Abstract
Phosphorus (P) remobilization in plants is required for continuous growth and development. The Arabidopsis (Arabidopsis thaliana) inorganic phosphate (Pi) transporter Pht1;5 has been implicated in mobilizing stored Pi out of older leaves. In this study, we used a reverse genetics approach to study the role of Pht1;5 in Pi homeostasis. Under low-Pi conditions, Pht1;5 loss of function (pht1;5-1) resulted in reduced P allocation to shoots and elevated transcript levels for several Pi starvation-response genes. Under Pi-replete conditions, pht1;5-1 had higher shoot P content compared with the wild type but had reduced P content in roots. Constitutive overexpression of Pht1;5 had the opposite effect on P distribution: namely, lower P levels in shoots compared with the wild type but higher P content in roots. Pht1;5 overexpression also resulted in altered Pi remobilization, as evidenced by a greater than 2-fold increase in the accumulation of Pi in siliques, premature senescence, and an increase in transcript levels of genes involved in Pi scavenging. Furthermore, Pht1;5 overexpressors exhibited increased root hair formation and reduced primary root growth that could be rescued by the application of silver nitrate (ethylene perception inhibitor) or aminoethoxyvinylglycine (ethylene biosynthesis inhibitor), respectively. Together, these data indicate that Pht1;5 plays a critical role in mobilizing Pi from P source to sink organs in accordance with developmental cues and P status. The study also provides evidence for a link between Pi and ethylene signaling pathways.
Phosphorus (P) is an essential macronutrient required for many physiological and metabolic processes. It is integral to several molecules such as nucleic acids, phospholipids, and ATP as well as to intermediates of signal transduction events (Schachtman and Shin, 2007; Rouached et al., 2010). To modulate P homeostasis, plants must balance P uptake, mobilization, and partitioning to various organs. Although P is abundant in nature, the bioavailability of utilizable inorganic phosphate (Pi) is often suboptimal for crop productivity (Marschner, 1995; Raghothama, 1999; Ticconi and Abel, 2004; Lin et al., 2009). Since Pi concentrations in soil solution rarely exceed 2 μm and cellular Pi concentrations are greater than 10 mm, plants must acquire Pi in roots against a steep concentration gradient (Mimura, 1999; Raghothama, 2000). Pi acquisition appears to be largely mediated by plasma membrane-localized high-affinity Pi transporters belonging to the PHOSPHATE TRANSPORTER1 (Pht1) family (Muchhal et al., 1996; Raghothama, 2000; Chiou et al., 2001; Poirier and Bucher, 2002). These proteins are characterized by 12 membrane-spanning domains that are similar to the yeast Pho84p high-affinity Pi transporter (Muchhal et al., 1996; Rausch and Bucher, 2002). In Arabidopsis (Arabidopsis thaliana), there are nine such Pht1 proteins with 60% to 95% sequence similarity, and their homologs have been identified in several crop species (rice [Oryza sativa], wheat [Triticum aestivum], potato [Solanum tuberosum], tomato [Solanum lycopersicum], and tobacco [Nicotiana tabacum]; Rausch and Bucher, 2002). Promoter-reporter fusions of Pht1 members in Arabidopsis demonstrated the Pi deficiency-induced expression of eight of the nine members in roots (Karthikeyan et al., 2002; Mudge et al., 2002). Further functional characterization of loss-of-function mutants of Pht1;1 and Pht1;4 validated their roles in Pi acquisition (Shin et al., 2004).
After Pi is transported into root epidermal cells, it is loaded into the xylem for distribution to shoot tissues (Poirier et al., 1991). Several studies have characterized mutants that are unable to mobilize Pi from source (older leaves) to sink (roots and younger leaves) organs (Delhaize and Randall, 1995; Versaw and Harrison, 2002; Aung et al., 2006; Chiou et al., 2006). Under long-term Pi deprivation, Pi is redistributed from older leaves toward sink organs (young leaves, growing roots, and developing seeds) by a process requiring its transfer to phloem vessels (Raghothama, 2000; Bucher et al., 2001). An important process in plant growth and development is efficient nutrient remobilization from older, senescing leaves in order to scavenge resources that may be limiting in nature or energetically costly to acquire (Leopold, 1961). In this context, it has been demonstrated that up to 78% of stored Pi is remobilized from older leaves in Arabidopsis (Himelblau and Amasino, 2001). Therefore, the translocation of Pi into sink tissues/cells is important for sustaining growth under low-Pi conditions. Chloroplast-localized Pht2;1, a low-affinity Pi transporter (Km of approximately 0.4 mm), has been shown to mediate Pi translocation within the aerial parts of Arabidopsis (Daram et al., 1999; Versaw and Harrison, 2002). Low-affinity transporters from barley (Hordeum vulgare; HvPht1;6) and rice (OsPht1;2) have also been implicated in Pi remobilization from leaves and Pi movement from root to shoot, respectively (Rae et al., 2003; Ai et al., 2009; Preuss et al., 2010). Despite these developments, little is known about the molecular mechanisms that govern Pi translocation and remobilization in higher plants.
Spatial expression patterns of Pht1 members in different tissue types and organs of Arabidopsis suggest their potential involvement not only in Pi acquisition but also in internal Pi distribution to metabolically active and growing parts of the plant (Karthikeyan et al., 2002, 2009; Mudge et al., 2002; Miller et al., 2009). Among the Pht1 members, Pht1;5 showed Pi deficiency-induced expression specifically in the phloem cells of older leaves, cotyledons, and flowers (Mudge et al., 2002). Genome-wide transcriptome analysis further corroborated the expression of Pht1;5 over the course of developmentally regulated senescence in the leaves of Arabidopsis (van der Graaff et al., 2006). However, the functional characterization of Pht1;5 and its potential role in Pi translocation/remobilization have not been elucidated. Here, we used loss-of-function mutants of Pht1;5 and transgenic lines overexpressing this gene in Arabidopsis to demonstrate its role in Pi mobilization between source and sink under different Pi regimes. We also provide evidence for a tangible link between Pi transporters and ethylene signaling.
RESULTS
Expression Profile of Pht1;5, and Isolation of pht1;5 T-DNA Insertion Mutants
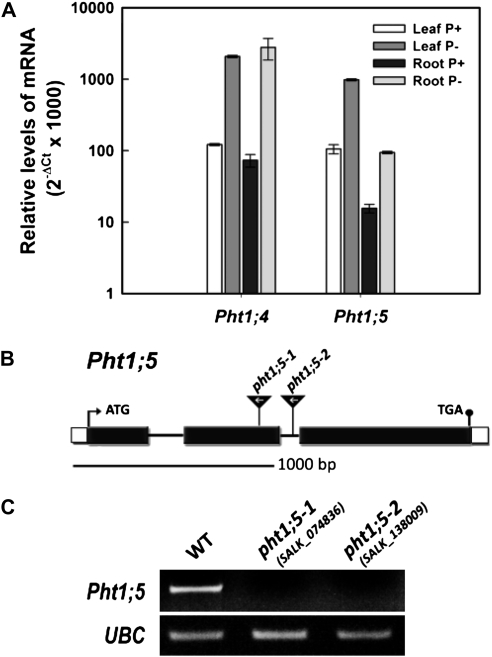
Previous studies have demonstrated the Pi starvation responsiveness of Pht1;5 expression in Arabidopsis (Mudge et al., 2002; Morcuende et al., 2007; Thibaud et al., 2010). To further determine the spatial expression pattern of Pht1;5, wild-type Arabidopsis plants were germinated hydroponically on half-strength Murashige and Skoog (MS) medium for 5 d and transferred to low Pi (10 μm; P−) and high Pi (1,250 μm; P+) for 7 d. Quantitative reverse transcription (qRT)-PCR analysis was used to measure Pht1;5 transcript levels in different tissues (roots and rosette leaves) grown under P+ and P− conditions. As shown in Figure 1A, Pi deficiency triggered increases in the abundance of Pht1;5 transcripts in both roots and rosette leaves. Consistent with earlier studies (Karthikeyan et al., 2002; Mudge et al., 2002; Shin et al., 2004), Pht1;4 transcript levels increased strongly in response to Pi deficiency in roots and rosette leaves (Fig. 1A). The data thus validated the fidelity of the growth condition used for elucidating the effect of Pi deficiency on the spatial expression profile of Pht1;5.
Figure 1.
