Abstract
Although phosphate (Pi) starvation signaling is well studied in Arabidopsis (Arabidopsis thaliana), it is still largely unknown in rice (Oryza sativa). In this work, a rice leaf tip necrosis1 (ltn1) mutant was identified and characterized. Map-based cloning identified LTN1 as LOC_Os05g48390, the putative ortholog of Arabidopsis PHO2, which plays important roles in Pi starvation signaling. Analysis of transgenic plants harboring a LTN1 promoter::β-glucuronidase construct revealed that LTN1 was preferentially expressed in vascular tissues. The ltn1 mutant exhibited increased Pi uptake and translocation, which led to Pi overaccumulation in shoots. In association with enhanced Pi uptake and transport, some Pi transporters were up-regulated in the ltn1 mutant in the presence of sufficient Pi. Furthermore, the elongation of primary and adventitious roots was enhanced in the ltn1 mutant under Pi starvation, suggesting that LTN1 is involved in Pi-dependent root architecture alteration. Under Pi-sufficient conditions, typical Pi starvation responses such as stimulation of phosphatase and RNase activities, lipid composition alteration, nitrogen assimilation repression, and increased metal uptake were also activated in ltn1. Moreover, analysis of OsmiR399-overexpressing plants showed that LTN1 was down-regulated by OsmiR399. Our results strongly indicate that LTN1 is a crucial Pi starvation signaling component downstream of miR399 involved in the regulation of multiple Pi starvation responses in rice.
Plants require large amounts of phosphate (Pi) to maintain growth and development (Raghothama, 1999). However, Pi is often a limiting factor for plants because of its low availability in the soil, which is mainly due to its low abundance and easy chelation with cations or organic compounds to form insoluble complexes (Misson et al., 2005). To cope with Pi starvation, plants have evolved multiple strategies to increase Pi availability (Raghothama, 1999). For example, changes in root architecture enable plants to increase surface contact with the soil and to acquire Pi more efficiently, and up-regulation of Pi transporter genes enhances Pi uptake and transport efficiency (Liu et al., 1998; Karthikeyan et al., 2002; Xiao et al., 2006). It is also thought that acid phosphatases are involved in Pi acquisition or recycling during Pi starvation (Duff et al., 1994; del Pozo et al., 1999), and acid phosphatase and RNase induction is a significant adaptive response to increase Pi availability (Duff et al., 1994; Trull and Deikman, 1998). In addition, Pi-starved plants can regulate multiple metabolic processes to reprioritize the utilization of internal Pi and maximize the acquisition of external Pi to adapt to low-Pi environments (Vance et al., 2003; Wasaki et al., 2003). Alteration in lipid metabolism, resulting in phospholipid degradation and galactolipid and sulfolipid synthesis, could increase intracellular Pi availability under Pi depletion (Gniazdowska et al., 1999; Misson et al., 2005). Gene chip analysis also revealed that genes related to phospholipid degradation and galactolipid and sulfolipid synthesis were up-regulated in rice (Oryza sativa) roots upon Pi deprivation (Wasaki et al., 2003). Moreover, genes responsible for nitrogen assimilation (i.e. nitrite reductase, Gln synthase, and Glu synthase) were repressed (Wu et al., 2003), and nitrate uptake was reduced by Pi starvation (Gniazdowska and Rychter, 2000), indicating that nitrogen assimilation is repressed under Pi depletion. In contrast, the uptake of several metallic elements, such as iron (Fe), aluminum, and calcium, was increased in response to Pi starvation (Misson et al., 2005). Recently, Pi starvation in rice was shown to increase Fe contents and influence the expression of Fe-responsive genes (Zheng et al., 2009), suggesting that enhanced capacity to absorb and scavenge Pi-complexing metals was an adaptive response to Pi starvation that could increase the availability of free Pi (Misson et al., 2005).
The Pi starvation signaling and regulatory system has been well studied in Arabidopsis (Arabidopsis thaliana; Bari et al., 2006; Schachtman and Shin, 2007), demonstrating that PHR1, SIZ1, PHOSPHATE-RESPONSIVE2 (PHO2), and miR399 are major players. PHR1, a MYB transcription factor, is a key regulator of Pi starvation signaling. PHR1 overexpression in Arabidopsis led to excessive Pi accumulation in shoots and activation of Pi starvation-induced gene expression (Rubio et al., 2001; Nilsson et al., 2007). SIZ1 is a small plant ubiquitin-like modifier (SUMO) E3 ligase that controls PHR1 sumoylation (Miura et al., 2005). miR399, the target gene of PHR1, is specifically induced by Pi starvation (Fujii et al., 2005) and can negatively regulate PHO2 expression through mRNA degradation (Chiou et al., 2006). Both miR399 overexpression and mutations in PHO2 resulted in overaccumulation of Pi in Arabidopsis shoots (Fujii et al., 2005; Chiou et al., 2006). Expression of several Pi starvation-induced genes such as Pht1;8, Pht1;9, AtIPS1, and AT4 was increased in pho2 mutants even under Pi-sufficient conditions, suggesting that PHO2 is involved in the regulation of Pi starvation responses (Bari et al., 2006).
Rice productivity is seriously affected by Pi availability, but the Pi starvation signaling pathway is still largely unknown in rice. Zhou et al. (2008) proposed that OsPHR2, the homolog of AtPHR1, is a significant regulator involved in Pi starvation signaling in rice. Wang et al. (2009) and Liu et al. (2010) further showed that OsSPX1 is involved in Pi homeostasis and suppresses the function of OsPHR2 in the regulation of OsPT2 expression. OsPHO2, the putative homolog of AtPHO2, has been shown to be involved in the Pi starvation signaling pathway mediated by OsSPX1-OsPHR2 (Bari et al., 2006; Wang et al., 2009; Liu et al., 2010). However, systematic work focusing on OsPHO2 protein function in the regulation of Pi starvation responses is still lacking. In this work, we identified a rice mutant that displayed leaf tip necrosis. Map-based cloning revealed that LEAVE TIP NECROSIS1 (LTN1) encodes a protein containing a ubiquitin-conjugating domain and is the AtPHO2 homolog (OsPHO2) in rice. The ltn1 mutant showed excessive Pi accumulation in shoots, which was caused by improved Pi uptake in the roots and increased Pi translocation from roots to shoots. The increased Pi uptake and transport in ltn1 was associated with increased expression of PHT genes. LTN1 was also involved in Pi-dependent root architecture alteration under Pi-deficient conditions. In addition to LTN1 participating in multiple Pi starvation responses, analysis of transgenic plants overexpressing OsmiR399 provided direct evidence that LTN1 was down-regulated by OsmiR399. All these results indicated that LTN1 is a crucial component of Pi starvation signaling and involved in regulating multiple Pi starvation responses in rice.
RESULTS
LTN1, Encoding a Protein Containing a Ubiquitin-Conjugating Domain, Is Responsible for Pi Overaccumulation in the ltn1 Mutant
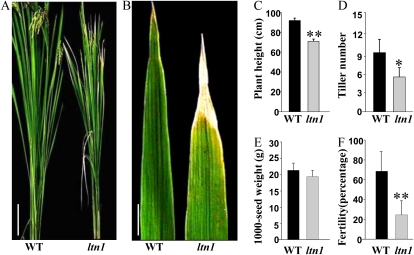
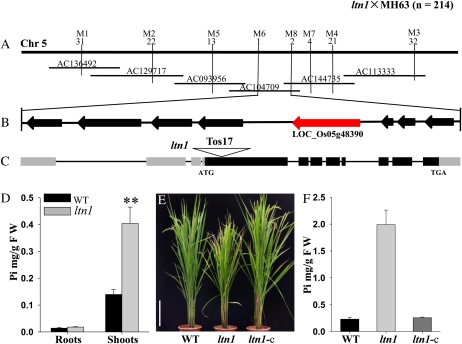
The ltn1 mutant was identified from a large-scale screening of our rice mutant population (Ma et al., 2009), which displayed leaf tip necrosis predominantly in mature leaves (Fig. 1, A and B). Significant alterations were also observed in ltn1 agronomic traits such as plant height, tiller number, and fertility (Fig. 1, C–F). LTN1 was initially mapped to rice chromosome 5 between markers M1 and M3 and further narrowed down to a 54-kb region between markers M6 and M8 (Fig. 2A). Within this 54-kb interval, eight predicted open reading frames (ORFs) were found (Fig. 2B). Sequence analysis demonstrated that a Tos17 transposon had inserted in LOC_Os05g48390, and no transcript of LOC_Os05g48390 was detected in the ltn1 mutant (Supplemental Fig. S1). Thus, LOC_Os05g48390 was a good candidate for the LTN1 gene. LTN1 was 7,946 bp long, contained 11 exons (Fig. 2C), and its 2,631-bp ORF encoded a predicted protein of 876 amino acids. LTN1 shared 58% amino acid sequence identity with Arabidopsis PHO2, which contains a ubiquitin-conjugating domain at the C terminus (Supplemental Fig. S2). LOC_Os05g48390 was previously proposed to be OsPHO2, the putative AtPHO2 homolog in rice (Bari et al., 2006), which is consistent with our results. Analysis of Pi contents revealed that Pi accumulated excessively in shoots of the ltn1 mutant (Fig. 2D), further demonstrating that LTN1 is the functional homolog of AtPHO2. To determine whether an excess of Pi was responsible for the leaf tip necrosis phenotype, wild-type plants were treated with different levels of Pi. When treated with extremely high amounts of Pi, wild-type plants also displayed excessive Pi accumulation and leaf tip necrosis (Supplemental Fig. S3). Furthermore, the degree of leaf necrosis in the ltn1 mutant was significantly lower under low-Pi conditions (Supplemental Fig. S4). Thus, excessive Pi accumulation leading to toxicity in the shoots was the probable reason for leaf tip necrosis in the ltn1 mutant. To further confirm that the LTN1 mutation was responsible for the ltn1 mutant phenotypes, genetic complementation was carried out by introducing a 9.4-kb LOC_Os05g48390 genomic fragment into the ltn1 mutant. Leaf tip necrosis and growth defects of the complemented ltn1 mutant were rescued (Fig. 2E), and leaf Pi contents decreased to wild-type levels (Fig. 2F), demonstrating that the insertion mutation in LTN1 was responsible for the Pi overaccumulation of the ltn1mutant. Taken together, these results indicated that LTN1 is the functional homolog of AtPHO2 in rice and is involved in the regulation of shoot Pi accumulation.
