Abstract
Background & objectives:
Imbalances in compactly regulated DNA repair pathways in the form of single nucleotide polymorphisms (SNPs) within vital DNA repair genes may result in insufficient DNA repair and increase in DNA breaks thus rendering the human system vulnerable to the debilitatory effects of grave diseases like cancers. The present study involves investigation of association of the non-synonymous SNP rs1052133 (C8069G/Ser326Cys) located in the exonic region of the gene human 8-oxoguanine DNA glycosylase (hOGG1) with the risk of squamous cell carcinomas of the head and neck (SCCHN).
Methods:
Case-control based genetic association study was performed among 575 (250 SCCHN cases and 325 normal healthy controls) sub-population cluster-matched (Indo-Europeans linguistic subgroup + Caucasoid morphological subtype) samples from the north Indian States of Uttar Pradesh and Uttarakhand using polymerase chain reaction followed by restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analysis.
Results:
Our results demonstrated statistically significant protective association for the heterozygous CG [Odds Ratio (OR) 0.6587, 95% Confidence Interval (CI) 0.4615 to 0.9402, P=0.0238], homozygous mutant GG (OR 0.2570, 95% CI 0.1070 to 0.6175, P=0.0013) and combined mutant CG + GG (OR 0.6057, 95% CI 0.4272 to 0.8586, P=0.0059) genotypes.
Interpretation & conclusions:
The results indicate that the polymorphism rs1052133 is strongly associated with SCCHN susceptibility and the mutant (G) allele might be a protective factor for SCCHN among north Indian subpopulations.
Keywords: BER, MAF, Odds ratio, OGG1, SCCHN, SNP
Base excision repair (BER) pathway is the most pro-active DNA repair pathway that corrects DNA modifications which arise either spontaneously or from attack by reactive chemicals i.e., removal of DNA damage involving structurally non-distorting and non-bulky lesions like oxidized and ring-saturated bases, alkylated and deaminated bases, apurinic/apyrimidinic (AP) sites and some types of mismatches1. Discrepancies in the compactly regulated BER pathway in the form of polymorphic variations/SNPs within vital DNA repair genes may result in reduction in activity causing insufficient DNA repair, genomic instability and increase in DNA breaks thus rendering the human system vulnerable to the debilitatory effects of critical diseases like cancers2–5. The gene human 8-oxoguanine DNA glycosylase (hOGG1 or OGG1) (HUGO Gene ID 4968; OMIM ID 601982; Gene length 17638bp; Chromosomal location 3p26.2) generates a 345 amino acid OGG1 protein that is an important member of the BER pathway involved in the repair of one of the most mutagenic lesions among base damages, i.e., 8-oxoguanine or 8-hydroxyguanine (8-hydroxy-2′-deoxyguanine, 8-OH-dG) present in DNA in its alternative tautomeric form. 8-Oxoguanine have been shown to cause base-pairing with adenine and result in G:C/T:A transversions in repair-deficient bacteria and yeast5. Among the validated sequence variants (SNPs) within OGG1 described in sequence databases, the SNP rs1052133, a C to G change at gene location 8069 located in Exon 7 of the Plus strand resulting in an amino acid change of Pro332Ala in transcript variant ENST00000302003, Ser326Cys in transcript variant ENST00000344629 and Pro119Ala change in transcript variant ENST00000339542 has been studied most frequently showing association between OGG1 genotypes and enzyme activity in a number of in vivo or in vitro studies, although the results have been inconsistent6. The rs1052133 variants show differences in DNA repair activity which has been implicated in increased risk of various cancers7–9. Interestingly, one study found that the rs1052133 wild type allele (C)-containing OGG1 has a seven-fold higher activity for repairing 8-oxoguanine than the rs1052133 mutant allele (G)-containing OGG12, although two other studies could observe no association between OGG1 genotypes and enzyme activity10,11. Genetic association studies on the polymorphism rs1052133/Ser326Cys (OGG1) have been conducted in various cancers associated with a wide range of risk ratios8,9,13–19 although, so far no such study has been undertaken to assess the association of OGG1 SNPs with the risk of SCCHN among the Indian population despite the fact that the India has the largest human genetic diversity among comparable global regions, second only to Africa20,21.