Spatial expression pattern of Pht1;5 and isolation of pht1;5 loss-of-function mutants. A, qRT-PCR analysis of the expression of Pht1;4 and Pht1;5 in different organs under different Pi regimes. Wild-type and mutant plants were raised hydroponically under aseptic conditions on half-strength MS medium for 5 d and transferred to P+ or P− medium for 7 d. B, Schematic representation of the Pht1;5 gene and the location of T-DNA inserts (triangles) in the different pht1;5 alleles. White and black boxes represent untranslated regions and exons, respectively. C, RT-PCR analysis of Pht1;5 expression in the wild type (WT) and pht1;5 mutants. Seedlings were grown hydroponically under aseptic conditions on half-strength MS medium for 5 d and transferred to P− medium for 7 d. Whole seedlings were harvested. Transcript abundance was determined using primers specific to Pht1;5 and UBC genes.
To determine the role of Pht1;5 in the acquisition and mobilization of Pi, a reverse genetics approach was employed. Two homozygous mutants (SALK_074836 [pht1;5-1] and SALK_138009C [pht1;5-2]) were identified with T-DNAs inserted at 919 bp (exon) and 1,020 bp (intron), respectively, downstream of the translation start site of the Pht1;5 gene (Fig. 1B). Genetic analysis revealed that the pht1;5-1 T-DNA segregated as a single insertion locus (data not shown), whereas estimation of the T-DNA copy number of pht1;5-2 (a confirmed homozygous insertion line; Arabidopsis Biological Resource Center) via qPCR revealed the presence of approximately five and seven copies of nptII and T-DNA left border compared with pht1;5-1 (Supplemental Table S2). RT-PCR was performed on 7-d-old Pi-starved wild-type, pht1;5-1, and pht1;5-2 seedlings for determining the levels of Pht1;5 expression (Fig. 1C). An amplified product corresponding to Pht1;5 was detected in the wild type, whereas no amplification products were detected in pht1;5-1 or pht1;5-2. Furthermore, qRT-PCR analyses of Pi-starved roots and shoots found no detectable amplification for Pht1;5 in these mutants (data not shown). These results confirmed the identification of two independent loss-of-function mutants for Pht1;5.
Loss-of-Function Mutation of Pht1;5 Results in Altered Pi Allocation between Shoot and Root
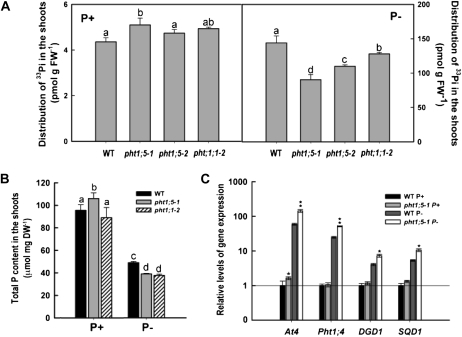
During Pi deficiency, the levels of Pht1;5 transcripts increased appreciably in both roots and rosette leaves (Fig. 1A). Therefore, a role for Pht1;5 in translocating Pi between root and shoot was hypothesized. To test this, radiolabeled 33Pi was used to compare the rates of root-to-shoot Pi translocation among the wild type, pht1;5-1, and pht1;5-2. A loss-of-function mutant (pht1;1-2) of high-affinity transporter Pht1;1, which affects both Pi acquisition and allocation (Shin et al., 2004), was included for comparison. Under P+ conditions, the distribution of 33Pi in the shoots was comparable among the wild type, pht1;5-2, and pht1;1-2 and was marginally higher for pht1;5-1 (Fig. 2A). However, under P− conditions, both pht1;5 mutants showed 35% to 40% lower 33Pi distribution to the shoots relative to the wild type. Although 33Pi distribution in pht1;1-2 was also significantly (P < 0.05) lower than in the wild type, it was significantly (P < 0.05) higher than in the pht1;5 mutants (Fig. 2A). These results suggest a more pronounced role for Pht1;5 in allocating Pi to the shoots during Pi deprivation. To determine whether the attenuated shoot Pi translocation rates observed in the mutants correlated with a change in P accumulation, we compared the total shoot P content of the wild type, pht1;5-1, and pht1;1-2 (Fig. 2B). Pi deficiency resulted in (P < 0.05) significant reductions in the total shoot P accumulation in all genotypes. Although the P content in the P+ shoots of pht1;5-1 was higher than in the wild type, under P− conditions, both mutants (pht1;1-2 and pht1;5-1) accumulated about 20% less P in their shoots compared with the wild type (Fig. 2B). Shoot P measurements in leaves and floral stalks from Pi-starved pht1;5-1 plants show similar decreases compared with the wild type when grown hydroponically under greenhouse conditions (Supplemental Fig. S1). The decrease in shoot P content in pht1;5-1 was amended by complementing the mutant with Pht1;5 cDNA (Supplemental Fig. S1). These data support a role for Pht1;5 in facilitating the mobilization of Pi between root and shoot independently of, or in conjunction with, Pht1;1. Whether altered Pi mobilization in the shoots of the pht1;5 mutant has a commensurate effect on Pi starvation-response (PSR) gene expression was determined via qRT-PCR analysis of several PSR genes (Misson et al., 2005; Shin et al., 2006). Under P+ conditions, other than a marginal increase in At4 transcripts in the pht1;5-1 mutant, the relative transcript levels for the PSR genes tested (At4, Pht1;4, DGD1, and SQD1) were similar between pht1;5-1 and the wild type (Fig. 2C). In contrast, the transcript levels for all the genes tested were significantly higher in the Pi-deprived shoots of pht1;5-1 compared with the wild type. These data corroborate that a more aggravated Pi starvation response is being experienced by the shoots of pht1;5-1 compared with the wild type due to the disruption of Pi mobilization from root to shoot during Pi deficiency.
Figure 2.
Pht1;5 loss of function affects Pi distribution in shoots. Wild-type (WT) and mutant plants were raised hydroponically under aseptic conditions on half-strength MS medium for 5 d and transferred to P+ or P− medium for 7 d. A, Distribution of 33Pi in the shoots (n = 4 replicates of approximately 50 seedlings each). B, Total P content in the shoots (n = 4 replicates of 25 seedlings each). For A and B, values are means, and different letters indicate means that differ significantly (P < 0.05). Error bars indicate se. C, qRT-PCR analyses of relative expression of Pi starvation-response genes in the shoots of wild-type and pht1;5-1 plants. At4g26410 was used as an internal control, and the values, normalized to wild-type P+ levels, are relative quantification ± maximum/minimum values of two independent biological replicates run in triplicate. Asterisks indicate 1.5-fold or greater (*) and 2.0-fold or greater (**) changes compared with the wild type. DW, Dry weight; FW, fresh weight.
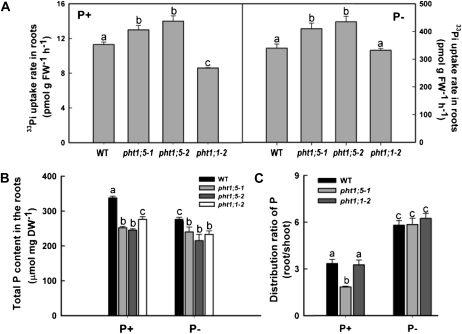
To gain further insight into a possible role for Pht1;5 in the maintenance of Pi homeostasis, 33Pi uptake rate in the roots was evaluated for wild-type, pht1;5-1, pht1;5-2, and pht1;1-2 seedlings grown under P+ and P− conditions (Fig. 3A). Independent of the Pi regime, there were significant (P < 0.05) increases (approximately 10%–20%) in the 33Pi uptake rates (i.e. net accumulation of 33Pi) in the roots of pht1;5-1 and pht1;5-2 compared with that of the wild type. On the contrary, there was a significant (P < 0.05) decline (approximately 30%) in root Pi uptake for pht1;1-2 compared with the wild type when grown under Pi-replete conditions, which is consistent with an earlier study (Shin et al., 2004). Despite differential 33Pi uptake rates observed for the pht1;1 and pht1;5 mutants under variable Pi conditions, the total P content in the roots of these mutants under both P+ and P− conditions was very similar and significantly (P < 0.05) lower compared with the wild type (Fig. 3B). The total P content data for roots (Fig. 3B) and shoots (Fig. 2B) of the wild type, pht1;5-1, and pht1;1-2 were used to calculate root-shoot distribution ratios of P (Fig. 3C). Although Pi deficiency exerted no significant (P < 0.05) differences on the ratio among the wild type and the mutants, pht1;5-1 showed a significant (P < 0.05) decline in the ratio compared with the wild type and pht1;1-2 when grown under Pi-replete conditions. Considering that Pht1;5 is expressed strongly in shoot tissues during Pi-replete conditions (Fig. 1A), these data suggest that Pht1;5 is involved in the mobilization of Pi from shoots to roots during high-Pi conditions.
