Abstract
Abscisic acid (ABA) regulates plant development and is crucial for plant responses to biotic and abiotic stresses. Studies have identified the key components of ABA signaling in Arabidopsis (Arabidopsis thaliana), some of which regulate ABA responses by the transcriptional regulation of downstream genes. Here, we report the functional identification of rice (Oryza sativa) ABI5-Like1 (ABL1), which is a basic region/leucine zipper motif transcription factor. ABL1 is expressed in various tissues and is induced by the hormones ABA and indole-3-acetic acid and stress conditions including salinity, drought, and osmotic pressure. The ABL1 deficiency mutant, abl1, shows suppressed ABA responses, and ABL1 expression in the Arabidopsis abi5 mutant rescued the ABA sensitivity. The ABL1 protein is localized to the nucleus and can directly bind ABA-responsive elements (ABREs; G-box) in vitro. A gene expression analysis by DNA chip hybridization confirms that a large proportion of down-regulated genes of abl1 are involved in stress responses, consistent with the transcriptional activating effects of ABL1. Further studies indicate that ABL1 regulates the plant stress responses by regulating a series of ABRE-containing WRKY family genes. In addition, the abl1 mutant is hypersensitive to exogenous indole-3-acetic acid, and some ABRE-containing genes related to auxin metabolism or signaling are altered under ABL1 deficiency, suggesting that ABL1 modulates ABA and auxin responses by directly regulating the ABRE-containing genes.
Abscisic acid (ABA) is an important plant hormone that affects many aspects of plant growth and developmental processes, including cell division, seed maturation and germination, seedling development, stomata opening, and leaf senescence, and plays a crucial role in plant responses to stresses. ABA serves as an endogenous signal to initiate the adaptive responses when plants are challenged by abiotic stress (Zhu, 2002) and biotic stress (Adie et al., 2007).
Insights into the signal transduction of ABA have unfolded dramatically in the past few years and revealed an unanticipated complexity. In Arabidopsis (Arabidopsis thaliana), several candidate receptors have been identified by their high-affinity binding to ABA. Recently, a 14-member family of intracellular ABA receptors, named PYR/PYL/RCAR (Ma et al., 2009; Pandey et al., 2009; Park et al., 2009; Santiago et al., 2009), was identified. However, no ABA receptor in the monocot rice (Oryza sativa) has been reported yet.
Genetics studies have helped to identify the key components of ABA signaling. Five ABA-insensitive members (ABI1–ABI5) have been identified by genetic screens, as the deficiency of corresponding genes results in the insensitive responses of seed germination to exogenous ABA. Transcriptome studies by DNA chip analysis revealed the dramatic changes in the expression of thousands of genes after ABA treatment and indicated the crucial roles of transcriptional regulation in ABA signaling (Hoth et al., 2002; Seki et al., 2002). ABI3, ABI4, and ABI5 are all transcription factors (VIVIPAROUS1 [VP1], APELATA2, and basic region/leucine zipper motif [bZIP] types, respectively) and are involved in the regulation of ABA-mediated gene expression (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Arabidopsis ABI4 and maize (Zea mays) ZmABI4 bind the dehydration-responsive element (DRE) that acts as a second cis-element or “coupling element” to assist with ABA-controlled gene expression (Niu et al., 2002; Narusaka et al., 2003). ABI3 represents an accessory enhancer of transcription and forms a complex with ABI5 to regulate the expression of downstream genes (Nakamura et al., 2001). Arabidopsis ABI5 and rice bZIP transcription factor TRAB1 interact with ABA-responsive elements (ABREs; ACGT-containing “G-boxes” in the promoter region; Hattori et al., 2002) to activate a series of downstream events. The activities of ABI5 and TRAB1 are regulated by phosphorylation (Kagaya et al., 2002), and ABI5 can be stabilized by blocking ABI FIVE-BINDING PROTEIN-mediated protein degradation through the 26S proteasome (Lopez-Molina et al., 2003).
Comprehensive analyses employing microarrays are very helpful for studying molecular mechanisms, especially when the functional clues are limited. Several groups have tried to analyze the gene expression profiling using the Arabidopsis abi5 mutant or ABI5-overexpressing seedlings, including dry seeds (Nakabayashi et al., 2005; Nakashima et al., 2009) and seedlings after ABA treatment at multiple time points (Hur, 2007). However, the detailed mechanisms of ABI5 function are still insufficient.
Plant bZIP transcription factors contain a basic region that binds DNA and a Leu zipper dimerization motif to create an amphipathic helix and regulate multiple processes, especially stress responses (Kang et al., 2002). In Arabidopsis, there are 75 bZIP members that can be divided into 10 groups according to the sequence similarity of the basic region (Jakoby et al., 2002). There are seven members of group A (AtbZIP39/ABI5, AtbZIP36/ABF2/AREB1, AtbZIP38/ABF4/AREB2, AtbZIP66/AREB3, AtbZIP40/GBF4, AtbZIP35/ABF1, and AtbZIP37/ABF3), and studies reveal that most of them are involved in ABA or stress signaling (Choi et al., 2000; Finkelstein and Lynch, 2000; Uno et al., 2000; Lopez-Molina et al., 2001). In addition, these members are designated as ABRE-binding factors (ABFs) or ABA-responsive element binding proteins (AREBs) because they can bind different ABRE-containing promoters (Choi et al., 2000; Uno et al., 2000).
There are 89 potential nonredundant bZIP transcription factor genes in rice (Nijhawan et al., 2008). We are particularly interested in the VI family (14 members) because they are highly homologous to the Arabidopsis group A subfamily (ABI5 subfamily). Until now, only three members of this subfamily have been functionally characterized. TRAB1 was identified by a yeast two-hybrid screen for isolating the protein interacting with VP1/ABI3 (Hobo et al., 1999). TRAB1 is localized to the nucleus and activated by ABA-dependent phosphorylation (Kagaya et al., 2002; Kobayashi et al., 2005). OsABI5, a transcriptional activator that can bind to ABRE (G-box), is involved in ABA signal transduction and regulates fertility and stress responses (Zou et al., 2007, 2008). OsbZIP23 functions as a transcriptional activator to positively regulate the ABA responses and hence increase the stress resistance in rice (Xiang et al., 2008). Whether other members of this subfamily have similar functions is still unknown.
Studies have shown similar inhibitory effects of ABA in germination and root growth of Arabidopsis and rice, although the sensitivities are much different, especially in seed germination. Considering that most studies at present focus on the ABA functions in stress responses in Arabidopsis, and that the conservation and specificity of rice ABA signaling remain largely unknown, studies on rice ABA signaling will be very valuable to illustrate the functional mechanism of ABA. Here, we report the functional identification of a rice bZIP transcriptional factor, ABI5-Like1 (ABL1), which is involved in rice ABA signaling and stress responses by directly binding to ABRE-containing genes, especially WRKY family genes. In addition, ABL1 suppresses auxin signaling by targeting ABRE-containing genes related to auxin metabolism or signaling, revealing the central role for ABL1 in the ABA and auxin responses.
RESULTS
ABL1 Is Expressed in Various Tissues and Is Induced by Hormones and Stress Conditions
A rice bZIP transcription factor, bZIP46 (LOC_Os6g10880), which belongs to subfamily VI of the rice bZIP family and has a close relationship with Arabidopsis ABI5, was identified and designated as ABL1. A comparison of the protein sequence revealed that several members of this subgroup, including ABL1, contain conserved motifs that have been identified in members of Arabidopsis ABI5 and rice TRAB1 (Hobo et al., 1999). Besides the conserved basic Leu zipper motif, ABL1 also contains phosphorylation sites, including the potential casein kinase II phosphorylation site (S/TxxD/E, where x represents any amino acid) and a Ca2+-dependent protein kinase phosphorylation site (R/KxxS/T), implying the possible regulation of ABL1 by various protein kinases.
