Abstract
The successful transmission of chromosomes during mitosis and meiosis relies on the establishment and subsequent release of cohesion between replicated chromatids. Cohesion is mediated by a four-subunit structural maintenance of chromosome complex, called cohesins. REC8 is a key component of this meiotic cohesion complex in most model organisms studied to date. Here, we isolated and dissected the functions of OsREC8, a rice (Oryza sativa) REC8 homolog, using two null Osrec8 mutants. We showed that OsREC8 encodes a protein that localized to meiotic chromosomes from approximately meiotic interphase to metaphase I. Homologous pairing and telomere bouquet formation were abnormal in Osrec8 meiocytes. Furthermore, fluorescent in situ hybridization experiments on Osrec8 meiocytes demonstrated that the mutation eliminated meiotic centromeric cohesion completely during prophase I and also led to the bipolar orientation of the kinetochores during the first meiotic division and accordingly resulted in premature separation of sister chromatid during meiosis I. Immunolocalization analyses revealed that the loading of PAIR2, PAIR3, OsMER3, and ZEP1 all depended on OsREC8. By contrast, the presence of the OsREC8 signal in pair2, pair3, Osmer3, and zep1 mutants indicated that the loading of OsREC8 did not rely on these four proteins. These results suggest that OsREC8 has several essential roles in the meiotic processes.
Meiosis is a process that accomplishes the requisite halving of the chromosome number by one single round of DNA replication, followed by two successive rounds of chromosome segregation. This process enables the production of haploid gametes from diploid sporocytes. The proper pairing, recombination, and segregation of homologous chromosomes are critical to meiosis and sexual reproduction (Kleckner, 1996; Zickler and Kleckner, 1999). Chromosome segregation in the second meiotic equational division resembles that in mitosis during which sister centromeres are segregated to opposite poles of the cell. However, in the initial reductional divisions, sister centromeres remain attached to each other as they are separated from their homologs (Miyazaki and Orr-Weaver, 1994).
Cohesion between sister chromatids is established during S-phase, and its maintenance and regulation is performed by the cohesin protein complex whose subunits are often referred to as cohesins (Jessberger, 2002; Hagstrom and Meyer, 2003; Uhlmann, 2004; Revenkova and Jessberger, 2006). The establishment and dissolution of sister chromatid cohesion are essential for accurate chromosome segregation during both mitosis and meiosis. In mitosis, cohesion is released all at once along the sister chromatids. By contrast, meiotic divisions require that sister chromatid cohesion be released in two steps: firstly from chromosome arms during meiosis I and secondly from centromeres during meiosis II. The homologous chromosomes are pulled to opposite poles of the cell at the first meiotic division, whereas the sister chromatids segregate only at the second meiotic division (Lee and Orr-Weaver, 2001). A successful first meiotic division requires the association of homologous chromosomes as bivalents during prophase I, the monopolar attachment of sister kinetochores at metaphase I, and the preservation of centromere cohesion at anaphase I when arm cohesion is released (Hauf and Watanabe, 2004; Ishiguro and Watanabe, 2007).
In Saccharomyces cerevisiae, mitotic cohesin is composed of four proteins: Scc1/Mcd1/Rad21, Scc3/Irr1, Smc1, and Smc3 (Guacci et al., 1997; Michaelis et al., 1997). The same complex was identified from Xenopus laevis extracts, except that it has at least two variants of Scc3, SA1 and SA2, each of them from a distinct complex (Losada et al., 1998, 2000). These proteins create a large ring structure that is presumed to entrap the sister chromatids (Gruber et al., 2003; Haering et al., 2004). However, the meiotic cohesin complex has been replaced by the meiosis-specific subunit REC8 in S. cerevisiae and Schizosaccharomyces pombe (Klein et al., 1999; Parisi et al., 1999; Watanabe and Nurse, 1999). The mutation of Rec8 of yeast disrupts cohesion at both chromosome arms and centromeres, resulting in reduced recombination, alterations in synaptonemal complex formation, and premature separation of sister chromatids at meiosis I (Klein et al., 1999; Watanabe and Nurse, 1999; Nasmyth, 2001; Kitajima et al., 2003). In recent years, homologs of yeast REC8 have been identified in mice (MmRec8; Bannister et al., 2004), humans (HsRec8; Parisi et al., 1999), Caenorhabditis elegans (CeRec8; Pasierbek et al., 2001), Arabidopsis (Arabidopsis thaliana; SYN1; Bai et al., 1999), and maize (Zea mays; AFD1; Golubovskaya et al., 2006). Depletion of this protein leads to defects in meiosis. In C. elegans, depletion of REC8 using RNA interference (RNAi) resulted in the formation of univalents and chromosome fragmentation at diakinesis (Pasierbek et al., 2001). In Arabidopsis, syn1/dif1 plants exhibit defects in meiotic chromosome cohesion and condensation that lead to the fragmentation of chromosomes and the formation of polyads (Bhatt et al., 1999; Cai et al., 2003). Moreover, the protection of centromeric Rec8 at anaphase I appears to be mediated by the Sgo1 protein identified in S. cerevisiae (Katis et al., 2004) and widely conserved across different species (Hamant et al., 2005; Kitajima et al., 2005; Watanabe, 2005; Wang et al., 2011b).
Kinetochore orientation provides a strong bias for the sister kinetochores to bind microtubules emanating from the same spindle pole. However, the role of Rec8 in kinetochore orientation at meiosis I remains ambiguous. Studies in S. cerevisiae showed that at least in this organism, Rec8 is not obligatory for monopolar orientation (Tóth et al., 2000). By contrast, SYN1 is necessary not only to maintain centromere cohesion at anaphase I but also for the monopolar orientation of kinetochores during the first meiotic division in Arabidopsis (Chelysheva et al., 2005). In maize, AFD1/REC8 controls homologous pairing through its role in axial element elongation and the subsequent distribution of the recombination machinery (Golubovskaya et al., 2006).
