Abstract
The seed oil content in oilseed crops is a major selection trait to breeders. In Arabidopsis (Arabidopsis thaliana), LEAFY COTYLEDON1 (LEC1) and LEC1-LIKE (L1L) are key regulators of fatty acid biosynthesis. Overexpression of AtLEC1 and its orthologs in canola (Brassica napus), BnLEC1 and BnL1L, causes an increased fatty acid level in transgenic Arabidopsis plants, which, however, also show severe developmental abnormalities. Here, we use truncated napin A promoters, which retain the seed-specific expression pattern but with a reduced expression level, to drive the expression of BnLEC1 and BnL1L in transgenic canola. Conditional expression of BnLEC1 and BnL1L increases the seed oil content by 2% to 20% and has no detrimental effects on major agronomic traits. In the transgenic canola, expression of a subset of genes involved in fatty acid biosynthesis and glycolysis is up-regulated in developing seeds. Moreover, the BnLEC1 transgene enhances the expression of several genes involved in Suc synthesis and transport in developing seeds and the silique wall. Consistently, the accumulation of Suc and Fru is increased in developing seeds of the transgenic rapeseed, suggesting the increased carbon flux to fatty acid biosynthesis. These results demonstrate that BnLEC1 and BnL1L are reliable targets for genetic improvement of rapeseed in seed oil production.
Canola (Brassica napus) is a major oil-producing crop, and the global production of rapeseed oil was over 22 million tons during 2009 to 2010, which is the third largest source of the vegetable oil supply (http://usda.mannlib.cornell.edu/usda/ers/89002/Table47.xls). Canola oil is of high nutritional value with high concentrations of unsaturated C18 fatty acids (FAs; >60%) and a low level of erucic acid (C22:1; <1%), an undesirable FA in edible oil (Harvey and Downey, 1964; Dupont et al., 1989). Moreover, vegetable oil is a suitable source for biodiesel fuels and important raw materials in industry (Ohlrogge, 1994; Thelen and Ohlrogge, 2002). The demand for vegetable oil is sharply increased in recent years, and the increase of the oil content is thus a major challenge for genetic improvement of oilseed crops.
Vegetable oil is mainly synthesized during the maturation phase of the seed and is the major energy reserve for later growth and development prior to the full establishment of photosynthetic capacity of the seedling. The storage oil in seeds mainly consists of triacylglycerol (TAG) synthesized from glycerol-3-P and FAs (Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Voelker and Kinney, 2001). In higher plants, biosynthesis of FA and lipids has been well characterized by biochemical and molecular studies (Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Harwood, 1996; Beisson et al., 2003). FA is de novo synthesized mainly from acetyl-CoA catalyzed in plastids by a series of enzymes, of which acetyl-CoA carboxylase (ACCase) and the FA synthase (FAS) multienzyme complex are the most critical enzymes. ACCase, a rate-limiting enzyme of FA synthesis, catalyzes the formation of malonyl-CoA from acetyl-CoA. Subsequently, FAS catalyzes the transfer of the malonyl moiety of malonyl-CoA to acyl-carrier protein (ACP) by adding two carbons to the growing chain, eventually resulting in the formation of C16:0 and C18:0 acyl-ACP, which are then released from the FAS complex and transferred into the cytoplasm (Slabas and Fawcett, 1992; Ohlrogge and Jaworski, 1997; Voelker and Kinney, 2001; Sasaki and Nagano, 2004). In the cytoplasm, FA dehydrogenase (FAD) catalyzes the formation of unsaturated FA, and FA elongase (FAE) sequentially adds two-carbon units to the growing acyl chain to form long-chain FAs. Finally, TAG synthesis is initiated in the endoplasmic reticulum by glycerol-3-P acyltransferase, and the reaction is sequentially completed by lysophosphatidic acid acyltransferase and diacylglycerol acyltransferase ( Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Voelker and Kinney, 2001).
Biosynthesis of FA and TAG is tightly linked to photosynthesis and carbohydrate metabolism, which provide carbon source for FA synthesis. In rapeseed, Suc is mainly produced in the silique wall during the seed-filling stage, which is transported into developing seeds, and is then converted into hexose UDP-Glc and Fru. In the cytosol and the plastid of the embryo cells, hexose is further converted into acetyl-CoA, a precursor of FA biosynthesis (King et al., 1997; Baud et al., 2002; Hill et al., 2003; Schwender et al., 2003). Using isolated plastids from developing rapeseed seeds, it has been found that key cytosolic metabolites, including Glc-6-P, malate, phosphoenol pyruvate, and pyruvate, are imported into the plastid and then converted into acetyl-CoA through the glycolytic pathway to initiate de novo synthesis of FAs, of which pyruvate and Glc-6-P cause the highest rate of FA biosynthesis (Kang and Rawsthorne, 1994; Rawsthorne, 2002). Thus, the increased carbon flux is necessary for a higher rate of FA biosynthesis.
During the past years, significant efforts have been made to manipulate key FA synthetic genes in various species using the transgenic technology. Transgenic studies with transgenes encoding key enzymes or enzyme subunits resulted in altered levels of lipids at varying degrees (Shorrosh et al., 1995; Zou et al., 1997; Jako et al., 2001), and in some cases the oil content was reduced (Dehesh et al., 2001). It is somewhat expected that the manipulation of single genes may not be an efficient approach because FA synthesis is a highly coordinated process involved in not only FA synthetic genes but also carbon metabolism genes, especially the control of carbon flux from glycolysis to FA biosynthesis (Broun et al., 1999; Thelen and Ohlrogge, 2002; Jaworski and Cahoon, 2003; Cahoon et al., 2007; Weselake et al., 2009). Therefore, genetic manipulations of key regulatory genes, likely represented by transcription factors or protein kinases, should be able to overcome the bottlenecks of the entire pathway of FA synthesis (Ohlrogge and Jaworski, 1997; Ruuska et al., 2002; Cahoon et al., 2007; Mu et al., 2008).
In Arabidopsis (Arabidopsis thaliana), several genes encoding embryo-specific transcription factors have been shown to play important roles in the regulation of FA biosynthesis. The best-understood examples are LEAFY COTYLEDON1 (AtLEC1), LEC1-LIKE (AtL1L), LEC2, and WRINKLED1 (WRI1; Focks and Benning, 1998; Lotan et al., 1998; Stone et al., 2001; Kwong et al., 2003; Cernac and Benning, 2004; Mu et al., 2008). WRI1, encoding an AP2/EREBP transcription factor, regulates a subset of genes involved in glycolysis and the incorporation of Suc into TAG (Focks and Benning, 1998; Cernac and Benning, 2004). LEC2 encodes a plant-specific B3-type transcription factor and directly regulates WRI1, which, in turn controls a subset of late glycolytic genes and FA synthetic genes (Stone et al., 2001; Baud et al., 2007). AtLEC1, encoding an NF-YB-type transcription factor and acting in a pathway independent from that of LEC2 (Lotan et al., 1998; Cernac et al., 2006), positively regulates WRI1 and a large repertoire of FA synthetic genes and several glycolytic genes (Baud et al., 2007; Mu et al., 2008). Overexpression of these genes results in an increased level of FAs in the transgenic Arabidopsis plants, accompanied by up-regulated expression of key FA synthetic genes as well as glycolytic genes (Stone et al., 2001; Kwong et al., 2003; Cernac and Benning, 2004; Baud et al., 2007; Mu et al., 2008). However, in most, if not all cases, overexpression of these transcription factor genes causes a variety of developmental abnormalities and even lethality (Lotan et al., 1998; Stone et al., 2001; Cernac and Benning, 2004; Wang et al., 2007; Mu et al., 2008; Shen et al., 2010). These detrimental effects render it difficult to directly utilize these genes in genetic improvement of oil-producing crops. A recent study showed seed-specific expression ZmWRI1, a WRI1-like gene of maize (Zea mays), enhanced oil accumulation in transgenic maize without detectable abnormalities. However, expression of ZmLEC1 under similar conditions severely affected growth and development of the resulting transgenic maize plants (Shen et al., 2010). Similar results were obtained by constitutive overexpression of the ZmWRI1 gene in the transgenic maize plants (Pouvreau et al., 2011).