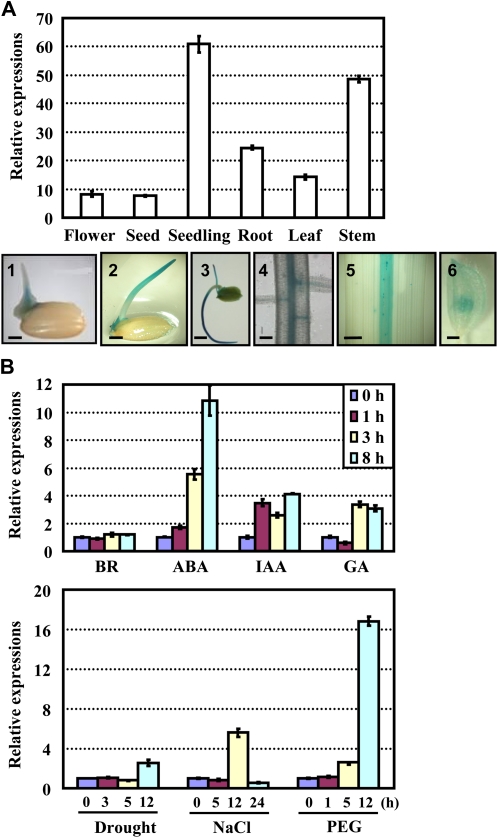
An analysis of the expression pattern by quantitative real-time reverse transcription (qRT)-PCR analysis or promoter-reporter gene (GUS) fusion studies revealed that ABL1 is expressed in various tissues, with a relatively higher expression in leaves and stems (Fig. 1A, top panel). Further detailed histochemical analyses of the GUS activities of pABL1:GUS lines showed that ABL1 is mainly expressed in the coleoptiles and primary roots at the seedling stage (Fig. 1A, bottom panels 1–3), particularly in the root vascular bundles (Fig. 1A, bottom panel 4). ABL1 is mainly expressed in the midvein of the leaves of adult plants (Fig. 1A, bottom panel 5), suggesting the possible role of ABL1 throughout the development process, primarily in the vegetative growth stage.
Figure 1.
Expression pattern analysis of ABL1. A, qRT-PCR analysis of ABL1 expression in various tissues (top panel). Seven-day-old seedlings were used to harvest the samples of the seedling (shoot and leaf) and root (primary and adventitious). Leaf and stem samples were harvested from the plants before heading. The bottom panel shows the results of promoter-GUS fusion studies, which revealed the expression of ABL1 in the seed (1), seedling (2 and 3), primary root of the seedling (4), leaf (5), and flower (6). Bars = 2 mm (1, 2, 5, and 6), 5 mm (3), and 50 μm (4). B, qRT-PCR analysis of ABL1 expression under ABA, BR, IAA, or GA treatment (top panel) or under drought, high-salinity, or PEG treatment (bottom panel). Twelve-day-old seedlings under different treatments were used for the analysis, and the expression of ABL1 without treatment was set at 1.0. Biological replicates of the experiments were performed, and the data are presented as averages of three independent experiments.
Some Arabidopsis bZIP transcription factor-encoding genes, AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5, exhibit ABA-, drought-, and high salinity-inducible expression (Fujita et al., 2005), and expression of rice bZIP23 is induced by ABA or under different stresses such as drought and salinity (Xiang et al., 2008), which is consistent with the hypothesis that ABA stimulates the expression of target transcription factors. To explore the possible involvement of ABL1 in the regulation of ABA and abiotic stress-related genes, the expression of ABL1 under exogenous hormones (ABA, brassinosteroid [BR], indole-3-acetic acid [IAA], and gibberellic acid [GA]) and abiotic stress conditions was examined by qRT-PCR. The results showed that ABL1 was strongly induced by ABA treatment and slightly stimulated by IAA and GA but not BR (Fig. 1B, top panel). Similarly, ABL1 was induced by drought, high salinity, and polyethylene glycol (PEG; Fig. 1B, bottom panel), and the most dramatic induction was under the 12-h PEG treatment. These results are consistent with the previous reports by microarray hybridization indicating that ABL1 is up-regulated by dehydration and salt but not cold (Nijhawan et al., 2008).
Deficiency of ABL1 by T-DNA Insertion Results in Suppressed ABA Responses
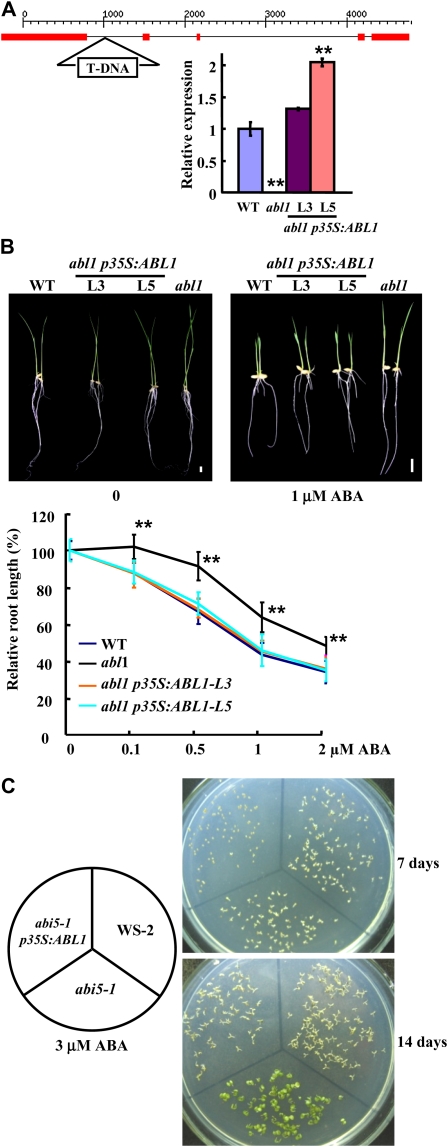
A search of the Shanghai T-DNA insertion population (http://ship.plantsignal.cn; Fu et al., 2009) resulted in the identification of a putative mutant line in which the insertion is located at the first intron of ABL1 (Fig. 2A, top panel). An analysis by PCR amplification confirmed the single-copy integration of T-DNA (Supplemental Fig. S1) and the deficiency of ABL1 in the homozygous lines by qRT-PCR analysis (Fig. 2A, bottom panel), indicating that abl1 is a knockout mutant.
Figure 2.
The deficiency of ABL1 caused by a T-DNA insertion resulted in suppressed ABA responses. A, The schematic exon-intron structure of the ABL1 gene and the position of the T-DNA insertion (first intron). Red boxes represent the exons (top panel). The bottom panel shows qRT-PCR analysis of the ABL1 expression in homozygous mutant and transgenic lines with complementary expression of ABL1 (L3 and L5). The expression of ABL1 in the wild type (WT) was normalized to 1.0. RNAs extracted from rice leaves were used for analysis, and the rice ACTIN gene was used as the internal control. B, The phenotypic observation (top panel; bars = 1 cm) and calculation (bottom panel) of root growth revealed that abl1 had suppressed responses to the ABA treatment, whereas abl1 seedlings with complemented expression of ABL1 (representative lines L3 and L5) had normal responses to the ABA treatment. The root lengths of 6-d-old seedlings (after germination) were measured, and the relative root lengths were calculated (the root lengths of untreated seedlings were set at 100%). The data are presented as averages ± sd (n > 20). A statistical analysis with a one-tailed Student’s t test indicates significant differences (** P < 0.01). C, ABL1 rescues the ABA insensitivity of Arabidopsis abi5-1. The seeds of Ws-2, abi5-1, and transgenic abi5-1 lines carrying p35S:ABL1 were germinated on half-strength MS medium supplemented with 3 μm ABA for 7 or 14 d.
The Arabidopsis abi5 mutant is insensitive to exogenous ABA, and the deficiencies of rice ABI5 and bZIP23, which belong to the same subfamily of ABL1, also display insensitive responses to ABA (Carles et al., 2002; Xiang et al., 2008; Zou et al., 2008). Similarly, phenotypic observation and statistical analysis of root and seedling growth of the wild type and abl1 under ABA treatment showed that the deficiency of ABL1 results in suppressed responses to exogenous ABA. abl1 shows only a 9% inhibition of the root length under the 0.5 μm ABA treatment, whereas the wild type has an approximately 30% inhibition (Fig. 2B). Unlike in Arabidopsis, both phenotypic observation and calculation of seed germination frequency revealed that there is no significant difference of abl1 in ABA-inhibited seed germination when compared with the wild type.