In the present study, the nonsynonymous SNP rs1052133 located in the exonic region of the gene OGG1 was analyzed using polymerase chain reaction followed by restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analysis for association with the risk of squamous cell carcinomas of the head and neck (SCCHN) in a subpopulation cluster-matched (Indo-Europeans linguistic subgroup + Caucasoid morphological subtype) case-control based genetic association study.
Material & Methods
Case and control sample collection: Blood samples (2 ml each) were collected from consecutive patients following written informed consent from 250 SCCHN patients, after histopathological and cytological confirmation, from different parts of north India undergoing treatment at Lucknow Cancer Institute (LCI), Lucknow, Uttar Pradesh, India between September 2005 and October 2008. Subpopulation cluster-matched (linguistic and morphological clusters) normal healthy volunteers (n=325) with no diagnosed case of any cancer from the northern States of India (Uttar Pradesh & Uttarakhand) were included as controls for cancer association studies. They were contacted personally and blood samples were drawn following informed consent. All the case and control subjects inducted in this study belonged specifically to the Caucasoid morphological subtype22 and Indo-European linguistic group23 of north India. A questionnaire was filled by each subject providing information on race, ethnicity, education, religion, marital status, first-degree family history, history of benign disease, etc. Information on tumour subtype, grading and stage of cancer was obtained from medical records of the patients. The study was approved by the Institutional Medical Ethics Committee of Central Drug Research Institute (CDRI), Lucknow.
DNA isolation: DNA was isolated from blood samples of SCCHN cases and normal healthy controls using QIAamp DNA Blood Midikit (Qiagen Inc., Chatsworth, CA, USA) following manufacturer's protocol, quantitated using spectrophotometer (Genequant pro, Amersham Biosciences, Pittsburg, PA, USA) and stored at -20°C.
Selection of gene and SNP for cancer association study: The gene OGG1 was chosen for the study mainly on the basis of its crucial role in the DNA repair process, especially the base excision repair (BER) pathway. Reference sequences of the gene and information on coding regions (CDS) were retrieved from NCBI's (National Center for Biotechnology Information) sequence databases (www.ncbi.nlm.nih.gov). The nonsynonymous SNP rs1052133 (OGG1) was selected for study on association with the risk of SCCHN based on available literatures on genetic association studies and the potential functional importance of the SNP in vital pathways, especially the DNA repair pathway, thereby serving as critical medium- or low- penetrance candidate for cancers.
Primer designing and synthesis: The oligos 5’ CTTCCACCTCCCAACACTGTCAC 3’ and 5’ GTGCCTGGCCTTTGAGGTAGTC 3’ were designed as forward and reverse primers, respectively using the PrimerSelect module of Lasergene v6.0 software (DNASTAR Inc., USA) for cancer genetic association study with the SNP rs1052133 (OGG1). The primer sequences were verified using NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), UCSC BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) (Fig.) and UCSC in-silico PCR (http://genome-mirror.duhs.duke.edu/cgi-bin/hgPcr) to eradicate the possibility of amplification of any non-specific DNA sequences and synthesized commercially.
Fig. 1.
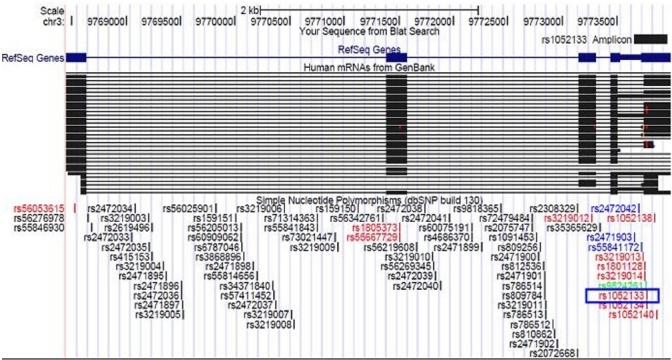
Pictorial representation of the human OGG1gene depicting exons, introns, alternatively spliced OGG1 mRNA sequences, SNPs in gene region and the PCR amplicon (rs1052133_Amplicon) used for the current study. [Figure generated using uCSC genome browser; the SNP investigated in the present study (rs1052133) is enclosed by a blue box].
