Abstract
Barley (Hordeum vulgare) and wheat (Triticum aestivum) possess significant phytase activity in the mature grains. Maize (Zea mays) and rice (Oryza sativa) possess little or virtually no preformed phytase activity in the mature grain and depend fully on de novo synthesis during germination. Here, it is demonstrated that wheat, barley, maize, and rice all possess purple acid phosphatase (PAP) genes that, expressed in Pichia pastoris, give fully functional phytases (PAPhys) with very similar enzyme kinetics. Preformed wheat PAPhy was localized to the protein crystalloid of the aleurone vacuole. Phylogenetic analyses indicated that PAPhys possess four conserved domains unique to the PAPhys. In barley and wheat, the PAPhy genes can be grouped as PAPhy_a or PAPhy_b isogenes (barley, HvPAPhy_a, HvPAPhy_b1, and HvPAPhy_b2; wheat, TaPAPhy_a1, TaPAPhy_a2, TaPAPhy_b1, and TaPAPhy_b2). In rice and maize, only the b type (OsPAPhy_b and ZmPAPhy_b, respectively) were identified. HvPAPhy_a and HvPAPhy_b1/b2 share 86% and TaPAPhya1/a2 and TaPAPhyb1/b2 share up to 90% (TaPAPhy_a2 and TaPAPhy_b2) identical amino acid sequences. despite of this, PAPhy_a and PAPhy_b isogenes are differentially expressed during grain development and germination. In wheat, it was demonstrated that a and b isogene expression is driven by different promoters (approximately 31% identity). TaPAPhy_a/b promoter reporter gene expression in transgenic grains and peptide mapping of TaPAPhy purified from wheat bran and germinating grains confirmed that the PAPhy_a isogene set present in wheat/barley but not in rice/maize is the origin of high phytase activity in mature grains.
Phytases (myoinositol hexakisphosphate phosphohydrolase; EC 3.1.3.26 and EC 3.1.3.8) are phosphatases that initiate the sequential liberation of orthophosphate groups from phytate (myoinositol 1,2,3,4,5,6-hexakisphosphate). Hereby, phosphate, inositol phosphates, and inositol are provided for a range of cellular activities (Brinch-Pedersen et al., 2002). A number of enzymes with phytase activity are known from plants, animals, and microorganisms (Dvoráková, 1998). They are classified according to their catalytic mechanism as belonging to the histidine acid phosphatases (HAPs), purple acid phosphatases (PAPs), Cys phosphatases, or β-propeller phosphatases (Lei et al., 2007). Each group consists of several phosphatases, but only a few of them have phytase activity. In plants, only phytases belonging to the HAP and PAP groups have been described.
The HAPs constitute a large group of enzymes that share the catalytic mechanism as an N-terminal RHGXRXP motif and a C-terminal HD motif position together and form the active site (Lei et al., 2007). The PAPs are metallohydrolases that bind two metal ions in the active center. One of the ions is usually iron III, while the second metal in plant PAPs can be zinc, manganese, or iron II. The ions are responsible for the coloring of the enzyme (Vogel et al., 2006). PAPs with phytase activity appear to be restricted to plants.
Phytases are of particular importance during seed germination, where they mobilize phosphate from phytate, the major reserve of phosphorus in plant seeds, accounting for approximately 70% of the total phosphorus (Lott, 1984). Different plant species have developed various strategies for phytase-mediated degradation of phytate during germination. Among the cereals, barley (Hordeum vulgare), wheat (Triticum aestivum and Triticum durum), and rye (Secale cereale) synthesize and accumulate significant amounts of phytase during grain development as well as during germination, and the mature grains possess a significant level of preformed phytase activity (Eeckhout and De Paepe, 1994). The preformed phytase launches the first wave of phytate hydrolysis during early germination. Other cereals, like maize (Zea mays) and rice (Oryza sativa), possess little or virtually no preformed phytase activity in the mature grains and depend fully on de novo synthesis during germination (Eeckhout and De Paepe, 1994).
The spatial and temporal regulation of phytase biosynthesis in plant seeds has profound effects on phosphate bioavailability when dry grains are used as food and feed. Monogastric animals like pig, poultry, and human have little or no phytase activity in their digestive tracts and, in most cases, the preformed phytase potential of the mature grain is inadequate for phytate degradation. In consequence, most phytate is excreted, adding to the phosphate load on the environment in areas with intense livestock production. The low phosphate bioavailability in feed based on dry grain furthermore necessitates large-scale feed supplementation with rock phosphate. This practice is not sustainable, as phosphate is a nonrenewable resource that will be depleted within a few decades (Steen, 1998). To alleviate these problems, microbe-derived phytase is commonly added to the feed in areas with intense pig and poultry production (Brinch-Pedersen et al., 2002). Another strategy is to engineer plants for improved phytase activity in the seeds. Thus, increased phytase activities in transgenic soybean (Glycine max) and canola (Brassica napus) seeds reduced phosphorus secretion by 50% and 48% when fed to broilers and piglets, respectively (Denbow et al., 1998; Zhang et al., 2000).
Despite of their importance for basic plant processes and their significance for human and livestock nutrition, little is known about the molecular mechanisms regulating phytase formation during grain development and germination. However, several plant PAP phytases (PAPhys) have been purified to homogeneity or near homogeneity and biochemically characterized. Two phytases have been identified in mature grains of wheat (PHYI, approximately 66 kD; PHYII, approximately 68 kD), barley (P1 and P2, both 66 kD), and rice (F1, 66 kD; F2, 68 kD; Hayakawa et al., 1989; Nakano et al., 1999; Greiner et al., 2000). A wheat PHY sequence has been deposited in GenBank (AX298209), and a patent application describes it as a 66-kD PAPhy with the same temperature and pH optima as PHYI (Rasmussen et al., 2007). The first PAPhy gene described (GmPhy) was isolated from soybean and was observed to be expressed in the cotyledons of germinating seedlings (Hegeman and Grabau, 2001). In Medicago truncatula, a cDNA of a PAPhy has been isolated and found to be expressed in leaves and in roots as secreted enzymes contributing to the acquisition of organic phosphorus (Xiao et al., 2005). Finally, phytase activities have been detected in two Arabidopsis (Arabidopsis thaliana) proteins termed AtPAP15 and AtPAP23 (Zhu et al., 2005; Kuang et al., 2009).
Recently, the wheat and barley HAP genes TaPhyII and HvPhyII, encoding multiple inositol phosphate phosphatases, were cloned, and the proteins were expressed in Escherichia coli and biochemically characterized as phytases (Dionisio et al., 2007). A HAP phytase was identified and characterized in lily (Lilium longiflorum) pollen (Mehta et al., 2006). Maize has been reported to possess two genomic HAP-encoding sequences (PHYTI and PHYT2; Maugenest et al., 1997, 1999). Both maize genes were expressed preferentially in the rhizodermis, endodermis, and pericycle layers of the adult root.
In this study, we have cloned and characterized a series of PAP genes from wheat, barley, maize, and rice expressed during grain formation or germination. Two major PAP types, termed a and b, were identified. The genes were expressed in Pichia pastoris and the derived proteins shown to be efficient phytases. Promoter-reporter gene studies in transgenic wheat, peptide mapping, and expression analysis revealed that the genes and derived proteins expressed during grain formation preferentially are of the a type, while the b types preferentially are expressed during germination. This indicates that the PAP-derived phytase potential of a cereal grain comprises two different pools, one pool being synthesized and stored during grain filling and the other one being synthesized during germination.
RESULTS
Cloning of 12 Cereal PAP cDNAs
Databases were searched for the presence of wheat, barley, maize, and rice PAP sequences. Multiple alignments of the contigs allowed a common map of contigs (cluster) to be assembled. The clusters were subsequently used for the design of primers for the cloning of cDNAs for all isogenes. First-strand cDNA was synthesized from a pool of mRNAs isolated from developing and germinating grains. From wheat, two isogenes, TaPAPhy_a and TaPAPhy_b, were cloned, distinguished by different lengths of their open reading frames. For each isogene, two variants were found, differing by single nucleotide differences or insertions/deletions in the 3′ untranslated region. The four clones were named TaPAPhy_a1 (FJ973998), TaPAPhy_a2 (FJ973999), TaPAPhy_b1 (FJ974000), and TaPAPhy_b2 (FJ974001). In barley, three cDNAs, HvPAPhy_a (FJ974003), HvPAPhy_b1 (FJ974004), and HvPAPhy_b2 (FJ974005), were cloned. Two PAP sequences named ZmPAPhy_b (FJ974007) and OsPAPhy_b (HM0006823) were cloned from maize and rice, respectively. The open reading frames of the genes ranged from 1,611 to 1,653 bp and encoded proteins with 538 to 551 amino acids and predicted molecular masses from 57.2 to 59 kD (Supplemental Table S1). An additional cDNA was cloned from barley, HvPAP_c (FJ974006), due to its similarity to Arabidopsis PAP23, previously demonstrated to possess phytase activity (Zhu et al., 2005). Finally, wheat Ta_ACP (FJ974002) and maize PAP_c (FJ974008) were cloned for alignment purposes.