Here, we present that seed-specific expression of BnLEC1 and BnL1L, driven by two modified napin A (napA) promoters, significantly increases the oil content in the transgenic rapeseed seeds. The transgenic plants show normal growth and development without any detrimental effects on major agronomic traits. Consistent with these results, expression of a subset of genes involved in FA biosynthesis, glycolysis, and sugar metabolism is increased in developing seeds and the silique wall of the transgenic rapeseed. These results provide a practical approach for the genetic improvement of canola and possibly other oilseed crops as well.
RESULTS
Modification and Characterization of Seed-Specific Promoters for Transgenic Studies in Rapeseed
We previously reported that overexpression of AtLEC1 and AtL1L genes and their orthologs BnLEC1 and BnL1L from canola resulted in a dramatically increased level of FAs in the transgenic Arabidopsis plants (Mu et al., 2008). However, overexpression of these genes caused pleiotropic phenotypes and lethality in most transgenic lines (Mu et al., 2008). Because AtLEC1 and AtL1L are key regulators of embryogenesis and embryo maturation and are specifically expressed in embryos (Lotan et al., 1998; Kwong et al., 2003), it is expected that ectopic expression of these genes in vegetative tissues causes developmental abnormalities.
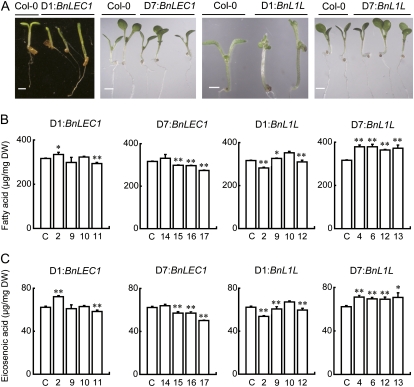
To explore the possible applications of these genes in genetic improvement of oilseed crops, we first tested if seed-specific expression of these genes could increase the FA level in transgenic Arabidopsis seeds. The BnLEC1 and BnL1L genes were placed under the control of the rapeseed storage protein 2S-1 promoter, which is also known as the napA promoter (hereafter referred to as the D1 promoter; see below for explanation). The resulting D1:BnLEC1 and D1:BnL1L transgenes were transformed into wild-type Arabidopsis plants (Columbia-0 [Col-0]). Analysis of multiple independent transgenic lines revealed that these transgenes caused severe abnormality after germination, and most of these plants died before flowering or did not produce seeds after flowering (Fig. 1A). Among the transgenic lines that showed relatively normal growth, the level of FAs and eicosenoic acid (C20:1), a marker for the accumulation of TAG in Arabidopsis, was marginally altered or reduced compared with the wild-type seeds (Fig. 1, B and C; Supplemental Fig. S1, A and B).
Figure 1.
Characterization of transgenic Arabidopsis plants carrying the BnLEC1 and BnL1L transgenes driven by seed-specific promoters. A, Seven-day-old wild-type (Col-0) and transgenic seedlings carrying different transgenes as indicated at the top. Seedlings shown at the left were germinated and grown in soil, and all others were germinated and grown on an agar plate. Note that D1:BnLEC1 and D1:L1L transgenic seedlings showed malformed cotyledons and the delayed initiation of true leaves. Bars = 1 mm. B, Total FA contents in the seeds harvested from Col-0 (C) and transgenic plants (transgenic line numbers are given below the graph) analyzed by GC-MS. Four independent transgenic lines were analyzed for each construct. Data presented are mean values of three experiments, and standard deviations are indicated by bars. See Supplemental Figure S1 for detailed analysis of FAs in these transgenic seeds. C, The accumulation of eicosenoic acid (C20:1; an FA maker for the formation of triacylglycerol in Arabidopsis) in seeds of Col-0 (C) and the transgenic plants (line numbers are given below the graph). Data presented are mean values of three experiments, and standard deviations are indicated by bars. In B and C, DW indicates dry weight. Asterisks indicate statistically significant differences compared with the control (Student’s t test: *P < 0.05; **P < 0.01).
We reasoned that seed-specific expression of the BnLEC1 and BnL1L genes at a reduced level might increase the FA level and minimize the developmental abnormalities observed in D1:BnLEC1 and D1:BnL1L transgenic plants. Previous studies on the napA promoter indicated that the sequences between −152 and +44 are essential for the seed-specific expression pattern in transgenic tobacco plants, and several cis-elements upstream from −152 were found to act as positive or negative regulatory motifs (Stålberg et al., 1993; Ellerström et al., 1996). We made similar napA promoter GUS reporter constructs that were stably transformed into Arabidopsis plants. For conciseness, we designated these mutant promoters as D2 through D8, whereas the wild-type napA promoter was referred to as D1 (Supplemental Fig. S2A). Consistent with the results obtained from the transgenic tobacco (Nicotiana tabacum) studies (Stålberg et al., 1993; Ellerström et al., 1996), the reporter gene assay in the transgenic Arabidopsis plants revealed the presence of a negative regulatory element upstream of −309 and multiple positive regulatory elements downstream of −309 (Supplemental Fig. S2B). We found that whereas deletions up to −270 (D4) and −250 (D5) maintained approximately 52% activity of the wild-type promoter, deletions of further downstream sequences (−230 or D6 and −211 or D7) caused the loss of the promoter activity more than 82% and 96%, respectively (Supplemental Fig. S2B). Histochemical analysis indicated that these truncated promoters retained the seed-specific expression pattern (Supplemental Fig. S3, A and B). Previous studies revealed that wild-type and the mutant napA promoters, including −1101 (D1), −309 (D2), −211 (D7), and −152 (D8), showed a similar expression pattern at different developmental stages of embryogenesis in the transgenic tobacco plants, although the expression level of the mutant promoters varied significantly. The promoter reporter constructs had barely detectable expression 5 to 10 d after pollination (DAP; globule to heart stages), followed by significant expression at 10 to 15 DAP (heart to torpedo stages), and reached the highest expression level 30 DAP (Stålberg et al., 1993; Ellerström et al., 1996). Consistent with these results, expression of D7:BnL1L (see below) in the transgenic Arabidopsis embryos was detectable around 7 to 10 DAP and had a higher expression level at 13 DAP (Supplemental Fig. S3C).
Based on these results, we placed BnLEC1 and BnL1L under the control of the D7 and D8 promoters, which were transformed into Arabidopsis plants. The resulting transgenic plants did not have any detectable abnormalities (Fig. 1A). Moreover, the level of FAs and C20:1 was increased in the D7:BnL1L transgenic seeds but was unaltered or reduced in the D7:BnLEC1 transgenic seeds (Fig. 1, B and C; Supplemental Fig. S1, C and D). Therefore, we decided to use D6 and D7 promoters to drive BnLEC1 and BnL1L for transgenic studies in rapeseed plants.