PCR standardization and amplification: Gradient PCR reaction was performed for standardization of DNA amplification condition and optimization of annealing temperature for the primer set (forward + reverse). Briefly, the primer set was used to amplify a standard DNA template at different annealing temperatures (with increment of approximately 2°C each) and the temperature at which highest amount of PCR product was formed (as visualised from agarose gel) was considered the optimum annealing temperature for further PCR reactions. Following standardization of annealing temperatures using Gradient PCR, PCR reactions were performed using the optimized annealing temperatures.
All PCR reactions were performed in 200 μl transparent PCR tubes on a peltier-based thermal cycler (PTC100, MJ Research, USA) using reagents from Fermentas Life Sciences (Fermentas, Lithuania) in a total reaction volume of 50 μl containing nearly 100 ng genomic DNA, 1.5 U Taq polymerase in 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, and 15 pmol (0.3 μM) of each primer. Thermal cycling conditions were as follows: initial denaturation step at 95°C for 10 min, 31 cycles of PCR consisting of denaturation at 94°C for 1 min, annealing at 63.2°C for 1 min and extension at 72°C for 1 min, followed by a final extension step at 72°C for 5 min. The reaction was held at 4°C. The PCR products were visualized by electrophoresis on 1.2 per cent agarose gel. For gel electrophoresis, 5 μl of the amplified product was mixed with 1 μl of 6X gel loading buffer (analytical grade water containing 30% glycerol, 0.25% bromophenol blue, 0.25% xylene cynole) and resolved on 1.2 per cent agarose gel in TAE buffer, at 80 volts for 2 h. 100 bp DNA markers (New England Biolabs, MA, USA) were run with the amplified products as reference. The amplicons (PCR products) were stored at 4°C.
RFLP analysis for cancer association study: The restriction enzyme SatI was selected for PCR-RFLP study on the coding SNP rs1052133 (OGG1) using SeqBuilder module of Lasergene 6.0 (DNAStar) and WATCUT (http://watcut.uwaterloo.ca/watcut/watcut/template.php), an online tool for SNP-RFLP analysis, and procured from Fermentas Life Sciences (Fermentas, Lithuania). The 302 bp PCR product was subjected to restriction digestion using SatI following optimum reaction conditions as per manufacturer's protocol. The digestion products were visualized by electrophoresis on 3 per cent agarose gel for RFLP analysis. The homozygous wild type (CC) genotype generated a single band of 302 bp upon restriction digestion while the homozygous mutant genotype (GG) and the heterozygous genotype (CG) produced two and three bands, respectively upon visualisation on agarose gel following restriction digestion using the enzyme SatI.
DNA resequencing: Bi-directional DNA sequencing was performed using ABI3100 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA) for validation of the results obtained through RFLP analysis.
Comparison with HapMap database: SNP data obtained on control samples from the north Indian subpopulations were compared with the data available on the other world populations available through the HapMap Genome Browser (HapMap Public Release #27) of Hapmap database24.
Comparison with SNP500Cancer database of the Cancer Genome Anatomy Project: The data obtained on control samples from the north Indian subpopulations for the purpose of cancer association studies were further compared with the data available on SNP500Cancer database of the Cancer Genome Anatomy Project. The SNP500Cancer database includes genomes of 102 individuals of self-described heritage consisting of 24 individuals of the African/African American heritage, 31 of Caucasian heritage, 23 of Hispanic heritage and 24 of Pacific Rim heritage25,26.
Prediction of consequences on protein structure and/or function: Non-synonymous single nucleotide polymorphisms (nsSNPs) may have considerable effect on the structure or function of the OGG1 protein. Therefore, the potential phenotypic implications of the nsSNP rs1052133 was evaluated using the prediction program PolyPhen (Polymorphism Phenotyping, http://www.bork.embl-heidelberg.de/polyphen) that merges a conservation score with physicochemical differences and structural features of the polymorphic variants to predict the functional importance of the amino acid substitution27–29.