Phytase Activity and Biochemical Properties of Cereal PAPhys
TaPAPhy_a1, TaPAPhy_b1, HvPAPhy_a, HvPAPhy_b2, ZmPAPhy_b, and OsPAPhy_b proteins were produced in P. pastoris and their enzyme kinetics determined. The P. pastoris PHO1 phosphatase was repressed by 0.1 m phosphate buffer, and nontransformed P. pastoris showed no detectable secretion or cell wall-associated phosphatase or phytase activity during 5 d of culture. Also, P. pastoris transformed with the empty vector (pPICZ_alpha A) showed no phytase activity. Predicted endoplasmic reticulum (ER) signal peptides and potential C-terminal membrane retention signals were excised from the expression constructs (for details, see Supplemental Table S2), and recombinant (r-) proteins were secreted with yields from 1.5 to 20 mg L−1.
After gel filtration, the recombinant proteins appeared in two overlapping peaks at approximately 165 ± 5 and approximately 75 ± 3.2 kD (Supplemental Fig. S1A). Proteins isolated from both peaks were active against para-nitrophenylphosphate (p-NPP) and phytate. Endoglycosylase H (Endo H) deglycosylation reduced the number of peaks to one 66-kD peak (Supplemental Fig. S1B), indicating that P. pastoris produces the PAPhy as a monomer with differential degrees of glycosylation.
It is known that binuclear metallohydrolases can lose their active site ions during purification and that this can negatively affect enzyme activity (Waratrujiwong et al., 2006). To use a highly active enzyme preparation for the biochemical studies, purified r-TaPAPhy_a1 and b1 were incubated with a range of metals before enzyme activity measurements (Table I). Most metals had no effect on enzyme activity. However, for r-TaPAPhy_a1, incubation with Mn2+ increased the specific activity by approximately 12-fold. For r-TaPAPhy_b1, only Fe2+ gave a significant increase in specific activity (approximately 5-fold). The biochemical experiments were performed using Mn2+-activated r-PAPhy_a and Fe2+-activated r-PAPhy_b. Using phytate as substrate, Km values for the a isoforms were 35 and 36 μm for r-TaPAPhy_a1 and r-HVPAPhy_a, respectively (Table II). For the b isoforms, values ranged from 45 (wheat b1) to 54 (rice) μm. The kcat/Km values for r-TaPAPhy_a1 and r-HVPAPhy_a were 796 and 722 × 104 s−1 m−1, respectively. The b isoforms ranged from 428 (rice) to 600 (wheat b1) × 104 s−1 m−1. A collection of phosphorylated compounds were further tested as substrate for r-TaPAPhy_b1. The affinities against these were all substantially lower than for phytate (Table II).
Table I. Specific phytase activity of purified r-TaPAPhy_a1 and b1 without and with bivalent metal ions.
The enzymes and metals were incubated for 10 min at room temperature before assaying for phytase activity.
| Enzyme | No Metal | FeSO4 (10 mm) + Ascorbic Acid (3 mm) | FeCl3 (10 mm) | CaCl2 (10 mm) | MnSO4 (10 mm) |
| r-TaPAPhy_a1 activity (μmol phosphorus min−1 mg−1) | 18.45 ± 1.49 | 27.65 ± 2.45 | 17.45 ± 1.56 | 19.34 ± 2.34 | 223.5 ± 4.68 |
| Relative activity (%) | 100 | 150 | 95 | 105 | 1,210 |
| r-TaPAPhy_b1 activity (μmol phosphorus min−1 mg−1) | 36.24 ± 2.12 | 216 ± 5.11 | 32.45 ± 3.56 | 35.45 ± 3.41 | 41.30 ± 4.78 |
| Relative activity (%) | 100 | 584 | 90 | 98 | 114 |
Table II. Kinetics parameters of r-PAPhy enzymes.
The specific activities were determined at 36°C, pH 5.0. r-TaPAPhy_b1 and phytate were used for reference. All data were determined in triplicate.
| Substrate and Protein | Km | Vmax | kcat | kcat/Km |
| μm | μmol min−1 mg−1 | s−1 | ×104 s−1m−1 | |
| Phytate | ||||
| r-TaPAPhy_a1 | 35 ± 6.8 | 223 ± 9.4 | 279 | 796 |
| r-TaPAPhy_b1 | 45 ± 3.4 | 216 ± 12.4 | 270 | 600 |
| r-HvPAPhy_a | 36 ± 4.2 | 208 ± 6.8 | 260 | 722 |
| r-HvPAPhy_b2 | 46 ± 7.3 | 202 ± 9.9 | 253 | 550 |
| r-ZmPAPhy_b | 48 ± 6.9 | 198 ± 13.5 | 248 | 517 |
| r-OsPAPhy_b | 54 ± 8.2 | 185 ± 11.7 | 231 | 428 |
| r-TaPAPhy_b1 | ||||
| p-NPP | 1,917 ± 32.5 | 496 ± 11.0 | 620 | 32.34 |
| Fru-1,6-bisP | 921 ± 21.8 | 36 ± 3.5 | 45 | 4.89 |
| Phosphoenolpyruvate | 1,793 ± 33.4 | 256 ± 8.4 | 320 | 17.85 |
| Glycerol-1-phosphate | 1,650 ± 42.1 | 653 ± 14.3 | 816 | 49.47 |
| Pyrophosphate | 343 ± 12.8 | 22 ± 3.1 | 28 | 8.13 |
| Glc-6-P | 2,675 ± 64.3 | 22 ± 7.2 | 28 | 1.04 |
| Fru-1-P | 2,314 ± 54.7 | 24 ± 4.4 | 31 | 1.33 |
| Phospho-Ser | 2,560 ± 48.2 | 15 ± 2.2 | 19 | 0.75 |
| Adenosine triphosphate | 1,046 ± 132.3 | 111 ± 4.2 | 139 | 13.26 |
| Adenosine diphosphate | 1,377 ± 142 | 66 ± 3.2 | 83 | 6.03 |
| r-TaPAPhy_b1, phytate control | 100% | |||
| Pyridoxal-5-phosphate | 6% ± 1.2% | |||
| ortho-Carboxyphenyl phosphate | 45% ± 4.6% | |||
| Naphthyl phosphate | 68% ± 7.4% | |||
| Adenosine monophosphate | 11% ± 2.6% | |||
| Guanosine triphosphate | 210% ± 7% | |||
| Fru-6-P | Not detectable |
With phytate as substrate, the pH optimum was determined to 5.5 ± 0.14 for r-TaPAPhy_a1 and 5.0 ± 0.2 for r-TaPAPhy_b1 (Supplemental Fig. S2A). The pH stability range was investigated from pH 1 to 13. After 30 min at pH ≤ 2.8, both enzymes lost 95% of their initial activity, whereas 35% activity was retained at pH ≥ 12.5. Between pH 3.5 and 10, preincubation of the enzymes caused no enzyme activity loss. The temperature optimum curves were quite broad, with optima at 55°C ± 1.8°C and 50°C ± 2°C for r-TaPAPhy_a1 and r-TaPAPhy_b1, respectively (Supplemental Fig. S2B). Based on Arrhenius plots, the activation energies for phytate hydrolysis were calculated to 118.2 kJ mol−1 for r-TaPAPhy_a1 and 88.55 kJ mol−1 for r-TaPAPhy_b1.
The effects of metal ions on the r-TaPAPhy_b1 phytase activity were tested for several ions, in this case without activation by FeSO4 (Supplemental Fig. S3). Ferrous iron caused a strong induction of enzyme activity already at 0.03 mm FeSO4, whereas ferrous iron and manganese at concentrations above 5 mm caused phytate precipitation. The inhibition constants (Ki) of MoO42+, VO43+, Zn2+, Cu2+, and F− were 6, 20, 25, 78, and 1,245 μm, respectively. The Ki of phosphate was 7.2 mm when tested with p-NPP as substrate.
Phylogenetic and Structural Analysis of the PAPhy Proteins
The wheat PAPhy proteins shared 88% to 97% identity, with the largest difference between TaPAPhy_a1 and TaPAPhy_b2 (Supplemental Table S3). The barley PAPhys shared from 86% to 99% identity. Between wheat and barley PAPhys, the identity ranged from 87% between TaPAPhy_a1 and HvPAPhy_b2 to 93% between TaPAPhy_a2 and HvPAPhy_a. The sequence identities between the wheat and barley PAPhys and PAPhy from soybean (AAK49438), rice OsPAPhy (ADG07931), maize (ZmPAPhy_b; ACR23335) and Arabidopsis (AtPAP15; AAN74650) ranged from 64% for soybean GmPAPhy to 85% for rice OsPAPhy.