Rice (Oryza sativa) is a model organism for monocots that represents another excellent system for studying the molecular mechanism of meiosis in high plants. The rice genome has four Rad21-like genes that share very low identity at polypeptide levels, and each is present as a single copy. Phylogenetic analysis demonstrates that OsRad21-4 is a rice homolog of yeast REC8, and the functional analysis of OsRad21-4 by RNAi suggests that OsRad21-4 is essential for efficient meiosis (Zhang et al., 2006). In this study, we investigated the function of OsREC8 by characterizing null mutants of Osrec8 and demonstrated its involvement in meiotic division. Our results suggest that OsREC8 is required for chromosome axis building and bouquet formation, as well as homologous chromosome pairing, synapsis, and recombination. Furthermore, OsREC8 is also necessary for meiotic centromere cohesion and monopolar orientation of kinetochores at meiosis I. Thus, OsREC8 plays several crucial roles in these meiotic processes that are critically different from mitosis.
RESULTS
Morphological Characterization of a Sterile Mutant
We identified one completely sterile mutant from a japonica rice variety, Yandao 8, induced by 60Co γ-ray radiation. The mutant showed a normal phenotype in the vegetative stage and could not be distinguished from its wild type based on plant morphology (Fig. 1, A and B). However, after flowering, we found that all the mature pollen grains of the mutant plant were empty and shrunken (Fig. 1, C and D). Moreover, when homozygous plants were pollinated with wild-type pollens, the mutant flowers could not set any seeds, suggesting that the mutation also affected megagametogenesis.
Figure 1.
Characterization of the Osrec8-1 mutant phenotype. A, Comparison of a wild-type plant (left) and an Osrec8-1 mutant plant (right). B, Comparison of a wild-type panicle (left) and an Osrec8-1 mutant panicle (right). C, Fertile pollen grains stained with 1% I2-KI solution in a wild-type plant. D, Completely sterile pollen grains stained with 1% I2-KI solution in an Osrec8-1 plant. Bars = 50 μm.
Map-Based Cloning of the Mutated Gene Causing a Sterile Phenotype
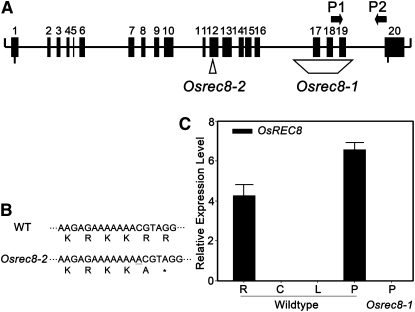
To isolate the mutated gene, we generated a large population by crossing heterozygous plant with Guangluai4, an indica rice variety. A total of 892 F2 segregates showing the complete sterile phenotype were used for map-based cloning. Linkage analysis mapped the gene on the long arm of chromosome 5, which was further delimited to a 62-kb region between the newly developed InDel markers AC120988-77 and AC120988-139. Sequencing analysis uncovered a truncated open reading frame of one candidate gene, Os05g50410, in the 62-kb region. A 1,390-bp deletion induced complete loss of three exons in AY371049 (GenBank; Fig. 2A), which encodes the rice OsRad21-4 protein, an ortholog of yeast Rec8 protein (Zhang et al., 2006). Thus, we named the mutant here Osrec8-1 and deduced that the rice Os05g50410 gene was responsible for its sterile phenotype. We also obtained another allele showing the same defects as Osrec8-1 from the progeny of rice tissue culture from a japonica rice variety, Nipponbare. Sequencing analysis revealed that this mutant contains a redundant nucleotide (A) that produces a premature termination codon (TAG) in exon 12 of AY371049 (Fig. 2, A and B) and was consequently designated as Osrec8-2. As Osrec8-1 showed severe mutant phenotype from the cytological analysis, we selected it for all subsequent experiments described below.
Figure 2.
OsRec8 encodes a rice OsRad21-4 protein. A, Position of mutated sites in Osrec8-1 and Osrec8-2 and gene-specific primers P1 and P2. B, Partial sequence of OsRec8 in the wild type and Osrec8-2 mutant. In Osrec8-2, a redundant A mutation in exon 12 introduces a stop codon (asterisk). C, Real-time PCR analysis of the OsREC8 gene. PCR was performed with first-strand cDNAs synthesized with total RNA from roots (R), culms (C), leaves (L), and panicles (P) of the wild-type plant and Osrec8-1 panicles.
By quantitative reverse transcription-PCR, we showed that the OsREC8 gene is expressed in roots and panicles of rice (Fig. 2C). Thus, OsREC8 expression is not meiosis specific, although the mutant phenotype is limited to reproductive organs. We did not detect any OsREC8 transcripts in the deletion allele Osrec8-1, just as expected.
Abnormal Meiosis Was Detected in Osrec8 Mutants
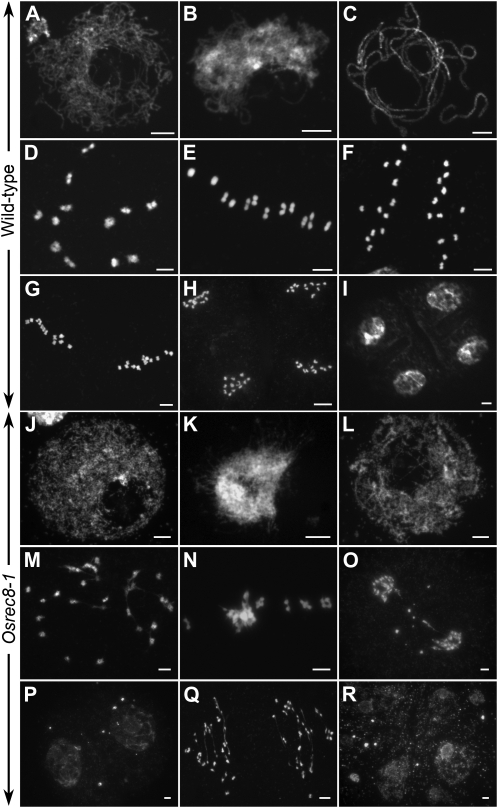
To determine how the OsREC8 mutation affects meiosis, we investigated the meiotic chromosomes in different stages of pollen mother cells (PMCs) from both the wild type and Osrec8-1 (Fig. 3). In the wild type, individual chromosomes appeared as thin threads at leptotene (Fig. 3A). During chromosome condensation, the homologous chromosomes came closer together at zygotene (Fig. 3B) and then became tightly associated and fully synapsed at pachytene (Fig. 3C). The chromosomes condensed further, and 12 bivalents were visible during diakinesis (Fig. 3D). The bivalents aligned on the equatorial plate in an ordered pattern at metaphase I (Fig. 3E). Subsequently, the homologous chromosomes separated and migrated in opposite directions at anaphase I (Fig. 3F). During the second meiotic division, the sister chromatids of each chromosome separated from each other, an event that was similar to that in mitosis and resulted in the formation of four daughter cells with 12 chromosomes each (Fig. 3, G–I).