Generation and Characterization of Transgenic Rapeseed Plants Carrying BnLEC1 and BnL1L Transgenes
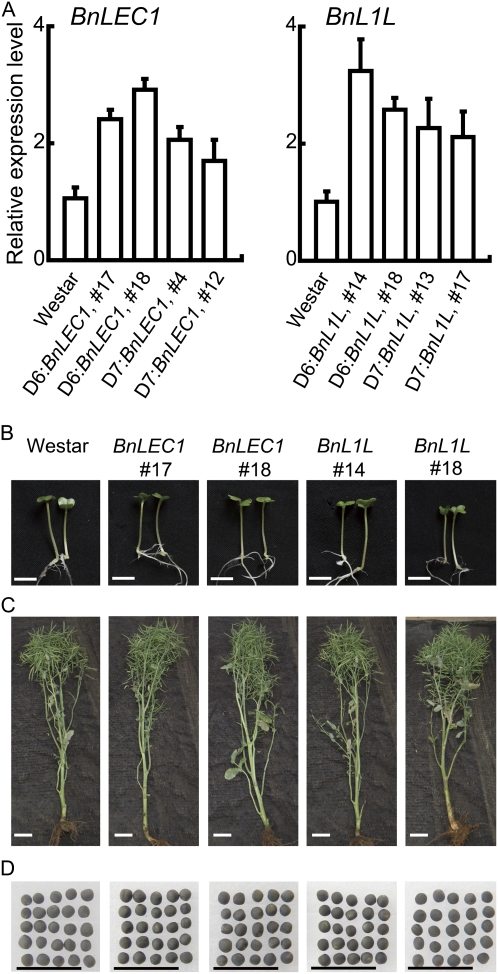
Four expression vectors, D6:BnLEC1, D7:BnLEC1, D6:BnL1L, and D7:BnL1L were constructed and transformed into Westar, a most commonly used variety, by the floral dip method (see “Materials and Methods”). Multiple independent transgenic lines were obtained and were initially analyzed (see below). Quantitative reverse transcription (qRT)-PCR analysis revealed the expression of BnLEC1 and BnL1L in developing seeds of these transgenic plants was 2- to 3-fold higher than that in nontransgenic Westar plants, and the expression level in developing seeds of the D6 promoter-derived transgenics was slightly higher than that in D7 promoter-derived transgenics (Fig. 2A). No expression of BnLEC1 and BnL1L was detected in leaves and the silique wall (Supplemental Fig. S4, A and B). This result indicates that the BnLEC1 and BnL1L transgenes are specifically expressed in developing seeds at a level higher than that of the respective endogenous genes.
Figure 2.
Characterization of transgenic BnLEC1 and BnL1L rapeseed plants. A, qRT-PCR analysis of BnLEC1 and BnL1L expression in developing seeds (35 DAP) collected from the Westar and transgenic plants as indicated at the bottom. The relative expression level of each gene was normalized using BnACTIN2 as an internal control. Data presented are mean values of three experiments, and standard deviations are indicated by bars. The expression level of each gene in Westar is set as 1.0. B, Six-day-old rapeseed seedlings germinated and grown on an agar plate. Bars = 1 cm. C, Seven-month-old rapeseed plants grown under the field condition. Bars = 10 cm. D, Seeds of Westar and the transgenic plants. Bars = 1 cm. B to D, Genotypes of plants/seeds in each column are shown on the top of B. All constructs shown are driven by the D6 promoter. T3 homozygous transgenics were used in all experiments.
Multiple independent transgenic lines were followed for three generations (T1 through T3), and no developmental abnormalities were observed throughout the entire life cycle (Fig. 2, B–D). A detailed analysis of major agronomic traits of the field-grown T3 homozygous plants revealed that the transgenic plants were phenotypically similar to Westar plants in most categories, including 1000-seed weight and the seed yield (Supplemental Table S1). Because AtLEC1 acts upstream of ABSCISIC ACID INSENSITIVE3, a key regulator of seed maturation and seed germination (Koornneef et al., 1984; Kagaya et al., 2005), and also because seed-specific expression of ZmLEC1 causes reduced seed germination (Shen et al., 2010), we compared the germination rate of the transgenic seeds with Westar seeds. No difference of the germination rate was found between Westar and the transgenic seeds (Supplemental Fig. S5). Taken together, these results indicate that seed-specific expression of BnLEC1 and BnL1L driven by the D6 and D7 promoters does not have detectable detrimental effects on plant growth and development.
Analysis of the Seed Oil Content in T1 and T2 Transgenic Rapeseed Plants Carrying the BnLEC1 and BnL1L Transgenes
For each construct, multiple independent T1 transgenic lines (grown during the 2008 to 2009 season in the Yangtze River Valley, southern China) were analyzed for the seed oil content. When screened by near infrared reflectance spectroscopy (NIR), 35 out of the 60 analyzed transgenic lines had an increased seed oil level by 5% to 21% higher than the nontransgenic Westar (Supplemental Table S2). To more precisely measure the oil content, we further analyzed the seed oil content by the standard Soxhlet extraction method in representative transgenic lines with an increased oil level higher than 5% as determined by NIR. Among the analyzed 22 lines, the seed oil content increased by 6% to 22% than Westar as assayed by the Soxhlet method (Supplemental Table S3), which were comparable with the results obtained by the NIR method.
T2 plants derived from representative T1 transgenic seeds were grown during the summer of 2009 in a high altitude area (approximately 2,200 m) in northwestern China, where most rapeseed cultivars have a higher level of the seed oil than that grown in the low altitude areas in the Yangtze River Valley (Fu et al., 2009, and refs. therein). Seeds from individual plants of homozygous transgenic lines were harvested, and the seed oil content was analyzed by NIR. The seed oil content in most plants increased at varying degrees, up to 24%, although a few plants (three out of the 80 analyzed plants) had a lower oil level than the nontransgenic control (Supplemental Table S4). These results suggest that overexpression of BnLEC1 and BnL1L at an appropriate level is able to increase the seed oil content in a manner independent from the growth locations.
Increased Oil Production in the BnLEC1 and BnL1L Transgenic Rapeseed Seeds
To more accurately analyze the field performance of these transgenic plants, we grew T3 homozygous plants during the 2009 to 2010 season in southern China, and detailed results obtained from two representative lines of each construct are presented below. As mentioned before, these T3 homozygous plants grew indistinguishable from Westar plants and had no alterations in major agronomic traits (Fig. 2, B–D; Supplemental Table S1). Seeds collected from 20 to 30 plants were used for the measurement of the oil content by the Soxhlet method. Under the experimental condition, the oil content of nontransgenic Westar seeds was 35.57%. The seed oil content in the transgenic seeds was 36.36% to 42.72%, increasing by 2.22% to 20.10% than the nontransgenic Westar seeds (Table I). Together with the data presented in Supplemental Tables S2 to S4, these results demonstrate that seed-specific expression of BnLEC1 or BnL1L at an appropriate level is able to increase oil production in the seed of the transgenic canola plants without any detectable abnormalities in plant growth and development.
Table I. Oil content in T3 transgenic rapeseed seeds (% dry weight).
Mature seeds were used for the analysis of the oil content by the Soxhlet method. Numbers refer to the transgenic line numbers. Asterisks indicate statistically significant differences compared with the control (Student's t test: *P < 0.05; **P < 0.01).
| Genotype | Oil Content | Percentage of Increase |
| Westar | 35.57 ± 0.23 | – |
| D6:BnLEC1 | ||
| 17 | 39.93 ± 0.93* | 12.26 |
| 18 | 42.72 ± 0.46** | 20.10 |
| D7:BnLEC1 | ||
| 4 | 36.36 ± 0.45 | 2.22 |
| 12 | 39.24 ± 0.53* | 10.32 |
| D6:BnL1L | ||
| 14 | 40.57 ± 0.31** | 14.06 |
| 18 | 41.28 ± 0.22** | 16.05 |
| D7:BnL1L | ||
| 13 | 38.82 ± 0.01 | 9.14 |
| 17 | 39.73 ± 0.03* | 11.70 |
When compared with that of the nontransgenic Westar control, the 1000-seed weight and the seed yield per plant remained unaltered (Supplemental Table S1), indicating that the increased oil production in the transgenic seeds was not penalized by the reduced size and number of the seed.
Nutritional Quality of the BnLEC1 and BnL1L Transgenic Canola Seeds
The nutritional quality of oilseed rape is mainly determined by the FA balance and the protein level. Whereas the FA balance determines the quality of edible oil, the protein content is a major contributor to the meal energy value for feed (Nesi et al., 2008). Analysis of the FA composition by gas chromatography-mass spectrometry (GC-MS) revealed that most of the examined FA species (except C18:1) were accumulated at a higher level in transgenic seeds than in the wild type (Table II). The most apparently increased FAs were unsaturated C18:2 and C18:3, whereas the C18:1 level was reduced. The C16:0 and C18:0 levels were increased at a lesser degree (Table II).
Table II. Relative FA contents in wild-type (Westar) and transgenic seeds (mol %).