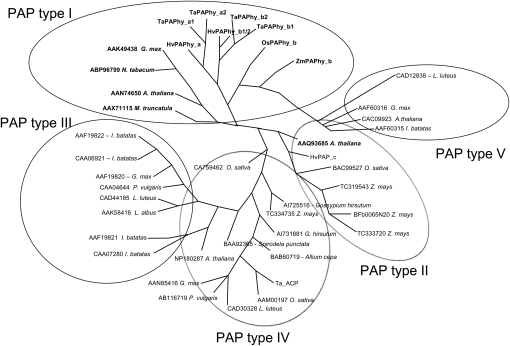
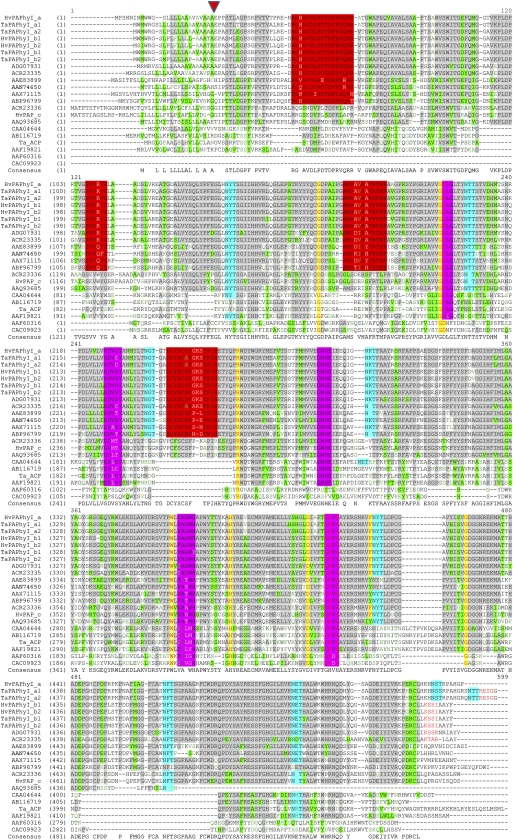
A phylogenetic tree of the protein sequences of the cloned wheat, barley, maize, and rice PAP genes, together with known and putative plant PAPhys and a large collection of existing GenBank and public ESTs of plant PAP sequences, is shown in Figure 1. The proteins group in five clades. Type I contains the PAPhy group, including TaPAPhy, HvPAPhy, ZmPAPhy, and OsPAPhys and the known PAPhys from M. truncatula (Xiao et al., 2005), Nicotiana tabacum (Lung et al., 2005), soybean (Hegeman and Grabau, 2001), and Arabidopsis (Zhang et al., 2008). Type II PAPs mainly consist of proteins that contain a signal peptide for chloroplast entry as predicted by ChloroP 1.1 (Emanuelsson et al., 1999) and one with a predicted ER signal peptide (i.e. AAQ93685). All type III PAPs are relatively short (470–490 amino acids) and have ER signal peptides typical of dimeric secreted or vacuolar PAPs (i.e. AAW29950 and CAA07280). Type III PAPs are induced by phosphorus starvation (Lu et al., 2008). Type IV contains the monomeric PAPs with either ER (CAD30328) or mitochondria signal peptides (TaPAP_c; ACR23330 and AAM00197). Type V comprises the small (about 35 kD) mammal-like PAPs (i.e. CAC09923). Alignment of the PAPhy protein sequences and representatives from the five clades of PAPs revealed that all possessed the characteristic PAP metalloesterase seven metal-binding residues (D, D, Y, N, H, H, H). These are contained in a conserved pattern of five consensus motifs [DxG/GDx2Y/GNH(E,D)/Vx2H/GHxH; Sträter et al., 1995). In addition to these sequences, all PAPs with phytase activity except Arabidopsis (AAQ93685) shared the following four consensus motifs: (1) R-G-(H/V/Q/N)-A-(V/I)-D-(L/I)-P-(D/E)-T-D-P-(R/L)-V-Q-R-(R/N/T); (2) S-(V/I)-V-(R/Q)-(Y/F)-G; (3) A-M-S-X-X-(H/Y)-(A/Y/H)-F-(R/K)-T-M-P; and (4) D-C-Y-S-C-(S/A)-F-X-X-X-T-P-I-H (Fig. 2). The potential involvement of the signature in a phytate-binding motif needs to be explored. However, the motifs may facilitate the identification of new PAPhys.
Figure 1.
Phylogenetic radial tree of plant PAP protein sequences with phytase activity (boldface) and without known phytase activity. Multiple alignment (ClustalW) and a parsimony algorithm were applied using the program Plylyp (http://evolution.genetics.washington.edu/phylip.html).
Figure 2.
Multiple alignments (ClustalW) of selected PAPs with or without known phytase activity. Gray shading, partial similarity; yellow shading, full similarity; green shading, weak similarity; purple-pink shading, PAP motifs; red shading, predicted PAPhy motifs; red letters, potential C-terminal ER-retention signals; cyan shading, potential N-linked glycosylation sites. The alignment includes all PAPhys represented in Figure 1 and at least two representatives from each of the five PAP types. A predicted signal peptide cleavage site is indicated by an arrowhead approximately 20 amino acids from the N termini for some of the PAPs. GenBank protein accession numbers are as follows: HvPAPhy_a, ACR23331; TaPAPhy_a1, ACR23326; TaPAPhy_a2, ACR23327; HvPAPhy_b1, ACR23332; HvPAPhy_b2, ACR23333; TaPAPhy_b1, ACR23328; TaPAPhy_b2, ACR23329; rice PAPhy_b, ADG07931; maize PAPhy_b, ACR23335; soybean PAPhy_b, AAE83899; Arabidopsis PAP15, AAN74650; M. truncatula PAPhy, AAX71115; Nicotiana tabacum PAPhy, ABP96799; maize PAP_c (type IV), ACR23336; HvPAP_c, ACR23334; Arabidopsis PAP_c (type IV), AAQ93685; kidney bean PAP group type III, CAA04644; kidney bean PAP type IV, AB116719; Ta_ACP, ACR23330; Ipomoea batatas PAP group type III, AAF19821; soybean PAP type V, AAF60316; Arabidopsis PAP type V, CAC09923.
A 21- to 22-amino acid N-terminal ER signal peptide was predicted for all HvPAPhy, TaPAPhy, ZmPAPhy, and OsPAPhy isoforms, indicating either vacuolar localization or secretion (Supplemental Table S1).
TaPAPhy_a1 and a2 contain nine potential N-linked glycosylation sites. Eight sites are found in HvPAPhy_a, HvPAPhy_b1, HvPAPhy_b2, and OsPAPhy_b, whereas TaPAPhy_ b1, TaPAPhy_b2, and ZmPAPhy_b contain seven potential sites (Supplemental Table S1). Thus, all appear to be heavily glycosylated.
Temporal and Spatial Expression of PAPhy
TaPAPhy and HvPAPhy expression in wheat and barley, respectively, was measured by quantitative reverse transcription (qRT)-PCR in developing grains at 15, 21, and 35 d postanthesis (DPA) and in grains after 2, 4, and 6 d of germination (Fig. 3). The developing grains were dissected into three fractions: (1) embryo; (2) endosperm squeezed out from the grain; and (3) a seed coat fraction consisting of the testa and pericarp together with the aleurone. The germinating grains were divided into three fractions: (1) early primary leaf; (2) early primary root; and (3) the residual fraction consisting of the germinated grain minus the primary root and leaf. In both species, a type isogenes were predominantly expressed during grain development, in particular in the embryo and seed coat at 15 and 21 DPA (Fig. 3). The a isogenes showed higher expression during grain development than the b isogenes. Limited expression of the a isogenes was observed during germination. In contrast, high levels of b isogene expression were seen in the early germinating grain, though not in the primary leaf and root.
Figure 3.
Expression of the wheat (A) and barley (B) PAPhy isogenes. Developing grains at 15, 21, and 35 DPA were dissected into three fractions, embryo (EM), endosperm (EN), and seed coat (SC), containing pericarp and aleurone. Germination grains were examined at 2, 4, and 6 d after germination (DAG) and were dissected into three fractions, early primary leaves (L), early primary root (R), and a fraction consisting of the germinated grain minus the primary leaf and root (S). Data represent averages of three biological repeats each in three technical repeats. Relative expression units have been transformed to expression fold (log2) relative to α2-tubulin expression (0-fold expression).
To provide additional support for the differential expression of the TaPAPhy_a and b isoforms, promoters from the TaPAPhy_a1 and TaPAPhy_b1 isogenes were isolated from a genomic library. Sequence comparison up to −474 bp upstream the ATG site showed only 31% identity, thus strongly supporting the differential expression of the isoforms. Moreover, TaPAPhy_a1-GUS and TaPAPhy_b1_GUS promoter-reporter gene constructs were introduced into transgenic wheat. In TaPAPhy_a1 transgenes, analysis of the developing seeds revealed a clear and distinct GUS staining in the scutellum and in the seed coat layers (Fig. 4, F and G), thus supporting the qRT-PCR expression data and the results obtained by peptide mapping (see below). No GUS staining was present in the endosperm of the TaPAPhy_a1-GUS transgenes. TaPAPhy_b2 caused no visible GUS staining in developing wheat grains, and no detectable staining was present in the negative control (data not shown).