To confirm that the altered ABA response of abl1 is indeed due to the deficiency of ABL1, a complementation study was performed. The full-length cDNA of ABL1 under the cauliflower mosaic virus 35S promoter was transformed into the abl1 mutant. An analysis of the transgenic lines with the complementary expression of ABL1 (Fig. 2A, bottom panel) revealed normal ABA responses in seedling and root growth under the ABA treatment (Fig. 2B), demonstrating the effects of ABL1 in mediating ABA response and signaling.
The full-length cDNA of ABL1 was transformed into the Arabidopsis abi5-1 mutant to examine the conserved function of ABA signaling in rice and Arabidopsis. The analysis of transgenic Arabidopsis lines (homozygous lines in the T3 generation) expressing ABL1 (Supplemental Fig. S2) showed that, in comparison with the fact that abi5-1 can grow well and is insensitive to ABA treatment, the ABA inhibition of seed germination and seedling growth under 3 μm ABA were rescued by expressing ABL1 (Fig. 2C), indicating that rice ABL1 has a function that is conserved from Arabidopsis ABI5.
ABL1 Localizes to the Nucleus and Binds to the ABRE cis-Element
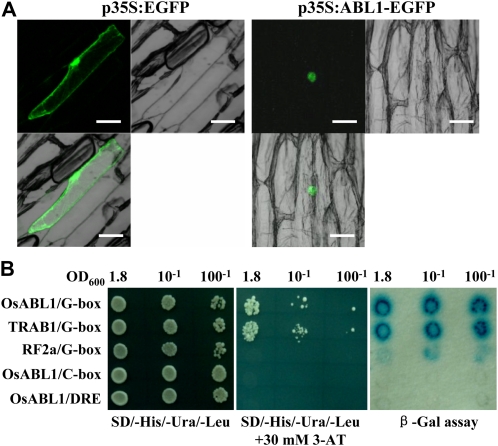
A nuclear localization signal is detected in ABL1. Observation of the green fluorescence by transient expression of the ABL1-GFP fusion protein in onion (Allium cepa) epidermal cells revealed the specific nuclear localization of ABL1 (Fig. 3A), implying a role of ABL1 as a transcription factor.
Figure 3.
ABL1 localizes to the nucleus and binds G-box/ABRE elements. A, Nuclear green fluorescence after transient expression of ABL1 fused to EGFP in onion epidermal cells revealed that ABL1 was localized to the nucleus. Onion cells expressing EGFP alone (left panel) were used as the control. Bars = 100 μm. B, The yeast-based one-hybrid assay showed the binding specificity of ABL1 to the ABRE/G-box element. The yeast strain YM4271 was used, and transformed yeast cells were spotted with serial dilutions of cells (optical density at 600 nm [OD600]) on SD/-His/-Ura/-Leu selective medium for 3 d. Proteins TRAB1 (BAA83740) and RF2a (AF005492) were used as positive and negative controls, respectively; ABRE/C-box and DRE element were used as the site mutation of ABRE/G-box. Transformants of different concentrations were dropped onto SD/-His/-Ura/-Leu + 30 mm 3-AT or selected by β-Gal activity.
Previous studies have demonstrated the binding activity of the rice bZIP VI subfamily members to the ABRE cis-element containing a 5′-CACGTG-3′ core sequence (G-box; Xiang et al., 2008; Zou et al., 2008). We then examined the specificity of the binding activity of ABL1 to the G-box cis-element with a yeast one-hybrid system. As shown in Figure 3B, a yeast strain (YM4271) transformed with the G-box and expressing the ABL1 fusion protein can grow on the synthetic dextrose (SD) medium (-His/-Ura/-Leu, supplemented with 30 mm 3-aminotriazole [3-AT]), and a β-Gal activity test of selected transformants indicated that ABL1 binds to the G-box to activate the expression of the reporter gene. Further analysis with site mutation of the G-box (ABRE/C-box or DRE element) indicated the defective growth of yeast cells and deficiency of β-Gal activity, confirming that OsABL1 specifically binds to the ABRE/G-box. These results suggest that ABL1 might act as a transcription factor by binding to the ABRE cis-element (G-box).
ABL1 Is Involved in ABA and Stress Responses by Regulating the ABRE (G-Box)-Containing Genes
To study the functional mechanism of ABL1 and identify the potential targets, the genome-wide expression profile was analyzed by DNA chip hybridization using roots as the sample material, as the phenotype of insensitive response to ABA was found notably in roots. Total RNAs of roots of 14-d-old abl1 and wild-type seedlings under ABA treatment (100 μm, 4 h) were used, and biological replicates were performed for the hybridization. Analysis of the Pearson’s correlation coefficient for the replicates showed that they were highly correlated (greater than 0.98), indicating good reproducibility of the chip hybridization. With a threshold of 1.5-fold change (false discovery rate [FDR] P < 0.01), a total of 373 genes (excluding different probes of the same transcript) were down-regulated (Supplemental Table S1) and 304 genes were up-regulated (Supplemental Table S2) in the ABL1-deficient plant.
Because ABL1 binds to the ABRE (G-box) cis-element to regulate gene expression, the cis-elements in the promoter regions of the altered genes were analyzed in detail to further identify the targets whose promoters might be bound by ABL1. The DNA sequence up to 3,000 bp upstream of the start codon ATG of each selected gene was extracted, and the analysis showed that several ABRE core motifs were enriched in the regulated genes (Table I), particularly in the down-regulated ones. These results indicate that ABL1 may bind the ABRE (G-box) elements of the promoters of these target genes to activate their transcription. Although some ABRE (G-box) motifs are detected as enriched in the up-regulated genes, the majority of the motifs are not the most representative putative binding elements of this bZIP subfamily (Nijhawan et al., 2008).
Table I. Cis-elements that have been demonstrated to be involved in ABA responses are enriched in down- or up-regulated genes under ABL1 deficiency (FDR P < 0.06).
| Cis Element | P | Description | Sequence |
| Down-regulated genes | |||
| ACGTTBOX | 8.33E-05 | T-box, ACGT element | AACGTT |
| ACGTABOX | 8.04E-04 | A-box, ACGT element, sugar | TACGTA |
| DPBFCOREDCDC3 | 2.05E-03 | Dc3, LEA class gene, ABA, ABI5 | ACACNNG |
| ABRERATCAL | 2.07E-03 | ABRE, calcium | MACGYGB |
| ABRELATERD1 | 2.57E-02 | ABRE, etiolation, ERD | ACGTG |
| ACGTATERD1 | 3.16E-02 | ACGT, etiolation, ERD | ACGT |
| CE3OSOSEM | 4.98E-02 | CE3, ABA, VP1, TRAB1, bZIP | AACGCGTGTC |
| HY5AT | 5.95E-02 | bZIP, HY5, stimulus response | TGACACGTGGCA |
| Up-regulated genes | |||
| ACGTABOX | 2.16E-02 | A-box, ACGT element, sugar | TACGTA |
| RYREPEATBNNAPA | 2.31E-02 | RY repeat, RY/G box | CATGCA |
| ABRE3HVA1 | 2.91E-02 | ABRE, ABA, HVA1 | GCAACGTGTC |
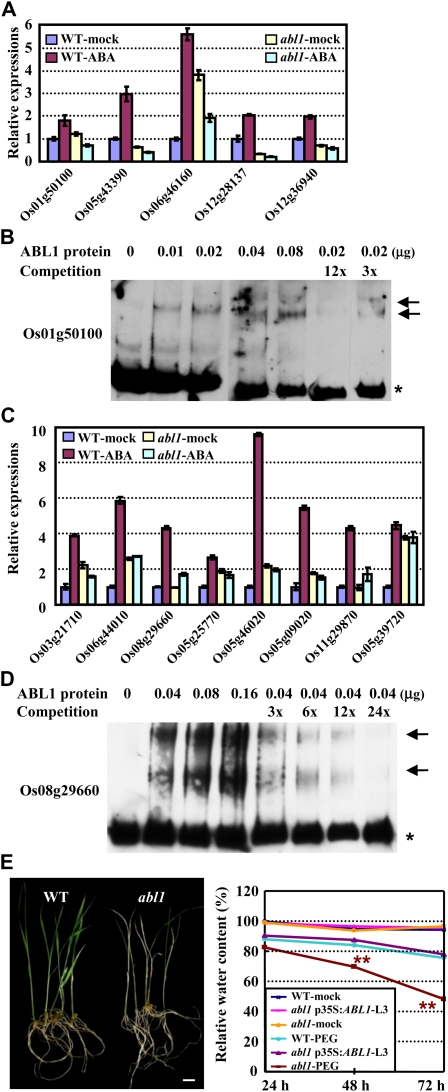
Furthermore, several down-regulated genes that contain at least four G-box elements were selected for qRT-PCR analysis in the abl1 mutant and the wild type in the presence or absence of ABA. As shown in Figure 4A, all five of the selected genes are differentially up-regulated after ABA treatment in the wild type, while they are uniformly greatly reduced in abl1 in comparison with the wild type, although the absolute values of the fold changes showed some variations for some genes between the microarray and qRT-PCR analyses. Based on the results that ABL1 binds to the ABRE cis-element, these observations further suggested that the down-regulated G-box-containing genes were potential target genes of ABL1. Indeed, gene LOC_Os01g50100 was selected and proved to be directly bound by ABL1 by an electrophoretic mobility shift assay (EMSA) using a promoter region containing at least two ABRE core cis-elements (ACGT; Fig. 4B), confirming that ABL1 could specifically and directly bind to the ABRE core cis-element (ACGT)-containing segment of promoters of regulated genes.
