodrow and Mott, 1989). This relaxation time was used as an indicator of the time necessary to complete light activation of Rubisco (Fig. 5B). Although the relaxation times were marginally increased by the expression of sorghum RbcS, there was no statistical difference between transgenic and nontransgenic rice, indicating that Rubisco activase in rice can mediate the activation of Rubisco-containing sorghum RbcS at a rate comparable to rice Rubisco. As described above, Rubisco content was significantly higher in transgenic rice compared with nontransgenic rice (Fig. 4). A reduction in Rubisco activation state was also observed in transgenic rice that overexpressed endogenous rice Rubisco (Suzuki et al., 2007). In addition, the increase in inactive Rubisco content presumably slowed the apparent rate constant for Rubisco activation (Woodrow et al., 1996). Therefore, the observed slight decrease in Rubisco activation state and marginal increase in relaxation time are considered to result from increased Rubisco content in transgenic rice, but not from a partial inability of Rubisco activase to interact with Rubisco-containing sorghum RbcS.
Figure 5.
Rubisco activation in transgenic rice. A, Activation of Rubisco under different light intensities. Rubisco activity in rice leaves was measured after 30-min irradiation of PPFD of 250 or 1,000 μmol m−2 s−1. Activation level was calculated as ratio of initial activity to total activity of Rubisco. B, Relaxation time for light activation of Rubisco after a step increase in light intensity. The CO2 assimilation rate was measured after sudden increase in light intensity from PPFD of 60 to 1,800 μmol m−2 s−1 for 20 min and the relaxation time for Rubisco activation was calculated as described by Woodrow and Mott (1989). NT, Nontransgenic rice; SS5, SS16, transgenic rice that express sorghum RbcS. Data represent means ± sd of five biological replicates. One-way ANOVA: *P < 0.05.
Effects of Sorghum RbcS on Photosynthesis
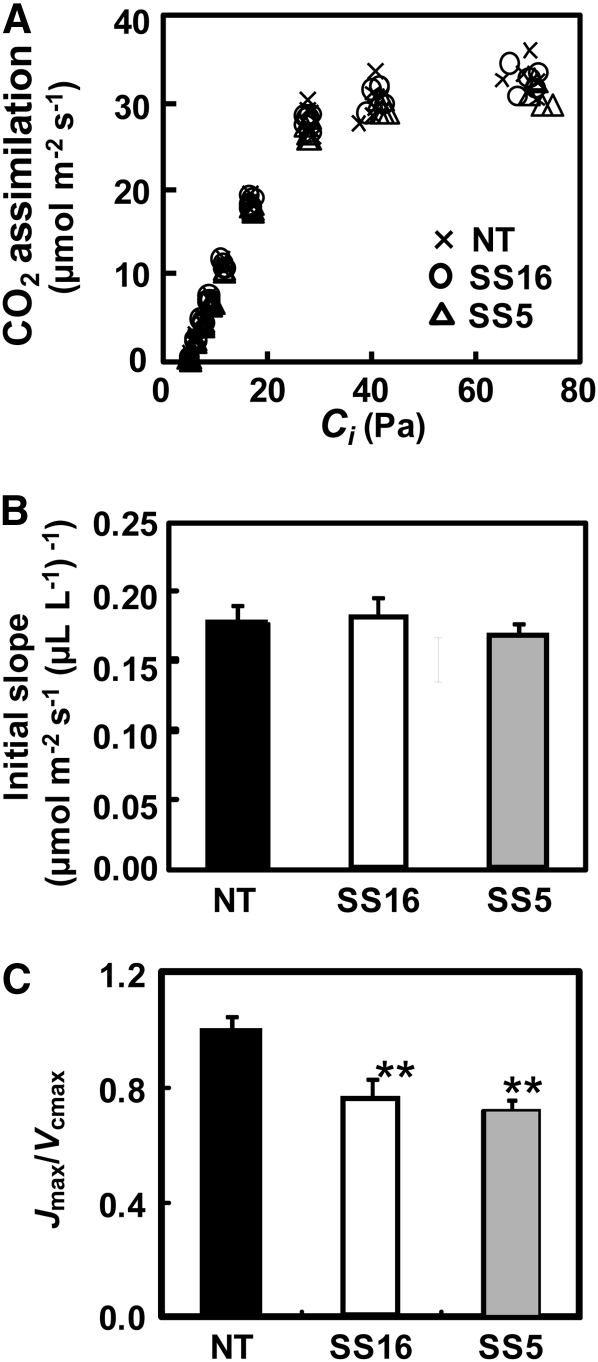
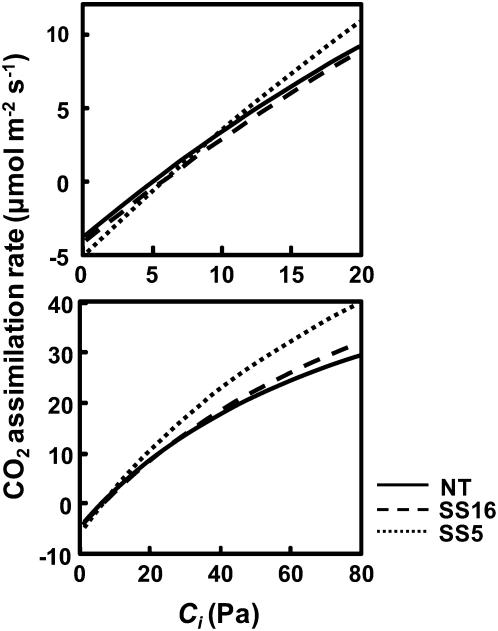