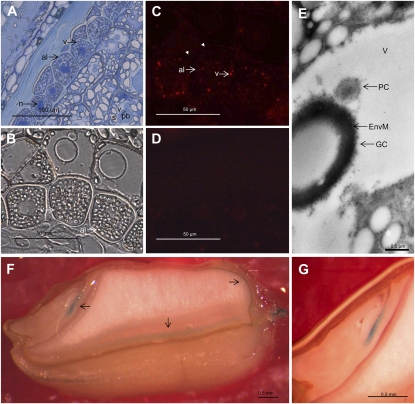
Figure 4.
Light (A–D, F, and G) and immunoelectron (E) microscopy analysis of the localization of PAPhy in the developing wheat grain, approximately 18 DPA. A, Toluidine blue-stained semithin cross-section of endosperm, aleurone, and pericarp tissues. B, Differential interference contrast microscopy with indications of the aleurone vacuoles. C, Immunofluorescence detection of PAPhy in 1-μm-thick sections. The aleurone vacuoles are clearly labeled, while there is no fluorescence from any other compartment of the cell, the apoplast (arrowheads), or other cell types. D, Immunofluorescence of a 1-μm-thick section incubated with secondary antibody only. There is virtually no background from the secondary antibody. E, Immunoelectron microscopy analysis showing an aleurone vacuole with gold labeling of protein crystalloid. F, Transgenic wheat grain transformed with a TaPAPhy_a1-GUS construct and showing GUS activity in the embryo and the seed coat fraction (arrows). G, GUS activity is restricted to the embryo scutellum. al, Aleurone; EnvM, globoid crystal-enveloping membrane; GC, globoid crystal; n, nucleus; pb, protein body; PC, protein crystalloid; s, starch; v, vacuole.
In addition, mature grains were analyzed to assess the presence of long-lived PAPhy transcripts. In both barley and wheat, mature grains possessed transcripts primarily of the a type isogenes, while there was a low content of the b types (Supplemental Fig. S4). This further supports the conclusion that the PAPhy genes in both barley and wheat are differentially expressed, the a type being expressed preferentially during grain development while the b type is expressed during germination.
Localization of TaPAPhy in the Grain
Western blotting of protein from mature wheat, barley, maize, and rice grains confirmed the presence of significant amounts of preformed PAPhy in wheat and barley (data not shown). Only very faint bands were seen in mature maize and rice grains. All the PAPhys were predicted to be either secreted or localized in the vacuole (Supplemental Table S1). To reveal the subcellular localization, immunofluorescence was performed on sections of fixed and embedded 18-DPA wheat grains (Fig. 4). Distinct labeling was seen in the vacuoles of the aleurone layer (Fig. 4C). There were no indications for the presence of larger amounts of TaPAPhy in other cell compartments or the apoplast. No signal was detected in the endosperm, and the secondary antibody caused no labeling of grain proteins. Electron microscopy provided a more detailed image of the distribution of TaPAPhy in the wheat aleurone vacuole (Fig. 4E). At least two types of inclusions are found in the aleurone cell protein storage vacuoles: I, the globoid crystal, which is surrounded by a characteristic enveloping membrane; and II, the protein crystalloid (Bethke et al., 1998). Abundant gold probes were found in the protein crystalloid of the aleurone vacuole.
Identification of Wheat Phytase in Mature and Germinating Grains by Tandem Mass Spectrometry Analysis
The phytase localized in the aleurone vacuole was purified from wheat bran, and phytase de novo synthesized during germination was purified from wheat grains germinated for 6 d. Reduced and alkylated samples of native and Endo H-deglycosylated TaPAPhys were subjected to four types of proteolytic digestion and tandem mass spectrometry (MS/MS) sequencing. Chymotryptic and tryptic digestions identified a number of peptides (Supplemental Fig. S5). Unique peptides confirmed the presence of TaPAPhy_a1, a2, and b1/b2 but did not distinguish the b isoforms in wheat bran (Supplemental Fig. S5A), whereas TaPAPhy_a1, a2, b1, and b2 were all distinguished in germinated wheat (Supplemental Fig. S5B). The relative concentration of these isoforms can be estimated by the abundance of unique peptides by the exponentially modified protein abundance index (emPAI) score (http://www.matrixscience.com/help/quant_empai_help.html). In wheat bran, the a isoforms dominated, with approximately 54% TaPAPhy_a1 and 35% TaPAPhy_a2 contributions to the total score. TaPAPhy_b1/b2 accounted for 12% of the score in wheat bran. In germinating grain, the b isoforms dominated, with 18% for TaPAPhy_b1 and 53% for TaPAPhy_b2, whereas TaPAPhy_a1 accounted for approximately 14% and TaPAPhy_a2 for 16%.
DISCUSSION
The first demonstration of plant PAPs as phytases was done in soybean (Hegeman and Grabau, 2001). Several studies have confirmed this. However, unraveling of a full plant phytase gene complement within the very large group of PAPs has so far been complicated due to the lack of motifs that could help distinguishing the phosphatases with phytase activity from the very large group of nonphytase phosphatases. Another complicating factor for compiling the plant phytase complement is that in a single plant species, the total PAPhys activity in developing and germinating grains is derived from a number of PAPhy isoforms with similar or very similar molecular mass and properties. In cereals, this is well known from barley, which synthesizes two 67-kD phytases (P1 and P2), and from rice, where 66-kD (F1) and 68-kD (F2) PAPhys have been purified (Hayakawa et al., 1989; Greiner et al., 2000). To achieve a detailed understanding of the PAPhy complement, the individual genes need to be isolated, their protein product properly characterized biochemically, and their temporal and spatial expression pattern clarified. Previous studies with PAPs from kidney bean (Phaseolus vulgaris) and soybean have suggested P. pastoris as a potential system for recombinant plant PAP production (Penheiter et al., 1998). In this study, P. pastoris was successfully established as a system for the production of functional enzymes of individual wheat, barley, maize, and rice PAPhy isogene candidates. The candidates were subsequently confirmed as being significant phytases, with significantly higher affinity against phytate than the multiple inositol phosphate phosphatase wheat and barley phytases described previously (Dionisio et al., 2007). Moreover, with a specific activity against phytate of approximately 200 μmol min−1 mg−1, the cereal PAPhys have the potential to compete with most bacterial and fungal phytases in hydrolyzing phytate (for detailed comparisons, see http://www.brenda-enzymes.org/php/result_flat.php4?ecno=3.1.3.8). This study thus combines the necessary molecular and biochemical techniques for the identification and evaluation of individual PAPhys candidates.
The PAPhy Clade
Phylogenetic analysis comprising 43 PAPs grouped the wheat, barley, maize, and rice PAPhy proteins together with a collection of plant PAPs with known phytase activity. The only example of a PAP with phytase activity that did not group in the PAPhy clade was the Arabidopsis PAP23 (Zhu et al., 2005), which grouped in the PAP type II clade. A common trait of the PAPhy group is the sharing of four consensus motifs. The potential roles of these motifs needs to be unraveled; however, in this study, they strongly facilitated the identification of wheat, barley, maize, and rice PAPhy candidates. The exact mechanism of PAPhy-mediated phytate hydrolysis remains elusive. Soybean PAPhy is proposed to be a homodimer (Hegeman and Grabau, 2001). However, in cereals, PAPhys purified from grains (Nakano et al., 1999; Greiner et al., 2000) and the current r-PAPhys were monomeric and still fully active phytases.
PAPhy_a and PAPhy_b during Grain Development and Germination
PAPhy genes have previously been described in soybean, M. truncatula, and Arabidopsis (Hegeman and Grabau, 2001; Xiao et al., 2005; Zhu et al., 2005; Kuang et al., 2009). In this study, the molecular and biochemical characteristics are described for four additional wheat, three barley, one maize, and one rice PAPhy genes. This has allowed a much more detailed understanding of the significance of PAPhy genes in the phytate metabolism of the cereal grain. Our findings after evaluation of P. pastoris-produced r-PAPhy reveal that all four species possess PAPhy genes, encoding fully functional phytases with very similar enzyme kinetics (Table II). The affinities of the r-PAPhys against phytate were higher than for any other physiologically important substrate tested and underline the importance of the PAPhys as phytases. However, in wheat and barley, the PAPhys comprised two similar gene families, termed PAPhy_a and PAPhy_b. Variants of PAPhys, termed P1 and P2, have previously been identified in barley (Greiner et al., 2000). P2 was reported as the sole phytase contributing to the preformed phytase activity of the dry grain, whereas P1 was active during germination. In this study, qRT-PCR analyses showed that in barley and wheat, PAPhy_a isogenes were predominantly expressed during grain development. In contrast, HvPAPhy_b and TaPAPhy_b genes were expressed mainly during germination and very little during grain development. In agreement with this, the RNA stored in the mature grain was derived from the PAPhy_a genes. In mature maize and rice grains, only PAPhy genes with the closest homology to the b type were identified. This is in agreement with the almost complete lack of preformed phytase activity in mature grains of these species. Promoter-GUS studies in transgenic wheat confirmed that TaPAPhy_a is expressed during development, in the scutellum and seed coat layers. TaPAPhy_b caused no visible GUS staining during grain development. Moreover, peptide mapping of PAPhy purified from wheat bran and from germinating grain confirmed that preformed TaPAP_a variants were abundant (88.3%) in bran, whereas TaPAP_b variants were predominant (70.4%) during germination and, therefore, de novo synthesized. Promoter isolations revealed that a and b isoform expressions are driven by different promoters.