Figure 1.
Phenotypes of field-grown wild-type and ltn1 mutant plants. A and B, Growth performance of whole plants (A) and leaf tip necrosis of ltn1 mutant and wild-type (WT) plants (B) grown in the field for 3 months. Bars = 20 cm in A and 1 cm in B. C to F, Agronomic traits (plant height, tiller number, 1,000-seed weight, and fertility) of wild-type (black bars) and ltn1 mutant (gray bars) plants. Error bars indicate sd (n = 15). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01.
Figure 2.
Map-based cloning of the LTN1 gene. A, Preliminary and fine mapping. The LTN1 locus was preliminarily mapped to rice chromosome 5 (Chr 5) between markers M1 and M3. The gene was further delimited to a 54-kb genomic region between markers M6 and M8 within the bacterial artificial chromosome clone AC104709. The number of recombinants is marked corresponding to the molecular markers. B, The 54-kb interval has eight predicted ORFs, and the candidate gene is marked in red. C, LTN1 gene structure. Gray shading, untranslated region; black shading, ORF region; triangle, Tos17 transposon insertion. D, Pi concentration in 2-week-old seedlings of wild-type (WT; black bars) and ltn1 mutant (gray bars) plants grown under Pi-sufficient conditions (16 mg L−1). FW, Fresh weight. Error bars indicate sd (n = 5). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test: ** P < 0.01. E, Growth performance of wild-type, ltn1 mutant, and ltn1-complemented (ltn1-c) plants grown in the field for 3 months. Bar = 20 cm. F, Leaf Pi concentration in 3-month-old field-grown wild-type (black bar), ltn1 mutant (gray bar), and ltn1-complemented (dark gray bar) plants. Error bars indicate sd (n = 5).
LTN1 Expression Profiling
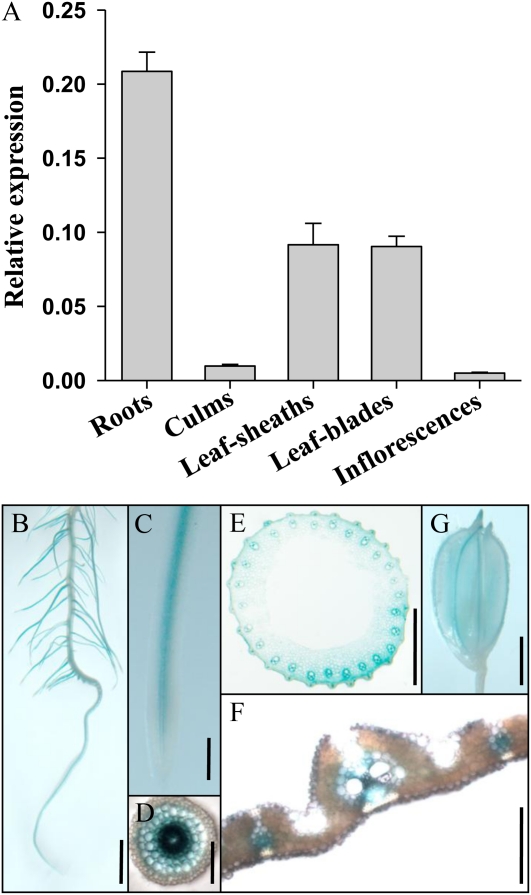
To investigate the LTN1 expression profile, total RNA was extracted from roots, culms, leaf blades, leaf sheaths, and inflorescences, and quantitative reverse transcription (qRT)-PCR analyses were performed. LTN1 was ubiquitously expressed in all tissues, with relatively higher levels in roots (Fig. 3A). To determine the spatial expression pattern of LTN1, a 4.7-kb fragment upstream of the LTN1 ATG was fused to the GUS reporter gene and transformed into wild-type rice. Histochemical analysis of LTN1PRO:GUS transgenic plants revealed that GUS activity was preferentially detected in the vascular tissues of all tissues examined, such as roots, culms, leaves, and grain husks (Fig. 3, B–G). The highest GUS activity was detected in roots, which was consistent with the qRT-PCR analysis. Pi is mainly acquired by roots and transported through vascular tissues, coinciding with LTN1 expression, suggesting that the LTN1 protein is involved in Pi uptake and transport.
Figure 3.
LTN1 expression profile. A, Relative LTN1 expression levels in the roots, culms, leaf sheaths, leaf blades, and inflorescences. Error bars represent sd (n = 3). B to G, GUS staining of LTN1PRO::GUS transgenic plant roots (B–D), culms (E), leaf blades (F), and grain husks (G), showing cross-sections in D, E, and F. Bars = 5 mm (B), 1 mm (C–F), and 2 mm (G).
The ltn1 Mutant Exhibits Increased Uptake and Translocation of Pi and Impaired Pi Remobilization
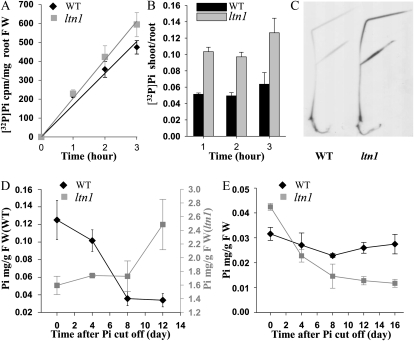
A Pi uptake experiment was performed to explore whether Pi overaccumulation in ltn1 shoots was due to increased Pi uptake and translocation from roots to shoots. ltn1 roots exhibited higher [32P]Pi uptake activity when cultured in hydroponic culture containing [32P]orthophosphate (Fig. 4A). The [32P]Pi shoot-root ratio was greater in ltn1 mutant than in wild-type plants, indicating that Pi translocation from roots to shoots was also increased in the ltn1 mutant (Fig. 4, B and C). Therefore, increased Pi uptake in roots and Pi transport from roots to shoots led to Pi overaccumulation in ltn1 shoots. In wild-type plants, the Pi contents in older leaves decreased over time when Pi was removed from the culture medium (Fig. 4D). However, no reduction in Pi levels was observed during Pi deficiency in the ltn1 mutant (Fig. 4D), suggesting that Pi remobilization out of ltn1 leaves was impaired. Pi contents were further analyzed in roots of ltn1 mutant and wild-type plants under Pi starvation conditions. In wild-type plants, Pi contents initially decreased and then recovered (Fig. 4E), suggesting that Pi can be retranslocated from shoots to roots to compensate for the loss of Pi under Pi starvation conditions. In contrast, Pi contents in ltn1 roots kept decreasing at all the time points analyzed (Fig. 4E), indicating that Pi retranslocation from shoots to roots was also impaired in the ltn1 mutant. In addition, the Pi distribution pattern in aerial plant parts was also altered in the mutant. In wild-type plants, Pi levels were highest in culms, followed by leaf sheaths, leaf blades, and inflorescences. In contrast, the highest Pi accumulation in the ltn1 mutant occurred in leaf blades, then leaf sheaths, culms, and inflorescences (Supplemental Fig. S5). These observations revealed that the Pi distribution pattern was altered in ltn1 aerial parts, suggesting that Pi transport into the leaf blades was enhanced during Pi distribution in the ltn1 shoots.
Figure 4.
Pi uptake and remobilization analysis in wild-type and ltn1 mutant plants. A, [32P]Pi uptake activity in roots of wild-type (WT; black diamonds) and ltn1 mutant (gray squares) plants. FW, Fresh weight. Error bars indicate sd (n = 3). B, Shoot-to-root ratios of the [32P]Pi taken up by wild-type (black bars) and ltn1 mutant (gray bars) plants from A. Error bars indicate sd (n = 3). C, Autoradiograph of [32P]Pi taken up by wild-type and ltn1 mutant plants. D and E, Pi concentration alteration in the fourth leaf (D) or roots (E) of wild-type (black diamonds) and ltn1 mutant (gray squares) plants after Pi cutoff. Error bars indicate sd (n = 5).