Figure 3.
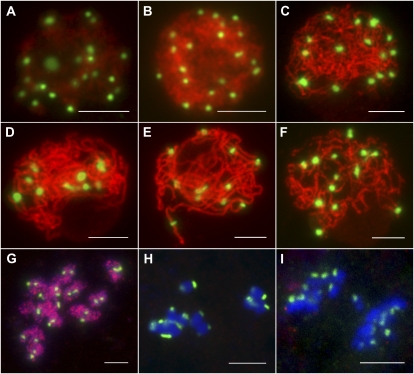
Meiosis in the wild type and the Osrec8-1 mutant. A to I, The wild type; J to R, the Osrec8-1 mutant. A and J, Leptotene. B and K, Zygotene. C and L, Pachytene. D and M, Diakinesis. E and N, Metaphase I. F and O, Anaphase I. G, Metaphase II. P, Telophase I. H and Q, Anaphase II. I, Tetrads. R, Polytrads. Bars = 5 μm.
In Osrec8-1, the chromosomal behavior at leptotene was similar to that in the wild type (Fig. 3J). The first detectable defects of chromosomal behavior appeared in zygotene. As shown in Figure 3K, sticky chromosomes were observed as a compact entangled mass with bright staining, and most regions of chromatins were highly condensed into agglomerations due to uneven condensation. During pachytene, the typical clear thick-threads were rarely observed, whereas the abnormal tangled chromosomes displayed many separated regions (Fig. 3L). Nevertheless, chromosome condensation and progression through the subsequent stages of meiosis occurred in Osrec8-1 PMCs. At diakinesis, 24 univalents were observed with some chromosome bridges (Fig. 3M). Intriguingly, most chromosomes were intertwined together and displayed a highly sticky phenotype when aligned on the equatorial plate at metaphase I (Fig. 3N). At anaphase I, we observed 20 to 24 chromosomes segregating toward the poles, suggesting there was premature separation of sister chromatids at the first meiotic division (Fig. 3O). In most cells, we also detected chromosome fragmentation, appearing as small dots in anaphase I and telophase I (Fig. 3, O and P). During meiosis II, we observed a large number of chromosome bridges that corresponded to prematurely released chromatids subjected to bipolar tension (Fig. 3Q). As a result, its meiotic products lacked the regular tetrahedral structure and were either asymmetric tetrads or polyads containing many micronuclei (Fig. 3R).
Osrec8 Mutants Show Defective Telomere Bouquet Formation
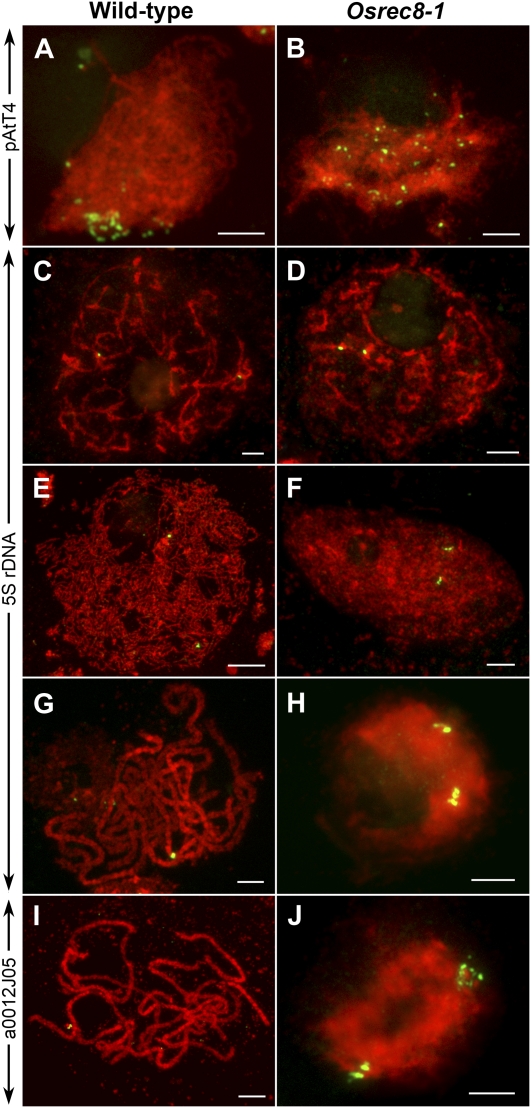
Telomeres attach to the inner nuclear envelope and cluster to form the so-called meiotic bouquet during meiotic prophase. Bouquet formation is a key event of a general repositioning of chromosomes and is thought to facilitate homologous chromosome pairing and synapsis (Harper et al., 2004; Zickler, 2006). To examine bouquet formation, we conducted fluorescence in situ hybridization (FISH) analysis using pAtT4 containing telomere-specific sequences as a probe. In the wild-type rice meiocytes, the telomeres attached to the nuclear envelope during leptotene and clustered in early zygotene (Fig. 4A). However, telomeres in the Osrec8-1 mutant meiocytes did not cluster to a certain region but were scattered throughout the nuclear volume (Fig. 4B), indicating that the bouquet formation was defective in the Osrec8-1 mutant.
Figure 4.
Chromosome behavior in male meiocytes in both the wild type and the Osrec8-1 mutant. A and B, Telomere bouquet analysis using FISH with the telomere-specific pAtT4 probe. C to J, Chromosome pairing and chromatid cohesion analyzed by FISH using both 5S rDNA and a0012J05 as probes, individually. C and D, Premeiotic interphase. E and F, Leptotene. G to J, Pachytene. The DAPI-stained chromosomes are in red. Bars = 5 μm.