Given the basic and applied potential of preformed and de novo-formed grain enzymes, surprisingly little is known about their synthesis, deposition, activation, and biochemical properties. One exception is β-amylases, known to be formed exclusively during grain filling (Zhang et al., 2006). Another exception is the lipoxygenases. In barley, they are synthesized in the embryo but are differentially regulated: lipoxygenase 1 is only formed during grain filling, while lipoxygenase 2 is synthesized exclusively during germination (Holtman et al., 1997; Rouster et al., 1998). This study demonstrates that for phytase in wheat and barley, PAPhy_a accounts for the synthesis of preformed phytase present in the mature grains. During germination, PAPhy is synthesized from the PAPhy_b genes. In maize and rice, where little or virtually no preformed phytase is present in the mature grain, only the PAPhy_b type has been identified. The highly conserved cDNAs of the PAPhy_a and PAPhy_b isogenes gave no indications of potential mechanisms regulating the differential expression of the PAPhy_a and PAPhy_b isogenes. However, in wheat, it was shown that TaPAPhy_a and TaPAPhy_b expressions are driven by different promoters.
TaPAPhy in the Wheat Grains
In small-grained cereals, approximately 90% of the grain phytate is accumulated in the aleurone layer and approximately 10% in the embryo (O’Dell et al., 1972). Almost all the phytate is present as phytin, a mixed salt (usually with K+, Ca2+, Mg2+, or Zn2+) that is deposited as globoid crystals in single membrane vesicles together with protein (Lott, 1984). This study indicates that wheat preformed PAPhy is localized in the vacuole protein crystal of the aleurone cell, close to its substrate phytin but not in the same type of inclusion. Previous studies on wheat myoinositol phosphate composition showed that grain phytin is not hydrolyzed during grain filling and storage (Brinch-Pedersen et al., 2003). The mechanism protecting phytin from hydrolysis during grain development and storage is not known; however, localization in different vacuolar inclusions may play a role. Another factor may be pH. r-TaPAPhy has close to zero activity at neutral pH, which is the approximate pH in the mature grain. In contrast, r-TaPAPhy is very active when the pH becomes acidic, which is the case when the vacuole becomes lytic during germination (Bethke et al., 1998).
CONCLUSION
In conclusion, wheat and barley, where preformed phytase activity is present in the mature grain, as well as maize and rice, with little or no preformed phytase activity in the mature grain, possess PAPhy genes encoding fully functional phytases with very similar enzyme kinetics. The PAPhy clade shares four consensus motifs that can be used for initial PAPhy identification followed by assaying after heterologous expression in P. pastoris. Preformed PAPhy in wheat grains is localized in the vacuole protein crystal of the aleurone layer, close to its substrate, phytate, but not in the same inclusion. For wheat and barley, PAPhy genes could be divided into two groups, termed PAPhy_a and PAPhy_b. In rice and maize, only the PAPhy_b type has been identified. Although HvPAPhy_a and HvPAPhy_b1/b2 share 86% identical amino acid sequence, and TaPAPhya1/a2 and TaPAPhyb1/b2 share up to 90% identity, PAPhy_a and PAPhy_b were differentially expressed during grain development and germination. In agreement with this, it was demonstrated in wheat that the a and b isogenes are driven by distinctly different promoters. TaPAPhy-promoter GUS studies in transgenic wheat and peptide mapping of TaPAPhy purified from bran and from germinating grains confirmed that preformed phytase activity in mature grains is constituted largely by the TaPAPhy_a isoforms, whereas phytases synthesized de novo during wheat grain germination are dominated by the TaPAPhy_b phytases.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Barley (Hordeum vulgare ‘Golden Promise’), wheat (Triticum aestivum ‘Bobwhite SH 98 26’), and maize (Zea mays genotype F7RR/RR; Uzarowska et al., 2009) were grown in the greenhouse according to Brinch-Pedersen et al. (2000). Indica rice (Oryza sativa ‘Himalaya’) was germinated on wet filter paper and leaves were harvested from approximately 5-cm plantlets. For qRT-PCR, grains were germinated on wet filter paper (daylight and room temperature).
Cloning, Sequencing, and Bioinformatics
Cloning primers (Supplemental Table S4) for wheat, barley, maize, and rice phosphatases and α-tubulin from barley and wheat were designed from sequence alignment of cDNA contigs. mRNA was isolated from germinating and developing grains, roots, and leaves using the Plant RNAeasy Kit (Qiagen) and the Dynabead T25 mRNA Isolation Kit (Invitrogen). First-strand cDNA was synthesized from a pool of mRNA from developing and germinating grains using oligo(dT)18N and SuperScript II-RT (Invitrogen). PCR on the single-strand cDNA was carried out by DNA polymerases Pfu Turbo (Promega) or Phusion (Finnzymes) using the following conditions: 95°C for 2 min and 36 cycles of 95°C for 1 min, 59°C for 1 min, and 72°C for 2 min. PCR products were cloned into the pCR Blunt vector (Invitrogen) or the EcoRV site of pBluescript II SK+ (Stratagene). Sequencing was carried out by Eurofins MWG Operon. Bioinformatics and sequence analyses were performed using the DNAstar (Lasergene) and VectorNTI 10 software (Invitrogen). SignalP version 3.0 was used for signal peptide predictions (Nielsen et al., 1997; Bendtsen, et al., 2004). Protein processing was predicted by TargetP version 1.1 (Emanuelsson et al., 2000). Potential phosphorylation sites were predicted by NetPhosK 1.0 (http://www.cbs.dtu.dk/services/NetPhosK/).
Expression in Escherichia coli for Antibody Production
TaPAPhy_b1 was selected for antibody production. The predicted signal peptide was excluded and an N-terminal His (His6) was included in the expression cassette (Supplemental Table S2). A new polylinker was introduced into the pET15b (Novagen) vector by annealing and ligating the upper 5′-TATGATCGATGAATTCAAGCTTGCGGCCGCCTCGAGG-3′ and lower 5′-ACTAGCTACTTAAGTTCGAACGCCGGCGGAGCTCCTAG-3′ oligonucleotides between the NdeI and BamHI sites of pET15b. The resulting vector contained the additional restriction sites ClaI, EcoRI, HindIII, NotI, and XhoI and was named pET15m. NdeI and HindIII sites were introduced to the 5′ and 3′ ends of TaPAPhy_b1 via PCR using the primers described in Supplemental Table S2 and Phusion DNA Polymerase (Finnzymes). The purified PCR product was digested and ligated into the NdeI and HindIII sites of pET15m. The constructs was verified by sequencing, transformed into E. coli strain Origami B pRARE 2 (DE3) pLysS (Novagen), which was grown in Overnight Express medium (Novagen), and induced with 0.2 mm isopropylthio-β-galactoside at 30°C for 6 h.
TaPAPhy_b1 was poorly soluble and had low phytase activity. Purification of recombinant proteins was carried out according to the pET System Manual, 10th edition (Novagen). The protein was dialyzed in 0.5 m Arg, 1 mm dithiothreitol (DTT), 50 mm Tris-HCl, and 1 mm EDTA, pH 7.5, using a 20-kD cutoff membrane. Protein concentrations were determined according to Bradford (1976).
Polyclonal antibodies was produced in rabbit by Agrisera (www.agrisera.com) using 1 mg of TaPAPhy_b1. Antiserum was affinity purified over an immobilized TaPAPhy_b1 column prepared with N-hydroxysuccinimidyl-activated Sepharose 4 Fast Flow (GE Healthcare). Affinity-purified antibody was specificity tested by preblocking it with TaPAPhy_b1 before western blotting and immunolocalization. Preblocked affinity-purified antibody gave no signal in western blotting or immunolocalization. Testing of the antibody on the recombinant PAPhys revealed that the specificity covered both a and b isoforms.