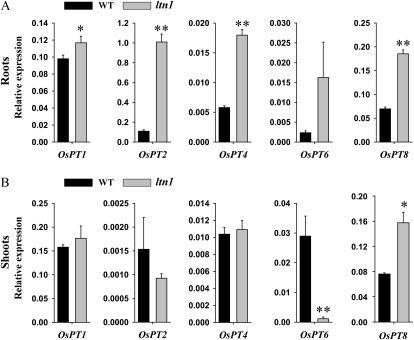
To determine whether the improved Pi uptake and transport in ltn1 were associated with Pi transporters, the expression of five members (OsPT1, OsPT2, OsPT4, OsPT6, and OsPT8) of rice high-affinity phosphate transporters (PHTs) was examined by qRT-PCR analysis. Under Pi-sufficient conditions, the transcript levels of four PHT members (OsPT1, OsPT2, OsPT4, and OsPT8) were increased significantly in ltn1 roots (Fig. 5A), whereas in shoots, only OsPT8 was up-regulated and OsPT6 was dramatically repressed (Fig. 5B). These results suggest that enhanced Pi uptake and transport in ltn1 roots probably correlated with up-regulation of OsPT1, OsPT2, OsPT4, and OsPT8 and that repression of OsPT6 resulted in impaired Pi distribution and mobilization in the shoot.
Figure 5.
qRT-PCR expression analysis of five members of the rice PHT gene family in roots (A) and shoots (B) of wild-type (WT; black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01.
LTN1 Is Involved in Root Architecture Alteration under Pi Starvation Conditions
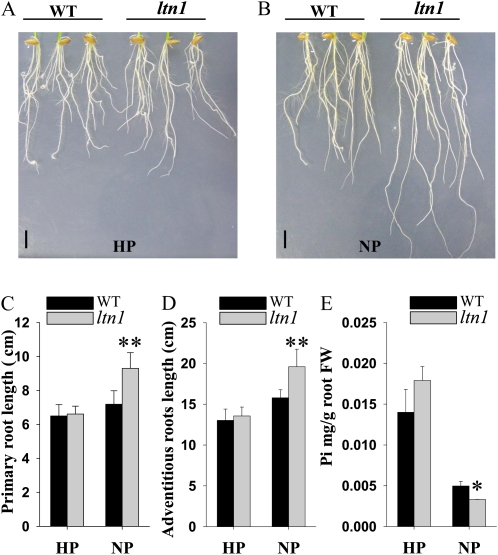
Pi starvation can alter root architecture, as indicated by the elongation of primary and adventitious roots in rice (Wissuwa, 2003; Yi et al., 2005). To investigate whether LTN1 is involved in root architecture alteration in response to Pi starvation, plants were grown under Pi-sufficient (16 mg L−1) or Pi-deficient (0 mg L−1) hydroponic culture conditions. Under Pi-sufficient conditions, no significant difference was observed in root architecture between wild-type and ltn1 mutant plants (Fig. 6A). However, under Pi-deficient conditions, the elongation of primary and adventitious roots was significantly enhanced in ltn1 mutant compared with wild-type plants (Fig. 6, B–D), indicating that the ltn1 mutant is more sensitive to Pi starvation. Consistently, Pi contents in ltn1 roots also decreased significantly compared with wild-type plants under Pi-deficient conditions (Fig. 6E). Thus, the hypersensitivity of the ltn1 mutant to Pi starvation might result from very low Pi contents in the roots under Pi starvation. These results suggested that LTN1 is involved in Pi-dependent root architecture alteration, possibly by affecting the Pi contents in roots under Pi starvation conditions.
Figure 6.
Effect of Pi availability on root architecture in wild-type and ltn1 mutant plants. A and B, Root performance of 10-d-old seedlings of wild-type (WT) and ltn1 mutant plants under Pi-sufficient (HP; 16 mg L−1; A) or Pi-deficient (NP; 0 mg L−1; B) conditions. Bars = 1 cm. C and D, The length of primary roots (C) and the three longest adventitious roots (D) of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient (16 mg L−1) or Pi-deficient (0 mg L−1) conditions. Error bars indicate sd (n = 10). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants under the same growth conditions as determined by Student’s t test analysis: ** P < 0.01. E, Pi contents in roots of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient (16 mg L−1) or Pi-deficient (0 mg L−1) conditions. FW, Fresh weight. Error bars indicate sd (n = 3). The asterisk indicates the significance of the difference between wild-type and ltn1 mutant plants under the same growth conditions as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05.
LTN1 Is Involved in Regulating Acid Phosphatase and RNase Activities
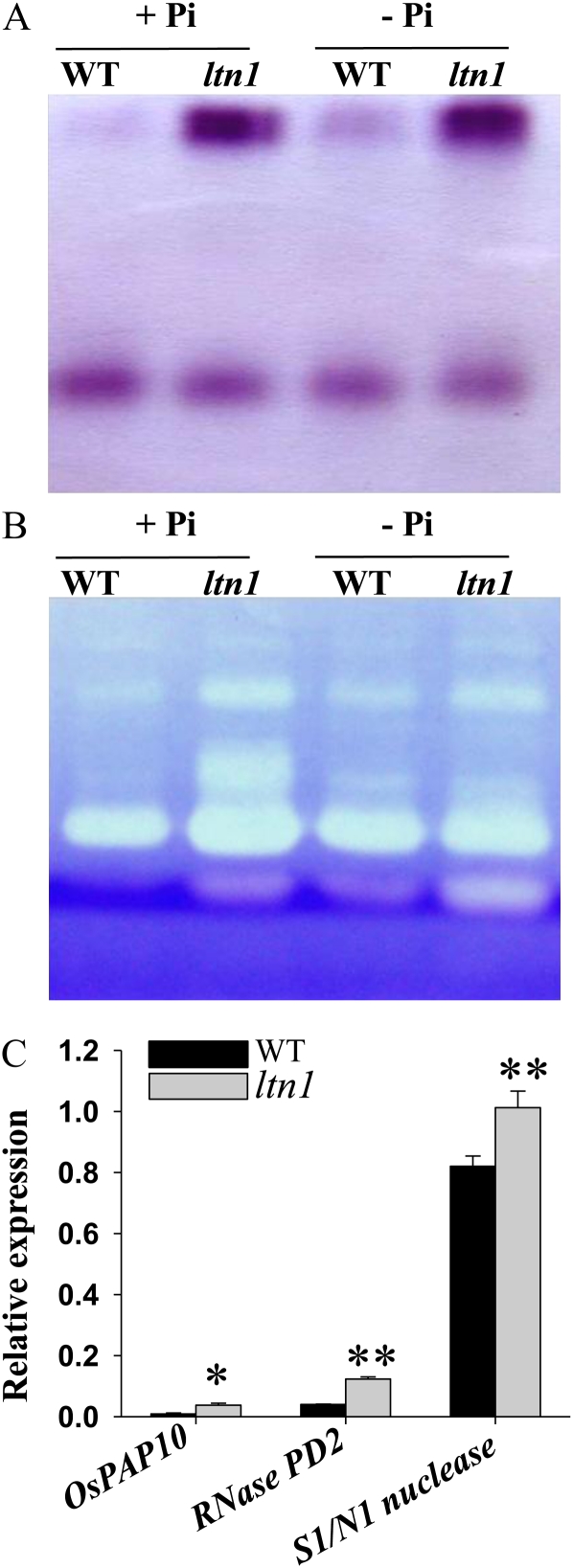
The stimulation of acid phosphatases and ribonucleases is a typical Pi starvation response in plants. To examine whether LTN1 was involved in this process, the intracellular acid phosphatase and RNase activities were compared between ltn1 mutant and wild-type plants. The activity of acid phosphatases and ribonucleases in shoots showed no significant difference between ltn1 mutant and wild-type plants (Supplemental Fig. S6). However, the activity of both types of enzymes in ltn1 roots was much higher than in wild-type plants (Fig. 7, A and B). Consistently, genes coding for Purple Acid Phosphatase10 (PAP10), RNase PD2, and S1/P1 nuclease were also up-regulated in ltn1 roots under Pi-sufficient conditions (Fig. 7C). These results demonstrated that ltn1 mutant plants can stimulate acid phosphatases and ribonucleases through activating the expression of related genes in roots.
Figure 7.
Analysis of acid phosphatase and RNase activities in wild-type and ltn1 mutant plants. A and B, Analysis of acid phosphatase (A) and RNase (B) activities in roots of wild-type (WT) and ltn1 mutant plants under Pi-sufficient (+Pi; 16 mg L−1) or Pi-deficient (−Pi; 0 mg L−1) conditions. C, Relative qRT-PCR expression analysis of genes encoding rice acid phosphatase (OsPAP10) and two nucleases (RNase PD2 and S1/N1 nuclease) in roots of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01.