Mature seeds were used for the analysis. Numbers refer to the transgenic line numbers. Asterisks indicate statistically significant differences compared with the control (Student’s t test: *P < 0.05; **P < 0.01).
| FA | Westar | D6:BnLEC1: 17 | D6:BnLEC1: 18 | D7:BnLEC1: 4 | D7:BnLEC1: 12 | D6:BnL1L: 14 | D6:BnL1L: 18 | D7:BnL1L: 13 | D7:BnL1L: 17 |
| C16:0 | 3.34 ± 0.15 | 3.44 ± 0.17 | 3.63 ± 0.13 | 3.60 ± 0.76 | 3.59 ± 0.13 | 3.65 ± 0.32 | 3.45 ± 0.17 | 2.95 ± 0.02* | 3.42 ± 1.11 |
| C18:0 | 1.12 ± 0.85 | 1.44 ± 0.08 | 1.57 ± 0.11 | 1.456 ± 0.50 | 1.74 ± 0.22 | 1.53 ± 0.11 | 1.71 ± 0.06 | 1.12 ± 0.10 | 1.50 ± 0.21 |
| C18:1 | 72.03 ± 0.97 | 65.32 ± 1.19** | 64.75 ± 0.72** | 68.35 ± 2.90 | 68.73 ± 1.00* | 71.66 ± 0.46 | 72.55 ± 1.15 | 70.05 ± 1.03 | 69.76 ± 0.93* |
| C18:2 | 16.22 ± 0.259 | 20.67 ± 0.12** | 18.73 ± 0.53** | 17.54 ± 0.89 | 16.53 ± 0.27 | 15.99 ± 0.20 | 15.02 ± 0.50* | 16.97 ± 0.16* | 18.36 ± 0.57** |
| C18:3 | 7.30 ± 0.27 | 9.14 ± 1.15* | 11.32 ± 1.08** | 9.06 ± 1.01* | 9.41 ± 0.71* | 7.16 ± 0.42 | 7.26 ± 0.43 | 8.91 ± 0.85* | 6.97 ± 0.41 |
Long-chain FAs (C20 and longer chain) were undetectable in Westar and the transgenic seeds. Among those long-chain FAs, erucic acid (C22:1) is an undesirable FA and is therefore a key factor for the quality of edible oil. Similar to most modern cultivars, Westar contains very low erucic acid content owing to a mutation in FAE1, a key gene in erucic acid biosynthesis (Han et al., 2001; Katavic et al., 2002). Similar to that of Westar, the erucic acid content was undetectable in the transgenic seeds, indicating that the increased oil content was not accompanied with an elevated level of this undesirable species in the transgenic seeds.
An additional criterion of the oil quality is the ratio of C18 unsaturated FAs. Both linoleic acid (C18:2) and linolenic acid (C18:3) are essential FAs for humans; therefore, a higher level of these two FA species in edible oil is nutritionally important. On the other hand, a higher level of C18:2 and C18:3, especially C18:3, affects oil stability, thus reducing the storage time (Nesi et al., 2008). Compared with that of Westar seeds, the relative level of C18 unsaturated FAs, indicated by the ratio of different species of these FAs, was slightly improved (a lower ratio of C18:1/C18:2 and C18:1/C18:3 and a higher ratio of C18:2/C18:3) or marginally altered in the transgenic seeds (Table III), indicating that the nutrition quality of the transgenic rapeseed oil remains unaltered. Lastly, the protein content was slightly decreased in the transgenic seeds (Table III), presumably owing to the increased FA level in the transgenic seeds. A similar observation was made in the transgenic maize carrying a seed-specific expressed ZmWRI1 transgene (Shen et al., 2010). Taken together, these results indicate that the nutritional quality remains unaltered in the BnLEC1 and BnL1L transgenic canola seeds.
Table III. Nutritional quality of the transgenic seeds.
| 18:1/18:2a | 18:1/18:3a | 18:2/18:3a | Protein | |
| % | ||||
| Westar | 4.44 | 9.59 | 2.22 | 24.52 ± 0.65 |
| D6:BnLEC1 | ||||
| 17b | 3.16 | 7.15 | 2.26 | 23.35 ± 0.92 |
| 18b | 3.46 | 5.72 | 1.65 | 23.01 ± 0.24*c |
| D7:BnLEC1 | ||||
| 4b | 3.90 | 7.54 | 1.94 | 24.10 ± 0.32 |
| 12b | 4.16 | 7.30 | 1.76 | 23.86 ± 0.36 |
| D6:BnL1L | ||||
| 14b | 4.48 | 10.01 | 2.22 | 23.20 ± 0.09 |
| 18b | 4.83 | 9.99 | 2.06 | 22.63 ± 0.34*c |
| D7:BnL1L | ||||
| 13b | 4.13 | 7.86 | 1.90 | 23.68 ± 0.76 |
| 17b | 3.80 | 10.01 | 2.63 | 23.81 ± 0.24 |
The ratio of different C18 unsaturated FAs is calculated according to data presented in Table II.
Numbers refer to the transgenic line numbers.
Asterisks indicate statistically significant differences compared with the control (Student’s t test: *P < 0.05).
Increased Expression of FA Synthetic Genes in Developing Seeds of the BnLEC1 and BnL1L Transgenic Plants
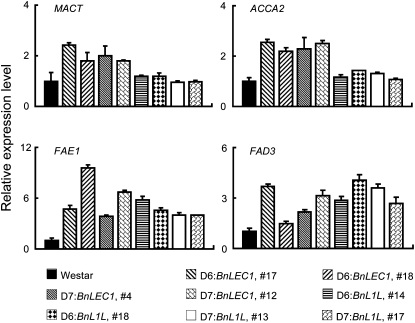
To explore the regulatory mechanism of BnLEC1 and BnL1L in the control of FA synthesis, we analyzed the expression level of several representative genes of the FA biosynthesis pathway in developing seeds of the transgenic plants. These genes include key genes in the condensation reaction (subunit A of ACCase or ACCA2 and malonyl-CoA:ACP transacylase), chain elongation reaction (FAE1), and desaturation reaction (ω-3 FA desaturase [FAD3]). The accumulation of oil in developing seeds of rapeseed occurs around 28 to 56 DAP, whereas the storage protein content in developing seeds remains almost constant at 35 DAP (Fowler and Downey, 1970; Gurr et al., 1972). We used materials (developing seeds and the silique wall) collected 35 DAP in all experiments described below unless otherwise specified.
qRT-PCR analysis revealed that expression of the FA synthetic genes was up-regulated at varying degrees (Fig. 3). Among these analyzed genes, expression of malonyl-CoA:ACP transacylase and ACCA2 was marginally increased the BnL1L transgenic seeds but displayed a higher level in the BnLEC1 transgenic seeds (Fig. 3). FAD3 catalyzes the conversion of C18:2 into C18:3. Expression of FAD3 was significantly induced in most transgenic lines, which likely ascribed to the increased level of C18:3 in the transgenic seeds (Tables II and III). We noticed that expression of FAE1, a key gene in erucic acid biosynthesis, was substantially increased in all the tested transgenic lines (4- to 11-fold; Fig. 3). Despite the elevated expression level of FAE1, the erucic acid level was not altered in the transgenic seeds, owing to a nonfunctional mutation in FAE1 of Westar (Han et al., 2001; Katavic et al., 2002).
Figure 3.
Expression of representative FA synthetic genes in developing seeds of the transgenic rapeseed plants. Expression of representative FA synthetic genes analyzed by qRT-PCR using total RNA prepared from developing seeds (35 DAP) collected from the Westar and transgenic plants. The relative expression level of each gene was normalized using BnACTIN2 as an internal control. Data presented are mean values of three experiments. Error bars in the graph represent standard deviations. The expression level of each gene in Westar is set as 1.0.