Figure 3.
Pht1;5 loss of function disrupts the distribution and assimilation of Pi in the roots. Wild-type (WT) and mutant plants were grown hydroponically on half-strength MS medium for 5 d and transferred to P+ or P− medium for 7 d. A, Rate of Pi uptake in the roots (n = 4 replicates of approximately 50 seedlings each). Seedlings grown under P+ and P− were transferred to respective medium supplemented with 33Pi for 2 h, and roots were analyzed for 33Pi uptake. B, Total P content in the roots (n = 4 replicates of 25 seedlings each). C, Distribution ratio of P between root and shoot (n = 4 replicates of 25 seedlings each). Histograms show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se. DW, Dry weight; FW, fresh weight.
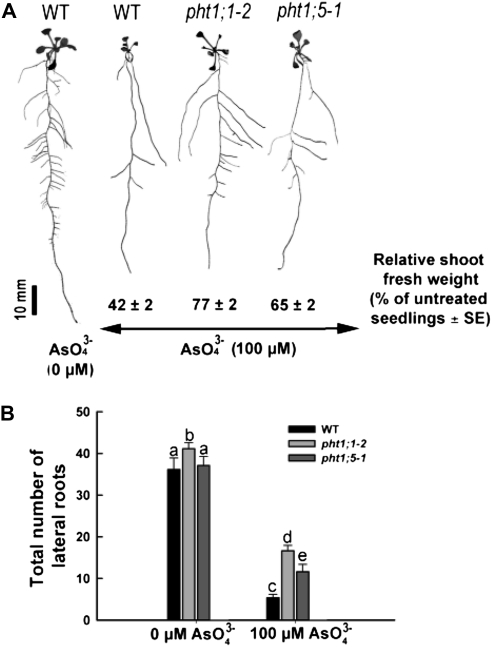
Arsenate (AsO43−) is an oxyanion structurally analogous to Pi and is taken up by roots via high-affinity Pi transporters (Asher and Reay, 1979; Shin et al., 2004; Catarecha et al., 2007). To gain further insight into the role of Pht1;5 in Pi acquisition, we compared the phenotypic responses of the pht1;1-2 and pht1;5-1 mutants with arsenate (Fig. 4). Although the toxic effect of arsenate was evident on the growth of both the mutants and the wild type, the degree of tolerance varied. The shoot fresh weight of the wild type grown on arsenate was 42% compared with that of untreated wild-type seedlings (Fig. 4A). The corresponding values for the pht1;1-2 and pht1;5-1 mutants were 77% and 65%, respectively, which indicated greater tolerance to arsenate by the mutants relative to the wild type. A similar trend of higher tolerance in the mutants was evident with respect to lateral root development (Fig. 4B). Together, these data indicate that loss of Pht1;5 confers weak tolerance to arsenate, suggesting that Pht1;5 could influence the acquisition of both Pi and arsenate.
Figure 4.
Loss of Pht1;5 results in moderate arsenate tolerance. Wild-type (WT) and mutant plants were grown on agar-solidified half-strength MS medium for 5 d and transferred to P+ Pi medium with and without 100 μm arsenate (AsO43−) for 7 d. A, Images of representative seedlings are shown (n = 20). Shoot fresh weights are presented relative to those of the respective seedlings grown in the absence of arsenate (n = 4 replicates of 20 seedlings each). B, Total number of lateral roots (n = 20). Histograms show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se.
Overexpression of Pht1;5 Affects the Distribution and Remobilization of Pi between Source and Sink
To further examine the in planta role of Pht1;5 in Pi allocation, we overexpressed the Pht1;5 coding region under the control of the ACTIN2 (ACT2) promoter in wild-type Arabidopsis. The ACT2 promoter imparts strong constitutive expression in vegetative tissues (An et al., 1996; Kandasamy et al., 2002). Two independently generated transgenic lines (5A and 11C) with relatively high levels of Pht1;5 expression compared with the wild type (Supplemental Fig, S2) were selected for phenotypic characterization. After 4 weeks of growth, the Pht1;5 overexpressors showed substantial increases in shoot biomass and leaf area compared with the wild type and pht1;5-1 (Fig. 5A). A more detailed phenotypic characterization was carried out on the wild type, pht1;5-1, and overexpressors that were grown for an additional 2 weeks. The Pht1;5 overexpressors exhibited significant (P < 0.05) increases in total leaf area (Fig. 5B), floral stalk thickness (Fig. 5C), and total leaf dry weight (Fig. 5D) compared with the wild type and pht1;5-1. On the other hand, pht1;5-1 initially displayed slower growth at the 4-week stage (Fig. 5A), but morphological parameters after 2 more weeks were only marginally affected compared with the wild type (Fig. 5, B and D). These phenotypic differences were observed under both short-day (Fig. 5) and long-day (data not shown) growth conditions.
Figure 5.
Pht1;5 overexpression increases shoot biomass. Seeds were germinated in potting mix in the greenhouse. A, Four-week-old wild-type (WT), pht1;5-1, and Pht1;5-overexpressor (5A and 11C) plants fertilized with half-strength Hoagland solution. B and C, Rosette leaves and floral stalks from 6-week-old plants were dissected and scanned at 200 dpi for documenting total leaf area (B) and stalk thickness (C). D, Rosette leaves were dried at 65°C for 24 h to determine the leaf biomass (n = 6 plants for each genotype). Histograms (B–D) show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se.
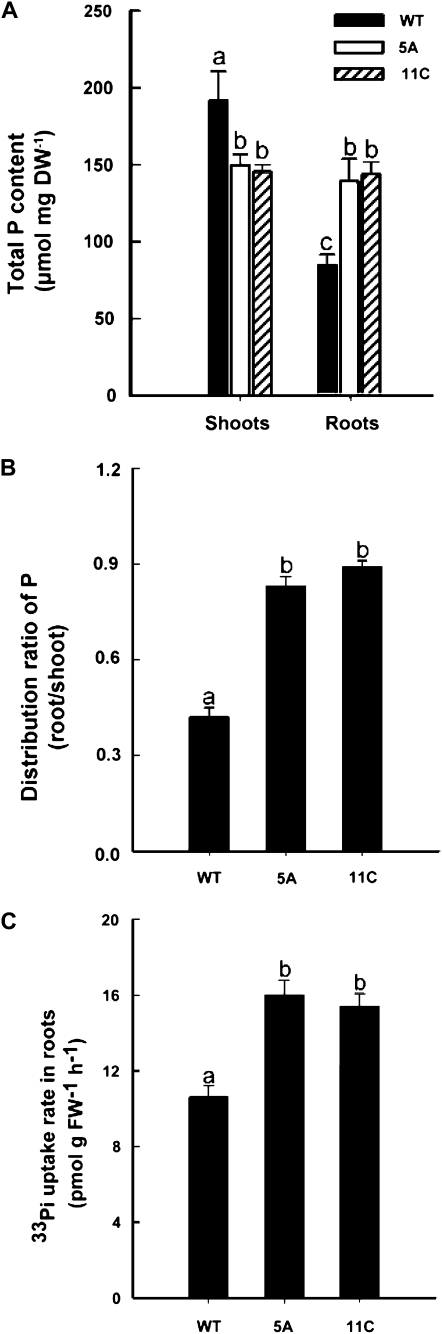
To examine the effect of Pht1;5 overexpression on P distribution and acquisition, the total shoot and root P contents and 33Pi root uptake rates of wild-type and Pht1;5 overexpressors were determined. Seedlings were grown on agar-solidified Pi-replete medium. Relative to the wild type, there was a significant (P < 0.05) decline in the total P content in the shoots of both overexpressors but an increase in total root P content (Fig. 6A). Interestingly, while the wild type showed higher (P < 0.05) shoot P content than root, there was no difference in P content between the two organs in the Pht1;5 overexpression lines. This suggests altered P distribution between source and sink in the overexpressors (Fig. 6B). It is plausible that Pht1;5-overexpression results in recycling of Pi from the shoots back to the roots. Elevated (approximately 60%) root 33Pi uptake rates in the Pht1;5 overexpressors compared with the wild type (Fig. 6C) further supported this notion. Although the 33Pi uptake rates in roots of the Pi-deprived Pht1;5 overexpressors were attenuated compared with the wild type, the P contents were comparable (data not shown). This indicates a differential effect of Pht1;5 overexpression on Pi homeostasis under different Pi regimes.