A Mutation in LTN1 Activates Multiple Metabolic Responses to Pi Starvation
Lipid Composition Is Altered in the ltn1 Mutant
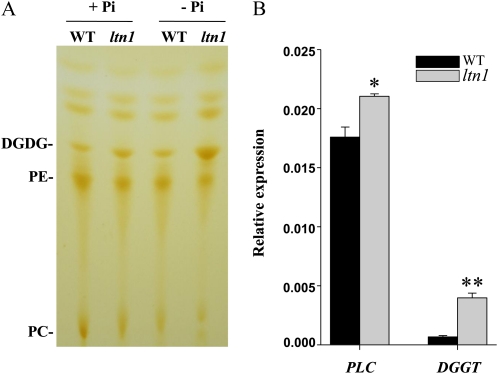
To investigate whether LTN1 regulates lipid metabolism, the lipid composition of ltn1 mutant and wild-type plants was analyzed. Total lipids were extracted from roots and shoots and separated by thin-layer chromatography (TLC). Two major phospholipids, phosphatidylcholine (PC) and phosphatidylethanolamine (PE), were decreased, while a galactolipid, digalactosyl diacylglycerol (DGDG), was increased in ltn1 roots (Fig. 8A). The same result was observed in roots of wild-type plants under Pi-deficient conditions (Fig. 8A). These results were further confirmed by analyzing the relative percentage of each lipid on the TLC plates (Table I). However, in ltn1 shoots, no alteration in lipid composition was seen compared with wild-type plants (Supplemental Fig. S7). Moreover, the genes coding for phospholipase C and diacylglycerol galactosyltransferase, which are responsible for phospholipid degradation and galactolipid synthesis, respectively, were up-regulated in ltn1 roots under Pi-sufficient conditions (Fig. 8B). These results indicate that the mutation in LTN1 triggered Pi starvation-responsive lipid composition alteration via up-regulation of genes coding for phospholipid degradation and galactolipid synthesis in roots.
Figure 8.
Lipid composition analysis in wild-type and ltn1 mutant plants. A, TLC analysis of lipid composition in roots of wild-type (WT) and ltn1 mutant plants under Pi-sufficient (+Pi; 16 mg L−1) or Pi-deficient (−Pi; 0 mg L−1) conditions. B, qRT-PCR expression analysis of genes encoding phospholipase C (PLC) and diacylglycerol galactosyltransferase (DGGT) in roots of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01.
Table I. Quantitative analysis of the relative percentage of lipid bands on the TLC plates (Fig. 8A) using ImageJ (http://rsb.info.nih.gov/ij/index.html) software.
The data are means ± sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants under the same growth conditions as determined by Student’s t test: * 0.01 ≤ P < 0.05, ** P < 0.01.
| Lipid | +Pi |
−Pi |
||
| Wild Type | ltn1 | Wild Type | ltn1 | |
| DGDG | 8.17 ± 2.15 | 14.05 ± 1.38* | 11.96 ± 2.32 | 65.81 ± 5.85** |
| PE | 35.91 ± 3.28 | 25.27 ± 4.23* | 24.87 ± 3.01 | 13.93 ± 3.96* |
| PC | 41.92 ± 3.33 | 24.21 ± 4.54* | 21.06 ± 0.75 | 12.80 ± 0.45** |
Nitrate Assimilation Is Repressed in the ltn1 Mutant
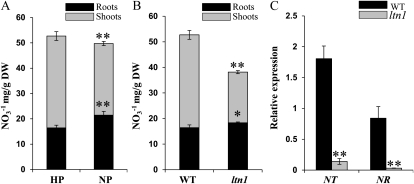
Pi deprivation increases nitrate accumulation in bean (Phaseolus vulgaris) roots and reduces nitrate levels in shoots, possibly by reducing both nitrate reductase activity in roots and nitrate transport from roots to shoots (Gniazdowska and Rychter, 2000). Similarly, rice also had increased nitrate levels in roots and decreased levels in shoots under Pi-deficient conditions (Fig. 9A). The nitrate contents were further analyzed in roots and shoots of the ltn1 mutant under Pi-sufficient conditions. The result showed that nitrate contents were increased in roots and decreased dramatically in shoots of the ltn1 mutant, with an overall decrease of total nitrate levels (Fig. 9B), which resembled the response of wild-type plants under Pi starvation conditions (Fig. 9A). The genes encoding a nitrate transporter and a nitrate reductase were repressed dramatically in ltn1 roots under Pi-sufficient conditions (Fig. 9C), indicating that the repression in nitrate assimilation in the ltn1 mutant probably resulted from reduced nitrate transport and nitrate reductase activity. These results demonstrated that a mutation in LTN1 might repress nitrate assimilation by affecting nitrate transport and nitrate reductase activity in roots.
Figure 9.
Nitrate contents and nitrate-related gene expression analysis in wild-type and ltn1 mutant plants. A, Nitrate concentration in roots (black bars) and shoots (gray bars) of wild-type plants under Pi-sufficient (HP; 16 mg L−1) or Pi-deficient (NP; 0 mg L−1) conditions. Error bars indicate sd (n = 3). DW, Dry weight. Asterisks indicate the significance of differences between wild-type plants grown under HP and NP conditions as determined by Student’s t test analysis: ** P < 0.01. B, Nitrate concentration in roots (black bars) and shoots (gray bars) of wild-type (WT) and ltn1 mutant plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01. C, qRT-PCR expression analysis of nitrate transporter (NT) and nitrate reductase (NR) genes in roots of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: ** P < 0.01.
Total Fe Contents Are Increased in the ltn1 Mutant
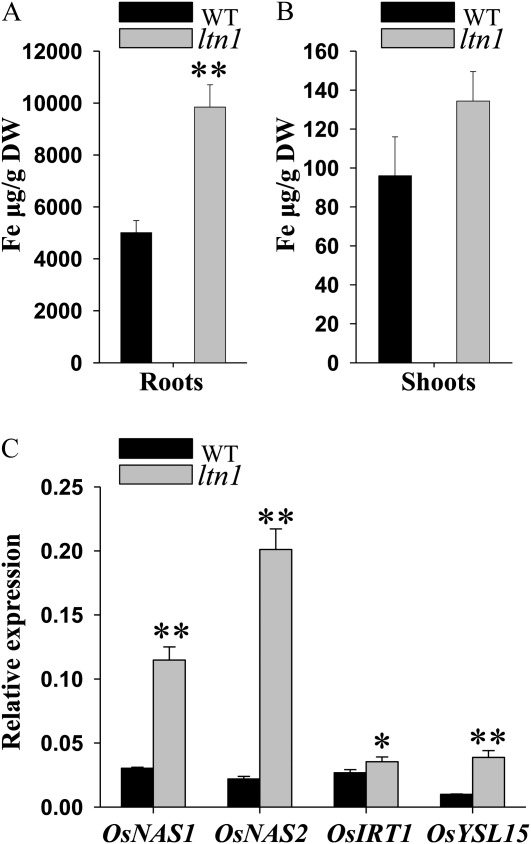
Total Fe contents were increased significantly in roots of the ltn1 mutant compared with wild-type plants under Pi-sufficient conditions, but not in ltn1 shoots (Fig. 10, A and B). In the ltn1 mutant, Fe-responsive genes such as OsNAS1, OsNAS2, OsIRT1, and OsYSL15 were also activated dramatically in roots under Pi-sufficient conditions (Fig. 10C), which correlated with increased Fe accumulation. These observations demonstrated that a mutation in LTN1 up-regulates Fe assimilation in rice, which activates Fe-responsive genes.
Figure 10.
Fe contents and Fe-responsive gene expression in wild-type and ltn1 mutant plants. A and B, Total Fe contents in roots (A) and shoots (B) of wild-type (WT; black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). DW, Dry weight. Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: ** P < 0.01. C, Expression of Fe-responsive genes in roots of wild-type (black bars) and ltn1 mutant (gray bars) plants under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test analysis: * 0.01 ≤ P < 0.05, ** P < 0.01.