Statistical analysis for determination of genetic association: Statistical analysis of data was performed using the computer softwares Statistical Package for the Social Sciences (SPSS) version 16.0 and GraphPad softwares, Prism 5.0 and Instat 3.0. Maximum likelihood estimates of descriptive statistics like allele and genotype frequencies were calculated. Hardy-Weinberg Equilibrium (HWE) was tested among cases and controls separately, comparing the observed genotype count with that of the expected, using Goodness-of-fit Chi square test. Crude and adjusted odds ratios (OR) and corresponding 95 per cent confidence intervals (CI) were estimated to ascertain association of individual genotypes with SCCHN. Effect of modification was assessed on a multiplicative scale by calculating ORs for the genotypes stratified by indicators like subtype and stage of cancer, etc. running separate models for each exposure category. All statistical tests were two-sided.
Results
Genotype results were successfully obtained among 302 healthy normal unaffected controls and among 235 SCCHN-affected cases for the SNP rs1052133 (OGG1).
ChisquareHWE for genotype distribution was 14.025 among controls for the loci rs1052133 (OGG1). The allele frequencies and genotype frequencies of the SNP rs1052133 (OGG1) among SCCHN cases and healthy control subjects are provided in Table I. Minor allele frequencies (MAFs) were 36.9 per cent among the controls and 28.1 per cent among SCCHN cases.
Table I.
Details of allele and genotype frequencies of the SNP rs1052133 (OGG1) observed in normal and SCCHN samples
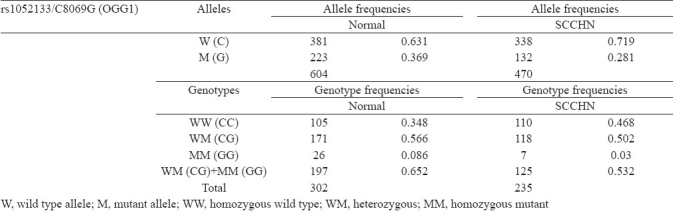
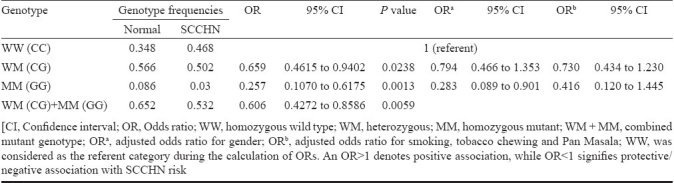
Subsequent analysis pertaining to the assessment of risks associated with individual mutant genotypes of the polymorphism rs1052133 (OGG1) covered in this investigation with regards to SCCHN risk depicted protective association with predisposition towards SCCHN for the heterozygous (CG) (OR 0.659, 95% CI 0.462 to 0.940), homozygous (GG) (OR 0.257, 95% CI 0.107 to 0.618) and combined mutant (CG + GG) (OR 0.606, 95% CI 0.427 to 0.859) genotypes. Results of the genetic association study between the polymorphism rs1052133 (OGG1) selected in the present study with SCCHN risk presented in terms of odds ratios of mutant genotypes is shown in Table II. Association of the selected SNP with SCCHN risk did not vary greatly with gender or tumour grading (data not shown).
Table II.
Representation of genetic association of the SNP rs1052133 in the gene OGG1 with the risk of SCCHN among north Indians determined in terms of odds ratios of mutant genotypes
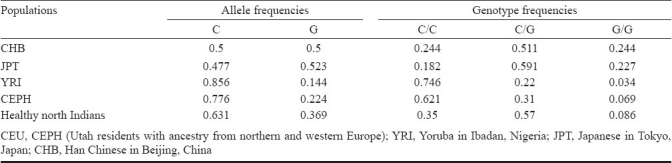
Comparison with HapMap database: Results of comparison between the genotype frequencies of rs1052133 in healthy north Indian volunteers and the world population data on the HapMap database are displayed in Table III. The frequencies of reference/wild type homozygote genotype (CC) were 0.244, 0.182, 0.746, 0.621, 0.35 among Han Chinese (CHB), Japanese (JPT), Yoruba (YRI) Central European (CEU) and healthy normal north Indian subjects, respectively.
Table III.