Expression of sorghum RbcS did not affect the photosynthesis rates at any tested CO2 partial pressures, including high CO2 conditions (Fig. 6A). The initial slope of the plot of CO2 assimilation rate versus Ci, an indicator of Rubisco-limited photosynthesis was 0.176, 0.180, and 0.166 in a nontransgenic line and transgenic lines SS16 and SS5, respectively (Fig. 6B). There was no statistical difference between these lines, confirming that the expression of high kcat Rubisco did not stimulate the rate of photosynthesis under Rubisco-limited low-CO2 conditions. The similar results were also reported for overexpression of native Rubisco in rice (Suzuki et al., 2007). Using the data of photosynthetic rate and kinetic properties of Rubisco, the maximum rate of ribulose 1,5-bisphosphate (RuBP) carboxylation (Vcmax) and the maximum rate of electron transport (Jmax) were estimated (Supplemental Table S1). The Vcmax was higher in transgenic lines than that in nontransgenic rice, whereas the Jmax did not significantly differ among them. The Jmax/Vcmax ratio was 0.99, 0.75, and 0.72 in nontransgenic rice, SS16 and SS5, respectively (Fig. 6C). These values were significantly lower in transgenic rice than that in nontransgenic rice, suggesting that the capacity of electron transport is not sufficient to support the capacity of RuBP carboxylation by Rubisco enhanced by sorghum RbcS. This imbalance might cause partial deactivation of Rubisco and could be a reason why increase in kcat does not enhance the photosynthesis rate in transgenic rice. Then, the RuBP saturated rate of photosynthesis was modeled according to von Caemmerer and Farquhar (1981) using kinetic parameters, contents, and activation levels of Rubisco obtained in this study (Fig. 7). The modeled photosynthetic rate was totally consistent with the measured photosynthetic rate (Fig. 6A) but slightly lower in transgenic rice than that in nontransgenic rice under low CO2 partial pressure up to 10 Pa. This finding confirms that the kinetic properties of Rubisco altered by the introduction of sorghum RbcS do not largely affect the photosynthetic rate under low-CO2 condition. In contrast, the photosynthetic rates above CO2 partial pressures of 50 Pa were significantly higher in transgenic rice. These results imply that the capacity of RuBP carboxylation under high CO2 is enhanced by the overexpression of sorghum RbcS, whereas the photosynthetic rate under high CO2 was not increased in transgenic rice because other factors that may determine the electron transport rate would limit the photosynthetic rate. To improve the photosynthetic rate, the enhancement of electron transport capacity should also be required in our transgenic rice.