Figure 4.
Suppressed expression of G-box/ABRE-containing genes, especially WRKY family members, in ABL1-deficient plants. A, qRT-PCR analysis of genes containing multiple G-box elements in the absence or presence of ABA (0.1 mm). Expression of the corresponding genes in the wild type (WT) in the absence of ABA was normalized to 1.0, and relative gene expression is shown. The experiments were biologically repeated, and the data are presented as averages of three independent experiments (as for C). B, EMSA confirms the binding of ABL1 to the promoter regions of a putative ABRE-containing gene (LOC_Os01g50100). The recombinant expressed ABL1 protein was used to test the binding capacity to the digoxigenin-labeled DNA fragment of the promoter region. Unlabeled DNAs (3× or 12×) were used as the cold competitor for assays. Free probes are indicated with a star, and shift bands are indicated with arrows. C, The G-box/ABRE-containing WRKY family genes were down-regulated in abl1 under the ABA treatment. Twelve-day-old wild-type or abl1 seedlings in the absence or presence of ABA (0.1 mm) were harvested and used for the qRT-PCR analysis. The expression of the corresponding genes in the wild type in the absence of ABA was set at 1.0. D, EMSA confirms the binding of ABL1 to the promoter regions of one ABRE-containing WRKY gene (LOC_Os08g29660). The experiments were performed as described in B. Unlabeled DNAs (3× to 24×) were used as the cold competitor for assays. E, abl1 was more sensitive to the PEG treatment (left panel; bar = 1 cm). Twenty-day-old wild-type and abl1 seedlings were exposed to 15% PEG for 10 d. The relative leaf water content after PEG treatment for 3 d was measured, revealing that abl1 was hypersensitive to drought stress in comparison with the wild type. Complementary expression of ABL1 rescued the suppressed growth of abl1 (right panel). Seven-day-old seedlings were exposed to 20% PEG for 3 d. Statistical analysis by one-tailed Student’s t test indicated significant differences (** P < 0.01).
Interestingly, the Gene Ontology (GO) process analysis presented a notable difference between the up- and down-regulated genes. Over one-third (P < 0.2) of the down-regulated genes are annotated as being involved in responses to abiotic or biotic stresses, including cold, nitrogen starvation, and water deprivation, or the hormones salicylic acid, jasmonic acid, and ABA (Supplemental Table S3). In contrast, the up-regulated genes showed diverse functions, and fewer of them are involved in the responses to stresses, although some stress pathways are still enriched, including oxidative stress and defense responses (Supplemental Table S4). These results suggest that ABL1 plays a critical role in ABA signaling and ABA-triggered stress responses. In addition, although some GO stress response processes were enriched in both the down- and up-regulated genes under ABL1 deficiency, it is reasonable to suppose that the regulation of these genes was an indirect effect of ABL1.
ABL1 Is Involved in Stress Responses by Regulating ABRE (G-Box)-Containing WRKY Family Genes
Many plant transcription factors serve as targets of other upstream factors to elaborate developmental regulation or signal magnification. Analysis of the microarray data showed that there were 32 transcription factor-encoding genes among the down-regulated genes and only nine among the up-regulated genes. Surprisingly, eight WRKY family genes were uniformly and significantly down-regulated. To clarify the activating effects of ABL1 on these WRKY genes, the expression of these WRKY genes in abl1 and the wild type in the absence or presence of ABA was verified. As shown in Figure 4C, under ABA treatment, all eight WRKY genes were dramatically up-regulated in wild-type plants, while this stimulation was significantly suppressed in abl1, implying that ABL1 mediates the stimulation of these WRKY genes by ABA. Further EMSA analysis of the binding activity of ABL1 to the promoter region of putative target WRKY genes confirmed, as expected, the direct binding of ABL1 to the promoter region of one WRKY gene, LOC_Os08g29660 (Fig. 4D).
Interestingly, the analysis of the promoter regions of these eight WRKY family genes showed that they also contain multiple ABRE cis-elements. Previous studies showed that the transcription of WRKY genes was strongly and rapidly up-regulated in response to wounding, pathogen infection, or abiotic stresses in numerous plant species (Eulgem et al., 2000), strongly supporting the assumption that ABL1 may mediate ABA stimulation of these WRKY genes through direct activation. In addition, it is also noted that genes containing the cis-elements possibly directly bound by WRKY transcription factors were enriched in the down-regulated genes in ABL1-deficient plants (Table II), which suggests that ABL1 triggered a chain of WRKY-regulated transcriptional events.
Table II. Cis-elements possibly directly bound by WRKY transcription factors are enriched in down-regulated genes under ABL1 deficiency (FDR P < 0.02).
| Cis Element | P | Description | Sequence |
| WBOXATNPR1 | 5.46E-03 | NPR1, WRKY18, disease resistance, W-box | TTGAC |
| WBOXNTCHN48 | 9.80E-03 | W-box, WRKY | CTGACY |
| WBBOXPCWRKY1 | 1.10E-02 | W-box, WRKY | TTTGACY |
| WBOXNTERF3 | 1.63E-02 | W-box, ERF3, wounding | TGACY |
| WBOXHVISO1 | 1.66E-02 | Sugar, WRKY | TGACT |
Additionally, we examined the tolerance of abl1 mutant plants to high salinity and PEG treatment. The results showed that the ABL1 deficiency did not result in obvious growth differences under various concentrations of salt (150, 180, or 200 mm), whereas abl1 plants had a much lower resistance to the PEG (15%) treatment (grew slowly and weak, and more leaves turned yellow; Fig. 4E, left panel). Further measurement of the leaf relative water content (according to Guo and Qian, 2003) under PEG treatment showed the hypersensitivity of the abl1 mutant to drought stress in comparison with the wild type (reduced relative water content), whereas the complementary expression of ABL1 rescued the suppressed growth of abl1 under PEG treatment (Fig. 4E, right panel). These findings suggested a primary role for ABL1 in the abiotic stress response.