Sister Chromatids Are Loosely Connected, But Centromeric Cohesion Cannot Be Maintained in Osrec8 Meiocytes
To determine whether sister chromatid cohesion is well established and maintained in Osrec8-1 mutant meiocytes, meiotic chromosomes around prophase I were probed together with 5S rDNA and a0012J05, a BAC clone located in the long arm of chromosome 8. The rice genome contains a single 5S rDNA locus located on the short arm of chromosome 11 (Zhang et al., 2005a). In wild-type meiocytes, the two unpaired 5S rDNA signals were visible at premeiotic interphase G2 (Fig. 4C). At leptotene, they could still be constantly detected (Fig. 4E). However, at pachytene, a single bright signal representing the paired 5S rDNA loci was observed in each cell (Fig. 4G). However, in Osrec8-1 mutant meiocytes, there were also two 5S rDNA signals located on the two unpaired chromosome 11 at premeiotic interphase G2 (Fig. 4D), which were similar to that in the wild type. At leptotene, the two 5S rDNA signals were also unpaired in Osrec8-1, while there were three different cases at this stage. In the first case, each 5S rDNA signal was a single dot, just like that in the wild type (35 out of 78 PMCs). In the second case, only one 5S rDNA signal showed a single dot, but the other showed two adjacent dots (23 out of 78 PMCs). In the last case, the two 5S rDNA signals exhibited two adjacent dots (20 out of 78 PMCs; Figure 4F). From zygotene to pachytene, we found the two 5S rDNA signals on chromosome 11 were always separated, each of them with two adjacent signal dots (Fig. 4H). Those observations indicated that homologous chromosomes did not pair in Osrec8-1; nonetheless, its sister chromatid cohesion could not be maintained well after the onset of leptotene. The same result was observed when using a0012J05 as the FISH probe. We suspected the sister chromatid cohesion was pretty much disrupted in Osrec8-1 mutant meiocytes, but the two sister chromatids were still connected by other unknown cohesins or interactions.
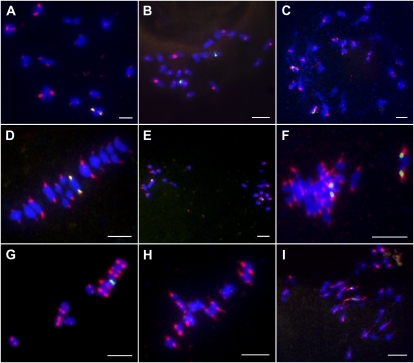
Meiotic centromeric cohesion was also evaluated by FISH analysis using CentO as the probe containing rice centromere-specific tandem repetitive sequences. The meiocytes at three different stages in meiosis, including diakinesis, metaphase I, and metaphase II, were investigated in the wild type, the Osrec8-1 mutant, and the pair2 mutant. In the wild type, 24 chromosomes were paired into 12 bivalents at diakinesis. There were two separated CentO signals of each bivalent, corresponding to the centromeres of two homologous chromosomes. However, for a single chromosome, the two centromeres of the duplicated chromatids were held together, showing centromere cohesion was well preserved at diakinesis (Fig. 5A). The meiotic centromeric cohesion could be well preserved at metaphase I (Fig. 5D) but was lost before metaphase II in the wild type (Fig. 5G). However, in Osrec8-1, we could not observe 12 bivalents at diakinesis during the lacking of homologous chromosomes pairing. We found there were two separated CentO signals of most univalents, which corresponded to the centromeres of two sister chromatids. The two centromeres of the replicated chromatids did not cohere, indicating that the centromere cohesion of sister chromatids had been lost before diakinesis (Fig. 5C). Moreover, we found the two CentO signals of each chromosome were always separated and pulled out by the microtubes emanating from different spindle poles during metaphase I (Fig. 5F).
Figure 5.
Dual-color FISH of the centromere probe CentO and 5S rDNA to chromosomes in wild-type, pair2, and Osrec8-1 meiocytes. Hybridization signals of CentO are in red and 5S rDNA are in green. Chromosomes were stained with DAPI (blue). A to C, Diakinesis in the wild type, pair2, and Osrec8-1, respectively. D to F, Metaphase I. G to I, One of the daughter cell at metaphase II. Bars = 5 μm.
To reveal whether the meiotic centromere cohesion defect in Osrec8-1 was caused by univalent formation, we also examined the centromere cohesion status in pair2 meiocytes. At diakinesis, only 24 univalents could be detected due to lack of homologous chromosome pairing, which were similar to those observed in Osrec8-1. However, there was only one CentO signal of each univalent in contrast to the two separated CentO signals of each single chromosome in Osrec8-1 meiocytes (Fig. 5B), indicating that centromere cohesion was retained at diakinesis, although univalent formation was abnormal in pair2. Similar to that in the wild type, the meiotic centromeric cohesion of sister chromatids in pair2 were maintained well at metaphase I (Fig. 5E) but were completely lost before metaphase II (Fig. 5H).
OsREC8 Is Required for the Monopolar Orientation of Kinetochore in Meiosis I
The success of meiosis relies on the monopolar orientation of sister kinetochores at metaphase I until the onset of anaphase I (Hauf and Watanabe, 2004; Chelysheva et al., 2005). Cytological observations on meiocytes of Osrec8-1 showed that sister chromatids prematurely segregated toward the opposite poles from metaphase I to anaphase I transition. To accurately investigate whether OsREC8 plays a critical role in monopolar orientation, we monitored the kinetochore behavior from metaphase I to anaphase I transition using FISH probed with centromere-specific tandem repetitive sequence CentO in both wild-type and Osrec8-1 meiocytes. In wild-type meiocytes, two individual CentO signals denoting the centromeres of each pair of bivalents were clearly observed at metaphase I when 12 pairs of bivalents aligned on the equatorial plate (Fig. 5D). This observation indicated that centromeric cohesion was preserved, and the two sister chromatids of each chromosome could migrate to the same pole as a unit. In other words, a pair of sister kinetochores must be attached to microtubules emanating from the same spindle pole. In S. cerevisiae, a protein complex (the monopolin) is necessary for establishing this monopolar attachment (Tóth et al., 2000; Rabitsch et al., 2003). In fact, this monopolar orientation is only retained in reductional division when the bivalents are aligned on the equatorial plate in metaphase I. At the metaphase II/anaphase II transition, each kinetochore of a pair of sister chromatids binds microtubules facing the opposite spindle pole, accompanied by the loss of sister chromatid cohesion at the centromeres and leading to the faithful segregation of sister chromatids. As shown in Figure 5G, two separated red signals corresponding to the centromeres of replicated sister chromatids of each single chromosome were visible at metaphase II in wild-type rice meiocytes.