Figure 6.
Photosynthetic rate of transgenic rice plants. A, Dependence on Ci of photosynthetic CO2 assimilation rate. The CO2 assimilation was measured at PPFD of 1,500 μmol m−2s−1, leaf temperature of 27°C, 21% O2, vapor pressure deficit of 1.0 to 1.2 kPa, and CO2 partial pressures of 4.5 to 88 Pa. Uppermost fully expanded leaves at vegetative growth stage (40–42 d after transplanting) of five independent plants for each lines were used for the experiments. B, Initial slope of CO2 assimilation rate as a function of Ci. The slope was calculated by the data of Figure 6A. C, The Jmax/Vcmax ratio. Vcmax and Jmax were estimated by the data of Figure 6A and kinetic parameters obtained in this study (summarized in Supplemental Table S1) using the equation of von Caemmerer and Farquhar (1981). NT, Nontransgenic rice; SS5, SS16, transgenic rice that express sorghum RbcS. One-way ANOVA: **P < 0.01.
Figure 7.
The CO2 assimilation rate as a function of Ci modeled with kinetic parameters, contents, and activation state of Rubisco. The CO2 assimilation rates at 28°C of nontransgenic rice and transgenic rice that express sorghum RbcS (SS5 and SS16) were calculated using the kinetic parameters, Rubisco content, and Rubisco activation level at PPFD of 1,000 μmol m−2 s−1 obtained in this study (summarized in Supplemental Table S1). However, Ko was assumed to be constant among plants and used the value of 410 μm determined in rice (Makino et al., 1985).
CONCLUSION
This study shows that the chimeric incorporation of sorghum RbcS significantly increases the kcat of Rubisco in transgenic rice (Table I). However, the photosynthetic rate was not increased in transgenic rice (Fig. 6). It is proposed that the increase in both kcat and content of Rubisco could lead to excess activity of the enzyme for photosynthesis. In general, Rubisco does not limit photosynthesis under high-CO2 conditions (von Caemmerer and Quick, 2000), where Rubisco will be in excess of that required to support photosynthesis, particularly in our transgenic rice. It has been reported that a specific reduction of Rubisco content by antisense inhibition of RbcS expression in rice stimulated the rate of photosynthesis under elevated CO2 (Makino et al., 1997). In that report, it was estimated that 65% of Rubisco in nontransgenic rice would be optimum for photosynthesis under elevated CO2. If a plant acquired high kcat Rubisco, as in our transgenic rice, a much smaller amount of Rubisco would be sufficient to support photosynthetic flux. This could reallocate more Rubisco nitrogen to the other photosynthetic proteins determining the electron transport capacity, leading to enhancement of nitrogen-use efficiency and photosynthesis.
Many attempts have previously been made to improve the kinetic properties of Rubisco by genetic manipulation and have not succeeded (Andrews and Whitney, 2003). We propose that the introduction of RbcS of a high kcat Rubisco from a closely related species is a better approach to improve Rubisco kinetic properties and photosynthesis in C3 plants. Many agriculturally important crops such as rice, wheat (Triticum aestivum), potato (Solanum tuberosum), and soybean (Glycine max) are classified as C3 plants. The strategy shown here in rice could be expanded to include other important C3 crops and contribute to a stable food supply in the future.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Sorghum (Sorghum bicolor ‘Tentaka’) was grown in the experimental field at Kobe University. Rice (Oryza sativa ‘Nipponbare’) was grown in soil under natural sunlight conditions in a temperature-controlled growth chamber on a 28°C day/23°C night cycle. For analyses of SDS-PAGE and Rubisco kinetics, leaves were sampled at 10:30 am to 11:30 am on sunny days. For the determination of Rubisco, soluble protein, and chlorophyll, leaves were harvested after measurement of gas-exchange rate. After sampling, the leaves were immediately frozen in liquid nitrogen and stored at −80°C until required.