Increased Expression of Glycolytic and Suc Photoassimilation Genes in Developing Seeds and the Silique Wall of the BnLEC1 Transgenic Plants
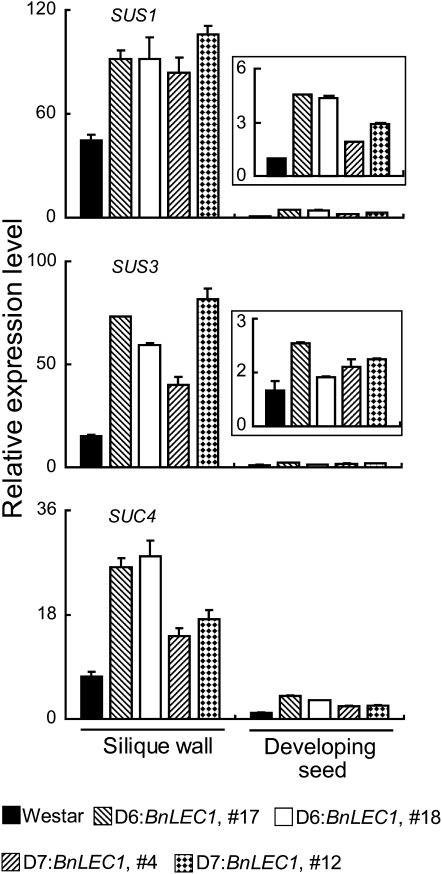
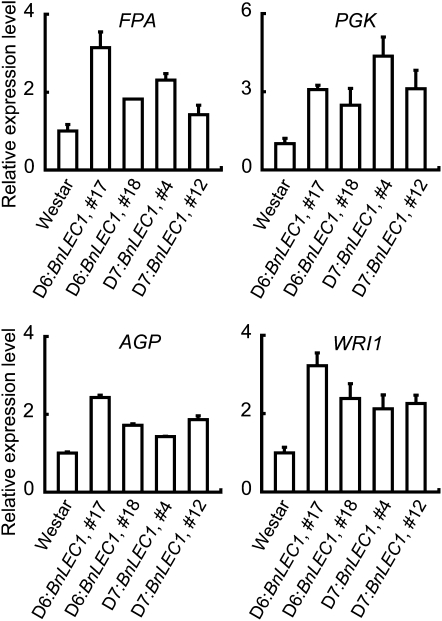
The observation that seed-specific expression of BnLEC1 and BnL1L caused the increased accumulation of oil in the transgenic seeds prompted us to ask if the expression of genes involved in Suc photoassimilation and glycolysis is altered. In developing seeds of the BnLEC1 transgenic plants, expression of SUC SYNTHASE1 (SUS1), SUS3, and SUC TRANSPORTER4 (SUC4) was increased compared with the Westar control (Fig. 4; see also below). Expression of SUC1, however, did not show significant alterations (Supplemental Fig. S6). In the glycolysis pathway, expression of Fru-bisphosphate aldolase (FPA) and phosphoglycerate kinase (PGK) was significantly increased (Fig. 5). Other tested glycolytic genes, including hexose kinase, glyceraldehyde-3-P dehydrogenase, and pyrophosphate-dependent phosphofructokinase, did not show an apparently altered expression level in developing seeds of the BnLEC1 transgenic plants (Supplemental Fig. S6). Consistent with these results, expression of canola WRI1 (BnWRI1), an ortholog of the Arabidopsis WRI1 gene that plays a critical role in regulating glycolysis and lipid biosynthesis (Focks and Benning, 1998; Cernac and Benning, 2004; Baud et al., 2007), was also increased in developing seeds of the BnLEC1 transgenic plants (Fig. 5). In addition, we also found expression of ADP-Glc pyrophosphorylase (AGP), a key gene regulating starch synthesis, was increased (Fig. 5). AGP catalyzes the conversion of Glc1P to ADPGlc that is then incorporated into starch granule in developing seeds (Kang and Rawsthorne, 1994; da Silva et al., 1997). A reduced AGP activity caused a decreased accumulation of starch as well as lipids in seeds of rapeseed, indicating a positive correlation between starch synthesis and the accumulation of lipids (Periappuram et al., 2000; Vigeolas et al., 2004). Taken together, these results suggest that seed-specific expression of BnLEC1 up-regulates a subset of genes that are positively correlated to FA synthesis, including those involved in Suc metabolism, glycolysis, and starch synthesis.
Figure 4.
Expression of Suc metabolism genes in developing seeds and the silique wall of the transgenic BnLEC1 rapeseed plants. Expression of SUS1, SUS3, and SUC4 in developing seeds and the silique wall of Westar, D6:BnLEC1, and D7:BnLEC1 transgenic plants (35 DAP) analyzed by qRT-PCR. The relative expression level of each gene was normalized with BnACTIN2. Data presented are mean values of three experiments, and standard deviations are indicated by bars. The expression level of each gene in developing seeds of Westar is set as 1.0. Insets in the panels of SUS1 and SUS3 are enlarged views of the expression patterns in developing seeds.
Figure 5.
Expression of glycolytic genes and BnWRI1 in developing seeds of the BnLEC1 transgenic plants. Expression of FPA, PGK, AGP, and BnWRI1 in developing seeds (35 DAP) of Westar and the BnLEC1 transgenic plants analyzed by qRT-PCR. The relative expression level of each gene was normalized using BnACTIN2 as an internal control. Data presented are mean values of three experiments, and standard deviations are indicated by bars. The expression level of each gene in developing seeds of Westar is set as 1.0.
During seed development in rapeseed, the silique wall is the major photoassimilation tissues, from which sugars are imported into developing seeds and then used for the synthesis of FAs (King et al., 1997). Thus, we next asked if expression of the carbohydrate metabolism genes was altered in the silique wall. All the tested glycolytic genes (FPA, PGK, glyceraldehyde-3-P dehydrogenase, and pyrophosphate-dependent phosphofructokinase) showed a similar expression level in the silique wall of Westar and the transgenic plants (Supplemental Fig. S7). Note that two of these four genes, FPA and PGK, showed an increased expression level in developing seeds of the transgenic plants (Fig. 5), suggesting that expression of the glycolytic genes is regulated by different mechanisms in developing seeds and in the silique wall. In contrast with that of the glycolytic genes, the Suc metabolism genes (SUS1, SUS3, and SUC4) showed an increased expression level in the silique wall of the transgenic plants than that in Westar plants (Fig. 4). Note that SUS1, SUS3, and SUC4 had a significantly higher expression level in the silique wall than in developing seeds (7- to 44-fold; see Fig. 4), consistent with the notion that the silique wall is the primary source of photoassimilation. Despite a higher basal expression level in the silique wall, the increased expression level of the each gene was 2- to 5-fold higher in the transgenics than that in Westar (Fig. 4). Therefore, in the BnLEC1 transgenic plants, whereas expression of Suc metabolism genes was increased in both developing seeds and the silique wall, expression of the glycolytic genes was up-regulated only in developing seeds.
Metabolism of Sugars in the Seed and the Silique Wall of the BnLEC1 and BnL1L Transgenic Rapeseed Plants
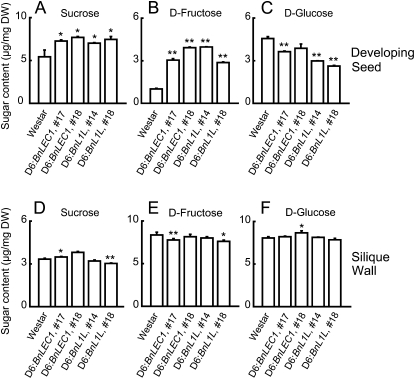
Soluble sugars, including Suc, Glc, and Fru, are the primary carbon source for lipid synthesis in developing seeds, which are largely synthesized in the silique wall and then transported into developing seeds of oilseed rape (King et al., 1997). In BnLEC1 transgenic seeds, an increased accumulation of lipids, accompanied by an elevated expression level of a subset of sugar metabolism genes, may be directly linked to sugar metabolism. To test this possibility, we analyzed the major soluble sugar contents (Suc, Glc, and Fru) in developing seeds and in the silique wall by GC-MS. In developing seeds of nontransgenic Westar, Suc accumulated at a higher concentration (5.4 μg per mg dry weight) than Fru and Glc (1.0 and 4.6 μg per mg dry weight, respectively; Fig. 6, A–C). In the silique wall, however, a reverse pattern was observed, with a higher level of Fru and Glc (8.4 and 8.1 μg per mg dry weight, respectively) and a lower level of Suc (3.3 μg per mg dry weight; Fig. 6, D–F). Our data are largely similar to the results obtained in a previous study using the enzymatic method (King et al., 1997).