Figure 6.
Overexpression of Pht1;5 results in altered Pi homeostasis. A and B, Wild-type (WT) and Pht1;5 overexpressor (5A and 11C) plants were grown on agar-solidified half-strength MS medium for 5 d and transferred to P+ for 7 d. A, Total P content in the tissues indicated (n = 5 replicates of 20 seedlings each). B, P distribution between root and shoot (n = 5 replicates of 20 seedlings each). C, Rate of 33Pi uptake in the roots. Seedlings grown initially under P+ medium were transferred to the same medium supplemented with 33Pi for 2 h, and roots were analyzed for 33Pi uptake (n = 4 replicates of approximately 50 seedlings each). Histograms show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se. DW, Dry weight; FW, fresh weight.
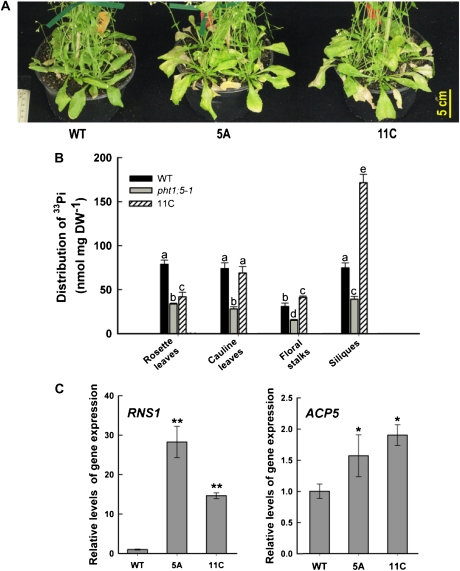
Pht1;5 overexpressors that were grown to maturity under greenhouse conditions consistently displayed chlorosis in older leaves that subsequently senesced earlier than those of the wild type (Fig. 7A). To gain insight into this premature senescence, 33Pi tracer studies were carried out to compare the Pi accumulation among a Pht1;5overexpressor (11C), the pht1;5-1 mutant, and the wild type (Fig. 7B). Compared with the wild-type, 33Pi accumulation in the Pht1;5 overexpressor was significantly (P < 0.05) lower in rosette leaves, comparable in cauline leaves, marginally higher in floral stalks, and approximately 2-fold higher in siliques. The 33Pi distribution in pht1;5-1 tissues was consistently and significantly (P < 0.05) lower relative to the wild type. These data suggest increased mobilization of Pi in the Pht1;5 overexpressor from source (i.e. rosette leaves) to sink (i.e. siliques), which may be attributed to enhanced Pi-scavenging mechanisms that facilitate the release of Pi from bound sources such as nucleic acids (Green, 1994; Raghothama, 1999; Misson et al., 2005). qRT-PCR analysis was used to quantify the transcript levels of RNase (RNS1; Bariola et al., 1994) and phosphatase (ACP5; del Pozo et al., 1999) genes involved in Pi remobilization in leaves of 4-week-old Pht1;5 overexpressors grown under Pi-replete conditions. As anticipated, there were increases in the relative transcript levels of RNS1 (14- to 28-fold) and ACP5 (1.5- to 2-fold) in the Pht1;5 overexpressors compared with the wild type (Fig. 7C). These molecular data support a role for Pht1;5 in facilitating the remobilization of Pi from senescing to metabolically active parts of the plant.
Figure 7.
Pht1;5 overexpressors exhibit premature senescence and increased Pi remobilization. A, Eight-week-old wild-type (WT) and Pht1;5 overexpressor (5A and 11C) plants showing senescent rosette leaves. B, Wild-type, pht1;5-1, and 11C plants were grown on agar-solidified half-strength MS medium for 10 d and transferred to well-aerated tubes with one-fifth-strength Hoagland nutrient solution supplemented with 100 μm Pi. After 10 d, roots were immersed in nutrient solution supplemented with 33Pi for 2 h. Roots were rinsed with unlabeled nutrient solution, and plants were transferred back to the nutrient solution for 7 d before scintillation counting (n = 5). Histogram bars represent means, and different letters indicate means that differ significantly (P < 0.05). Error bars indicate se. C, qRT-PCR analyses showing increased expression of genes involved in Pi remobilization in Pht1;5 overexpressors. Three-week-old plants were grown hydroponically in modified half-strength Hoagland nutrient solution for 7 d and transferred to high-Pi medium for 7 d. At4g26410 was used as an internal control, and the values, normalized to wild-type levels, are relative quantification ± maximum/minimum values of two independent biological replicates run in triplicate. Asterisks indicate 1.5-fold or greater (*) and 2.0-fold or greater (**) changes compared with the wild type. DW, Dry weight,
Pht1;5 Overexpression Alters Root Hair Development and Primary Root Growth in Association with Ethylene Signaling
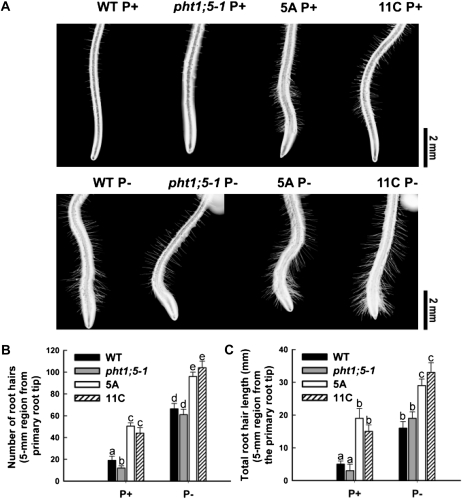
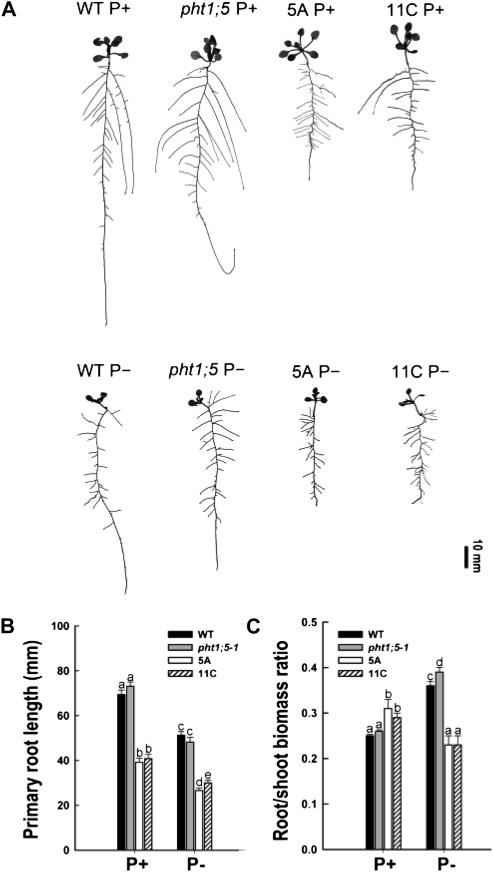
To study whether differential nutrient partitioning between root and shoot affects root development in the Pht1;5 overexpression lines, different root traits of the overexpressors were compared with those of the wild type and the pht1;5-1 mutant. Among the root traits examined, increased root hair proliferation is an early response of plants to Pi deficiency (Bates and Lynch, 1996; Ma et al., 2001; Jain et al., 2007b). Irrespective of the Pi regime, the Pht1;5 overexpressors showed significant (P < 0.05) increases in both the number and length of root hairs compared with the wild type and pht1;5-1 (Fig. 8). A second Pi deficiency-induced root response is a reduction in primary root growth due to premature cell differentiation (López-Bucio et al., 2002, 2003; Sánchez-Calderón et al., 2005; Jain et al., 2007a). Under P+ and P− conditions, there were significant (P < 0.05) reductions in the primary root length of both Pht1;5 overexpressors compared with the wild type and pht1;5-1 (Fig. 9, A and B). Pi deficiency-mediated root proliferation aimed at enhancing Pi mining capacity results in an increased root-shoot biomass ratio (Hermans et al., 2006; Hammond and White, 2008). Analysis of this trait revealed a significant (P < 0.05) reduction in the Pht1;5 overexpressors relative to the wild type and pht1;5-1 under P− conditions, whereas a reverse trend was observed under P+ conditions (Fig. 9C). Together, these data indicate accentuated Pi deficiency responses of Pht1;5 overexpressors under variable Pi conditions.