Plasmid Construction and Transformation of Rice
The full-length cDNA of sorghum RbcS (accession no. AB564718) was amplified by RT-PCR using gene-specific primers SbRbcS-XbaI-F and SbRbcS-SacI-R (Supplemental Table S2). PCR products were digested with XbaI and SacI and cloned into the binary vector pIG121Hm (a generous gift from Prof. K. Nakamura, Nagoya University, Japan) containing the rice chlorophyll a/b-binding protein promoter. The construct was introduced into rice via Agrobacterium-mediated gene transfer. Antibiotic-resistant transgenic rice plants were regenerated and grown in soil. The level of transgene expression was screened by SDS-PAGE after staining with Coomassie Brilliant Blue R-250. Transgenic lines in which the next generation exhibited a segregation ratio of around 1:3 were selected. These progeny with the highest expression level were presumed to be homozygous and used for further analysis.
Electrophoresis and Immunoblotting
Leaf tissues were homogenized using a cold mortar and pestle in extraction buffer containing 50 mm HEPES-KOH, 10 mm MgCl2, 1 mm EDTA, 5 mm dithiothreitol (DTT), 0.01 mm leupeptin, 1 mm phenylmethylsulfonylfluoride, 10% (w/v) glycerol, and 5% (w/v) polyvinylpolypyrrolidone, pH 7.4, with a small amount of quartz sand. The homogenate was centrifuged at 15,000g for 5 min at 4°C and the supernatant was collected as a total soluble protein extract. SDS-PAGE and immunoblotting were performed as described previously (Masumoto et al., 2010). After SDS-PAGE, the gels were stained with Coomassie Blue or subjected to immunoblotting using antisera raised against the rice Rubisco (Fukayama et al., 1996). BN-PAGE was carried out using a precast gradient gel (NativePAGE Novex 3%–12% Bis-Tris gel, Life Technologies) according to the manufacturer’s instructions.
RT-PCR Analyses
Total RNA was isolated from uppermost fully expanded leaves using the RNeasy plant mini kit (Qiagen). The first-strand cDNA was synthesized from 1.2 μg of total RNA with oligo(dT)18 and random hexamer as primers using PrimeScript II first-strand cDNA synthesis kit (Takara). Semiquantitative RT-PCR was performed using Quick Taq HS DyeMix (Toyobo) with 5% dimethyl sulfoxide and primers listed in Supplemental Table S2. The DNA polymerase was first activated at 94°C for 2 min, and PCR was carried out for 25 cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 68°C, followed by a final extension step for 7 min at 68°C. Quantitative RT-PCR was performed essentially as described by Suzuki et al. (2009a) using gene-specific primers listed in Supplemental Table S2. PCR was carried out using SYBR Premix Ex Taq GC (Takara) and Thermal Cycler Dice TP800 (Takara) according to the manufacturer’s instruction. Expression of the actin gene (Rac1, AB047313) was examined as an internal control.
Rubisco Activity
Leaf tissues were homogenized in extraction buffer containing 100 mm Bicine-NaOH, 1 mm EDTA, 5 mm MgCl2, 2 mm NaH2PO4, 0.4% (w/v) bovine serum albumin, 5 mm DTT, 4 mm amino-n-caproic acid, and 0.8 mm benzamidine, pH 8.0, using a chilled mortar and pestle with a small amount of quartz sand. The homogenate was then centrifuged at 15,000g for 2 min at 4°C. The supernatant was used for the determination of Rubisco activity and Rubisco catalytic site.