In Osrec8-1, we could not clearly detect the 24 centromere signals resembling that of the wild type during metaphase I and metaphase II. Due to defective homologous pairing, for each univalent, two separate red signals corresponding to the centromeres of sister chromatids were found at the metaphase I/anaphase I transition, suggesting that sister chromatid kinetochores separated under the pulling force of the bipolar microtubules at the first meiotic division when OsREC8 was deficient. We failed to detect 48 sister chromatid centromere signals accurately because of the sticky nature of most chromosomes (Fig. 5F). The bipolar attachment of sister chromatid kinetochores at metaphase I in Osrec8-1 meiocytes was confirmed by FISH using 5S rDNA probe, which localized very closely to the centromere region of rice chromosome 11. Four 5S rDNA signals, each two migrating to the opposite poles, were typically observed at metaphase I in Osrec8-1, rather than only two 5S rDNA signals in wild-type meiocytes. This result suggested that OsREC8 was required for the monopolar orientation of the two sister chromatid kinetochores at metaphase I. Interestingly, we could detect some overt thin thread-like signals when the kinetochore of a single sister chromatid was dragged from bipoles at the metaphase II/anaphase II transition (Fig. 5I). Thus, in Osrec8 mutants, sister kinetochores have a bipolar instead of a monopolar orientation, transforming the first meiotic division into a mitotic one.
OsREC8 Localized to Meiotic Chromosomes from Approximately Interphase to Metaphase I
The distribution and chronology of the OsREC8 protein were investigated by immunofluorescent localization on wild-type microsporocytes using a polyclonal antibody against OsREC8 protein raised in a rabbit. The OsREC8 signal appeared in the nucleus beginning at approximately the meiotic interphase. Meiocytes at interphase displayed diffusing chromatin and OsREC8 labeling (Fig. 6A). During early leptotene, a strong OsREC8 signal was associated with the condensing chromatin (Fig. 6B). As meiocytes proceeded through leptotene to pachytene, the OsREC8 labeling was distributed over most of the chromosomes (Fig. 6, C–E). As the chromosome began to condense during diplotene, the OsREC8 signal faded and then shifted from chromosomes into nucleoplasm after diakinesis (Fig. 6, F and G). The OsREC8 signal disappeared completely as cells proceeded through metaphase I (Fig. 6H) and was no longer observed in chromosomes after the onset of anaphase I (Fig. 6I). Therefore, OsREC8 proteins were detectable in meiocytes from approximately interphase to metaphase I, and the results from our immunolocalization studies suggested that OsREC8 was localized preferentially along the entire length of the meiotic chromosomes.
Figure 6.
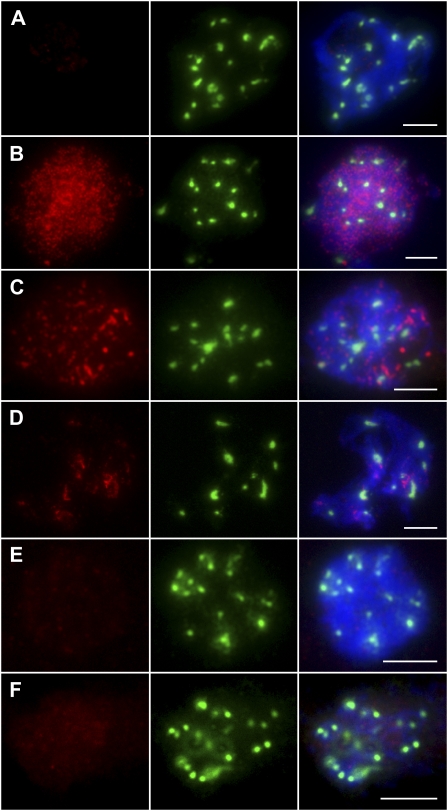
Immunofluorescent localization of OsREC8 (red) on meiotic spreads of wild-type rice chromosomes counterstained with DAPI (blue). The robust CENH3 signal (green) denoted these cells were rice PMCs. A, Interphase. B, Leptotene. C, Zygotene. D, Early pachytene. E, Pachytene. F, Diplotene. G, Diakinesis. H, Metaphase I. I, Anaphase I. Bars = 5 μm.
OsREC8 Is Essential for Localization of PAIR2, PAIR3, OsMER3, and ZEP1 But Is Not Required for OsAM1
As the deletion in Osrec8-1 resulted in the alteration of OsREC8 transcripts, it was important to determine whether the mutation also affected the synthesis of OsREC8 protein. As expected, OsREC8 was not detected in the meiocytes of the Osrec8-1 mutant by immunostaining (Fig. 7A), indicating that Osrec8-1 is a null mutant. To further investigate whether the depletion of OsREC8 also affected the distribution of OsAM1, PAIR2, PAIR3, OsMER3, and ZEP1, Osrec8-1 meiocytes were immunostained with antibodies against these five proteins. During leptotene and early zygotene in Osrec8-1, OsAM1 localized to the chromatin, forming punctuated foci, which resembled that in wild-type meiocytes (Fig. 7B). PAIR3 is an axis-associated protein in rice. In the Osrec8 mutant, PAIR3 localization was severely disrupted, and only a few discrete PAIR3 signals were observed on the chromosomes (Fig. 7D). Similar to PAIR3, PAIR2 was also shown as diffuse punctuated signals in Osrec8-1, and most of them did not localize to the chromatin correctly in early prophase I (Fig. 7C). Nevertheless, almost no OsMER3 signals or very faint ZEP1 signals were visible in the Osrec8-1 prophase I meiocytes (Fig. 7, E and F). These immunodetection experiments implied that OsREC8 is required for normal localization of PAIR2, PAIR3, MER3, and ZEP1, whereas the normal loading on the prophase I chromosomes of OsAM1 does not depend on OsREC8.