Figure 6.
Analysis of Suc, Fru, and Glc levels in developing seeds and the silique wall of the BnLEC1 and BnL1L transgenic plants. The content of Suc, Fru, and Glc in developing seeds (A–C) and the silique wall (D–F) collected from Westar and the BnLEC1 and BnL1L transgenic plants analyzed by GC-MS. All materials were collected at 35 DAP. Results are mean values of three experiments, and standard deviations are indicated by bars. DW, Dry weight. Asterisks indicate statistically significant differences compared with the control (Student’s t test: *P < 0.05; **P < 0.01).
In developing seeds of the BnLEC1 and BnL1L transgenic plants, the level of Suc and Fru was higher than that of the nontransgenic Westar, whereas the accumulation of Glc was reduced in developing seeds of the transgenic plants (Fig. 6, A–C). In particular, the Fru content in developing seeds of the transgenic plants was 3- to 4-fold higher than that in the nontransgenic Westar plants (Fig. 6B). These results suggest that more Suc and Fru are synthesized in developing seeds or imported from the source tissues of the transgenic plants. In the silique wall, these analyzed soluble sugars did not show apparent alterations in the transgenics (Fig. 6, D–F).
DISCUSSION
The oil content is the most important agronomic trait in the oilseed crops, which has been subjected to extensive selection during breeding. In this study, we show that conditional overexpression of BnLEC1 and BnL1L under the control of two modified seed-specific promoters significantly increases the oil content in the transgenic rapeseed seeds. Moreover, conditional overexpression of these two transgenes does not have detectable detrimental effects on plant growth and development, and all the analyzed major agronomic traits of the transgenic plants were similar to that of the nontransgenic control. Lastly, the nutrition quality of the transgenic seed, including the compositions of FAs and the protein content of the seed, is comparable with the nontransgenic control. Taken together, these results demonstrate that seed-specific overexpression of BnLEC1 and BnL1L at an appropriate level substantially increases the seed oil content of the transgenic oilseed rape without detectable negative effects on other major agronomic traits.
The manipulation of key regulatory genes in the FA biosynthesis pathway is of great interests in the efforts for genetic improvement of oilseed crops. Unfortunately, overexpression of these genes causes a pleiotropic phenotype and even a lethal phenotype, largely owing to that these genes also play a critical role in embryogenesis and seed maturation (Lotan et al., 1998; Stone et al., 2001; Kwong et al., 2003; Cernac and Benning, 2004; Wang et al., 2007; Mu et al., 2008; Shen et al., 2010). This detrimental effect renders it impractical for the use of these genes in molecular breeding by conventional overexpression. Two recent studies showed that overexpression of ZmWRI1 was able to increase the oil content without affecting seed germination, seedling growth, and grain yield in transgenic maize (Shen et al., 2010; Pouvreau et al., 2011). However, overexpression of ZmLEC1 with two seed-specific promoters reduces seed germination and leaf growth, although the transgenic seed contains a higher oil level than the control (Shen et al., 2010).
How do we explain the different results obtained from the transgenic ZmLEC1 maize and the transgenic BnLEC1 rapeseed? We notice that D6 and D7 promoters used in this study display <18% and 4% of the activity of the napA promoter in transgenic Arabidopsis plants, which is comparable to the observation made in transgenic tobacco plants (Stålberg et al., 1993; Ellerström et al., 1996). It is reasonable to assume that a similar activity is present in transgenic rapeseed seeds. Indeed, the expression level of BnLEC1 in developing seeds of the transgenic plants is approximately 2- to 3-fold higher than the endogenous BnLEC1 level, which appears to be tolerable for normal growth and development of rapeseed plants. In the transgenic maize, it remains unclear about the expression level of the ZmLEC1 transgene, especially in the comparison with the endogenous expression level (Shen et al., 2010). Nevertheless, the developmental abnormalities observed in the transgenic maize plants is likely owing to a higher expression level the ZmLEC1 transgene as observed in Arabidopsis (Lotan et al., 1998; Mu et al., 2008). Thus, the control of the expression level of a LEC1 or LEC1-like transgene at an appropriate level (e.g. 4% to 18% of the napA promoter activity under our assay conditions) is critical for the successful utilization of this class of genes in transgenic studies. Moreover, it will be interesting to test the tolerable limit of a further increased expression level of a LEC1 or LEC1-like transgene in rapeseed, at which the transgene allows the increase of the oil content with minimal negative effects on plant growth and development.
In Arabidopsis, AtLEC1 has been shown as a master regulator of FA biosynthesis, of which more than 50% FA synthetic genes are up-regulated in the AtLEC1-overexpressing transgenic plants. In addition, expression of a large number of genes related to carbohydrate metabolism is also increased in AtLEC1-overexpressing plants (Mu et al., 2008). In agreement with this observation, the increased oil content in the BnLEC1 and BnL1L transgenic canola seeds is accompanied by the increased Suc content and the increased glycolytic activity. This notion is supported by two lines of evidence. First, expression of a subset of genes involved in Suc synthesis and glycolysis is increased in developing seeds of the BnLEC1 transgenic plants, whereas several genes related to Suc synthesis and transport, but not the glycolytic genes, are up-regulated in the silique wall. These distinctive expression patterns of these genes support the view of an increased Suc photoassimilation capacity in the silique wall or the source tissues and, in parallel, an increased glycolytic activity in developing seeds or the sink tissues. Second, the content of Suc and Fru is increased in developing seeds of the transgenic plants but not in the silique wall. The relatively unaltered level of Suc in the silique wall may have resulted from rapid transport of Suc from the silique wall into developing seeds, which, in turn, may act as a driving force to promote Suc synthesis in the source tissues. Although the precise mechanism of this phenotype related to Suc synthesis and transport in the siliques wall remains elusive, these observations may provide an attractive model to analyze the source-sink network in plants. Nevertheless, these observations suggest that the increased seed oil content in the BnLEC1 and BnL1L transgenic plants is likely directly related to the increased carbon flux to the FA biosynthesis pathway.
Finally, LEC1 belongs to the HAP3 or the CCAAT-binding factor family, which is highly conserved in eukaryotes. Our preliminary studies suggest that whereas overexpression of the AtLEC1 and AtL1L genes in other plant species, including rapeseed, is capable of stimulating the accumulation of lipids, overexpression of LEC1-like genes from other plant species can also significantly enhance FA biosynthesis in the transgenic Arabidopsis plants. In a more extended view, in mammalian cells, the CCAAT-binding factors directly bind to the FAS promoter and play an important role in the regulation of FAS gene expression (Rangan et al., 1996; Roder et al., 1997; Schweizer et al., 2002). Thus, this class of transcription factors is evolutionarily and functionally conserved in the regulation of FA biosynthesis. Along with these observations, our results demonstrate that BnLEC1 and BnL1L are reliable targets for genetic improvement of oilseed rape and that the approach used here is technically practical in molecular breeding, which should be applicable to other oilseed crops as well.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Plants of oilseed rape (Brassica napus ‘Westar’) and all related transgenic plants were grown under the field conditions. In the Yangtze River Valley area, seeds were sown usually in late September or early October and harvested around late May. In the northwestern region, seeds were sown in late March or early April and harvested in late September or early October. When grown under the tissue culture condition, seeds were surface sterilized with 75% ethanol for 2 min, followed by 0.1% mercury chloride sodium for 10 min, and then rinsed four times with sterile water. Sterilized seeds were sown on agar plates (1× MS salts, 3% Suc, and 0.8% agar, pH 5.7; Murashige and Skoog, 1962) and then cultured at 22°C under a 16/8-h light/dark cycle.
Plants of Arabidopsis (Arabidopsis thaliana; the Col-0 accession) were grown under continuous white light at 22°C in soil or on MS agar (0.5× MS salts, 3% Suc, and 0.8% agar, pH 5.7) as previously described (Mu et al., 2008).