ABL1 Modulates the Auxin Responses
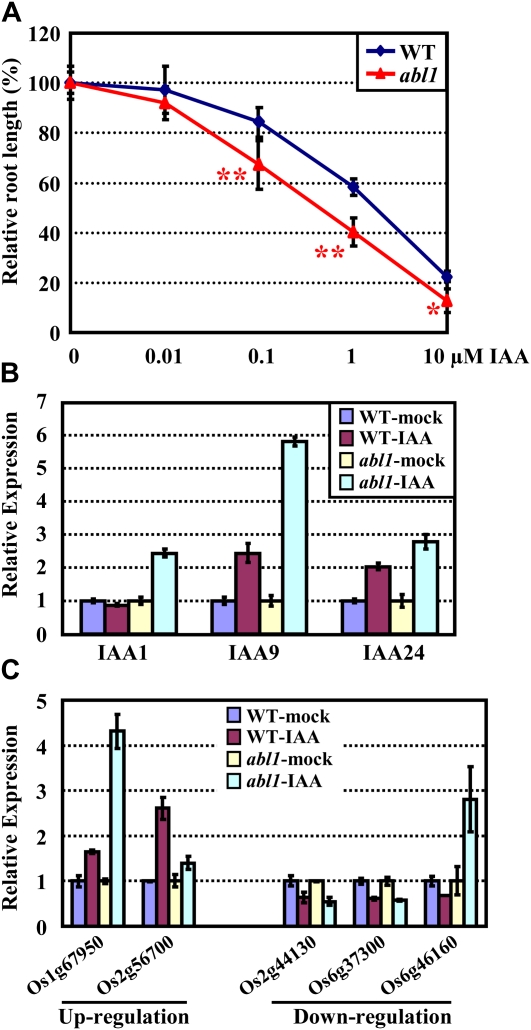
It is interesting that ABL1 is induced by exogenous IAA (Fig. 1B), which suggests a possible role of ABL1 in mediating the cross talk of ABA and IAA. The examination of IAA sensitivity by measuring the primary root length under different concentrations of IAA showed that abl1 exhibits much more inhibition, indicating a hypersensitive response of abl1 to exogenous IAA (Fig. 5A). Consistently, examination of the transcription of three auxin-responsive genes, IAA1, IAA9, and IAA24 (genes with pronounced auxin stimulation of Aux/IAA), in abl1 confirmed the significantly enhanced expression under exogenous IAA and hence the increased auxin responses (Fig. 5B).
Figure 5.
ABL1 is involved in the IAA responses. A, The measurement and calculation of the relative root elongation reveal the hypersensitive responses of abl1 to exogenous auxin. Five-day-old wild-type (WT) and abl1 seedlings (after germination) grown on medium supplemented with various concentrations of IAA were analyzed. The root length without IAA treatment was set at 100%. The data are presented as averages ± sd (n > 20). Statistical analysis by one-tailed Student’s t test indicated significant differences (* P < 0.05, ** P < 0.01). B, qRT-PCR analysis showed that the IAA-responsive marker genes (IAA1, IAA9, and IAA24) were increased to a higher level in abl1 under the IAA treatment than those of the wild type. Twelve-day-old wild-type and abl1 seedlings were treated with IAA (10 μm) for 6 h and then harvested for analysis. The expression of the corresponding genes of the wild type and abl1 in the absence of IAA was set at 1.0. The experiments were repeated, and the data are presented as averages of three independent experiments (as for C). C, The altered genes in the abl1 mutant that share a similar response to the IAA treatment (up- or down-regulation) were selected, and their expression in the absence or presence of IAA (10 μm) was examined by qRT-PCR analysis.
DNA chip hybridization analysis (this study and a public database of the Gene Expression Omnibus [GEO; http://www.ncbi.nlm.nih.gov/geo/]; GSE5167) of the genes with altered expression under ABL1 deficiency showed that some altered genes shared a similar regulation to the IAA treatment, and most of the genes contain the concentrated ABRE (G-box) cis-elements in their promoter regions (Table III). Two up-regulated and three down-regulated auxin-related genes that contain multiple ABRE cis-elements were then selected for analyzing the transcription under IAA treatment. The results showed the same tendency after IAA treatment both in the wild type and the abl1 mutant, except for one gene (LOC_Os06g46160), and some genes presented more significant increases in abl1 (Fig. 5C), confirming the hypersensitive response of abl1 to exogenous IAA.
Table III. Genes changed in the abl1 mutant and presenting similar responses to IAA treatment.
Genes containing at least three ABRE cis-elements in the promoter region are marked with asterisks. Numbers indicate the regulation ratio.
| Gene | Ratio in abl1 | IAA Regulation | Description |
| Up-regulated | |||
| LOC_Os07g36430 | 1.8 | 5.9 | Expressed protein |
| LOC_Os02g56700* | 1.8 | 5.3 | Dehydrogenase, putative |
| LOC_Os06g12500 | 1.6 | 2.9 | Membrane-associated DUF588 domain-containing protein, putative |
| LOC_Os01g67950* | 1.9 | 2.2 | Ubiquitin family protein, putative |
| LOC_Os10g18870* | 1.7 | 2.2 | DIRIGENT, putative |
| Down-regulated | |||
| LOC_Os06g37300* | −2.4 | −2.0 | Cytochrome P450, putative |
| LOC_Os06g46160* | −2.1 | −2.1 | Expressed protein |
| LOC_Os07g03590* | −1.8 | −2.1 | SCP-like extracellular protein |
| LOC_Os09g34160* | −2.1 | −2.2 | Resistance protein, putative |
| LOC_Os11g03290* | −2.1 | −2.3 | Nucleoside-triphosphatase, putative |
| AK059453 | −1.8 | −2.4 | |
| LOC_Os01g68740* | −1.9 | −2.4 | Keratin, type I cytoskeletal 9, putative |
| LOC_Os06g33100* | −2.3 | −3.4 | Peroxidase precursor, putative |
| LOC_Os02g44130* | −2.1 | −5.3 | ZOS2-14, C2H2 zinc finger protein |
In addition, a detailed GO process analysis of the altered genes in abl1 showed that six genes are related to IAA, and five of them have enriched ABRE cis-elements (Table IV). However, not all of the regulated genes contained ABRE cis-elements, implying that indirect regulation of ABL1 in IAA signaling might exist.
Table IV. Genes changed in the abl1 mutant and related to IAA by GO process analysis.
Genes containing at least three ABRE cis-elements in the promoter region are marked with asterisks. Numbers indicate the regulation ratio.
| Gene | Ratio | Description | Function |
| LOC_Os03g62060* | 2.2 | Hydrolase, putative | Auxin metabolic process |
| LOC_Os04g56690* | 2.0 | OsSAUR23, auxin-responsive SAUR gene family member | Response to auxin stimulus |
| LOC_Os05g04820 | 1.6 | MYB family transcription factor, putative | Response to auxin stimulus |
| LOC_Os01g57470* | −1.6 | EF hand family protein, putative | Response to auxin stimulus |
| LOC_Os02g37000* | −1.6 | Mitochondrial prohibitin complex protein 1, putative | Response to auxin stimulus |
| LOC_Os07g14610* | −1.7 | IAA-amino acid hydrolase ILR1-like 6 precursor, putative | Auxin metabolic process |
DISCUSSION
Crucial Roles of Rice ABL1 in ABA and Stress Response
Although ABL1 could partially rescue the insensitive response to ABA of the Arabidopsis abi5-1 mutant, a functional difference exists in Arabidopsis ABI5 and rice ABL1. Seeds of abl1 do not show altered germination under ABA treatment, which is particularly obvious in Arabidopsis abi5. The tissue expression pattern of Arabidopsis ABI5 is much more abundant in developing siliques than in vegetative tissue, while ABL1 is transcribed throughout the developmental processes primarily in the vegetative phase. This indicates that Arabidopsis ABI5 and rice ABL1 still have functional diversity that may be achieved by different regulatory networks.
Most members of the rice bZIP subfamily VI (10 of 14) are involved in ABA signaling and abiotic stress responses (Xiang et al., 2008; Zou et al., 2008), especially stress-related AREB/ABF factors of both rice and Arabidopsis (Yoshida et al., 2010). Phylogenetic analysis showed that ABL1 is the closest homolog to rice bZIP23 and TRAB1 (a functionally unknown OsbZIP72) and Arabidopsis AREB1, AREB2, and ABF3 (Supplemental Fig. S3). The three Arabidopsis AREBs have been proven to interact with SRK2D/SnRK2.2, a SnRK2 protein kinase that was identified as a regulator of AREB1. A previous study showed that all these rice homologs require ABA for full activation as Arabidopsis AREB (Yoshida et al., 2010), suggesting that rice ABL1, bZIP23, and bZIP72 (TRAB1) may function downstream of SnRK2 to activate the target genes in ABA signaling.