Figure 8.
Pht1;5 overexpression leads to increases in root hair number and length. Wild-type (WT), pht1;5- 1, and Pht1;5 overexpressor (5A and 11C) plants were grown on agar-solidified half-strength MS medium for 5 d and transferred to P+ or P− medium for 2 d. A, Primary root tips of the representative seedlings showing root hairs (n = 10). B and C, Numbers (B) and total lengths (C) of root hairs in a 5-mm section from the primary root tip. Histograms show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se.
Figure 9.
Pht1;5 loss of function and overexpression affect RSA. Wild-type (WT), pht1;5-1, and Pht1;5 overexpressor (5A and 11C) plants were grown on agar-solidified half-strength MS medium for 5 d and transferred to P+ or P− medium for 7 d. A, RSA of the representative seedlings (n = 12). B, Primary root length (n = 12). C, Root-shoot biomass ratio recorded on a dry weight basis (n = 5 replicates of 20 seedlings each). For B and C, histograms show means, and different letters indicate means that differ significantly (P < 0.05). Error bars represent se.
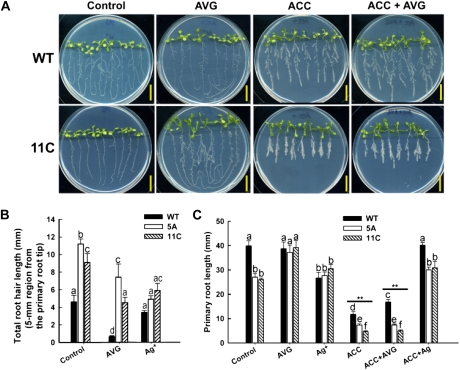
Modulation of root traits has been shown to be a manifestation of complex interactions among signaling pathways of Pi deficiency and phytohormones such as auxin and ethylene (Williamson et al., 2001; López-Bucio et al., 2002; Nacry et al., 2005; Jain et al., 2007a; Lei et al., 2011). Several lines of evidence indicate that ethylene stimulates auxin biosynthesis, and synergistic interactions between them modulate root growth and root hair development (Osmont et al., 2007; Stepanova and Alonso, 2009). Since overexpression of Pht1;5 caused a dramatic inhibition of primary root growth and alteration in root hair development under both P+ and P− conditions, its root responses toward ethylene precursors/inhibitors were investigated. Wild-type and Pht1;5 overexpressor (5A and 11C) plants were grown on half-strength MS medium for 5 d and then transferred to P+ supplemented with ethylene precursor (1 μm 1-aminocyclopropane-1-carboxylic acid [ACC]), biosynthesis inhibitor (0.2 μm aminoethoxyvinylglycine [AVG]), perception inhibitor (20 μm silver nitrate [AgNO3]), or simultaneous application of ACC + AVG and ACC + AgNO3 (Fig. 10). The effects of these treatments on root hair development and primary root length were documented after 1 and 7 d, respectively. Treatment with AVG resulted in significant (P < 0.05) reductions in total root hair length of the wild type and the Pht1;5 overexpressors (Fig. 10B). However, AgNO3 treatment resulted in a substantial decrease in total root hair length for the Pht1;5 overexpressors but only a marginal decrease for the wild type (Fig. 10B). A similar phenomenon was observed with the impact of AVG on primary root growth. Treatment with AVG had no effect on wild-type primary root length, but it dramatically increased (P < 0.001) primary root growth in the overexpressors, resulting in lengths comparable to the wild type (Fig. 10, A and C). The AgNO3 treatment significantly (P < 0.05) reduced the primary root length of the wild type but did not impose any detrimental effect on the root length of the Pht1;5 overexpression lines (Fig. 10C). Although medium supplemented with ACC or ACC + AVG exerted strong (P < 0.001) inhibitory effects on the primary root growth of all the genotypes tested, the Pht1;5 overexpressors displayed augmented sensitivity toward ACC compared with the wild type (Fig. 10C). Together, these results implicate ethylene signaling in modulating the primary root and root hair phenotypes of Pht1;5 overexpression lines.
Figure 10.
Ethylene signaling and biosynthesis inhibitors rescue the root phenotype of Pht1;5 overexpressors. Wild-type (WT) and Pht1;5 overexpressor (5A and 11C) plants were grown on agar-solidified half-strength MS medium for 5 d and then transferred to P+ medium (control) or the same medium supplemented with the inhibitors shown for 7 d. A, Phenotypes of wild-type and 11C seedlings grown in the presence of the indicated treatments. Scale = 20 mm. B, Total root hair length in a 5-mm section from the primary root tip (n = 10). C, Primary root length under the indicated treatments (n = 16). Histograms show means, and different letters indicate means that differ significantly (P < 0.05). Asterisks represent means that differ significantly (** P < 0.001) compared with the untreated control sets.
DISCUSSION
Plants possess multiple Pi transport systems, regulated at both high and low affinities, that facilitate Pi uptake from the rhizosphere and subsequently distribute it to cells and subcellular compartments. The movement of Pi inside the plant is an intricate and complex process, since source and sink relationships are constantly changing depending on the rate of growth, stage of development, and Pi availability (Raghothama, 1999; Bucher et al., 2001; Miller et al., 2009). The data presented here demonstrate that Pht1;5 is required for the normal mobilization of Pi between source and sink tissues.
Pht1;5 Influences Shoot Pi Status by Regulating Pi Distribution between Root and Shoot
Among the members of the Pht1 family, Pht1;5 was shown to be expressed in cotyledons and hypocotyls of germinating seedlings, floral buds, older leaves at the onset of senescence, and Pi-starved roots (Mudge et al., 2002). Our experiments corroborated these results and also showed that Pht1;5 is expressed in leaves under both P+ and P− conditions (Fig. 1A). Functional inactivation of this gene in pht1;5 mutant seedlings (pht1;5-1 and pht1;5-2) grown under Pi-deficient conditions led to low shoot P content, despite also resulting in an increased root Pi uptake rate (Figs. 2 and 3). Shoots of mature Pi-starved pht1;5-1 plants showed a similar reduction in P content compared with the wild type (Supplemental Fig. S1). Decreased shoot P content of pht1;5 mutants correlated with the up-regulation of several PSR genes (At4, Pht1;4, DGD1, and SQD1; Fig. 2C). Together, these results indicate that the loss of Pht1;5 affects the mobilization of Pi between root and shoot during Pi deficiency. Pht1;5 may play a role similar to Pht1 transporters from rice (OsPht1;2 and OsPht1;6) and barley (HvPht1;6), which have been implicated in Pi mobilization (Rae et al., 2003; Ai et al., 2009). Interestingly, Pht1;5, OsPht1;2, and HvPht1;6 are all expressed in stele cells in the root during Pi deficiency, consistent with them playing a role in root-to-shoot mobilization of Pi (Mudge et al., 2002; Rae et al., 2003; Ai et al., 2009).
Since Pht1;5 is predominantly expressed in shoot tissues during Pi-replete conditions (Mudge et al., 2002; Fig. 1A), Pht1;5 may play a role in Pi transfer from shoot to root. Our data demonstrate that the pht1;5-1 mutant accumulates more P in shoots and less in roots compared with the wild type (Figs. 2 and 3). Also under Pi-replete conditions, the root Pi uptake rates for the pht1;5 mutants were 10% to 20% higher than for the wild type (Fig. 3A). This likely results from enhanced uptake capacity by other Pi starvation-induced Pht1 transporters. In a previous study, loss of Pht1;4 showed similar increases in shoot P accumulation and root Pi uptake under Pi-replete conditions (Shin et al., 2004). This suggests that Pht1;4 and Pht1;5 may have partially overlapping functions during high-Pi conditions. Notably, Pht1;4 expression has been detected in the root stele and in shoot tissues under Pi-replete conditions (Karthikeyan et al., 2002; Mudge et al., 2002; Misson et al., 2004; Fig. 1A), consistent with a role for Pht1;4 in Pi mobilization. A key difference between the loss of Pht1;4 and Pht1;5 is the impact on root Pi uptake during Pi deficiency. pht1;4 mutants experience a drop in root Pi uptake during Pi deficiency (Shin et al., 2004), whereas pht1;5 mutants exhibit an increase (Fig. 3A). This supports the notion that Pht1;4, but not Pht1;5, contributes significantly to root Pi acquisition. Nevertheless, loss of Pht1;5 results in moderate tolerance to arsenate (Fig. 4), suggesting that Pht1;5 has a role in influencing the root acquisition of arsenate and, by inference, of Pi.