All rapeseed plant materials (leaves, developing seeds, and the silique wall) were collected from the field-grown plants, immediately frozen in liquid nitrogen, and then stored at −80°C until the use.
Plasmid Construction
To generate napA:GUS reporter constructs, the napA promoter region was PCR amplified from genomic DNA prepared from Westar leaves using primers D1F and D1B (Supplemental Table S6 for sequences of all primers used in this study). This promoter fragment (D1; Supplemental Fig. S2) was 1,178 bp, including 44 bp of 5′-untranslation region and the sequence encoding the first 11 amino acid residues coding sequence (33 bp). In all reporter constructs, the first 11 amino acid residues were fused in frame to the GUS coding sequences. The mutant promoters (D2 through D8; Supplemental Fig. S2) were made by the PCR-based cloning method using different combinations of specific primer pairs (D2F through D8F as forward primers and D1B as a reverse primer in all reactions; Supplemental Table S6). The resulting fragment was digested with HindIII and BamHI (embedded in the PCR primers) and then inserted into the same sites of pBI121 (Clontech).
D1, D6, D7, and D8 promoters also cloned into the pCAM2300 vector carrying a kanamycin selectable marker, and the genomic DNA fragments containing BnLEC1 and BnL1L coding sequences (Mu et al., 2008) were inserted downstream of these promoters, respectively. Note that, the sequences encoding the first 11 amino acid residues of Napin protein were removed by PCR using appropriate forward primers (D1F, D6F, D7F, and D8F) and the backward primer D2B.
All constructs were verified by extensive restriction digestion and DNA sequencing analysis.
Generation of Transgenic Plants
All constructs were transformed into Agrobacterium tumefaciens strain GV3101, which were then used for the transformation. Transformation of Arabidopsis plants was carried out as described (Clough and Bent, 1998).
Transformation of rapeseed plants (D6:BnLEC1, D7:BnLEC1, D6:BnL1L, and D7:BnL1L) were carried out with a modified floral dip method (Clough and Bent, 1998). Briefly, agrobacteria cultures carrying a target construct were collected by centrifugation and then resuspended in a solution containing 0.5× MS salts, 3% Suc, 0.1% Silwet L-77, 2 ng/L 6-benzyladenine, and 8 mg/L acetosyringone. Field-grown rapeseed plants at the flowering stage (approximately 5 months old) were used for the transformation. The head of a flowering plant was bent downward and dipped into a beaker containing the agrobacterial culture for 1 to 2 min with gentle agitation, and the treated plant head was loosely wrapped with a vegetable parchment paper. The plant was treated as described above every other day 3 times and then continued to grow until maturation. Seeds harvested from the transformed plant were surface sterilized and sown on the MS medium containing 180 mg/L kanamycin to screen for putative transformants. The putative transformants were identified upon the initiation of the first pair of green true leaves, usually after cultured on the selective medium for 3 to 4 weeks. Roots of the putative transformants were removed with a razor, and the aerial part was transferred on the root regeneration medium (1× MS, 3% Suc, 0.5 mg/L α-naphthyl acetic acid, 0.5 mg/L indole-3-acetic acid, and 0.8% agar, pH 5.7) by inserting the hypocotyl tip into the medium. After cultured for additional 3 to 4 weeks until the regeneration of new roots, the putative transformants were transferred into soil, and leaf materials were collected for PCR detection of the transgene. The transformation efficiency is usually 0.05% to 0.1%, and 16 to 21 primary transformants for these four constructs were obtained and used in subsequent studies.
Field Experiments and Agronomic Trait Investigation
For field experiments, Westar and the transgenic plants (30 plants each genotype) were grown in two-row plots at a density of 40 × 20 cm per plant. Analysis of agronomic traits was carried out as described (Qiu et al., 2006). Data were collected from the analysis of at least 10 plants of Westar and the transgenic lines.
Analysis of Lipids and Proteins
The seed oil content was analyzed by the Soxhlet extraction method and the NIR method, respectively. For the Soxhlet method, 2 g seeds were oven-dried overnight at 80°C and then cooled to the room temperature in a desiccator. The dried seeds were grinded into fine powder and then transferred into a preweighed bag made with Whatman 3M filter papers (Weight A) and sealed. The sample was oven-dried for 6 to 8 h until reaching constant weight (Weight B). After cooling to room temperature in a desiccator, the bagged sample was transferred into a Soxhlet tube and extracted with petroleum ether (boiling point under 50°C) for 24 h. After the extraction, the bagged sample was dried in a hood to evaporate remaining petroleum ether, cooled in a desiccator, and then weighed (Weight C). The oil content (%) was calculated by the formula (B − C)/(B − A) × 100%. For each sample, triplicates were performed and mean values of the triplicates were used to calculate the oil content.
NIR analysis of the oil content was carried out using a Foss NIRSystems Series-5000 NIR spectrophotometer according to the WinISI III manual instructions for routine analysis (Foss-tecator Infrasoft International). Approximately 4 to 5 g seeds of each sample were scanned in a 36-mm inner diameter ring cup, and reflectance spectra (log 1/R) from 400 to 2,500 nm were recorded at 2-nm intervals.
Analysis of the FA level was performed by GC-MS as previously described (Mu et al., 2008). The protein content was determined by the Kjeldahl method as described (Daun and DeClercq, 1994).
Analysis of the Sugar Content
Sample preparation and analysis of sugars were performed as previously described with minor modifications (Broeckling et al., 2005). Briefly, 6 mg of lyophilized fine powder prepared from rapeseed developing seeds or the silique wall were extracted with 1.5 mL chloroform. The sample was then vortexed and incubated at 37°C for 45 min. In order to separate polar (sugars) from nonpolar metabolites, 1.5 mL of HPLC-grade water containing 25 μg mL−1 ribitol (as an internal polar standard) was then added to the sample. The mixture was shaken up and then incubated at 37°C for 45 min. One milliliter of the aqueous supernatants was transferred into 1.5-mL Eppendorf tubes after centrifugation at 3,000g for 30 min at 4°C and then dried in a lyophilizer (ALPHA 1-2 LD plus). The dried samples were resuspended in 50 μL of methoxyamine hydrochloride (15 mg mL−1) in pyridine with brief sonication and incubation for 1 h at 50°C. One microliter of the sample was applied for GC-MS (Agilent 7890A GC coupled to 5975C MS) analysis at a 10:1 split ratio after silylation with 50 μL of MSTFA +1% TMCS (Sigma-Aldrich) for 1 h at 50°C. The GC-MS program started with 60°C for 1 min and then ramped at 10°C/min to 325°C and held for 10 min; injector and inlet temperature were set at 230°C and 290°C, respectively. Separation was performed on a HP-5 MS column (30 m × 0.25 mm × 0.25 μm) with a constant flow of 1.1 mL/min helium. The MS scan mass-to-charge range was from 50 to 600. The sugars were identified based on the retention time and MS information of authentic standards, and the quantification was performed by an external standard method according to the total ion chromatograms of the target compounds.
Analysis of GUS Activity
Histochemical and fluorimetric analysis of the GUS activity was performed as described (Jefferson et al., 1987).
Analysis of Gene Expression by qRT-PCR
Total RNA was prepared using Plant RNA Mini Kit (Watson Biotechnologies) according to the manufacturer’s instructions. qRT-PCR analysis was carried out as described previously (Deng et al., 2010). Briefly, 1 to 2 μg of total RNA was used for the synthesis of the first-strand cDNA using oligo(dT) as a primer and SuperScript II reverse transcriptase (Invitrogen). After being treated with RNase H, the diluted RT reaction was used as a template for qPCR using ExTaq DNA polymerase (Takara Biotechnology) and the fluorescent intercalating dye iQ SYBR Green Super mix (Bio-Rad). The reaction was run on a C1000 Thermal Cycler (Bio-Rad). Relative expression of the target genes was analyzed with the Δ-Δ Ct method using 26S rRNA (experiments in Supplemental Fig. S4B) or BnACTIN2 (all other experiments) as an internal control. All primers used in the RT-qPCR analyses are listed in Supplemental Table S6.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU371726 (BnLEC1) and EU371727 (BnL1L).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of fatty acids in transgenic Arabidopsis seeds.