Expression in Pichia pastoris
r-TaPAPhy_a1, r-TaPAPhy_b1, r-HvPAPhy_a, r-HvPAPhy_b2, r-OsPAPhy_b, and r-ZmPAPhy_b were produced extracellularly in P. pastoris using pPICZ_alpha A (Invitrogen) in fusion with the alpha Mating Factor and driven by the alcohol oxidase promoter. The nucleotides downstream of the ATG start codon coding for predicted signal peptides of 20 to 21 amino acids and the seven C-terminal residues that might target for the vacuole were excluded from all constructs. C-terminal His6 was included in all expression constructs. Cloning primers are given in Supplemental Table S2. PCR products were digested and ligated into pPICZ_alpha A or pPICZ_alpha A (NdeI). The latter is a derivative of the first but with an additional NdeI site downstream of the EcoRI site in pPICZ_alpha A. Positive clones were identified after sequencing (Eurofins MWG Operon) and were linearized by SacI. After heat inactivation of SacI, 10 μg of DNA was used for electroporation (1.8 kV, 25 μF, 200 Ω) of P. pastoris strain KM71H. Cells were left 3 h at 30°C before plating on yeast peptone dextrose solid medium containing 100 μg mL−1 zeocin. After 3 d of incubation at 30°C, colonies were transferred to fresh yeast peptone dextrose solid medium with 100 μg mL−1 zeocin. Colonies were PCR screened for the correct insert and tested for the production of secreted protein in feed-batch shaking cultures. Positive clones were grown in buffered minimal glycerol (1% yeast nitrogen base, 1% casaminoacids, 100 mm phosphate buffer, pH 6.0, 2% glycerol, 50 μm ZnSO4, 450 μm FeSO4, 150 μm MnSO4, 200 μm MgCl2, and 200 μm CaCl2) for 24 h, followed by induction with 1% methanol and added at daily intervals thereafter.
Cultures were grown under continuous shaking (330 rpm) at 30°C and buffered each day to pH 5.5 with 1 m NaOH. PAPhy proteins were detected in the medium after 1 d using anti-His6 (Qiagen) or TaPAPhy_b antibodies.
Recombinant proteins were purified from the supernatant after centrifuging the cultures (6,000g, 10 min). Dialysis (30-kD cutoff; Vivaspin 500; Sartorius) removed the phosphate buffer. After adjusting the pH to 8.0, the dialyzed broth was passed through a Q-Sepharose (GE Healthcare) column, and the recombinant phytase eluted with a 165 mm NaCl pulse. Nickel nitrilotriacetic acid Sepharose (Qiagen) chromatography was performed, and the phytase was eluted with 250 mm imidazole containing 350 mm NaCl. The protein was concentrated using Vivaspin columns and dialyzed with Microcon YM-30 (Millipore). A fraction of the eluate was deglycosylated with Endo H glycosidase (New England Biolabs) according to the manufacturer’s instructions, omitting the denaturation step. For more details on the purification, see Supplemental Table S5.
The r-PAPhy molecular mass was determined according to Andrews (1964) using an ÄKTA FPLC device (GE Healthcare). Calibration standards were aprotinin (6.5 kD), ribonuclease A (13.7 kD), carbonic anhydrase (29 kD), ovalbumin (43 kD), conalbumin (75 kD), aldolase (158 kD), ferritin (440 kD), and blue dextran (2,000 kD). Each standard (5 mg) was passed through the column, and molecular mass values were calculated according to a calibration curve.
Western Blotting
The protein was fractionated by 4% to 12% SDS-PAGE and blotted to nitrocellulose membranes using a semidry blot apparatus as described by the manufacturer (Hoefer). Polyclonal antibody against E. coli TaPAPhy_b1 was used in a 1:200 dilution and secondary goat anti-rabbit IgG conjugated with alkaline phosphatase in a 1:5,000 dilution.
Biochemical Characterization of Recombinant Phytases
Phytase activity was measured according to Engelen et al. (1994). For substrate specificities, the method of Greiner et al. (1998) was used. Specific activity of recombinant phytase was calculated after protein determination with Coomassie Brilliant Blue R-250 using bovine serum albumin as a standard. Standard phosphatase activity, using p-NPP as a substrate, was assayed in a final volume of 1 mL, 0.1 m acetate buffer, pH 5.0, measuring the A405 and determining the end-point activity using an extinction coefficient of 18.6 mmol cm−1 according to the Lambert-Beers law.
For pH optimum determination, the following buffers were used: pH 1 to 3.5, 100 mm Gly/HCl; pH 3.5 to 5.5, 100 mm Na-acetate/NaOH; pH 5.5 to 7, 100 mm MES (Na salt)/Tris/HCl; pH 7 to 9, 100 mm Tris/HCl; and pH 9 to 10, 100 mm Gly/NaOH. Two micrograms of enzyme was incubated for 10 min in both the pH and temperature optima experiments. Phytate (from rice; Sigma P-3168) and p-NPP were both used in 2 mm and 10 mm final concentrations.
Enzyme kinetics was carried out at the pH optimum and 36°C. Metal sensitivity and Ki were determined at pH 5.5, the pH at which maximum inhibition rates were obtained. Metal ions were preincubated with 2 mm phytate at room temperature, pH 5.5, and thereafter incubated at 36°C. After 10 min, 1 μg of recombinant enzyme was added and the incubation was continued for 10 min. Kinetic calculations were performed using Sigmaplot software. Enzyme metal-activating tests were performed after incubating Vivaspin-concentrated protein in 10 mm FeSO4 + 3 mm ascorbic acid, 10 mm FeCl3, 10 mm CaCl3, or 10 mm MnSO4 for 10 min at room temperature.
Real-Time RT-PCR
Developing and germinating grains were dissected with a microscope. New sterile scalpels and tweezers were used for each tissue type, and the dissected tissues were checked carefully to ensure minimum contamination with adjacent tissue. Using a modified extraction buffer (100 mm Tris-HCl, pH 7.5, 500 mm LiCl, 10 mm EDTA, 1% lithium dodecyl sulfate, and 50 mm DTT), total RNA was isolated using the Plant RNAeasy Kit (see manufacturer’s instructions; Qiagen). A DNase I treatment (RNase free; Roche) was performed by incubating with 20 units of DNase I, at 37°C for 30 min, in a final concentration of 40 mm Tris-HCl, pH 7.5, 6 mm MgCl2, 2 mm CaCl2, and 100 mm NaCl.
cDNA was synthesized from 2 μg of total RNA using oligo(dT)18N and SuperScript II (Invitrogen). qRT-PCR was performed on a Sequence Detection System (Applied Biosystem 9700HT) using SYBR Green master mix (Amersham Biosciences). Primers distinguishing each PAPhy isogene (Supplemental Table S6) were designed using the Primer Premier software (Premier Biosoft International). The specificity of each primer pair was tested by PCR amplifying the cDNA clones, followed by cloning and sequencing of the PCR products. The optimal cDNA quantity (1 μL; dilution 1:10) was determined by using a dilution series.
The relative expression levels of the PAPhy isogenes were normalized against the expression of the wheat (DQ435659) and barley (Y08490) α2-chain tubulins. The expression data were normalized according to the REST algorithm using the REST2005 software (Pfaffl et al., 2002). The relative expression units for each isogene were finally transformed to expression fold, defined as the log2 of relative expression units.
Subcellular Localization of TaPAPhy
Light and immunoelectron microscopy of developing wheat grains were performed using polyclonal rabbit anti-TaPAPhy antibody and the procedures already described (Brinch-Pedersen et al., 2006).
Purification of Phytase from Wheat Bran and Germinating Wheat Grains
The bran from 1 kg of wheat grains (cv Bobwhite) was dissolved in 1 L of buffer A (0.1 m acetate buffer, pH 5.5, 1 mm CaCl2, 1 mm phenylmethylsulfonate fluoride, 5 mm benzamidine, 50 μm tosyl-lysil-chloromethylchetone, and 0.1% Nonidet P-40), heated to 40°C under stirring (200 rpm) before 5,000 units of xylanase (Sigma X2753), 2,300 units of β-glucanase (Fluka 74385), and 500 units of phospholipase D (Sigma P0515) were added. Stirring at 40°C was continued for 3 h before centrifugation (6,000g, 30 min). Proteins were (NH4)2SO4 precipitated (60% saturation), and the pellet was resuspended in 50 mL of buffer B (20 mm acetate, pH 4.3, 0.1 mm CaCl2) and dialyzed (10-kD cutoff) against 10 L of water for 12 h. Dialyzed proteins were subjected to SP-Sepharose chromatography (50-mL bed size; GE Healthcare) in buffer B and eluted with a linear gradient of NaCl (0–0.5 m). Fractions with phytase activity were pooled and dialyzed as described above, although against buffer C (50 mm Tris-HCl, pH 7.5), and fractionated by Q-Sepharose chromatography (50-mL bed size; GE Healthcare) using a linear gradient of NaCl (0–0.5 m). Phytase-active fractions were pooled, subjected to concanavalin A-Sepharose chromatography (20-mL bed size; GE Healthcare), and eluted by buffer C including 0.25 m NaCl and 0.2 m β-d-glucopyranoside. Fractions with phytase activity were pooled and concentrated using Vivaspin cartridges (10-kD cutoff).