Figure 7.
Immunofluorescent localization of OsREC8, OsAM1, PAIR2, PAIR3, OsMER3, and ZEP1 antibodies (red) on meiotic spreads of Osrec8-1 chromosomes counterstained with DAPI (blue). Robust CENH3 signals (green) denoted that these cells were rice PMCs. A, OsREC8 at zygotene stage. B, OsAM1 at early zygotene. C, PAIR2 at zygotene. D, PAIR3 at zygotene. E, OsMER3 at zygotene. F, ZEP1 at zygotene. Bars = 5 μm.
OsREC8 Loading Is Independent of PAIR2, PAIR3, OsMER3, and ZEP1 But Relies on OsAM1
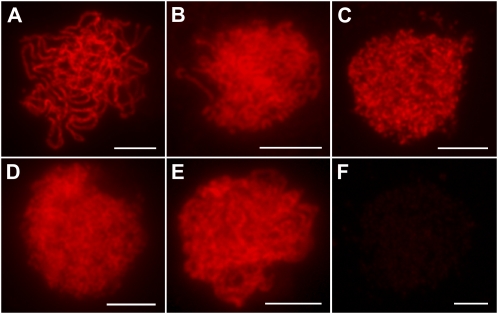
PAIR2, PAIR3, OsMER3, and ZEP1 are known to play crucial functions in axial element installation, recombination, and synaptonemal complex formation. As the localization of these important meiotic proteins was found to be affected by the absence of OsREC8 in the experiments above, we also carried out immunodetection of OsREC8 in pair2, pair3, Osmer3, and zep1 mutants to further characterize the relationship between these proteins. We found that OsREC8 proteins were normally loaded on the chromosomes and behaved in a similar manner to that in wild-type meiocytes at prophase I (Fig. 8). However, only very faint OsREC8 signals were detected in the Osam1 mutant. These results implied that the normal loading of OsREC8 is independent of PAIR2, PAIR3, OsMER3, and ZEP1 but relies on OsAM1.
Figure 8.
Immunofluorescent localization of OsREC8 in wild-type and different mutant prophase I meiocytes. A, The wild type. B, pair2. C, pair3. D, Osmer3. E, zep1. F, Osam1. Bars = 5 μm.
DISCUSSION
OsREC8 Is Essential for Chromatid Cohesion in Rice Meiosis
In all model organisms used in the study of meiosis, the cohesion sister chromatid cohesion 1 is mostly replaced by its meiotic equivalent, REC8, allowing the cohesin complex to fulfill their meiotic functions. During anaphase of the first meiotic division, the dissociation of REC8 from chromosome arms releases the arm cohesion, allowing homologous chromosomes to segregate, whereas centromeric cohesion is maintained until anaphase II owing to the protection of REC8 from cleavage by SGO1, which was isolated in yeast first and later in many other organisms (Rabitsch et al., 2003; Marston et al., 2004).
In this study, sister chromatids in Osrec8 mutants separated at anaphase I, indicating that OsREC8 is absolutely necessary in rice for maintenance of cohesion at centromeres at anaphase I. Actually, we found that centromeric cohesion was already lost before metaphase I, such as at diakinesis in Osrec8-1 meiocytes. These data suggest that OsREC8 is essential for the meiotic centromeric cohesion from its establishment at about interphase to its dissociation before anaphase I.
We found the sister chromatid cohesion could not be maintained well after the onset of leptotene and suspected the sister chromatid cohesion was pretty much disrupted in Osrec8-1 mutant meiocytes. However, we could not observe the complete loss of cohesion during late prophase I as observed in Arabidopsis switch1 mutants (Mercier et al., 2001) or C. elegans rec8/spo11 double mutants (Pasierbek et al., 2001). In Osrec8-1 meiocytes, a pair of sister chromatids of each univalent aligned on the equatorial plate, attached by microtubules emanating from the opposite pole at metaphase I. Since centromeric cohesion was absent in Osrec8 mutants, we proposed that the chromosomal arms of sister chromatids were still held by other cohesins or interactions. In fact, the two replicated chromatids are already held together by cohesin elements once entering the G2 stage of meiotic interphase. We suspect that those established cohesion might already perform their function before OsREC8 is installed onto meiotic chromosomes as meiosis starts. Of course, other chromosome interactions might also assist in holding sister chromatids together in the absence of OsREC8, such as sister crossovers or interlocks (Wang et al., 2009). Therefore, OsREC8 is essential for chromatid cohesion in rice meiosis.
OsREC8 Is Required for the Monopolar Orientation of the Two Sister Chromatid Kinetochores at Metaphase I
Kinetochores are complexes of proteinaceous structures, which are assembled on chromosomes and represent the major point of contact between spindle microtubules and centromeres. In contrast to back-to-back orientation of sister kinetochores in mitosis, kinetochore geometry in meiosis I is side-by-side arrangement of sister kinetochores (Hauf and Watanabe, 2004). Although Rec8 is thus far known to be conserved in all eukaryotes, whether its function in monopolar orientation of sister kinetochores is conserved has not been determined. It has been shown in S. pombe that Rec8 is required for kinetochores to be oriented to the same pole (Watanabe and Nurse, 1999; Yokobayashi et al., 2003). However, the replacement of Rec8 by Scc1 in S. cerevisiae preserves the monopolar orientation, showing that at least in this organism, Rec8 is not obligatory for monopolar attachment of kinetochores (Tóth et al., 2000). In the absence of maize or Arabidopsis REC8, sister chromatids apparently establish bipolar attachment, suggesting that REC8 in maize and Arabidopsis is also required for monopolar orientation (Chelysheva et al., 2005; Golubovskaya et al., 2006). Here, we show that sister chromatids in Osrec8 mutants prematurely separate at anaphase I, indicating that OsREC8 is absolutely necessary for monopolar orientation of sister kinetochores at metaphase I in rice. In Osrec8-1, that all the univalents align on the metaphase I plate and are subjected to a bipolar tension leading to a mitotic-like division demonstrates that OsREC8 disruption leads to a bipolar orientation of kinetochores in rice. In fission yeast, a meiosis-specific kinetochore protein, Moa1, physically interacts with REC8 and enables cohesion-mediated monopolar attachment at meiosis I (Yokobayashi and Watanabe, 2005). The S. cerevisiae kinetochore-associated protein MAM1 is also essential for monopolar attachment and controls orientation of the sister chromatid kinetochores during meiosis I (Tóth et al., 2000). However, in rice, whether OsREC8 is sufficient for monopolar orientation of the sister kinetochores and how OsREC8 affects kinetochore behavior during meiosis I remain to be further elucidated.