Rubisco in the extract was activated by preincubation with 15 mm MgCl2 and 10 mm NaHCO3 on ice for 15 to 20 min. Then, Rubisco activity was determined at 28°C using [14C] NaHCO3 by assaying the incorporation of 14C into acid-stable products, as described previously (Ishikawa et al., 2009). Rubisco catalytic site concentrations were determined by measuring the stoichiometric binding of [14C] carboxyarabinitol-1,5-bisphosphate (CABP) as described previously (Ishikawa et al., 2009). The kcat of Rubisco (mol mol−1 s−1) was calculated as the ratio of Rubisco activity to Rubisco catalytic sites. For determination of Km for CO2 of Rubisco (Kc), Rubisco activities were measured at six different [14C]NaHCO3 concentrations (0.5–15 mm). In this experiment, 1.0 W-A units of carbonic anhydrase were added to the reaction mixture. Kc was calculated from the Hanes-Woolf plot.
Photosynthetic Rate
Gas exchange of the uppermost full expanded leaf was measured using an open gas-exchange system (GFS-3000, Waltz). Measurements were performed at a leaf temperature of 27°C, 21% O2, a PPFD of 1,500 μmol quanta m−2 s−1, and a leaf-to-air vapor pressure difference of 1.0 to 1.2 kPa. The response curve of CO2 assimilation was obtained with a single leaf by changing the ambient CO2 partial pressures from 4.5 to 88 Pa.
Sc/o was determined by measurement of the gas-exchange rate (Laisk, 1977). The measurements were performed at five different ambient CO2 partial pressures (4.0–10 Pa) with two different light intensities (PPFD of 125 and 500 μmol m−2 s−1). The CO2 compensation point independent of photorespiration (Γ*) and the day respiration (Rd) was estimated by the intersection of these two regression lines. Sc/o was calculated by the following equation:
where O is the intercellular partial pressures of O2 (21 kPa). The RuBP saturated rates of CO2 assimilation (A) were calculated by the following equation (von Caemmerer and Farquhar, 1981):
where [Rubisco] is the catalytic site content of Rubisco. Kc and Ko are the Michaelis constants of Rubisco for CO2 and O2, respectively. Bunsen absorption coefficients were used to convert Kc and Ko values from concentration to partial pressure. The maximum rate of RuBP carboxylation (Vcmax) was calculated by the model of von Caemmerer and Farquhar (1981) using the initial slope of the CO2 response curve. The maximum rate of electron transport (Jmax) was calculated from the following equation, substituting the rate of CO2 assimilation rate at Ci > 60 Pa (As) as described in Suzuki et al. (2009a):
The CO2 partial pressure in the chloroplast (Cc) was obtained by the following equation:
where gw is the CO2 transfer conductance. The gw of 5 mol CO2 m−2 s−1 Pa−1 reported with rice was used for the calculation (Makino et al., 1994).
Relaxation time for Rubisco activation following an increase in PPFD was measured basically according to Woodrow and Mott (1989). Gas exchange of the uppermost fully expanded leaf was measured at a leaf temperature of 27°C, 25 Pa CO2, 21% O2, and a leaf-to-air vapor pressure difference of 1.0 to 1.2 kPa using an open gas-exchange system (LI-6400, Li-Cor). The leaves were exposed to saturating level of PPFD (1,800 μmol m−2 s−1) for 30 min, and placed at low PPFD (60 μmol m−2 s−1) for 45 min. The gas-exchange rate was recorded at 5-s interval following an increase in PPFD from 60 to 1,800 μmol m−2 s−1. Assimilation rates were normalized to an intercellular CO2 partial pressure (Ci) of 25 Pa and plotted as the natural logarithm of the difference between the final steady-state rates and rates at each time. The relaxation time for Rubisco activation was determined by the reciprocal of the slope of second linear phase that occurred from 0.5 to 5.0 min after increasing the light intensity (Woodrow and Mott, 1989). The relaxation time reflects the time required to complete light activation of Rubisco.