LTN1 Is Down-Regulated by OsmiR399
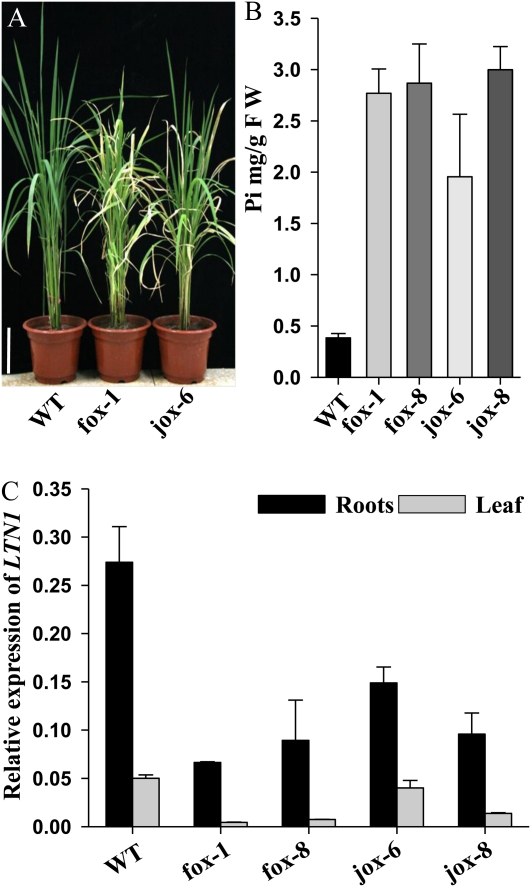
It was previously shown that four members (a, d, f, and j) of OsmiR399 were induced by Pi starvation (Bari et al., 2006). Five OsmiR399 binding sites were identified in the 5′ untranslated region of LTN1 (OsPHO2), suggesting that LTN1 may be down-regulated by OsmiR399. To confirm this, DNA fragments of OsmiR399f and OsmiR399j precursors under the control of the rice ACTIN1 promoter were constructed and transformed into wild-type rice. The transgenic plants constitutively expressing OsmiR399 displayed leaf tip necrosis and had other ltn1 mutant phenotypes (Fig. 11A). Pi levels increased in almost all the transgenic plants. Four transgenic lines (fox-1, fox-8, jox-6, and jox-8) were selected for further analysis. While leaf Pi concentration increased significantly (Fig. 11B), LTN1 transcript levels decreased dramatically in both roots and leaves under Pi-sufficient conditions (Fig. 11C). This indicated that overexpression of OsmiR399 (f and j) down-regulated LTN1 by degrading its transcript, which resulted in overaccumulation of Pi in rice leaves and a leaf tip necrosis phenotype. These results provided strong evidence that LTN1 is involved in Pi starvation signaling and functions downstream of OsmiR399 in rice.
Figure 11.
Phenotypical analysis, Pi contents, and LTN1 expression in wild-type and OsmiR399-overexpressing transgenic plants. A, Phenotypes of wild-type (WT), OsmiR399f-overexpressing (fox-1), and OsmiR399j-overexpressing (jox-6) plants grown in the greenhouse for 2 months under Pi-sufficient conditions. Bar = 20 cm. B, Leaf Pi concentration in wild-type, OsmiR399f-overexpressing transgenic (fox-1 and fox-8), and OsmiR399j-overexpressing transgenic (jox-6 and jox-8) plants grown in the greenhouse for 2 months under Pi-sufficient conditions. FW, Fresh weight. Error bars indicate sd (n = 3). C, Relative LTN1 expression in roots (black bars) and leaves (gray bars) of wild-type, OsmiR399f-overexpressing, and OsmiR399j-overexpression 2-week-old seedlings under Pi-sufficient conditions (16 mg L−1). Error bars indicate sd (n = 3).
DISCUSSION
A Mutation in LTN1 Results in Overaccumulation of Pi in Shoots
In Arabidopsis, mutations in PHO2 led to increased Pi uptake and transport and Pi overaccumulation in shoots (Dong et al., 1998; Aung et al., 2006), implying that PHO2 plays a pivotal role in regulating Pi uptake and transport. Hence, the investigation of PHO2 function in rice will provide useful information to develop improved crops with enhanced efficiency of Pi use. In this work, we identified the rice mutant ltn1, which showed leaf tip necrosis and growth defects in most agronomic traits. Map-based cloning identified LTN1 as LOC_Os05g48390, which encodes a ubiquitin-conjugating domain-containing protein with 58% amino acid sequence identity to Arabidopsis PHO2. Based on the conserved gene structure, Bari et al. (2006) previously proposed that the putative rice homolog of AtPHO2 was LOC_Os05g48390, which is consistent with our results. The ltn1 mutant also overaccumulated Pi in the shoots, implying that LTN1 and AtPHO2 proteins have conserved functions in regulating Pi uptake and transport. Genetic complementation further confirmed that the mutation in LTN1 was responsible for Pi overaccumulation in ltn1 shoots. Thus, LTN1 is the functional homolog of AtPHO2 and is involved in the regulation of Pi accumulation in rice.
LTN1 Regulates Pi Uptake and Transport by Controlling Expression of PHT Genes
Pi is acquired by roots and then transported through vascular tissues to the shoots (Schachtman et al., 1998). Expression analysis of LTN1 revealed that the gene was expressed more highly in roots than in other organs. Promoter::GUS analysis further showed that the LTN1 promoter was predominantly active in the vascular tissues of all organs tested. This expression pattern suggests that LTN1 may participate in Pi uptake and transport. Consistently, Pi uptake experiments with [32P]Pi showed that the ltn1 mutant had greater [32P]Pi uptake activity in roots and could translocate more [32P]Pi from roots to shoots. These results indicate that both Pi uptake in roots and Pi translocation from roots to shoots were stimulated in the ltn1mutant, resulting in Pi overaccumulation in the mutant shoots. The mobile Pi molecule can be remobilized from old to young leaves and can be retranslocated from shoots to roots to compensate for the Pi decrease in roots under Pi deprivation (Mimura et al., 1996; Jeschke et al., 1997; Chiou et al., 2006). In wild-type plants, Pi contents in old leaves decreased over time during Pi deprivation, indicating that Pi can be mobilized out of the old tissues under Pi starvation. However, Pi mobilization was impaired in the ltn1 mutant, since a Pi decrease in old leaves was not observed during Pi starvation. In our experiment, Pi concentrations in old leaves of the ltn1 mutant increased significantly at the last time point, which can be explained by a dramatic decrease in leaf fresh weight during senescence. Under Pi deficiency, Pi contents in the roots of wild-type plants initially decreased and then clearly recovered, which may be caused by Pi translocation from shoots to roots. But in the ltn1 mutant, Pi contents in the roots kept decreasing over time during Pi starvation and no recovery was observed, suggesting that Pi translocation from shoots to roots was defective in the ltn1 mutant. These results are consistent with the observed effects of the pho2 mutant in Arabidopsis, indicating that the function of PHO2 proteins in regulating Pi uptake and transport is highly conserved between Arabidopsis and rice.
Pi is first acquired by roots and then transported to and distributed in shoots (Schachtman et al., 1998). Analysis of Pi contents in different organs revealed that Pi distribution in shoots was also altered in the ltn1 mutant. In wild-type plants at the flowering stage, the culms contained the highest amount of Pi, followed by leaf sheaths, leaf blades, and inflorescences, whereas in the ltn1 mutant, all shoot organs showed increased Pi accumulation, with the most dramatic Pi overaccumulation in leaf blades. Pi transport into leaf blades seemed to be enhanced during Pi distribution in ltn1 shoots, suggesting that LTN1 plays a significant role in regulating Pi distribution in the shoots.
Pi transporters are directly responsible for Pi acquisition and transport in plants (Harrison et al., 2002; Misson et al., 2004; Seo et al., 2008), and plants generally up-regulate the expression of Pi transporters to enhance Pi uptake and transport efficiency under Pi starvation (Liu et al., 1998; Karthikeyan et al., 2002; Xiao et al., 2006). To determine if the increased Pi uptake and transport in the ltn1 mutant was dependent on the Pi transporter system, the expression of five members of the PHT gene family was compared between ltn1 mutant and wild-type plants. Under Pi-sufficient conditions, OsPT1, OsPT2, OsPT4, and OsPT8 were significantly induced in ltn1 roots compared with wild-type plants, whereas OsPT6 was dramatically repressed in ltn1 shoots. The differential response of PHT genes between roots and shoots in ltn1 was associated with Pi uptake and transport in the ltn1 mutant. Increased expression of OsPT1, OsPT2, OsPT4, and OsPT8 in roots may be responsible for the greater Pi uptake activity in roots and Pi transport from roots to shoots in the ltn1 mutant. The dramatic repression of OsPT6 in shoots may lead to defective Pi distribution and mobilization in ltn1 shoots. Taken together, these results strongly demonstrate that LTN1 is involved in the regulation of Pi acquisition and transport by controlling the expression of PHT genes.