Based on previous studies showing that rice bZIP23, TRAB1, and ABI5 also function in stress-related responses, it is supposed that the common stress-related effects of rice ABL1 are masked in the abl1 mutant due to the redundant functions and expression of other homologs such as OsbZIP23. Examination of the expression levels of other homologs of ABL1 in abl1 mutant and wild-type plants under the absence or presence of exogenous ABA revealed that although the ABL1 homologs are down-regulated in the abl1 mutant, their expression is restored to the normal expression level under ABA treatment (Supplemental Figure S4), which indicated that the observed phenotype related to stress in the abl1 mutant is mainly owing to the deficiency of ABL1. Generation of multiple mutants of genes including rice bZIP23, ABL1, TRAB1, and bZIP72 might help to study their functional redundancy and synergetic regulation, as has been done in Arabidopsis using the triple mutant areb1 areb2 abf3 (Yoshida et al., 2010). Whether these rice AREBs function cooperatively as master transcription factors to regulate the ABRE-dependent ABA signaling and to be involved in drought stress tolerance need further study.
ABL1 Regulates Abiotic Stress Responses by Modulating the Expression of ABRE-Containing Genes, Especially the WRKY Family Genes
Analysis of the transcriptome of rice seedlings exposed to cold, drought, high salinity, or application of ABA revealed that many transcription factors are involved in ABA-mediated stress responses (Rabbani et al., 2003). ABL1 localizes to the nucleus, and the suppressed expression of ABRE-containing genes under ABA treatment in abl1 plants indicates a primary role for ABL1 in ABA-triggered transcriptional regulation. In addition, GO analysis revealed that most of the altered genes in abl1 plants were closely correlated to the response to abiotic stresses, implying the important role of ABL1 in ABA-mediated stress responses.
It has been reported that overexpression of rice bZIP proteins such as OsbZIP23 resulted in enhanced responses to abiotic stresses, and further comparison of the up- or down-regulated genes in abl1 and OsbZIP23-overexpressing plants revealed the specific characteristics of ABL1. There were 795 or 318 up-regulated and 1,017 or 390 down-regulated genes, respectively, in OsbZIP23-overexpressing or abl1 plants, while only six and seven genes were in common in up- and down- regulated genes (Supplemental Table S5). The very limited regulated genes in common suggested that the function of ABL1 in transcriptional regulation in ABA responses is different from other bZIP family members. Compared with that of OsbZIP23-overexpressing plants (Xiang et al., 2008), there is only one lipid transfer protein-encoding gene, and no dehydrin family and late embryogenesis abundant gene is down-regulated in abl1, although many genes related to stress response are down-regulated. Meanwhile, many more stress-related regulatory factors were down-regulated in abl1 than OsbZIP23-overexpressing plants, including protein kinase and transcription factors, suggesting the differential regulation between ABL1 and OsbZIP23 in stress responses.
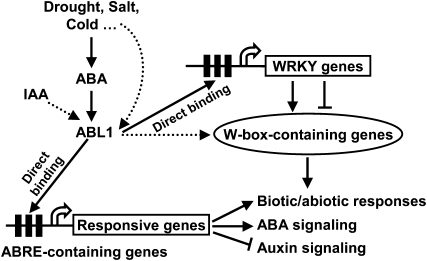
The WRKY gene family encodes a large group of transcription factors, and the WRKY domain can bind to the W-box or sugar-responsive cis-elements in the promoter regions of target genes to regulate their transcription (Rushton et al., 1995; Sun et al., 2003). Interestingly, a series of ABRE-containing WRKY family genes, which are involved in abiotic stress responses, are transcriptionally regulated by ABL1, and their induced expression under ABA treatment is evidently suppressed in ABL1-deficient plants. WRKY genes are up-regulated by multiple stress conditions and hormone treatments (Supplemental Table S6; Ramamoorthy et al., 2008), which is consistent with ABL1 being a transcriptional activator that is greatly up-regulated by drought, salinity, and IAA treatments and indicates that ABL1 regulates the abiotic stress responses by modulating the expression of ABRE-containing WRKY family genes (Fig. 6).
Figure 6.
A hypothetical working model of ABL1 function in stress response and auxin/ABA cross talk. ABL1 expression is stimulated by various environmental stresses, which may be mediated by ABA, and then activates the expression of a series of ABRE (G-box)-containing genes, especially those involved in ABA signaling and stress responses through direct binding to the ABRE motifs. In addition, ABL1 stimulates the expression of ABRE-containing WRKY members, which differentially regulate the downstream W-box-containing genes to regulate the biotic/abiotic responses and ABA signaling. Meanwhile, ABL1 is induced by IAA and negatively regulates auxin signaling by regulating the expression of ABRE (G-box)-containing genes related to auxin metabolism or signaling.
Most of the down-regulated genes in abl1 are stimulated by abiotic stress conditions. An analysis of the rice gene profile under drought, salinity, and cold conditions (microarray data were obtained from the public database under GEO accession no. GSE6901) and a comparison with the altered genes in abl1 showed that 58% of down-regulated genes (Supplemental Table S7) and 49% of up-regulated genes (Supplemental Table S8) in abl1 are also regulated under stress conditions (at least one of the three conditions). A further analysis of the promoter region of these commonly altered genes revealed the presence of the W-box cis-element, which is particularly enriched in both the up- and down-regulated genes with higher fold changes. Considering the role of WRKY transcription factors serving as either transcriptional activators or suppressors, it is thus assumed that under abiotic stresses, ABL1 is up-regulated (possibly mediated by ABA) and stimulates the downstream ABRE-containing genes to regulate the responses or ABA signaling. Importantly, ABL1 stimulates a series of ABRE-containing WRKY genes that activate or suppress the W-box-containing genes to promote stress responses (Fig. 6).
ABL1 Mediates the Cross Talk of ABA and Auxin
Previous studies revealed the cross talk of ABA and auxin on different levels. PGGT I encodes the β-subunit of protein geranylgeranyltransferase type I and negatively regulates ABA signaling in guard cells. The deficiency of PGGT I results in increased lateral root formation in response to exogenous auxin (Johnson et al., 2005), indicating synergistic cross talk of ABA and auxin. Detailed studies on the transcriptional profiling of genes assigned to the ABA and auxin biosynthetic pathways by GO annotation revealed the complex interaction of ABA and auxin signaling with ambiguous outcomes (Nemhauser et al., 2006).
Cross talk between the ABA- and auxin-dependent responses takes place mainly during seed germination and early seedling development, in which the ABA-dependent repression of growth is potentiated by auxin (Ni et al., 2001; Nakabayashi et al., 2005; Liu et al., 2007). Recent studies showed that ABA and auxin have synergic functions in repressing embryonic axis elongation in Arabidopsis (Belin et al., 2009), which involves the transcriptional repression of the Aux/IAA genes, AXR2/IAA7 and AXR3/IAA17, by ABI5 and ABI3 binding to the particular ABRE elements. The hypersensitive response to IAA treatment in root length by ABL1 deficiency (Fig. 5A) was somehow in contrast to the ABA repression of embryonic axis elongation in Arabidopsis through enhanced IAA signaling. However, when considering the physiological effects, there is a consistent tendency for ABA to suppress embryo axis elongation or seedling growth by regulating auxin signaling. ABI5 stimulates auxin signaling to suppress embryo axis elongation, and ABL1 suppresses auxin signaling and hence seedling growth, revealing an interesting and complex cross talk between ABA and auxin. In addition, auxin-induced lateral root formation is completely suppressed by ABA in 35S-VP1/abi3-6 plants, indicating that VP1 mediates the interaction between ABA and auxin signaling (Suzuki et al., 2001). Considering the interaction of TRAB1 and VP1, we ponder whether the role of ABL1 in auxin signaling was related to ABI3 orthologs in rice. It is assumed that the cross talk of ABA and auxin, especially the suppression of auxin by ABA, is correlated to specific growth and developmental processes such as embryo axis elongation or seedling growth. ABL1 could directly or indirectly target the ABRE-containing genes that encode the proteins involved in the auxin response, metabolism, or signaling, providing new clues to cross talk between ABA and auxin signaling and functions (Fig. 6).