Soluble Protein, Rubisco, and Chlorophyll
Leaf tissues were homogenized in extraction buffer containing 50 mm HEPES-KOH, 5 mm MgCl2, 1 mm EDTA, 5 mm DTT, 4 mm amino-n-caproic acid, 0.8 mm benzamidine, 0.05% (v/v) Triton X-100, 5% (w/v) glycerol, and 0.1% (w/v) polyvinylpolypyrrolidone, pH 7.4, using a chilled mortar and pestle with a small amount of quartz sand. The homogenate was then centrifuged at 15,000g for 2 min at 4°C. The supernatant was used for the determination of Rubisco and total soluble proteins. Rubisco was determined using [14C] CABP and calculated by assuming 6.5 molecules of CABP bound per molecule of Rubisco (Butz and Sharkey, 1989). Total soluble protein was determined by the method of Bradford (1976) with bovine serum albumin as the standard. For determination of chlorophyll, a portion of homogenate was taken from the mortar and extracted with 80% (v/v) acetone. Chlorophyll was determined spectrophotometrically as described in Porra et al. (1989).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB047313 (Rac1) and AB564718 (sorghum RbcS).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the amino acid sequences of RbcS from rice, sorghum, spinach, and Chlamydomonas.
Supplemental Figure S2. Structural comparison of RbcS from rice and sorghum.
Supplemental Table S1. Photosynthetic parameters used for calculation of Jmax/Vcmax ratio and modeling the photosynthetic rate.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We would like to thank Prof. Makoto Matsuoka (Nagoya University, Japan) for the generous gifts of vectors.
References
- Andersson I, Backlund A. (2008) Structure and function of Rubisco. Plant Physiol Biochem 46: 275–291 [DOI] [PubMed] [Google Scholar]
- Andrews T, Whitney SM. (2003) Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch Biochem Biophys 414: 159–169 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Butz ND, Sharkey TD. (1989) Activity ratios of ribulose-1,5-bisphosphate carboxylase accurately reflect carbamylation ratios. Plant Physiol 89: 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR. (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78: 9–19 [DOI] [PubMed] [Google Scholar]
- Evans JR, Austin RB. (1986) The specific activity of ribulose-1,5-bisphosphate carboxylase in relation to genotype in wheat. Planta 167: 344–350 [DOI] [PubMed] [Google Scholar]
- Fukayama H, Tsuchida H, Agarie S, Nomura M, Onodera H, Ono K, Lee B-H, Hirose S, Toki S, Ku MSB, et al. (2001) Significant accumulation of C4-specific pyruvate, orthophosphate dikinase in a C3 plant, rice. Plant Physiol 127: 1136–1146 [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Uchida N, Azuma T, Yasuda T. (1996) Relationships between photosynthetic activity and the amounts of Rubisco activase and Rubisco in rice leaves from emergence through senescence. Jpn J Crop Sci 65: 296–302 [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. (2010) Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in chlamydomonas. J Biol Chem 285: 19833–19841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzoff TP, Zhu G, Bohnert HJ, Jensen RG. (1998) Chimeric Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase containing a pea small subunit protein is compromised in carbamylation. Plant Physiol 116: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum O, Evans JR, Chow WS, Andrews TJ, Conroy JP, von Caemmerer S. (2005) Faster Rubisco is the key to superior nitrogen-use efficiency in NADP-malic enzyme relative to NAD-malic enzyme C4 grasses. Plant Physiol 137: 638–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Fukayama H. (2009) Screening of high kcat Rubisco among Poaceae for improvement of photosynthetic CO2 assimilation in rice. Plant Prod Sci 12: 345–350 [Google Scholar]
- Karkehabadi S, Peddi SR, Anwaruzzaman M, Taylor TC, Cederlund A, Genkov T, Andersson I, Spreitzer RJ. (2005) Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 44: 9851–9861 [DOI] [PubMed] [Google Scholar]
- Knight S, Andersson I, Brändén CI. (1990) Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution: subunit interactions and active site. J Mol Biol 215: 113–160 [DOI] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. (2008) The biochemistry of Rubisco in Flaveria. J Exp Bot 59: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Laisk A. (1977) Kinetics of Photosynthesis and Photorespiration in C3 Plants (in Russian). Nauka, Moscow, pp 1–198 [Google Scholar]