LTN1 Is Involved in Root Architecture Alteration under Pi Starvation
Root architecture alteration is one of the most important adaptive responses of plants to Pi starvation. In Arabidopsis, low Pi availability can increase lateral root density and length and reduce primary root growth mediated by reduced cell elongation (Williamson et al., 2001; López-Bucio et al., 2002). By characterization of the low phosphorus insensitive (lpi) mutants, Sánchez-Calderón et al. (2006) provided genetic support that phosphorus is an important signal to regulate root development and showed that LPI1 and LPI2 are required for Pi starvation-responsive root architecture alteration in Arabidopsis. Svistoonoff et al. (2007) also showed that LPR1 and LPR2, which encode multicopper oxidases, are involved in Pi starvation-triggered primary root inhibition. More recently, Pérez-Torres et al. (2008) demonstrated that lateral root development in response to Pi starvation is mediated by modulating auxin sensitivity via the auxin receptor TIR1, further revealing the molecular mechanism of lateral root development alteration in response to Pi starvation in Arabidopsis. Although Pi starvation-induced root architecture alteration has been well studied in Arabidopsis, it is far from well understood in rice. Different from Arabidopsis, both primary and adventitious root elongation is a typical root architecture alteration in response to Pi deprivation in rice (Wissuwa, 2003; Yi et al., 2005). To investigate whether LTN1 is involved in this process, we analyzed the effect of Pi availability on root architecture alteration in wild-type and ltn1 mutant plants. Under Pi-sufficient conditions, no significant difference in root architecture was observed between wild-type and ltn1 mutant plants. However, the elongation of primary and adventitious roots was significantly enhanced in the ltn1 mutant under Pi starvation, which indicated that the ltn1 mutant is hypersensitive to Pi starvation. Consistently, Pi contents in ltn1 roots were much lower than in wild-type plants under Pi starvation conditions. The hypersensitivity of the ltn1 mutant to Pi starvation may be due to a more severe Pi starvation signal resulting from a lower Pi concentration in ltn1 roots under Pi deprivation. Root architecture alteration in the ltn1 mutant was only observed under Pi starvation conditions, which differs from the constitutive activation of other Pi starvation responses even in Pi-sufficient conditions in the ltn1 mutant. Possibly, LTN1 regulates Pi-dependent root architecture alteration indirectly by affecting the Pi concentration in roots under Pi starvation conditions.
LTN1 Is Crucial in Regulating Multiple Pi Starvation Responses in Rice Roots
Plants have evolved multiple strategies to increase Pi availability under low-Pi stress. These adaptive strategies include root morphology alteration, Pi transporter induction, acid phosphatase and nuclease production, and complex metabolic adjustments (Raghothama, 1999; Wasaki et al., 2003; Misson et al., 2005). Loss of function of LTN1 led to induction of PHT genes in ltn1 roots even under Pi-sufficient conditions, suggesting that a mutation in LTN1 probably mimics a Pi starvation signal and triggers the Pi starvation response. Thus, we further investigated whether LTN1 was a specific regulator only involved in Pi uptake and transport or a general regulator involved in regulating multiple Pi starvation responses.
Production of acid phosphatases and nucleases enables plants to release Pi from organic compounds under low-Pi conditions (Raghothama, 1999; Abel et al., 2002). Thus, the induction of acid phosphatases and nucleases is a significant adaptive response for plants under Pi deprivation. In our experiment, we found that the activity of both acid phosphatases and ribonucleases was increased significantly in ltn1 roots compared with wild-type plants under Pi-sufficient conditions. The induction of acid phosphatases and ribonucleases was also observed in Pi-deprived wild-type plants, but to a much lower extent than in ltn1 plants. This phenomenon may suggest that the stimulation of acid phosphatases and ribonucleases depends on the repression extent of LTN1. Genes encoding a rice acid phosphatase (OsPAP10) and two ribonucleases (RNase PD2 and S1/P1 nuclease) were also up-regulated, implying that LTN1 regulates the activity of acid phosphatases and ribonucleases through activating the expression of related genes in roots. This result correlates with the transcriptional response of OsPAP10 to Pi limitation (Zhou et al., 2008), suggesting that OsPAP10 is an important acid phosphatase in response to Pi starvation in rice. However, the functional RNase genes that are responsive to Pi starvation have not yet been reported in rice. Based on the significant increase in RNase activity and the up-regulation of two RNase genes in ltn1 roots, it is reasonable to propose that RNase PD2 and S1/P1 nuclease may be the functional ribonucleases involved in the Pi starvation response in rice.
Phosphorus is one of the most important mineral nutrients and participates in multiple metabolic processes. Thus, Pi starvation stress will affect multiple metabolic processes, including lipid metabolism, nitrogen assimilation, metallic element acquisition, and glycolysis, in plants (Wasaki et al., 2003; Wu et al., 2003; Misson et al., 2005). Altering metabolism enhances Pi recycling and represses Pi consumption, which enables plants to more efficiently adapt to low-Pi stress. When plants experience Pi starvation stress, Pi can be recovered from phospholipids, which will be substituted with galactolipids or sulfolipids. Analysis of lipid composition showed that two major phospholipids, PC and PE, were decreased while DGDG was increased in ltn1 roots compared with wild-type plants under Pi-sufficient conditions. This alteration is consistent with a modification of lipid composition induced by Pi starvation, indicating that LTN1 affects lipid composition in response to Pi starvation. In our experiment, the most dramatic alteration of lipid composition, indicated by PC and PE decrease and DGDG increase, occurred in ltn1 roots under Pi-deficient conditions. This phenomenon may be caused by the more severe Pi starvation signal resulting from the lower Pi concentration in ltn1 roots under Pi deprivation. In ltn1 roots, genes responsible for phospholipid degradation and galactolipid synthesis were up-regulated in Pi-sufficient conditions, implying that LTN1 alters lipid composition by regulating the expression of lipid metabolism genes.
Repression of nitrogen assimilation by Pi limitation allows plants to save more energy for Pi acquisition. In bean, Pi deprivation leads to increased root nitrate accumulation and suppresses shoot nitrate accumulation, possibly by repressing nitrate reductase activity and nitrate transport from roots to shoots (Gniazdowska and Rychter, 2000). A similar alteration in nitrate contents was observed in wild-type rice plants under Pi starvation conditions, indicating that rice also represses nitrate assimilation during Pi starvation. Our data further revealed that the nitrate contents in ltn1 roots and shoots were, respectively, higher and much lower than in wild-type plants under Pi-sufficient conditions. These changes in ltn1 nitrate contents resemble the Pi starvation-triggered alteration of nitrate contents in wild-type plants, demonstrating that LTN1 participates in the regulation of nitrogen assimilation during Pi starvation. Nitrate is usually acquired by roots and then transported into the shoots or reduced into nitrite for further use. Genes encoding nitrate transporter and nitrate reductase were dramatically repressed in ltn1 roots under Pi-sufficient conditions, implying that the altered nitrate contents in the ltn1 mutant probably resulted from repression of both nitrate transport from roots to shoots and nitrate reductase activity. The nitrate transporter identified here probably plays a significant role in transporting nitrate from roots to shoots. Thus, nitrate transporter repression may lead to defective nitrate transport from roots to shoots and result in increased root nitrate contents and decreased shoot nitrate contents in the ltn1 mutant. In addition, the dramatic nitrate reductase repression in roots affects the reduction of nitrate to nitrite and thereby also increases nitrate accumulation in ltn1 roots.
Increase in metallic element acquisition is another adaptive response to Pi starvation in plants (Wasaki et al., 2003; Misson et al., 2005). An enhanced capacity to absorb and scavenge Pi-complexing metals is considered to be an adaptive response to Pi starvation that could increase free Pi availability (Misson et al., 2005). Pi deprivation leads to increased Fe contents and activation of Fe-responsive genes in rice (Zheng et al., 2009). Total Fe contents were also increased in both roots and shoots of ltn1 mutants under Pi-sufficient conditions compared with wild-type plants. In association with the increased Fe contents in the ltn1 mutant, several genes involved in Fe assimilation, such as OsNAS1, OsNAS2, OsIRT1, and OsYSL15, were activated in ltn1 roots under Pi-sufficient conditions. These observations reveal that LTN1 loss of function mimics the Pi starvation signal and activates the expression of Fe-responsive genes to increase Fe acquisition. Although Fe contents were increased in both ltn1 roots and shoots, Fe accumulation and up-regulation of Fe assimilation-related genes were much more significant in roots than in shoots, suggesting that LTN1-regulated Fe acquisition occurs mainly in roots.
The acceleration of carbon supply for organic acid synthesis through glycolysis was also proposed to be enhanced in response to Pi starvation. It was demonstrated that the activities of glycolysis enzymes, such as phosphoenolpyruvate carboxylase (PEPC), phosphoenolpyruvate phosphatase, and pyruvate kinase (PK), are stimulated by Pi starvation in plants (Nanamori et al., 2004). To determine whether LTN1 is involved in glycolysis, the expression of several glycolysis-related genes and PEPC and PK activities were analyzed. Although the expression of glycolysis-related genes and the activities of PEPC and PK seemed to be up-regulated in ltn1 roots, the changes were not significant (Supplemental Fig. S8; Supplemental Table S1). Possibly, the alteration of glycolysis during Pi starvation is not significant in rice, or glycolysis is indirectly regulated by LTN1.
By characterization of the ltn1 mutant, we observed that most of the Pi starvation responses were activated even under Pi-sufficient conditions. Interestingly, the Pi starvation responses were mainly detected in ltn1 roots, indicating that LTN1 has a distinct function in regulating Pi starvation responses between roots and shoots. LTN1 seems to be a negative regulator of Pi starvation responses only in roots, which correlates with the relatively high LTN1 expression levels in roots. Pi toxicity in shoots results in serious damage to ltn1 growth and development, which probably interfered with the experimental results obtained from shoots. Thus, in this work, we mainly focused on the function of LTN1 in roots. Further work is needed to understand the function of LTN1 in shoots. Detailed analysis of LTN1-overexpressing plants will be helpful to elucidate the function of this gene in shoots.