For the most part, the two independent T-DNA insertion lines, pht1;5-1 and pht1;5-2, showed similar physiological traits (Figs. 2 and 3). However, distribution of 33Pi in the shoots of the mutants differed (Fig. 2A). This may be due to the presence of multiple T-DNA copies in pht1;5-2 as estimated via qPCR (Supplemental Table S2). Thus, pht1;5-1 was characterized more extensively in this study.
Additional evidence supporting the role of Pht1;5 in Pi mobilization came from an analysis of plants overexpressing Pht1;5. Under Pi-replete conditions, Pht1;5 overexpressors accumulated more P in roots, but less in shoots, relative to the wild type (Fig. 6B), which is the opposite trend as that observed for the pht1;5-1 mutant (Fig. 3C). Interestingly, though, despite reduced shoot P concentrations, the Pht1;5 overexpression lines showed increased shoot biomass (Fig. 5). Our results suggest that overexpression of Pht1;5 results in an imbalance of Pi supply and demand, thereby affecting root/shoot biomass allocation (Fig. 9) and overall plant growth (Figs. 5 and 6). In an earlier study, Lai et al. (2007) reported that increased meristematic activity results in Pi consumption and that the magnitude of organ growth activity specifies Pi demand. Therefore, it is plausible that an increase in growth and Pi remobilization in Pht1;5 overexpression lines depletes Pi reserves in the shoot at a faster rate. Together, these results suggest that Pht1;5 plays a role in modulating Pi distribution between shoot and root in accordance with Pi status. Differences in physiological responses between the wild type and the Pht1;5 overexpressors were less evident under Pi deficiency conditions (data not shown), which could be due to strong induction of Pht1;5 during Pi deprivation in the wild type (Fig. 1A; Supplemental Fig. S2).
Pht1;5 Facilitates Pi Remobilization from Source to Sink in Adult Plants
Pht1;5 is the only Arabidopsis Pht1 member known to be induced in leaves by developmentally regulated senescence (Mudge et al., 2002; van der Graaff et al., 2006; Arabidopsis eFP Browser [http://bbc.botany.utoronto.ca/efp/]). Interestingly, Pht1;5-overexpressors displayed premature leaf senescence (Fig. 7A). Radiotracer experiments of feeding 33Pi to mature plants revealed that the Pht1;5 overexpressors accumulated 2-fold higher levels of 33Pi in siliques relative to the wild type, while the accumulation in rosette leaves was lower (Fig. 7B). The Pht1;5 overexpressors also had increased transcript levels for RNS1 and ACP5 in rosette leaves (Fig. 7C). During Pi limitation and senescence, scavenging processes involving the activation of these genes facilitate the release of Pi from organic forms of P (phosphate esters/nucleic acids), thereby making it available for recycling (Bariola et al., 1994; Green, 1994; del Pozo et al., 1999). The localization of Pht1;5 in the phloem of older leaves substantiates its role in Pi remobilization (Mudge et al., 2002). Together, the data support a role for Pht1;5 in facilitating the remobilization of Pi released by scavenging processes from senescing source (rosette leaves) to metabolically active sink (siliques) organs. A quantitative trait locus mapping study for seed nutrient content performed using two recombinant inbred populations, Columbia × Landsberg erecta and Cape Verde Islands × Landsberg erecta, identified Pht1;5 as a putative locus that governs seed P content (Waters and Grusak, 2008). The role of Pht1;5 in loading Pi into seeds warrants further investigation.
The Regulation of Pht1;5 and Its interactions with Ethylene- and Senescence-Related Pathways
Modulation of root system architecture (RSA) to increase root surface area for enhancing Pi adsorption is a characteristic adaptive response of Pi-starved plants (Jain et al., 2007b; Schachtman and Shin, 2007; Rouached et al., 2010). Constitutive Pht1;5 overexpression caused increases in the number and length of root hairs as well as a reduction of primary root length under both P+ and P− conditions (Figs. 8 and 9). Several lines of evidence suggest that the modulation of Pi deficiency-induced changes in RSA results from cross-talk among sugar, nutrient, and phytohormone signaling (López -Bucio et al., 2003; Jain et al., 2007b; Rubio et al., 2009). Thus, the alteration of root growth in the Pht1;5 overexpressors could be regulated independently of the Pi regime. Previous studies have implicated ethylene in Pi deficiency-induced root hair formation and primary root elongation (Ma et al., 2001, 2003; Zhang et al., 2003; Lei et al., 2011). Ethylene has also been implicated in tolerance to potassium deprivation and iron signaling responses (Zaid et al., 2003; Shin and Schachtman, 2004; Jung et al., 2009). We found that treatment with AVG, an inhibitor of ACC synthase and ethylene biosynthesis, rescued the primary root phenotype of Pht1;5 overexpressors (Fig. 10, A and C), while AgNO3 treatment decreased the total length of root hairs of Pht1;5 overexpressors to near wild-type levels (Fig. 10B). Expression of PhPT1, a high-affinity Pi transporter in petunia (Petunia hybrida), was shown to be induced during senescence and in response to ethylene treatment (Chapin and Jones, 2009), highlighting a potential link among ethylene, senescence, and activation of Pi transporters. In a recent study, Lei et al. (2011) showed that the Pi deficiency-induced expression of a Pht1;4 promoter-luciferase reporter transgene was strongly enhanced by ACC but attenuated by AgNO3 treatment. Furthermore, the expression of several PSR genes (including Pht1;1 and Pht1;4) was induced and suppressed in null mutant alleles of CONSTITUTIVE TRIPLE RESPONSE1 and ETHYLENE INSENSITIVE2, respectively. These studies along with our results provide evidence for a tangible connection between Pht1 transporters and ethylene signaling. Another possible link between ethylene and Pht1;5 activity could be ethylene-dependent modulation of reactive oxygen species, which may regulate Pht1;5. A similar phenomenon was recently identified for the regulation of HAK5, which encodes a high-affinity potassium transporter (Jung et al., 2009). Since ethylene and auxin function synergistically in regulating several root traits (Schmidt and Schikora, 2001; López-Bucio et al., 2002; Ma et al., 2003), the potential role of auxin in influencing the exaggerated root hair development and reduced primary root growth of Pht1;5 overexpressors cannot be ruled out. However, the cross talk with ethylene signaling appears to be conditional, as evidenced by comparable apical hook formation and hypocotyl lengths of etiolated seedlings of the wild type, pht1;5 mutants, and Pht1;5 overexpressors (data not shown).
Loss-of-function/RNA interference (RNAi) mutants of PHO2, PHR1, and WRKY75 impact pathways that regulate Pi acquisition and distribution, thereby playing critical roles in systemic Pi responses (Delhaize and Randall, 1995; Rubio et al., 2001; Bari et al., 2006; Devaiah et al., 2007; Thibaud et al., 2010). Using qRT-PCR, we found that relative Pht1;5 transcript levels in loss-of-function pho2 and phr1 mutants were comparable to wild-type levels, but those in a WRKY75-RNAi line were reduced under both P+ and P− conditions (Supplemental Fig. S3). This supports a role for WRKY75 in the regulation of Pht1;5. Previously, WRKY75 was shown to positively regulate Pi acquisition and negatively regulate lateral root and root hair development in Arabidopsis (Devaiah et al., 2007). The presence of two WRKY box (W-box) cis-elements (TTGACC/T) in the Pht1;5 promoter further suggests regulatory control by WRKY transcription factors. WRKY proteins have also been implicated in defense responses and senescence-regulated processes (Eulgem and Somssich, 2007). For instance, WRKY6 is known to be a positive regulator of genes involved in senescence signaling and pathogen defense responses (Robatzek and Somssich, 2002). Overexpression or loss of function of WRKY6 resulted in alterations in the transcript abundance of Pht1;5, suggesting a role for WRKY6 in Pht1;5 regulation (Chen et al., 2009). Interestingly, WRKY6 has also been shown to modulate Pi distribution between root and shoot by negatively regulating the expression of PHO1, which encodes a protein involved in loading Pi into root xylem (Poirier et al., 1991; Chen et al., 2009). One characteristic feature of WRKY proteins is their ability to autoregulate their own promoter as well as cross-regulating other WRKYs (Rushton et al., 2010). It is possible that feed-forward or feedback regulation of these determinants could occur concomitantly with the alteration in shoot Pi levels due to the activity of Pht1;5. More experiments are required to dissect the molecular mechanisms that regulate Pht1;5 and its responses that trigger senescence-regulated processes.