Additionally, coordination of the GA- and ABA-dependent signals is starting to be unveiled in Arabidopsis (Piskurewicz et al., 2009; Okamoto et al., 2010). ABL1 could be up-regulated by GA (Fig. 1B), and analysis of the microarray data showed that some genes related to GA were altered, and all of them contained ABRE cis-elements in the promoter regions (Supplemental Table S9). One of the down-regulated genes possibly encodes GA ent-kaurene synthase, and another is possibly involved in converting ent-kaurene to GA12. Two genes whose products catalyzed the penultimate steps of GA activation were up-regulated in the abl1 mutant. According to the previous finding that GA influences GA metabolism by a feedback regulation and most of the GA20ox genes are down-regulated by applied bioactive GAs (GA1 and GA4; Hedden and Phillips, 2000), the down-regulation of early GA synthesis genes and the up-regulation of GA metabolism genes implies a hypersensitive GA response of the abl1 mutant and an antagonistic relationship between GA and ABA.
In summary, our studies demonstrate that ABL1 is involved in the ABA and stress responses and mediates the cross talk of ABA and auxin by directly regulating a series of ABRE-containing downstream genes, providing new insights into the ABA signaling and ABA-auxin interactions.
MATERIALS AND METHODS
Chemicals and Plant Materials
IAA (I2886), 24-epi-brassinolide (E1641), ABA (A1049), and GA (G7645) were purchased from Sigma-Aldrich.
For rice (Oryza sativa) growth, seeds were germinated in sterilized water and then grown in pots in a Phytotron with a 12-h-light (26°C)/12-h-dark (18°C) cycle.
To detect the transcription levels of relevant genes, rice wild-type (Zhonghua 11, japonica variety) and abl1 plants were grown in the greenhouse with a 12-h-light/12-h-dark cycle, and 12-d-old seedlings were treated with hormones (1 μm BR, 10 μm IAA, 10 μm GA, or 0.1 mm ABA) followed by sampling at 0, 1, 3, and 8 h. For abiotic stress treatment, 12-d-old seedlings were irrigated with 20% PEG 6000 (followed by sampling at 1, 5, and 12 h) and 200 mm NaCl solution (sampling at 0, 5, 12, and 24 h). Drought stress was achieved by leaving the intact seedlings in the air without water supply, and seedlings were sampled at 0, 3, 5, and 12 h. Total RNAs were extracted from total seedlings after treatment and used for further analysis.
Seeds of Arabidopsis (Arabidopsis thaliana) wild-type Wassilewskija (Ws-2; stock no. CS22659) and abi5-1 (stock no. CS8105), which has a mutation in the Arabidopsis ABI5 gene (AT2G36270), were obtained from the Arabidopsis Biological Resource Center at Ohio State University.
Subcellular Localization Studies of ABL1
The whole ABL1 coding region was amplified with primers ABL1-G-S (5′-GAAGATCTAATGGAGTTGCCGGCGGATG-3′; added BglII site is underlined) and ABL1-G-A (5′-GACTAGTGCATGGACCAGTCAGTGTTC-3′; added SpeI site is underlined) and subcloned into the binary vector pCAMBIA1302 (CAMBIA), resulting in a C-terminal fusion to GFP. The resultant construct, p35S:ABL1-EGFP, was sequenced to confirm the in-frame fusion of ABL1 and GFP and used for transient expression in onion (Allium cepa) epidermal cells. Transient expression of GFP fusion protein was performed with a model PDS-1000/He biolistic particle delivery system (Bio-Rad). Five micrograms of purified plasmids was coated with 0.6- to 1-μm gold particles, and bombardment was performed with the following parameters: 1,100-p.s.i. rupture disc, 27-inch helium vacuum, 6-cm distance from the stopping screen to the target tissues. After bombardment, onion epidermal tissue was incubated in Murashige and Skoog (MS) medium at 25°C in darkness for at least 24 h. Cell plasmolysis was achieved by incubating the onion epidermal tissue in Suc solution (1 m) for 10 min. The green fluorescence was observed by confocal laser scanning microscopy (FITC488; Zeiss LSM500) with an argon laser excitation wavelength of 488 nm GFP. Onion cells harboring vector pCAMBIA1302 and expressing unique EGFP were observed as a control.
RT-PCR and qRT-PCR Analysis
Total RNAs were extracted from 12-d-old seedlings of the wild type (Zhonghua 11) and the abl1 mutant using TRIzol reagent (Invitrogen) and reverse transcribed according to the manufacturer’s instructions (ReverTra Ace-a-first-strand synthesis kit; Toyobo). For RT-PCR analysis of ABL1 transcription, equal amounts of first-strand cDNAs were used as templates for PCR amplification using primers 5′-GCGGCAGAGGCGGATGAT-3′ and 5′-GTTCGTCGGAGGCAAAATCT-3′. The rice ACTIN gene (Os03g50890) was amplified by primers 5′-CCTTCAACACCCCTGCTATG-3′ and 5′-TGAGTAACCACGCTCCGTCA-3′ and used as an internal positive control for quantification of the relative amounts of cDNA.
For qRT-PCR analysis, the resultant first-strand cDNA was used as a template to quantitatively analyze gene expression using the Rotor-Gene real-time thermocycler R3000 (Corbett Research) with real-time PCR Master Mix (Toyobo). For the analysis, a linear standard curve was generated using a series of dilutions for each PCR product, and the levels of transcript in all unknown samples were determined according to the standard curve. The rice ACTIN gene was amplified and used as an internal standard to normalize the expression of ABL1 and the other tested genes. The primers used are listed in Supplemental Table S10.
Constructs and Rice Transformation
The full-length cDNA of ABL1 was isolated from KOME AK105312 (the Rice Genome Resource Center, http://www.rgrc.dna.affrc.go.jp/) by digestion with EcoRI and XbaI and subcloned into the binary vector pCAMBIA2300S precut with KpnI and XbaI. The resultant construct was transferred into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by electroporation and used for transformation of the abl1 mutant using immature embryos as materials.
Promoter-Reporter Gene Fusion Studies
A 1.9-kb genomic DNA fragment containing the promoter region of the ABL1 gene was amplified by PCR using primers 5′-CCCAAGCTTTATCCCTCTGTAACCAAACCAAAC-3′ (added HindIII site is underlined) and 5′-CGGGATCCTCCCACCTCAAAATCCTTCAAATC-3′ (added BamHI site is underlined) and then subcloned into modified vector pCAMBIA1300+pBI101.1 (Liu et al., 2003), resulting in the fusion of the ABL1 promoter and GUS reporter gene. The construct was then transformed into rice plants as described above, and approximately 30 independent transgenic lines were obtained. GUS activities were histochemically detected as described (Jefferson et al., 1987).
DNA-Binding Assay
The DNA-binding assay was performed according to the methods described by Zou et al. (2008). The yeast reporter strain YM4271, which carries the dual reporter genes HIS3 and lacZ, with a trimer of 27-bp DNA fragments including ABREs (G-box) composed as follows: (5′-agctAGCCACGTGTCGGACACGTGGCA-3′; G-box positions are underlined) upstream of the TATA element was used as an assay system. ABRE/C-box (5′-agctAGCGACGTCTCGGAGACGTCGCA-3′; site-mutated positions are underlined) and DRE (5′-agctAGCTACCGACATTCGGATACCGACATGCA-3′; site-mutated positions are underlined) elements were constructed as the site mutation and used to examine the specific binding activity of the ABRE/G-box. Lowercase letters indicate four nucleotides used for PCR amplification to ensure accuracy. The full-length cDNA of ABL1 was subcloned into the vector pAD-Gal4-2.1, and the resultant construct was then transformed into yeast reporter strain YM4271. The transformed cells were incubated on medium lacking His and supplemented with 30 mm 3-AT, a competitive inhibitor of the HIS3 gene product, for 3 d, and then cell growth was observed. β-Gal activity of selected different transformants was examined after 6 h.