Although in previous studies many genes were proposed to regulate multiple Pi starvation responses, the support for their regulatory functions was mainly inferred from expression analysis. We provide direct physiological evidence for Pi starvation responses in ltn1 roots, clearly indicating that LTN1 is involved in the regulation of multiple Pi starvation responses in roots. Additionally, the expression of related genes also correlates with the Pi starvation responses in ltn1 roots, indicating that LTN1 can influence the expression of related genes at the transcriptional level to regulate Pi starvation responses. These results not only provide further insight into possible LTN1 function but also lead to the identification of downstream genes responsible for Pi starvation responses in rice. Further work using transgenic plants is needed to elucidate the function of those genes identified in this work.
LTN1 Works Downstream of OsmiR399
In Arabidopsis, Pi starvation-induced miR399 can degrade PHO2 mRNA, indicating that PHO2 is involved in Pi starvation signaling downstream of miR399 (Fujii et al., 2005; Bari et al., 2006; Chiou et al., 2006). Based on the conserved gene structure of OsPHO2 (LTN1) with AtPHO2 and the induction of OsmiR399 (OsmiR399a, OsmiR399d, OsmiR399f, and OsmiR399j) under Pi starvation in rice, the miR399 regulatory mechanism on PHO2 was proposed to be conserved between Arabidopsis and rice (Bari et al., 2006). To verify LTN1 repression by OsmiR399, OsmiR399f- and OsmiR399j-overexpressing transgenic plants were generated. LTN1 repression, overaccumulation of Pi, and Pi toxicity were observed in both OsmiR399f-overexpressing (fox-1 and fox-8) and OsmiR399j-overexpressing (jox-6 and jox-8) plants, strongly suggesting that LTN1 was negatively regulated by OsmiR399 at the mRNA level. Previous work has shown that OsmiR399 is involved in Pi starvation signaling in rice and is under the control of OsPHR2 (Zhou et al., 2008). Thus, these results demonstrate that LTN1 functions downstream of OsmiR399 to mediate Pi starvation signaling in rice.
LTN1 Functions in Pi Starvation Signaling Transduction
As described above, a mutation in LTN1 results in typical Pi starvation responses such as up-regulation of PHT genes, induction of phosphatases and ribonucleases, and metabolic alterations, suggesting that this mutant mimics Pi starvation signaling and triggers multiple Pi starvation responses. LTN1 down-regulation by Pi starvation-induced OsmiR399 further verifies that LTN1 is involved in Pi starvation signaling in rice. Based on these observations, we conclude that LTN1 is a significant component of Pi starvation signaling in rice and participates in the regulation of a wide range of Pi starvation responses. Under Pi-sufficient conditions, LTN1 probably functions as a negative regulator to repress the expression of Pi starvation-responsive genes. When Pi starvation stress occurs, OsmiR399 is induced and LTN1 mRNA is degraded by OsmiR399. LTN1 repression leads to the activation of Pi starvation-responsive genes and turns on Pi starvation responses. Recently, OsPHO2 (LTN1) was shown to be involved in Pi starvation signaling mediated by OsSPX1-OsPHR2, as indicated by a significant up-regulation of OsSPX1 in ospho2 roots (Wang et al., 2009; Liu et al., 2010). The same results were also obtained with our ltn1 mutant (Supplemental Fig. S9). Although the relationship between OsPHO2 (LTN1) and OsSPX1-OsPHR2-OsPT2 explains Pi overaccumulation in the shoots of OsPHR2 overexpressing and OsSPX1 RNA interference plants and provides new insight into the Pi starvation signaling in rice, the detailed mechanisms of Pi overaccumulation in the ospho2 (ltn1) mutant and OsPHO2 (LTN1) protein function in the regulation of Pi starvation responses remain to be elucidated. The fact that enhanced Pi uptake and transport in the ltn1 mutant correlated with up-regulation of OsPT1, OsPT2, OsPT4, and OsPT8 provides physiological and molecular evidence for the mechanism of Pi accumulation in the ltn1 mutant. More importantly, besides Pi uptake and transport, LTN1 also participates in the regulation of a series of important Pi starvation responses, such as stimulation of phosphatase and RNase activities, lipid composition alteration, nitrogen assimilation repression, and increased metal uptake. Therefore, we can conclude that LTN1 is a pivotal regulator involved in multiple Pi starvation responses in rice.
However, there is still a big gap in explaining the correlation between the proposed LTN1 gene function and the observed phenotypes of the ltn1 mutant. LTN1 encodes a ubiquitin-conjugating enzyme (E2) that plays a key role in the ubiquitin-mediated protein degradation pathway, suggesting that the LTN1 protein probably functions as a negative regulator that can repress its target gene expression by protein degradation. Therefore, in ltn1 plants, the degradation of LTN1 target proteins would be blocked because of the LTN1 mutation, resulting in LTN1 target protein activation. Possibly, LTN1 target proteins function as positive regulators that can directly up-regulate the expression of PHT genes and Pi starvation-responsive genes, which leads to excessive accumulation of Pi in shoots, leaf tip necrosis, and constitutive activation of multiple Pi starvation responses in the ltn1 mutant. Thus, identification of LTN1 target proteins is necessary to explain how LTN1 works to regulate the downstream genes and the phenotype caused by the LTN1 mutation. Since the regulation of Pi starvation responses by LTN1 occurs mainly by controlling the expression of related genes, we propose that the targets of LTN1 are most likely transcription factors that can activate the expression of PHT genes and Pi starvation-responsive genes. Moreover, the typical ubiquitin-mediated protein degradation pathway usually includes ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3). Whether LTN1 cooperates with E3 or works as an E2-E3 dual-function enzyme to mediate the degradation of the target protein remains to be determined. Further work, such as yeast two-hybrid screening of LTN1 interaction proteins or ltn1 suppressor/high-Pi mutant screening, will be helpful to clarify these questions and contribute to the understanding of the Pi starvation signaling mediated by LTN1 in rice.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) ltn1 mutant and its parent cv Nipponbare were grown in the field or in a greenhouse with a 12-h day (30°C) and a 12-h night (24°C). For hydroponic culture, half-strength Hoagland medium was used. The Pi deprivation culture was half-strength modified Hoagland medium with the substitution of (NH4)H2PO4 with NH4NO3. For Pi treatment, wild-type plants were first grown under normal half-strength Hoagland medium for 10 d after germination, then transferred to half-strength Hoagland medium supplemented with very high Pi concentrations (160 mg L−1 or 320 mg L−1). The hydroponic culture was adjusted to pH 6.0 with 1 m NaOH before use, and the medium was changed every 3 d. The hydroponic cultured seedlings of ltn1 and wild-type Nipponbare were grown in an incubator with a 12-h-day (30°C)/12-h-night (28°C) photoperiod, approximately 200 μmol photons m−2 s−1 photon density, and approximately 70% humidity.
Map-Based Cloning of LTN1
The ltn1 mutants were crossed with Minghui 63, an indica variety. Plants showing leaf tip necrosis in the F2 progeny were selected for the genetic linkage analysis. Preliminary mapping was performed with the molecular markers distributed throughout the rice genome (McCouch et al., 2002). The fine-mapping sequence-tagged site primers were designed according to the DNA sequences of indica and japonica (http://www.ncbi.nlm.nih.gov). Primers used in the fine mapping are listed in Supplemental Table S2.
Complementation of the ltn1 Mutant
For complementation of the ltn1 mutant, a 9.4-kb DNA fragment containing the entire LTN1 gene was obtained by splicing together three DNA fragments from PCR amplifications. The primers used are listed in Supplemental Table S3. These three DNA fragments with overlapping sequences were individually inserted into the pMD18-T vector (Takara) to produce the 9.4-kb fragment. XhoI- and KpnI-recognizing sequences were added to the ends. The 9.4-kb fragment was digested with XhoI and KpnI and inserted into the binary vector pCAMBIA2300 to generate the resulting binary vector. The resulting binary and empty pCAMBIA2300 vectors were introduced into the ltn1 mutants using the Agrobacterium tumefaciens-mediated transformation method (Liu et al., 2007).
RNA Extraction, cDNA Preparation, and qRT-PCR
Total RNAs were extracted using the RNAVzol reagent (Vigorous). About 2 μg of the total RNAs treated with DNase I was used to synthesize the first-strand cDNA with oligo(dT)18 as primer. The product of first-strand cDNA was used as the template for the PCR. For qRT-PCR, SYBR Green I was added to the reaction system and run on a Chromo4 real-time PCR detection system (Bio-Rad) according to the manufacturer’s instructions. The data were analyzed with Opticon monitor software (Bio-Rad). Three replicates were performed for each gene. Rice ACTIN1 was used as the internal control in all analyses. The primers used for qRT-PCR are listed in Supplemental Table S4.
GUS Staining
Different parts from mature transgenic plants and seedlings were collected and used for histochemical detection of GUS expression. The collected samples were incubated in a solution containing 50 mm sodium phosphate buffer (pH 7.0), 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, 0.1% Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid at 37°C (Jefferson, 1989). Images were taken directly or with a stereomicroscope (SZX16; Olympus).