In conclusion, our data indicate that Pht1;5 plays a complex role in differential partitioning of Pi to plant organs, thereby influencing overall plant growth. Under Pi-replete conditions, Pht1;5 expression is limited to shoots and is modulated by developmental cues to promote Pi distribution to sink tissues, including roots of young seedlings and reproductive tissues in mature plants. Upon perception of low Pi, Pht1;5 expression is induced in both shoots and roots. Under these conditions, Pht1;5 is required for proper Pi translocation from root to shoot, likely via a role in loading Pi into root xylem. Pht1;5 also indirectly influences Pi acquisition by regulating shoot P status.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia, pht1;1-2 (Shin et al., 2004), pho2 (Delhaize and Randall, 1995), WRKY75 RNAi line (Devaiah et al., 2007), and phr1 (Bari et al., 2006) were used in this study. For studying growth at the seedling stage, seeds were sterilized in 70% (v/v) ethanol for 1 min followed by 50% (v/v) commercial bleach and 0.1% (v/v) Tween 20 for 10 to 15 min. After three to five washes with sterile water, seeds were stratified at 4°C for 2 to 3 d and suspended in 0.2% (w/v) agar. Seeds were germinated on half-strength MS medium and 1.5% (w/v) Suc, buffered to pH 5.7 with 0.5 mm MES. Depending on the experiment, seedlings were grown on meshes in standard Magenta (GA-7) boxes under aseptic hydroponic conditions as described (Jain et al., 2009) or vertically grown on petri plates with medium solidified with 1.2% (w/v) agar (Sigma A1296; lot no. 096K01581, with a P content of approximately 30 μm). Seedlings were then transferred to a growth room set to these conditions: 22°C, 16-h photoperiod, and average photosynthetically active radiation between 60 and 70 μmol m–2 s–1. After 5 d, uniformly grown seedlings with primary root lengths ranging between 15 and 25 mm were transferred to P+ (1.25 mm KH2PO4) or P− (0 mm KH2PO4) medium as described (López-Bucio et al., 2002; Jain et al., 2009). Although hydroponic conditions are best suited for studying nutrient starvation stress conditions, we observed that lack of added Pi was detrimental to seedling growth and led to eventual death before the completion of the experiment. Therefore the P− treatments were supplemented with 10 μm KH2PO4 for the seedlings to deplete during the course of the experiment. In the Pi-deprived medium, KH2PO4 was replaced by an equimolar amount of K2SO4.
Ethylene-responsive root growth assays on seedlings were performed using the ethylene precursor ACC with or without the addition of AVG and AgNO3, as described by Růzicka et al. (2007).
For phenotypic characterization of older developmental stages, greenhouse studies were conducted on plants that were germinated and grown on Premier Promix TA (Premier Horticulture). Once every week, plants were irrigated with modified half-strength Hoagland solution comprising 2.5 mm Ca(NO3)20.4H2O, 1.25 mm K2SO4, 1 mm MgSO40.7H2O, 250 μm KH2PO4, 0.13 μm KCl, 2.25 μm MnSO4·H2O, 12.5 μm H3BO3, 2.0 μm ZnSO40.7H2O, 0.25 μm CuSO40.5H2O, 0.1 μm Na2MoO40.2H2O, and 27.5 mg L−1 Fe-sequestrene and buffered to pH 5.7 with 0.5 mm MES.
Isolation of Mutant Lines
Putative T-DNA insertion lines in the Pht1;5 gene were identified by searching the SIGNAL-SALK database (http://signal.salk.edu/cgi-bin/tdnaexpress). Homozygous plants were isolated by PCR screening of seed populations (obtained from the Arabidopsis Biological Resource Center) using T-DNA left-border (LBa1, 5′-TGGTTCACGTAGTGGGCCATCG-3′), gene-specific right (RP, 5′-GTTTGAGCTTTTGAGACGGTG-3′), and gene-specific left (LP, 5′-ACCTTTGCTAATTTCTTGGCC-3′) primer combinations to confirm insertions in the Pht1;5 locus. The T-DNA insertion site was PCR amplified and sequenced at the Purdue University Genomics Core Facility. The homozygous mutants were designated pht1;5-1 (SALK_074836) and pht1;5-2 (SALK_138009C).
Construction of Pht1;5 Overexpression Lines
For overexpression of Pht1;5, the 1.6-kb Pht1;5 cDNA was reverse transcribed with SuperScript III (Invitrogen) from total RNA extracted from Pi-starved Arabidopsis seedlings, and the primers PHT5_S1 (5′-TACGTCGGGCCCCCATGGCGAAAAAAGGAAAAGAAGT-3′) and PHT5_A1629 (5′-TAGCTGAAGCTTGGATCCTCAAACCGGGACTTTTCTACCGGA-3′; restriction sites are underlined) were used for amplification. The flanking NcoI and BamHI restriction sites were used for cloning the Pht1;5 coding sequence into a constitutive, vegetative ACT2 promoter/terminator cassette (pACT2; Kandasamy et al., 2002) within the binary vector pCambia-1300. Wild-type Arabidopsis plants (ecotype Columbia) or the pht1;5-1 mutant were transformed using Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Hygromycin resistance was used for identifying homozygous T3 plant lines.
RNA Expression Analyses
Total RNA was isolated from ground plant tissues using the RNeasy Plant Mini Kit (Qiagen; www.qiagen.com). One microgram of total RNA was treated with RQ1 RNase-free DNase I (Promega; www.promega.com) and was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen; www.invitrogen.com). RT-PCR was performed on 2 μL of the cDNA using gene-specific primers. Thermal cycling consisted of an initial denaturation at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 90 s at 72°C, and a final 7-min extension at 72°C. qRT-PCR analysis was performed on an Applied Biosystems 7500 Real-Time PCR system using SYBR Green detection chemistry (Applied Biosystems; www.appliedbiosystems.com). UBC and At4g26410 (Czechowski et al., 2005) were used as reference genes, and relative expression levels of the genes were computed by the 2–ΔΔCt method of relative quantification (Livak and Schmittgen, 2001). In Figure 1A, transcript abundances of Pht1;4 and Pht1;5 across different tissues were calculated as 2–ΔCt. The primers used are listed in Supplemental Table S1.
Estimation of T-DNA Copy Number
T-DNA copy number in the genomes of the pht1;5 mutants was estimated via qPCR as described previously by Mason et al. (2002). QuickExtract Plant DNA Extraction Solution (Epicentre) was used to obtain genomic DNA. Reactions for two biological replicates per genotype were performed in triplicate on an Applied Biosystems 7500 Real-Time PCR system using SYBR Green detection chemistry (Applied Biosystems). Starting quantities for two regions of the SALK transgene, nptII and a portion near the left border, and two endogenous single-copy genes, ACT2 and Pht1;4, were determined by LinRegPCR software (Ruijter et al., 2009). For each transgene, a virtual calibrator (Mason et al., 2002) was created to use for normalization of the data to represent T-DNA copy number.
Total P Estimation
Total P content was estimated using a modification of the U.S. Environmental Protection Agency method 365.2. Approximately 50 mg of plant material was taken in preweighed glass vials (Kimble Chase Life Sciences), dried overnight at 70°C, weighed, and flamed to ash in a furnace. The samples were dissolved in 100 μL of concentrated HCl, and 10 μL of the digested sample was diluted with 790 μL of deionized water. To this, 200 μL of molybdenum blue reagent (5 n H2SO4, 4% [w/v] ammonium molybdate, and 10% [w/v] ascorbic acid) was added, and samples were incubated at 45°C for 20 min prior to spectrophotometric determination of P.