Wheat grains (cv Bobwhite) were surface sterilized as described elsewhere (Dionisio et al., 2007) and germinated on filter paper wetted with distilled water. At day 6, roots and leaves were removed and 50 g of grains was homogenized in 300 mL of buffer A. After centrifugation (6,000g, 15 min, 4°C), the supernatant was dialyzed against buffer C and purified by Q-Sepharose, SP-Sepharose, concanavalin A-Sepharose, and Superdex G 200. The insoluble homogenate was suspended in 150 mL of buffer A at 40°C and stirring. Xylanase (2,000 units; Sigma X2753), β-glucanase (1,000 units; Fluka 74385), and phospholipase D (500 units; Sigma-Aldrich P0515) were added, and the stirring at 40°C was continued for 3 h before centrifugation (6,000g, 30 min). The supernatant was dialyzed (10-kD cutoff) against 10 L of buffer B (20 mm acetate, pH 4.3, 0.1 mm CaCl2) for 12 h and purified following the procedure described for wheat bran phytase from the Q-Sepharose step.
Proteolytic Digestions
Purified phytase (10 μg) was reduced in 50 μL of buffer (8 m urea, 200 mm Tris, 20 mm EDTA, and 20 mm DTT) in an ultrasound bath for 10 min followed by 30 min of incubation at 25°C. Proteins were then alkylated by 14 μL of 0.5 m iodoacetic acid in 0.5 m Tris, pH > 8, for 30 min at 25°C in the dark and precipitated with 6 volumes of ice-cold ethanol overnight at −20°C. The pellets were dissolved in 20 μL of 50 mm NH4HCO3, pH 8.0, and digested by sequencing-grade modified bovine chymotrypsin (Princeton Separation) or modified sequencing-grade porcine trypsin (Promega) dissolved in 50 mm acetic acid. Samples were digested at 37°C at enzyme:substrate = 100:1 (w/w) for 30 min, and after addition of more protease (1:100), digestions were continued for 1 h and stopped with 5 μL of 5% formic acid. Digests were concentrated 10-fold by vacuum centrifugation and diluted with 20 μL of 5% formic acid prior to liquid chromatography-MS/MS or storage at −20°C.
Deglycosylation with Glycopeptidase A
Carboxymethylated phytase was prepared and precipitated as described above. The pellet was dissolved in 15 μL of 0.1 mm ammonium acetate, pH 5, and incubated with 60 milliunits of glycopeptidase A from almond (Prunus dulcis; Sigma-Aldrich) for 18 h at 37°C, dried by vacuum centrifugation, and digested with chymotrypsin as describe above.
Nano-Liquid Chromatography-Electrospray Ionization-MS/MS and Data Analysis
Aliquots of proteolytic digests were analyzed by nanoflow capillary HPLC interfaced directly to an electrospray ionization Q-time of flight MS/MS device (MicroTOFQ; Bruker Daltonics) as described elsewhere (Knudsen et al., 2008).
The lists of MS and MS/MS spectra from each proteolytic experiment were analyzed and searched by Mascot software version 2.2 (www.matrixsciences.com) against a wheat protein database, prepared by translation of available wheat EST sequences (Dana-Farber Cancer Institute Wheat Gene Index, release 12.0, July 24, 2008; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=wheat). Search parameters were as follows: enzyme, semichymotrypsin, allowing three missed cleavages; complete modification, carboxymethylated; partial modification, oxidized Met; peptide tolerance, 0.1 D. Settings for Lys-C, Asp-N, and trypsin were similar. Glycopeptides were extracted manually from the raw MS/MS spectra using DataAnalysis version 3.4 (Bruker Daltonics). Error-tolerant searches were performed to identify deglycosylated Asn residues converted to Asp residues in digests of glycopeptidase A-treated samples. The level of each PAPhy isoform was estimated according to the emPAI score (http://www.matrixscience.com/help/quant_empai_help.html).
Promoter-GUS Constructs
A wheat (cv Bobwhite) genomic library was generated using the Lambda Fix II/Xho I Partial Fill-In Vector Kit (Agilent Technologies-Stratagene Products). The initial library was titered, and the size was found to be 5 × 106 plaque-forming units (pfu), corresponding to 45,000 to 115,000 Mb or 2.8 to 7.2 times the size of the wheat genome. The library was amplified to a final titer of 3 × 106 pfu μL−1. The amplified library was plated on NZY agar plates at a density of 600 pfu cm−2. Library screening was performed via plaque lifts using Hybond N+ membranes and the procedure described by the manufacturer (GE Healthcare). The probe was 20 μCi 32P labeled by PCR using [α-32P]dCTP and the primers PAP ex3 Fw (5′-CTTGAGCCTGGGACGAAGT-3′) and PAP ex3 Rv (5′-GAGAAGGACCCGCTCTCC-3′) and a template consisting of a plasmid comprising a cDNA molecule whose nucleotide sequence encoded the TaPAPhy_b. The primers amplified a fragment of the cDNA molecule whose nucleotide sequence corresponds to the highly conserved third exon of the Triticeae PAPhy gene. The amplified sequence generated a DNA probe of 479 nucleotides in length. Unincorporated deoxyribonucleotide triphosphates were removed with an Illustra MicroSpin G-50 column (GE Healthcare). The probe was denatured by boiling followed by shock cooling in 500 μL of 10 μg μL−1 sonicated salmon sperm DNA.
From a positive λ-clone, a 474-bp fragment of the TaPAPhy_a1 promoter fragment was amplified using the primers TaPAPhy_a1-474FW (5′-AAGCTTCTAGGATCATTATGG-3′) and TaPAPhy_a1-1RV (5′-GGATCCTGACAGAATTGGAATGCCTT-3′). HindIII and BamHI restriction sites (boldface) were added to the 5′ and 3′ ends, respectively, of the amplified product. The promoter was HindIII and BamHI ligated upstream of the GUS-encoding uidA gene (Jefferson et al., 1987) of the pUC18-based pGUSN plasmid, which downstream of the uidA gene holds the nos terminator sequence of the Agrobacterium tumefaciens nopaline synthase gene (Bevan et al., 1983). The resulting plasmid was named pTaPAPhy_a1-GUS-N.
The TaPAPhy_b promoter was PCR amplified from the λ-clones using the forward primer 5′-GGTCTTAAUATTCTCCACGAAATAGTGCCTCA-3′ and the reverse primer 5′-GGCATTAAUCCCGATAGACGTTTGGTGC-3′. The amplified PAPhy_b2 promoter fragment was 1,380 bp. The promoter was inserted upstream of the GUS gene after digesting the PCR product with the User Enzyme Mix (New England Biolabs) and opening the pCAMBIA_GUS_35Sterm vector with the PacI and Nt.BbvCI enzymes according to Nour-Eldin et al. (2006). The resulting plasmid was named pTaPAPhy_b2-GUS-35Sterm.
Generation and Identification of Transgenic Plants
The pTaPAPhy_a1-GUS-N and pTaPAPhy_b2-GUS-35Sterm plasmids were introduced into immature embryos of wheat cv Bobwhite using the DuPont PDS 1000 helium biolistic system, as described already (Brinch-Pedersen et al., 1996). Selection, regeneration, and identification of transgenic wheat plants were performed as described by Brinch-Pedersen et al. (2000). Assaying for GUS activity was performed according to Jefferson and coworkers (1987).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TaPAPhy_a1 (FJ973998), TaPAPhy_a2 (FJ973999), TaPAPhy_b1 (FJ974000), TaPAPhy_b2 (FJ974001), Ta_ACP (FJ974002), HvPAPhy_a (FJ974003), HvPAPhy_b1 (FJ974004), HvPAPhy_b2 (FJ974005), HvPAP_c (FJ974006), ZmPAPhy_b (FJ974007), ZmPAP_c (FJ974008), and OsPAPhy_b (HM0006823).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Superdex G 200 gel filtration of P. pastoris-produced r-TaPAPhy_b1.
Supplemental Figure S2. pH (A) and temperature (B) profiles for r-TaPAPhy_a1 and r-TaPAPhy_b1.
Supplemental Figure S3. Metal inhibition of r-TaPAPhy_b1.
Supplemental Figure S4. TaPAPhy and HvPAPhy transcripts in dry grains of barley and wheat.
Supplemental Figure S5. Peptide mapping of purified phytases.
Supplemental Table S1. Molecular features of the wheat, barley, maize, and rice PAPhys, wheat Ta_ACP, and barley HvPAP_c.
Supplemental Table S2. Cloning primers, vectors, strains, and results of heterologous expression of PAPhy in E. coli and P. pastoris.
Supplemental Table S3. Phylogenetic distances between PAP and PAPhy proteins.
Supplemental Table S4. Oligonucleotides for cloning.
Supplemental Table S5. Purification levels and yield parameters of r-TaPAPhy a1 and b1.
Supplemental Table S6. Oligonucleotides for qRT-PCR.