Pi Concentration Determination and Pi Remobilization Assay
Pi measurement was carried out as described previously (Delhaize and Randall, 1995; Wang et al., 2009). For the Pi remobilization assay, seedlings were grown in half-strength Hoagland medium for 2 weeks and then transferred into the Pi deprivation conditions [half-strength Hoagland medium with the substitution of (NH4)H2PO4 with NH4NO3]. The fourth leaves and the roots were collected and weighed every 4 d. The Pi contents of samples were measured and analyzed.
Pi Uptake Assay
Two-week-old seedlings grown in half-strength Hoagland medium were used for the Pi uptake assay. The uptake buffer was half-strength Hoagland solution with the addition of KH232PO4 (Fu Rui). The Pi uptake assay was performed according to Chiou et al. (2006). Four seedlings were pooled as a biological sample and incubated in the uptake buffer. After 1, 2, and 3 h, reactions were stopped by rinsing the seedlings 10 times with half-strength Hoagland solution to remove the KH232PO4. Root and shoot samples were weighed and lysed in 500 μL of 30% hydrogen peroxide and 200 μL of perchloric acid at 70°C for 1 h. Scintillation cocktail was added to 140 μL of the mixture up to a final volume of 2 mL, and the radioactivity was measured by scintillation counting (Perkin-Elmer). Autoradiography was performed according to Ai et al. (2009). Rice seedlings were grown in 100 mL of half-strength Hoagland medium containing 5 μCi of KH232PO4 for 24 h with a 16/8-h day/night cycle with temperature settings of 28°C/22°C. The seedlings were washed to get rid of the KH232PO4 on the surface and then dried with filter paper. Autoradiographs of the radioactive seedlings were developed using Kodak X-Omat film (http://www.kodak.com).
Assay of Intracellular Acid Phosphatase and RNase Activities
Analysis of intracellular acid phosphatase activity was done according to Trull and Deikman (1998). Briefly, protein was extracted with a native buffer (pH 7.8, 50 mm phosphate-buffered saline, 1 mm EDTA, 1% [w/v] polyvinylpyrrolidone, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) from roots of 2-week-old seedlings. Before electrophoresis, the protein concentration was measured by the method of Bradford (1976), using bovine serum albumin as a standard. The protein was separated by SDS-PAGE (9%) at 4°C. Gels were washed with cold distilled water to remove the SDS. Gels were then shaken in 50 mm sodium acetate (pH 5.5)/10 mm MgCl2 twice for 15 min. Finally, the gels were stained with Fast Black K salt and β-naphthyl acid phosphate. For analysis of RNase activity, protein extraction and measurement were carried out as described above, and enzyme assays were according to the protocol described by Yen and Green (1991).
TLC Lipid Composition Assay
Total lipids were extracted from roots and shoots as described previously by O’Brien and Benson (1964). Lipid extracts were assayed on silica gel TLC plates (Merck) with an acetone:toluene:water (91:30:7, v/v) solvent system. The lipids were visualized with iodine vapor and recognized by cochromatography with known standard lipid samples.
Assay of PEPC and PK Activities
PEPC and PK activities were analyzed using 2-week-old seedlings grown in half-strength Hoagland medium as described by Nanamori et al. (2004).
Nitrate Concentration Measurement
Nitrate concentration was measured using a colorimetric method (Cataldo et al., 1975). The roots and shoots of 2-week-old seedlings grown in half-strength Hoagland medium were collected and ground to a powder in liquid nitrogen. The samples were dried at 70°C for 48 h in an oven. Samples (100 mg) were suspended in 10 mL of deionized water and incubated at 45°C for 1 h. The suspension was centrifuged at 5,000g for 15 min. Supernatant (0.2 mL) and 0.8 mL of 5% (w/v) salicylic acid in concentrated H2SO4 were mixed together. After 20 min, 19 mL of 2 n NaOH was added slowly into the mixture to raise the pH above 12. Samples were cooled to room temperature, and the absorbance was determined at 410 nm. Nitrate concentration was calculated according to standards containing NO3−1.
Total Fe Content Measurement
Roots and shoots of 2-week-old seedlings grown in half-strength Hoagland medium were dried at 70°C for 48 h in an oven. Approximately 80-mg samples were digested with 13 mL of concentrated HNO3 and 2 mL of 30% hydrogen peroxide at 140°C for 30 min. Digested solution volumes were made to 25 mL with deionized water. The total Fe contents were determined with an inductively coupled plasma spectrometer (Optima 2000 DV; PerkinElmer).
Statistical Analysis
The data were analyzed using Excel software (Microsoft) for average values, sd, and Student’s t test analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AU032431 (LTN1), AB052843.1 (RNase PD2), AK106059.1 (S1/P1 nuclease), AJ276277.2 (PLC), AK064148.1 (DGGT), AK068409 (NT), and AK102178 (NR).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. LTN1 transcript detection in wild-type and ltn1 mutant plants using semiquantitative RT-PCR.
Supplemental Figure S2. Protein alignment between Arabidopsis PHO2 and rice LTN1.
Supplemental Figure S3. Leaf performance and Pi contents of wild-type plants grown under very high-Pi conditions.
Supplemental Figure S4. Leaf performance and Pi contents of ltn1 mutant plants grown under low-Pi conditions.
Supplemental Figure S5. Pi distribution in different tissues of wild-type and ltn1 mutant plants.
Supplemental Figure S6. Analysis of acid phosphatase and RNase activities in shoots of wild-type and ltn1 mutant plants.
Supplemental Figure S7. TLC analysis of lipid composition in shoots of wild-type and ltn1 mutant plants.
Supplemental Figure S8. Expression analysis of glycolysis-related genes in roots of wild-type and ltn1 mutant plants.
Supplemental Figure S9. Expression analysis of OsSPX1 and OsPHR2 in wild-type and ltn1 mutant plants.
Supplemental Table S1. Analysis of PEPC and PK activities in roots of wild-type and ltn1 mutant plants.
Supplemental Table S2. Primers used for map-based cloning.
Supplemental Table S3. Primers used for the plasmid constructs.
Supplemental Table S4. Primers used for qRT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Prof. Huixia Shou for kindly providing the detailed method for Pi determination. We thank Prof. Hongqing Lin and Hongfei Liu for help in Fe content determination. We thank Dr. Michael Schläppi for editing the language of the manuscript.
References
- Abel S, Ticconi CA, Delatorre CA. (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cataldo D, Maroon M, Schrader L, Youngs V. (1975) Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Dong B, Rengel Z, Delhaize E. (1998) Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205: 251–256 [DOI] [PubMed] [Google Scholar]
- Duff S, Sarath G, Plaxton W. (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90: 791–800 [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Gniazdowska A, Rychter A. (2000) Nitrate uptake by bean (Phaseolus vulgaris L.) roots under phosphate deficiency. Plant Soil 226: 79–85 [Google Scholar]
- Gniazdowska A, Szal B, Rychter A. (1999) The effect of phosphate deficiency on membrane phospholipid composition of bean (Phaseolus vulgaris L.) roots. Acta Physiol Plant 21: 263–269 [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jeschke W, Kirkby E, Peuke A, Pate J, Hartung W. (1997) Effects of P deficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L.). J Exp Bot 48: 75–91 [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. (1998) Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol 116: 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164: 969–979 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Liu L, Zhu C, Sun C, Xu B, Fang J, Tang J, Luo A, Cao S, Li G, et al. (2009) Molecular analysis of rice plants harboring a multi-functional T-DNA tagging system. J Genet Genomics 36: 267–276 [DOI] [PubMed] [Google Scholar]
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res (Suppl) 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Mimura T, Sakano K, Shimmen T. (1996) Studies on the distribution, re-translocation and homeostasis of inorganic phosphate in barley leaves. Plant Cell Environ 19: 311–320 [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamori M, Shinano T, Wasaki J, Yamamura T, Rao IM, Osaki M. (2004) Low phosphorus tolerance mechanisms: phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar Mulato compared with rice. Plant Cell Physiol 45: 460–469 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- O’Brien JS, Benson AA. (1964) Isolation and fatty acid composition of the plant sulfolipid and galactolipids. J Lipid Res 5: 432–434 [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L. (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Reid RJ, Ayling SM. (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Seo HM, Jung Y, Song S, Kim Y, Kwon T, Kim DH, Jeung SJ, Yi YB, Yi G, Nam MH, et al. (2008) Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnol Lett 30: 1833–1838 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Trull MC, Deikman J. (1998) An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206: 544–550 [DOI] [PubMed] [Google Scholar]
- Vance C, Uhde-Stone C, Allan D. (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H. (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T. (2003) Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ 26: 1515–1523 [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M. (2003) How do plants achieve tolerance to phosphorus deficiency? Small causes with big effects. Plant Physiol 133: 1947–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Liu J, Dewbre G, Harrison M, Wang ZY. (2006) Isolation and characterization of root-specific phosphate transporter promoters from Medicago truncatula. Plant Biol (Stuttg) 8: 439–449 [DOI] [PubMed] [Google Scholar]
- Yen Y, Green PJ. (1991) Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol 97: 1487–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138: 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, et al. (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.