- Makino A, Mae T, Ohira K. (1985) Photosynthesis and ribulose-1,5-bisphosphate carboxylase/oxygenase in rice leaves from emergence through senescence: quantitative analysis by carboxylation/oxygenation and regeneration of ribulose 1,5-bisphosphate. Planta 166: 414–420 [DOI] [PubMed] [Google Scholar]
- Makino A, Nakano H, Mae T. (1994) Responses of ribulose-1,5-bisphosphate carboxylase, cytochrome f, and sucrose synthesis enzymes in rice leaves to leaf nitrogen and their relationships to photosynthesis. Plant Physiol 105: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Sakashita H, Hidema J, Mae T, Ojima K, Osmond B. (1992) Distinctive responses of ribulose-1,5-bisphosphate carboxylase and carbonic anhydrase in wheat leaves to nitrogen nutrition and their possible relationships to CO2-transfer resistance. Plant Physiol 100: 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Shimada T, Takumi S, Kaneko K, Matsuoka M, Shimamoto K, Nakano H, Miyao-Tokutomi M, Mae T, Yamamoto N. (1997) Does decrease in ribulose-1,5-bisphosphate carboxylase by antisense RbcS lead to a higher N-use efficiency of photosynthesis under conditions of saturating CO2 and light in rice plants? Plant Physiol 114: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto C, Miyazawa S, Ohkawa H, Fukuda T, Taniguchi Y, Murayama S, Kusano M, Saito K, Fukayama H, Miyao M. (2010) Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc Natl Acad Sci USA 107: 5226–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MAJ, Andralojc PJ, Mitchell RAC, Madgwick PJ, Keys AJ. (2003) Manipulation of Rubisco: the amount, activity, function and regulation. J Exp Bot 54: 1321–1333 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedelmann PE. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Portis AR, Jr, Li C, Wang D, Salvucci ME. (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Sage RF. (2002) Variation in the kcat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. J Exp Bot 53: 609–620 [DOI] [PubMed] [Google Scholar]
- Seemann JR, Badger MR, Berry JA. (1984) Variations in the specific activity of ribulose-1,5-bisphosphate carboxylase between species utilizing differing photosynthetic pathways. Plant Physiol 74: 791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM. (2008) The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414: 141–149 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Miyamoto T, Yoshizawa R, Mae T, Makino A. (2009a) Rubisco content and photosynthesis of leaves at different positions in transgenic rice with an overexpression of RBCS. Plant Cell Environ 32: 417–427 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi K, Yoshizawa R, Mae T, Makino A. (2009b) Differences in expression of the RBCS multigene family and rubisco protein content in various rice plant tissues at different growth stages. Plant Cell Physiol 50: 1851–1855 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ohkubo M, Hatakeyama H, Ohashi K, Yoshizawa R, Kojima S, Hayakawa T, Yamaya T, Mae T, Makino A. (2007) Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol 48: 626–637 [DOI] [PubMed] [Google Scholar]
- Tada Y, Sakamoto M, Matsuoka M, Fujimura T. (1991) Expression of a monocot LHCP promoter in transgenic rice. EMBO J 10: 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Farquhar GD. (1981) Some relationship between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Quick WP. (2000) Rubisco: physiology in vivo. Leegood RC, Sharkey TD, von Caemmerer S, eds, Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 85–113 [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ. (2001) Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J 26: 535–547 [DOI] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. (2011) Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol 155: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow IE, Kelly ME, Mott KA. (1996) Limitation of the rate of ribulosebisphosphate carboxylase activation by carbamylation and ribulosebisphosphate carboxylase activase activity: development and tests of a mechanistic model. Aust J Plant Physiol 23: 141–149 [Google Scholar]
- Woodrow IE, Mott KA. (1989) Rate limitation of non-steady-state photosynthesis by ribulose-1,5-bisphosphate carboxylase in spinach. Aust J Plant Physiol 16: 487–500 [Google Scholar]
- Wostrikoff K, Stern D. (2007) Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc Natl Acad Sci USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.