Supplementary Material
Acknowledgments
Lis Bagnkop Holte and Ole Bråd Hansen are thanked for excellent technical assistance and for taking good care of the plants. Novozymes is thanked for its support in generating antibodies.
References
- Andrews P. (1964) Estimation of the molecular weights of proteins by Sephadex gel-filtration. Biochem J 91: 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Swanson SJ, Hillmer S, Jones RL. (1998) From storage compartment to lytic organelle: the metamorphosis of the aleurone protein storage vacuole. Ann Bot (Lond) 82: 399–412 [Google Scholar]
- Bevan MW, Barnes WM, Chilton MD. (1983) Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res 11: 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Galili G, Knudsen S, Holm PB. (1996) Engineering of the aspartate family biosynthetic pathway in barley (Hordeum vulgare L.) by transformation with heterologous genes encoding feed-back-insensitive aspartate kinase and dihydrodipicolinate synthase. Plant Mol Biol 32: 611–620 [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Hatzack F, Sørensen LD, Holm PB. (2003) Concerted action of endogenous and heterologous phytase on phytic acid degradation in seed of transgenic wheat (Triticum aestivum L.). Transgenic Res 12: 649–659 [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Hatzack F, Stöger E, Arcalis E, Pontopidan K, Holm PB. (2006) Heat-stable phytases in transgenic wheat (Triticum aestivum L.): deposition pattern, thermostability, and phytate hydrolysis. J Agric Food Chem 54: 4624–4632 [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Olesen A, Rasmussen SK, Holm PB. (2000) Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an Aspergillus phytase. Mol Breed 6: 195–206 [Google Scholar]
- Brinch-Pedersen H, Sørensen LD, Holm PB. (2002) Engineering crop plants: getting a handle on phosphate. Trends Plant Sci 7: 118–125 [DOI] [PubMed] [Google Scholar]
- Denbow DM, Grabau EA, Lacy GH, Kornegay ET, Russell DR, Umbeck PF. (1998) Soybeans transformed with a fungal phytase gene improve phosphorus availability for broilers. Poult Sci 77: 878–881 [DOI] [PubMed] [Google Scholar]
- Dionisio G, Holm PB, Brinch-Pedersen H. (2007) Wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) multiple inositol polyphosphate phosphatases (MINPPs) are phytases expressed during grain filling and germination. Plant Biotechnol J 5: 325–338 [DOI] [PubMed] [Google Scholar]
- Dvoráková J. (1998) Phytase: sources, preparation and exploitation. Folia Microbiol (Praha) 43: 323–338 [DOI] [PubMed] [Google Scholar]
- Eeckhout W, De Paepe M. (1994) Total phosphorus, phytate-phosphorus and phytase activity in plant feedstuffs. Anim Feed Sci Technol 47: 19–29 [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen AJ, van der Heeft FC, Randsdorp PHG, Smit ELC. (1994) Simple and rapid determination of phytase activity. J AOAC Int 77: 760–764 [PubMed] [Google Scholar]
- Greiner R, Jany KD, Alminger ML. (2000) Identification and properties of myo-inositol hexakisphosphate phosphohydrolases (phytases) from barley (Hordeum vulgare). J Cereal Sci 31: 127–139 [Google Scholar]
- Greiner R, Konietzny U, Jany KD. (1998) Purification and properties of a phytase from rye. J Food Biochem 22: 143–161 [Google Scholar]
- Hayakawa T, Toma Y, Igaue I. (1989) Purification and characterization of acid-phosphatases with or without phytase activity from rice bran. Agric Biol Chem 53: 1475–1483 [Google Scholar]
- Hegeman CE, Grabau EA. (2001) A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol 126: 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman WL, Vredenbregt-Heistek JC, Schmitt NF, Feussner I. (1997) Lipoxygenase-2 oxygenates storage lipids in embryos of germinating barley. Eur J Biochem 248: 452–458 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen SK, Stensballe A, Franzmann M, Westergaard UB, Otzen DE. (2008) Effect of glycosylation on the extracellular domain of the Ag43 bacterial autotransporter: enhanced stability and reduced cellular aggregation. Biochem J 412: 563–577 [DOI] [PubMed] [Google Scholar]
- Kuang RB, Chan KH, Yeung E, Lim BL. (2009) Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol 151: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei XG, Porres JM, Mullaney EJ, Brinch-Pedersen H. (2007) Phytase: source, structure, and application. Polaina J, MacCabe AP, eds, Industrial Enzymes: Structure, Function and Applications. Springer, Dordrecht, The Netherlands, pp 505–530 [Google Scholar]
- Lott JNA. (1984) Accumulation of seed reserves of phosphorus and other minerals. Murray DR, ed, Seed Physiology. Academic Press, New York, pp 139–166 [Google Scholar]
- Lu K, Chai YR, Zhang K, Wang R, Chen L, Lei B, Lu J, Xu XF, Li JN. (2008) Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor Appl Genet 117: 963–975 [DOI] [PubMed] [Google Scholar]
- Lung SC, Chan WL, Yip W, Wang L, Yeung EC, Lim BL. (2005) Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilization. Plant Sci 169: 341–349 [Google Scholar]
- Maugenest S, Martinez I, Godin B, Perez P, Lescure AM. (1999) Structure of two maize phytase genes and their spatio-temporal expression during seedling development. Plant Mol Biol 39: 503–514 [DOI] [PubMed] [Google Scholar]
- Maugenest S, Martinez I, Lescure AM. (1997) Cloning and characterization of a cDNA encoding a maize seedling phytase. Biochem J 322: 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta BD, Jog SP, Johnson SC, Murthy PPN. (2006) Lily pollen alkaline phytase is a histidine phosphatase similar to mammalian multiple inositol polyphosphate phosphatase (MINPP). Phytochemistry 67: 1874–1886 [DOI] [PubMed] [Google Scholar]
- Nakano T, Joh T, Tokumoto E, Hayakawa T. (1999) Purification and characterization of phytase from wheat bran of Triticum aestivum L. cv. Nourin #61. Food Sci Technol Res 5: 18–23 [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MH, Jensen JK, Halkier BA. (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dell BL, de Boland AR, Koirtyohann SR. (1972) Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J Agric Food Chem 20: 718–721 [Google Scholar]
- Penheiter AR, Klucas RV, Sarath G. (1998) Purification and characterization of a soybean root nodule phosphatase expressed in Pichia pastoris. Protein Expr Purif 14: 125–130 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative Expression Software Tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SK, Sorensen MB, Johansen KS, inventors March 6, 2007. Polynucleotides encoding phytase polypeptides. U.S. Patent Application No. 7,186,817 B2 [Google Scholar]
- Rouster J, van Mechelen J, Cameron-Mills V. (1998) The untranslated leader sequence of the barley lipoxygenase 1 (Lox1) gene confers embryo-specific expression. Plant J 15: 435–440 [DOI] [PubMed] [Google Scholar]
- Steen I. (1998) Phosphorus availability in the 21st century: management of a non-renewable resource. Phosphate and Potassium 217: 25–31 [Google Scholar]
- Sträter N, Klabunde T, Tucker P, Witzel H, Krebs B. (1995) Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science 268: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Uzarowska A, Dionisio G, Sarholz B, Piepho HP, Xu ML, Ingvardsen CR, Wenzel G, Lübberstedt T. (2009) Validation of candidate genes putatively associated with resistance to SCMV and MDMV in maize (Zea mays L.) by expression profiling. BMC Plant Biol 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Spener F, Krebs B. (2006) Purple acid phosphatases. Messerschmidt A, Huber R, Poulas T, Wieghardt K, Cygel M, Bode M, eds, Handbook of Metalloproteins. John Wiley & Sons, Hoboken, NJ, pp 752–767 [Google Scholar]
- Waratrujiwong T, Krebs B, Spener F, Visoottiviseth P. (2006) Recombinant purple acid phosphatase isoform 3 from sweet potato is an enzyme with a diiron metal center. FEBS J 273: 1649–1659 [DOI] [PubMed] [Google Scholar]
- Xiao K, Harrison MJ, Wang ZY. (2005) Transgenic expression of a novel M. truncatula phytase gene results in improved acquisition of organic phosphorus by Arabidopsis. Planta 222: 27–36 [DOI] [PubMed] [Google Scholar]
- Zhang GP, Chen JX, Dai F, Wang JM, Wu FB. (2006) The effect of cultivar and environment on beta-amylase activity is associated with the change of protein content in barley grains. J Agron Crop Sci 192: 43–49 [Google Scholar]
- Zhang W, Gruszewski HA, Chevone BI, Nessler CL. (2008) An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146: 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZB, Kornegay ET, Radcliffe JS, Wilson JH, Veit HP. (2000) Comparison of phytase from genetically engineered Aspergillus and canola in weanling pig diets. J Anim Sci 78: 2868–2878 [DOI] [PubMed] [Google Scholar]
- Zhu HF, Qian WQ, Lu XZ, Li DP, Liu X, Liu KF, Wang DW. (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol Biol 59: 581–594 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.