Pi Uptake Assay
Pi uptake was performed as described previously (Shin et al., 2004) with minor modifications. Seedlings (approximately 50) were raised under aseptic conditions hydroponically on half-strength MS medium for 5 d and transferred to P+ (1.25 mm Pi) and P− (0.01 mm Pi) media for 7 d. The P+ and P− seedlings along with the meshes were transferred to petri plates (100 mm × 15 mm) containing P+ or P− nutrient solution supplemented with 0.15 μCi mL−1 33Pi. After 2 h, samples were incubated in ice-cold desorption medium (0.1 mm CaCl2, 5 mm MES, and 2 mm KH2PO4, pH 5.7) for 30 min. The samples were rinsed with desorption solution followed by distilled water and blotted dry before separating roots and shoots. After their fresh weights were recorded, tissues were dried overnight at 65°C, and 33Pi activity was measured by using a liquid scintillation counter (Tri-Carb 2810; Perkin-Elmer). The root 33Pi uptake rate was calculated as pmol Pi g−1 fresh weight h−1, and distribution was calculated as pmol Pi g−1 fresh weight.
For 33Pi feeding and tracing experiments, wild-type, pht1;5-1 mutant, and Pht1;5 overexpressor (11C) plants were grown on agar-solidified half-strength MS medium for 10 d and transferred to well-aerated tubes containing one-fifth-strength Hoagland nutrient solution containing 100 μm Pi. After 10 d, roots were immersed in the same nutrient solution supplemented with 0.15 μCi mL−1 33Pi for 2 h. Roots were desorbed with unlabeled Hoagland solution for 30 min and rinsed with tap water thoroughly before transferring back to the nutrient solution. Plants were allowed to grow for 7 d before harvesting different parts of the plant. Tissue samples were dried overnight at 65°C and weighed, and 33Pi activity was measured as mentioned above.
Analysis of RSA
Root traits were documented as described (Jain et al., 2007a). Briefly, the seedlings grown on agar-solidified medium were scanned at 600 dots per inch (dpi) using a desktop scanner (UMAX PowerLook III) and transferred to 70% (v/v) ethanol, facilitating preservation for subsequent detailed analyses. The scanned images were used for measuring the elongation of the primary root after transfer from half-strength MS medium to P+ and P− media. For documenting the number of first-order and higher order lateral roots and their lengths, seedlings stored in ethanol were gently spread on agar (1%, w/v) plates with a camel hair brush and scanned at 600 dpi. Ten to 15 scanned images of the seedlings per genotype for every treatment were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/).
Documentation of Root Hairs
Root hair numbers and lengths were recorded as described (Jain et al., 2007a). Briefly, seedlings were initially grown on half-strength MS medium with agar (1.2%, w/v) for 5 d and transferred to agar-solidified P+ and P− media. After 2 d or as mentioned, images of root hairs growing in the 5-mm region from the tip of the primary root were captured using a stereomicroscope (Nikon SMZ-U). Root hairs were measured for approximately 10 seedlings per genotype for every treatment using ImageJ software.
Statistical Analyses
Data were analyzed by one-way ANOVA (Figs. 2A, 3A, 5, B–D, and 6, B and C) or two-way ANOVA (Figs. 2B, 3, B and C, 4B, 6A, 7B, 8, B and C, 9, B and C, and 10, B and C), and Tukey’s honestly significant difference test was carried out for multiple comparisons using the SPSS 10 program (www.spss.com). Different letters represent means that were statistically different at P < 0.05 or as mentioned.
Pht1;5 corresponds to GenBank accession number NM_128843 and Arabidopsis Genome Initiative locus identifier At2g32830.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Total P content in the pht1;5-1 mutant complemented with Pht1;5 cDNA.
Supplemental Figure S2. Relative expression of Pht1;5 in two independent Pht1;5 overexpression lines.
Supplemental Figure S3. Relative expression of Pht1;5 in Pi signaling mutants.
Supplemental Table S1. Primers used for PCR analyses.
Supplemental Table S2. qPCR estimation of T-DNA copy number in the pht1;5 mutants.
Supplementary Material
Acknowledgments
We thank Debra M. Sherman and Chia-Ping Huang of the Purdue Life Sciences Microscopy Facility for technical assistance with the microscopy work and Prof. Richard Meagher (University of Georgia, Athens) for providing the Pht1;5 overexpression lines.
References
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Asher CJ, Reay PF. (1979) Arsenic uptake by barley seedlings. Aust J Plant Physiol 6: 459–466 [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19: 529–538 [Google Scholar]
- Bucher M, Rausch C, Daram P. (2001) Molecular and biochemical mechanisms of phosphorus uptake into plants. J Plant Nutr Soil Sci 164: 209–217 [Google Scholar]
- Catarecha P, Segura MD, Franco-Zorrilla JM, García-Ponce B, Lanza M, Solano R, Paz-Ares J, Leyva A. (2007) A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin LJ, Jones ML. (2009) Ethylene regulates phosphorus remobilization and expression of a phosphate transporter (PhPT1) during petunia corolla senescence. J Exp Bot 60: 2179–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-F, Li L-Q, Xu Q, Kong YH, Wang H, Wu W-H. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Aung K, Lin S-I, Wu C-C, Chiang S-F, Su CL. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Liu H, Harrison MJ. (2001) The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J 25: 281–293 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. (1999) Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11: 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Green PJ. (1994) The ribonucleases of higher plants. Annu Rev Plant Physiol Plant Mol Biol 45: 421–445 [Google Scholar]
- Hammond JP, White PJ. (2008) Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J Exp Bot 59: 93–109 [DOI] [PubMed] [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM. (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG. (2007a) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144: 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Poling MD, Smith AP, Nagarajan VK, Lahner B, Meagher RB, Raghothama KG. (2009) Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiol 150: 1033–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vasconcelos MJ, Sahi SV, Raghothama KG. (2007b) Molecular mechanisms of plant adaptation to phosphate deficiency. Janick J, , Plant Breeding Reviews, Vol 29 John Wiley & Sons, New York, pp 359–419 [Google Scholar]
- Jung JY, Shin R, Schachtman DP. (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB. (2002) Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan AS, Ballachanda DN, Raghothama KG. (2009) Promoter deletion analysis elucidates the role of cis elements and 5’UTR intron in spatiotemporal regulation of AtPht1;4 expression in Arabidopsis. Physiol Plant 136: 10–18 [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, D’Urzo MP, Damsz B, Raghothama KG. (2002) Regulated expression of Arabidopsis phosphate transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Thacker J, Li Y, Doerner P. (2007) Cell division activity determines the magnitude of phosphate starvation responses in Arabidopsis. Plant J 50: 545–556 [DOI] [PubMed] [Google Scholar]
- Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D. (2011) Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol 189: 1084–1095 [DOI] [PubMed] [Google Scholar]
- Leopold AC. (1961) Senescence in plant development: the death of plants or plant parts may be of positive ecological or physiological value. Science 134: 1727–1732 [DOI] [PubMed] [Google Scholar]
- Lin W-Y, Lin S-I, Chiou T-J. (2009) Molecular regulators of phosphate homeostasis in plants. J Exp Bot 60: 1427–1438 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Baskin TI, Brown KM, Lynch JP. (2003) Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol 131: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24: 459–467 [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants. Academic Press, London [Google Scholar]
- Mason G, Provero P, Vaira AM, Accotto GP. (2002) Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Shen Q, Xu G. (2009) Freeways in the plant: transporters for N, P and S and their regulation. Curr Opin Plant Biol 12: 284–290 [DOI] [PubMed] [Google Scholar]
- Mimura T. (1999) Regulation of phosphate transport and homeostasis in plant cells. Int Rev Cytol 191: 149–200 [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Thibaud M-C, Bechtold N, Raghothama K, Nussaume L. (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari RP, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P. (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS. (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58: 93–113 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. (2002) Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book; 1: e0024, doi/10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Gilliham M, Tyerman SD. (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW. (2003) Characterization of two phosphate transporters from barley: evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rausch C, Bucher M. (2002) Molecular mechanisms of phosphate transport in plants. Planta 216: 23–37 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE. (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y. (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J. (2009) Plant hormones and nutrient signaling. Plant Mol Biol 69: 361–373 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L. (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Schmidt W, Schikora A. (2001) Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol 125: 2078–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ. (2006) Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J 45: 712–726 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Alonso JM. (2009) Ethylene signaling and response: where different regulatory modules meet. Curr Opin Plant Biol 12: 548–555 [DOI] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ. (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters BM, Grusak MA. (2008) Quantitative trait locus mapping for seed mineral concentrations in two Arabidopsis thaliana recombinant inbred populations. New Phytol 179: 1033–1047 [DOI] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid H, El Morabet R, Diem HG, Arahou M. (2003) Does ethylene mediate cluster root formation under iron deficiency? Ann Bot (Lond) 92: 673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Lynch JP, Brown KM. (2003) Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J Exp Bot 54: 2351–2361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.