OsREC8 Establishes a Platform for Homologous Recombination and Synapsis during Early Prophase I
REC8 has been shown to be involved in homologous pairing, synapsis, or recombination during early prophase I for most model organisms used to date. In yeast, Rec8 loading is required for formation of synaptonemal complex and is necessary for recombination between homologs (Klein et al., 1999). In Arabidopsis, SYN1 plays essential roles in chromosome condensation and homologous pairing during meiosis (Bai et al., 1999; Cai et al., 2003). Chromosome synapsis at pachytene was defective when C. elegans REC8 was depleted by RNAi (Pasierbek et al., 2001). Interestingly, the absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis (Xu et al., 2004). In maize, AFD1 controls homologous pairing through its roles in axial element elongation and the subsequent distribution of the recombination machinery (Golubovskaya et al., 2006).
Here, Osrec8 mutants showed defective telomere bouquet formation and homologous pairing, indicating that OsREC8 occupies a critical role in regulating chromosome structure and functions during meiosis. To further dissect the functions of OsREC8 during early prophase I, we performed immunodetection of PAIR2, PAIR3, OsMER3, and ZEP1 in Osrec8 mutants and investigated the distribution of OsREC8 protein in pair2, pair3, Osmer3, and zep1 mutants. PAIR3 is an axis-associated protein in rice, and it localizes to the chromosome core during prophase I and associates with both unsynapsed axial elements and synapsed lateral elements (Wang et al., 2011a). PAIR2 plays an essential role in promoting homologous chromosome synapsis, and its distribution can well reflect the distribution of axial elements in early stages as a protein that associates with axial elements in rice (Nonomura et al., 2006). OsMER3, a ZMM protein, has an important role in the formation of COs and may directly participate in double strand break repair in rice (Wang et al., 2009). ZEP1, a transverse filament protein, is the central element of the synaptonemal complex (Wang et al., 2010). We found that PAIR2, PAIR3, OsMER3, and ZEP1 could not load on chromosomes normally in Osrec8, whereas OsREC8 could load on chromosomes normally in pair2, pair3, Osmer3, or zep1 mutants, implying that OsREC8 is a master regulator in axial element formation, homologous recombination, and synapsis by regulating its downstream proteins. Additionally, OsREC8 signals appeared on meiotic chromosomes at approximately interphase just prior to the progression of recombination and synapsis. Therefore, we infer that OsREC8 establishes a platform for homologous recombination and synapsis during early prophase I in rice.
In summary, the characterization of Osrec8 mutants in our study has helped to unravel the roles of OsREC8 in sister chromatid cohesion, kinetochore orientation, and several other important prophase I events. As one of the earliest meiosis-specific proteins to be emerged, OsREC8 plays a master role in faithful transition of mitosis into meiosis.
MATERIALS AND METHODS
Plant Materials
The 60Co γ-ray radiation-induced mutant Osrec8-1 was found in a japonica rice (Oryza sativa) variety, Yandao 8. As Osrec8-1 is completely sterile for both male and female gametes, OsRec8-1+/−was crossed with Guangluai4 to make mapping populations. F2 segregates showing the completely sterile phenotype were used for fine mapping. Another allele showing the same defects as Osrec8-1 was obtained from the progeny of rice tissue culture from a japonica rice variety, Nipponbare, and was designated as Osrec8-2. Yandao 8 was used as a wild-type control for all experiments. All plant materials were grown in the paddy fields.
Molecular Cloning of OsREC8
To fine map OsREC8, many sequence-tagged-site markers were developed based on sequence differences between indica variety 9311 and japonica variety Nipponbare according to the data published in http://www.ncbi.nlm.nih.gov. The candidates were delimited between AC120988-77 (forward, 5′-TGCATGAATTGGTGAGTAAA-3′; reverse, 5′-TTGCTTT ATGCCTTAAAGCT-3′) and AC120988-139 (forward, 5′-AGCATTATACGAAGGG CATG-3′; reverse, 5′-GTTAAGTCTGAAGTGAAGTTG-3′).
Real-Time PCR for Transcript Expression Assay
Total RNA was extracted from the stem, leaf, root, and panicle of Nipponbare. Real-time PCR analysis was performed using the Chromo4 real-time PCR instrument (CFD-3240; Bio-Rad) and SYBR Green I (Invitrogen). RT-PCR was performed using P1 (5′-TCCACTCGTACCTCAAGCTA-3′) and P2 (5′-GTTGCTAAAACGCATGCTTG-3′) for OsREC8 and UBQF (5′-CAAGATGATCTGCCGCAAATGC-3′) and UBQR (5′-TTTAACCAGTCCATGAACCCG-3′) for ubiquitin. The real-time PCR results were analyzed using Opticon Monitor analysis software 3.1 (Bio-Rad). The experiment had three replicates.