EMSA
Recombinant ABL1 protein was expressed in Escherichia coli and used in the EMSA. The coding region of ABL1 was amplified via PCR using primers ABL1-pET32a-S (5′-CATGCCATGGAGATGGAGTTGCCGGCGGAT-3′; added NcoI site is underlined) and ABL1-pET32c-A (5′-GGAATTCGCATGGACCAGTCAGTGT-3′; added EcoRI site is underlined) and then subcloned into pET-32a(+) (Novagen). The resultant construct was sequenced to confirm the in-frame fusion and then transformed into E. coli strain BL21 for expression of the recombinant proteins. The recombinant proteins were purified with nickel-nitrilotriacetic acid agarose (catalog no. 30210; Qiagen).
The DNA fragments of the promoter regions of LOC_Os01g50100 (positions −523 to −265 bp) and LOC_Os08g29660 (positions −1,023 to −872 bp) were amplified with the primers 5′-TTGACCGTTGGTATCTGCATCA-3′/5′-CGTACTGCTGTTCCTTTTCTGC-3′ and 5′-TAGAAATGGACGTAGATCTCAACGC-3′/5′-CCAAGATCCTGGCACCTACCT-3′ by using rice genomic DNA as the template, sequenced, subcloned, and PCR labeled with digoxigenin according to the manufacturer’s instructions (Roche).
Labeled DNA was incubated with or without unlabeled DNA and recombinant proteins (10–160 ng) in binding buffer (75 mm HEPES, 175 mm KCl, 5 mm EDTA, 40% glycerol, 5 mm dithiothreitol, and 1 mm MgCl2). The reactions were incubated at room temperature for 20 min, run on a 0.5× 5% polyacrylamide gel, followed by transmembrane, cross-linking, blocking, and antibody incubation, and then detected with disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate (product no. 11655884001; Roche) according to the manufacturer’s instructions.
Expression of ABL1 in the Arabidopsis abi5-1 Mutant and ABA Sensitivity Assay
The ABL1 cDNA was subcloned into the vector pCAMBIA1301, and the resultant construct was transformed into the Arabidopsis abi5-1 mutant by the A. tumefaciens floral dip method (Clough and Bent, 1998). T1 seeds from the infiltrated plants were screened on MS medium containing 25 μg L−1 hygromycin (Roche), and the homozygous lines (T3 generation) were used for ABA sensitivity analysis.
The seeds of Arabidopsis Ws-2, abi5-1 mutant, and transgenic abi5-1 plants expressing ABL1 were placed on half-strength MS medium supplemented with 3 μm ABA, chilled for 2 d at 4°C in darkness, grown at 22°C (16-h-light/8-h-dark cycle) for 7 or 14 d, and then observed.
GeneChip Analysis Using Affymetrix Array and Data Statistical Analysis
GeneChip rice genome array (Affymetrix) was used to study gene expression under ABL1 deficiency. abl1 and wild-type seedlings were grown in the Phytotron with a 12-h-light/12-h-dark cycle, and 14-d-old seedlings were treated in water with ABA (100 μm, 4 h). Total RNAs were extracted from roots using TRIzol reagent (Invitrogen Life Technologies). Total RNA (2 μg) was used as starting material for hybridization, and each sample had two biological repeats. Washing, staining, and scanning of chips were performed as described by the supplier’s protocol.
The hybridization signals were normalized using Affymetrix’s MAS5.0 methods from the Bioconductor suite (www.bioconductor.org) of tools for the statistical package R (http://www.R-project.org). All the data can be accessed at GEO under the accession number GSE18529. Least-square linear regression for each sample (Schmid et al., 2005) was carried out to check the replicate quality of the microarray. The r2 statistic of each sample was higher than 0.98. The Institute for Genome Research Multiple experiment viewer (Mev; version 4.0; http://www.tm4.org/mev.html) was used to identify the genes differentially expressed in abl1. FDRs for various P value thresholds were determined by the method of Benjamini and Yekutieli (2001). The changed genes in the abl1 mutant under the threshold of 1.5-fold change (FDR P < 0.01) were selected for further analysis. The expression data of GSE6901 were downloaded from the National Center for Biotechnology Information GEO.
GO Process Analysis
Up to date, the integrity of GO information in Arabidopsis is much better than in rice, so the rice genes were first identified by using BLASTp (E < 1e-5) as described by The Institute for Genome Research (http://www.tigr.org), and then the functions of genes in Arabidopsis were assigned to their rice homolog. To determine the significance of the overrepresentation of selected genes for each pathway in a GO catalog, a χ2 test was performed to test the null hypothesis or the alternative hypothesis, defined as H0:p0 = p1; 1H:p0 ≠ p1, where p0 = m/M and p1 = n/N (m is the number of selected genes mapped to the pathway, M is the number of genes on the microarray that are mapped to that particular pathway, n is the total number of selected genes mapped to all pathways, and N is the total number of genes on the microarray mapped to all pathways).
cis-Element Analysis
The DNA sequence up to 3,000 nucleotides upstream of ATG for each selected gene was extracted and used for cis-element analysis. Known cis-elements were analyzed by using the PLACE program (for Plant Cis-Acting Regulatory DNA Elements; http://www.dna.affrc.go.jp/PLACE/), and the ratio of each cis-element in regulated genes in the abl1 mutant to that in the whole genome was followed by performing a χ2 test. FDRs for various P value thresholds were determined as 0.06 (Table I) and 0.02 (Table II). If a known cis-element was enriched with a low FDR P value, this cis-element was selected.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the abl1 mutant.
Supplemental Figure S2. Expression analysis of ABL1 in the Arabidopsis abi5-1 mutant transformed with ABL1.
Supplemental Figure S3. Phylogenetic analysis of the rice VI bZIP subfamily members.
Supplemental Figure S4. The expression level of ABL1 homolog genes in the abl1 mutant.
Supplemental Table S1. Down-regulated genes in the abl1 mutant by GeneChip analysis.
Supplemental Table S2. Up-regulated genes in the abl1 mutant by GeneChip analysis.
Supplemental Table S3. The down-regulated genes in abl1 involved in abiotic and biotic responses by GO process analysis (P < 0.2).
Supplemental Table S4. The up-regulated genes in abl1 involved in abiotic and biotic responses by GO process analysis (P < 0.2).
Supplemental Table S5. Up- and down-regulated genes in common in abl1 mutant and OsbZIP23-overexpressing plants.
Supplemental Table S6. WRKY transcription factors stimulated by ABL1 are up-regulated under stress conditions and hormone treatments.
Supplemental Table S7. Down-regulated genes in the abl1 mutant and their expression under drought, high-salinity, and cold stress by GeneChip analysis.
Supplemental Table S8. Up-regulated genes in the abl1 mutant and their expression under drought, high-salinity, and cold stress by GeneChip analysis.
Supplemental Table S9. Genes changed in the abl1 mutant that are related to GA by GO process analysis.
Supplemental Table S10. Primers used for qRT-PCR analysis.
Acknowledgments
We thank Ms. Shu-Ping Xu for rice transformation.
References
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C, Megies C, Hauserová E, Lopez-Molina L. (2009) Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21: 2253–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188 [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu FF, Ye R, Xu SP, Xue HW. (2009) Studies on rice seed quality through analysis of a large-scale T-DNA insertion population. Cell Res 19: 380–391 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LB, Qian Q. (2003) The methods of evaluating the drought resistance of cultivated rice varieties in field. China Rice 2003: 26–27 [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH. (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Hur J. (2007) Functional genomics analysis of the Arabidopsis ABI5 bZIP transcription factors. PhD thesis. Texas A&M University, College Station [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Chary SN, Chernoff EA, Zeng Q, Running MP, Crowell DN. (2005) Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol 139: 722–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T. (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T. (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. (2007) Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW. (2003) Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J 36: 189–202 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR. (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Ni DA, Wang LJ, Ding CH, Xu ZH. (2001) Auxin distribution and transport during embryogenesis and seed germination of Arabidopsis. Cell Res 11: 273–278 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ. (2002) Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell 14: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, et al. (2010) Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 62: 39–51 [DOI] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Turecková V, Lacombe E, Lopez-Molina L. (2009) Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. (2008) A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol 49: 865–879 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez JA, Cutler SR, Rodriguez PL. (2009) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, Cocciolone S, McCarty DR. (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28: 409–418 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F. (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360: 307–313 [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F. (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683 [DOI] [PubMed] [Google Scholar]