Supplemental Figure S2. Analysis of the napA promoter activity in transgenic Arabidopsis plants.
Supplemental Figure S3. The napA mutant promoters maintain the seed-specific expression pattern in transgenic Arabidopsis plants.
Supplemental Figure S4. Expression of BnLEC1 and BnL1L in the silique wall and leaves of the transgenic rapeseed plants.
Supplemental Figure S5. Analysis of the germination rate of the transgenic BnLEC1 and BnL1L seeds.
Supplemental Figure S6. Expression of SUC1, HXK, GPDH, and PPK in developing seeds of the transgenic rapeseed plants.
Supplemental Figure S7. Expression of FPA, PGK, GPDH, and PPK in the silique wall of the transgenic rapeseed plants.
Supplemental Table S1. Analysis of major agronomic traits of the transgenic BnLEC1 and BnL1L rapeseed plants.
Supplemental Table S2. Analysis of the oil content in the T1 transgenic rapeseed seeds by near infrared reflectance spectroscopy.
Supplemental Table S3. Analysis of the oil content in the T1 transgenic rapeseed seeds by the Soxhlet method.
Supplemental Table S4. Analysis of the oil content in T2 transgenic rapeseed seeds by near infrared reflectance spectroscopy.
Supplemental Table S5. Fatty acid contents in wild-type (Westar) and the transgenic seeds.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Mian Xia for expert advice on the transformation of rapeseed plants, Sanyuan Tang for his efforts at the early stage of this project, and Yan Liang for critically reading the manuscript.
References
- Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al. (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW. (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56: 323–336 [DOI] [PubMed] [Google Scholar]
- Broun P, Gettner S, Somerville C. (1999) Genetic engineering of plant lipids. Annu Rev Nutr 19: 197–216 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM. (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10: 236–244 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C. (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- da Silva PMFR, Eastmond PJ, Hill LM, Smith AM, Rawsthorne S. (1997) Starch metabolism in developing embryos of oilseed rape. Planta 203: 480–487 [Google Scholar]
- Daun J, DeClercq D. (1994) Comparison of combustion and Kjeldahl methods for determination of nitrogen in oilseeds. J Am Oil Chem Soc 71: 1069–1072 [Google Scholar]
- Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. (2001) Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol 125: 1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, Yang W-C, Liang Y, Zuo J. (2010) Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22: 1232–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J, White PJ, Johnston KM, Heggtveit HA, McDonald BE, Grundy SM, Bonanome A. (1989) Food safety and health effects of canola oil. J Am Coll Nutr 8: 360–375 [DOI] [PubMed] [Google Scholar]
- Ellerström M, Stålberg K, Ezcurra I, Rask L. (1996) Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol Biol 32: 1019–1027 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C. (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Downey RK. (1970) Lipid and morphological changes in developing rapeseed, Brassica napus. Can J Plant Sci 50: 233–247 [Google Scholar]
- Fu S-X, Cheng H, Qi C. (2009) Microarray analysis of gene expression in seeds of Brassica napus planted in Nanjing (altitude: 8.9 m), Xining (altitude: 2261.2 m) and Lhasa (altitude: 3658 m) with different oil content. Mol Biol Rep 36: 2375–2386 [DOI] [PubMed] [Google Scholar]
- Gurr MI, Blades J, Appleby RS. (1972) Studies on seed-oil triglycerides. The composition of Crambé abyssinica triglycerides during seed maturation. Eur J Biochem 29: 362–368 [DOI] [PubMed] [Google Scholar]
- Han J, Lühs W, Sonntag K, Zähringer U, Borchardt DS, Wolter FP, Heinz E, Frentzen M. (2001) Functional characterization of β-ketoacyl-CoA synthase genes from Brassica napus L. Plant Mol Biol 46: 229–239 [DOI] [PubMed] [Google Scholar]
- Harvey B, Downey R. (1964) The inheritance of erucic acid content in rapeseed (Brassica napus). Can J Plant Sci 44: 104–111 [Google Scholar]
- Harwood JL. (1996) Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta 1301: 7–56 [DOI] [PubMed] [Google Scholar]
- Hill LM, Morley-Smith ER, Rawsthorne S. (2003) Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiol 131: 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC. (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Cahoon EB. (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6: 178–184 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kang F, Rawsthorne S. (1994) Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus L.). Plant J 6: 795–805 [Google Scholar]
- Katavic V, Mietkiewska E, Barton DL, Giblin EM, Reed DW, Taylor DC. (2002) Restoring enzyme activity in nonfunctional low erucic acid Brassica napus fatty acid elongase 1 by a single amino acid substitution. Eur J Biochem 269: 5625–5631 [DOI] [PubMed] [Google Scholar]
- King SP, Lunn JE, Furbank RT. (1997) Carbohydrate content and enzyme metabolism in developing canola siliques. Plant Physiol 114: 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang X-J, Zuo J. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nesi N, Delourme R, Brégeon M, Falentin C, Renard M. (2008) Genetic and molecular approaches to improve nutritional value of Brassica napus L. seed. C R Biol 331: 763–771 [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB. (1994) Design of new plant products: engineering of fatty acid metabolism. Plant Physiol 104: 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Periappuram C, Steinhauer L, Barton DL, Taylor DC, Chatson B, Zou J. (2000) The plastidic phosphoglucomutase from Arabidopsis. A reversible enzyme reaction with an important role in metabolic control. Plant Physiol 122: 1193–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Gendrot G, Py C, Pichon J-P, Rouster J, Paul W, Rogowsky PM. (2011) Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, Long Y, Liu J, Li R, Zhuang X, Wang Y, Tan X, Dietrich E, et al. (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114: 67–80 [DOI] [PubMed] [Google Scholar]
- Rangan VS, Oskouian B, Smith S. (1996) Identification of an inverted CCAAT box motif in the fatty-acid synthase gene as an essential element for modification of transcriptional regulation by cAMP. J Biol Chem 271: 2307–2312 [DOI] [PubMed] [Google Scholar]
- Rawsthorne S. (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Roder K, Wolf SS, Beck K-F, Schweizer M. (1997) Cooperative binding of NF-Y and Sp1 at the DNase I-hypersensitive site, fatty acid synthase insulin-responsive element 1, located at -500 in the rat fatty acid synthase promoter. J Biol Chem 272: 21616–21624 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y. (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Schweizer M, Roder K, Zhang L, Wolf SS. (2002) Transcription factors acting on the promoter of the rat fatty acid synthase gene. Biochem Soc Trans 30: 1070–1072 [DOI] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y. (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC. (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Roesler KR, Shintani D, van de Loo FJ, Ohlrogge JB. (1995) Structural analysis, plastid localization, and expression of the biotin carboxylase subunit of acetyl-coenzyme A carboxylase from tobacco. Plant Physiol 108: 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabas AR, Fawcett T. (1992) The biochemistry and molecular biology of plant lipid biosynthesis. Plant Mol Biol 19: 169–191 [DOI] [PubMed] [Google Scholar]
- Stålberg K, Ellerström M, Josefsson L-G, Rask L. (1993) Deletion analysis of a 2S seed storage protein promoter of Brassica napus in transgenic tobacco. Plant Mol Biol 23: 671–683 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4: 12–21 [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Möhlmann T, Martini N, Neuhaus HE, Geigenberger P. (2004) Embryo-specific reduction of ADP-Glc pyrophosphorylase leads to an inhibition of starch synthesis and a delay in oil accumulation in developing seeds of oilseed rape. Plant Physiol 136: 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ. (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52: 335–361 [DOI] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert KN, Lin Y. (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226: 773–783 [DOI] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PBE, Harwood JL. (2009) Increasing the flow of carbon into seed oil. Biotechnol Adv 27: 866–878 [DOI] [PubMed] [Google Scholar]
- Zou J, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. (1997) Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9: 909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.