Antibody Production
To generate the antibody against OsREC8, a 747-bp fragment of OsREC8 (amino acids 139–384) was amplified from AY371049.1 (GenBank) with primer OsREC8GST. After being cloned into the pGEM-T vector and sequenced, this fragment was ligated into the expression vector pGEX-4T-2 (Amersham) digested with EcoRI-XhoI. This expression vector was transformed into BL21 (DE3) and was induced by addition of 0.3 mm isopropylthio-β-galactoside to the culture medium. The OsREC8 fusion peptides, mainly expressed in the soluble fraction, were purified using the MagnetGST Protein Purification System (Promega). Polyclonal antibodies of OsREC8 were raised in both rabbits and mice.
Meiotic Chromosome Preparation
Young panicles of both the wild type and Osrec8-1 were harvested and fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1). Microsporocytes undergoing meiosis were squashed in an acetocarmine solution. Slides with chromosomes were frozen in liquid nitrogen. After removing the coverslips, the slides were dehydrated through an ethanol series (70%, 90%, and 100%). Chromosomes were counterstained with 4′,6-diamino-phenylindole (DAPI) in an antifade solution (Vector Laboratories). Chromosome images were captured under the Olympus BX61 fluorescence microscope with a microCCD camera.
FISH
FISH analysis was conducted as described (Zhang et al., 2005a). The pTa794 clone containing the coding sequences for the 5S rRNA genes was used as one FISH probe to monitor the short arm of chromosome 11. a0012J05, a BAC clone that located on the long arm of chromosome 8, was used as FISH probe to show the chromatid cohesion of chromosome arms. The rice centromere-specific tandem repetitive sequence CentO was used as the other FISH probe to monitor centromeric regions (Zhang et al., 2005b). Chromosomes and FISH signal images were captured under the Olympus BX61 fluorescence microscope with a microCCD camera.
Immunofluorescence
Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature (25°C). Anthers in the proper stage were squashed on a slide with phosphate-buffered saline solution and covered with a coverslip. After soaking in liquid nitrogen and removing the coverslip, the slide was dehydrated through an ethanol series (70%, 90%, and 100%) prior to being used in immunostaining. Slides were then incubated in a humid chamber at 37°C for 4 h in different antibody combinations diluted 1:500 in TNB buffer (0.1 m Tris-HCl, pH 7.5, 0.15 m NaCl, and 0.5% blocking reagent). After three rounds of washing in phosphate-buffered saline, Texas red-conjugated goat anti-rabbit antibody and fluorescein isothiocyanate-conjugated sheep anti-mouse antibody (1:1,000) were added to the slides. The chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories). Immunolocalization using antibodies against OsREC8, PAIR2, PAIR3, OsMER3, OsAM1, and ZEP1 were performed as previously described (Wang et al., 2009).
References
- Bai X, Peirson BN, Dong F, Xue C, Makaroff CA. (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. (2004) Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 40: 184–194 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Cai X, Dong F, Edelmann RE, Makaroff CA. (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116: 2999–3007 [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, et al. (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118: 4621–4632 [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. (2003) Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. (1997) Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell 8: 957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J. (2004) Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell 15: 951–964 [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Meyer BJ. (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4: 520–534 [DOI] [PubMed] [Google Scholar]
- Hamant O, Golubovskaya I, Meeley R, Fiume E, Timofejeva L, Schleiffer A, Nasmyth K, Cande WZ. (2005) A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr Biol 15: 948–954 [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. (2004) A bouquet of chromosomes. J Cell Sci 117: 4025–4032 [DOI] [PubMed] [Google Scholar]
- Hauf S, Watanabe Y. (2004) Kinetochore orientation in mitosis and meiosis. Cell 119: 317–327 [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Watanabe Y. (2007) Chromosome cohesion in mitosis and meiosis. J Cell Sci 120: 367–369 [DOI] [PubMed] [Google Scholar]
- Jessberger R. (2002) The many functions of SMC proteins in chromosome dynamics. Nat Rev Mol Cell Biol 3: 767–778 [DOI] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. (2004) Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol 14: 560–572 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. (2005) Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol 15: 353–359 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. (2003) Distinct cohesin complexes organize meiotic chromosome domains. Science 300: 1152–1155 [DOI] [PubMed] [Google Scholar]
- Kleckner N. (1996) Meiosis: How could it work? Proc Natl Acad Sci USA 93: 8167–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103 [DOI] [PubMed] [Google Scholar]
- Lee JY, Orr-Weaver TL. (2001) The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol 17: 753–777 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. (2004) A genome-wide screen identifies genes required for centromeric cohesion. Science 303: 1367–1370 [DOI] [PubMed] [Google Scholar]
- Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefèvre F, Pelletier G, Horlow C. (2001) SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. (1994) Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28: 167–187 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745 [DOI] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Eiguchi M, Suzuki T, Kurata N. (2006) PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J Cell Sci 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JH, Kohli J. (1999) Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol 19: 3515–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. (2003) Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4: 535–548 [DOI] [PubMed] [Google Scholar]
- Revenkova E, Jessberger R. (2006) Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma 115: 235–240 [DOI] [PubMed] [Google Scholar]
- Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K. (2000) Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell 103: 1155–1168 [DOI] [PubMed] [Google Scholar]
- Uhlmann F. (2004) The mechanism of sister chromatid cohesion. Exp Cell Res 296: 80–85 [DOI] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, et al. (2009) MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J Cell Sci 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Qin B, Li M, Cheng Z. (2011a) PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol Biol Cell 22: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tang D, Wang K, Shen Y, Qin B, Miao C, Li M, Cheng Z. (2011b) OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z. (2010) The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. (2005) Sister chromatid cohesion along arms and at centromeres. Trends Genet 21: 405–412 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. (1999) Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464 [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley M, Verschoor S, Inselman A, Handel MA, McKay MJ. (2004) A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep 5: 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. (2005) The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817 [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. (2003) Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol 23: 3965–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Yang Q, Bao W, Zhang Y, Han B, Xue Y, Cheng Z. (2005a) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tao J, Wang S, Chong K, Wang T. (2006) The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60: 533–554 [DOI] [PubMed] [Google Scholar]
- Zhang W, Yi C, Bao W, Liu B, Cui J, Yu H, Cao X, Gu M, Liu M, Cheng Z. (2005b) The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol 139: 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. (2006) From early homologue recognition to synaptonemal complex formation. Chromosoma 115: 158–174 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]