Comparison of allele frequencies and genotype frequencies of rs1052133 between healthy north Indian volunteers and the data obtained from HapMap project23
Comparison with SNP500Cancer database: The mutant allele frequency for rs1052133 (OGG1) was 0.225 among the controls samples included in the SNP500Cancer database, whereas the corresponding MAF value for the control samples included in the present study was 0.369.
Characterization of protein coding genetic variant: Bioinformatic analyses of the non-synonymous coding changes using the prediction program Polyphen demonstrated that the SNP rs1052133 was predicted to be possibly damaging for the transcript variant OGG1-203/ENST00000339542 (PSIC score difference: 1.800).
Discussion
Effects of population stratification in disease association studies will be negligible if cases and controls are drawn from the same cluster even if they do not belong to the same ethnic group which implies that studies involving sampling cases and controls from the same population cluster need no additional corrections or confounding effects of population stratification and would additionally increase the power of such association studies30. Therefore, the present study involving subpopulation-specific case-control based association of SCCHN risk of the non-synonymous SNP rs1052133 in OGG1 may prove to be a framework for future studies on the association of this SNP in a subpopulation specific manner.
Studies investigating cancer susceptibility of the polymorphism rs1052133 in the gene OGG1 have found a wide range of risk associated with cancers8,9,13–19,31. Among studies conducted globally on the association of SCCHN risk with rs1052133 (OGG1)6,7,13,32, an increased SCCHN risk was observed among individuals with the mutant genotypes32. Elahi et al7 observed a significantly increased risk of orolaryngeal cancer with the homozygous mutant genotype (OR 4.1, 95% CI 1.3-13.0), and a non significant increased risk with the heterozygous genotype (OR 1.6, 95% CI 1.0-2.6). On the other hand, no association was observed in the study by Gorgens et al33 for the heterozygous (CG) and homozygous mutant (GG) genotypes where the odds ratios were 1.040 and 1.082 respectively. In the present study, a statistically significant protective association was found between OGG1 mutant genotypes viz. heterozygous CG, homozygous mutant GG and combined mutant CG + GG genotypes and SCCHN risk among the north Indians. No other study has been conducted to investigate the association of SNPs in the gene OGG1 with the risk of SCCHN among Indian subpopulations. When compared to the world population data on the HapMap database, the genotype frequencies of rs1052133 in healthy north Indian volunteers was found to be in close agreement with those of Japanese (JPT) and Han Chinese (CHB) populations but very different from the Central European (CEU) and Yoruba (YRI) populations. Conversely, when compared to the SNP500Cancer data, no correlation could be drawn with the allele and genotype frequencies of SNPs among control samples included in the present study . This may be, largely because of the non random sampling methods of the SNP500Cancer database and more likely since the populations covered in the SNP500Cancer database of the Cancer Genome Anatomy Project did not consider the enormous diversity of the Indian population during sample collection25,26. These results reiterate the uniqueness of the Indian population with respect to the worldwide scenario and suggest that any epidemiological study intended to study case-control based genetic association in the north Indian population should be conducted with ethnically matched sub-population clusters from north India to minimize the effects of population stratification.
Our results strongly suggest that the polymorphism rs1052133 is strongly associated with predisposition to SCCHN with its variant genotypes having a protective effect on the occurrence of SCCHN among north Indian subpopulations. Although individual genotypes might have only a modest effect on cancer risk, but the cumulative effect of a large number of such predisposition polymorphisms might have a substantial cancer risk. Research with genetic models that will include multiple genotypes may thus be instrumental to significantly and reliably predict cancer risk.
This is perhaps the first study investigating the association of OGG1 polymorphisms with SCCHN risk among any Indian subpopulation. The data generated from this study may have wide-ranging applications for further epidemiological and public health related research on the Indian population. However, insufficient information and inadequate understanding on the functional aspects of SNP alleles have made it difficult to appropriately interpret these potentially meaningful differences that may be cancer-causing. Furthermore, a comprehensive study of all the polymorphisms in all known DNA repair genes and analysis of the joint effects of the risk variants should be more beneficial in providing extensive information about the variability among gene and pathway dysfunctions contributing towards higher cancer susceptibility. Therefore, comprehensive analysis of the effects of functionally important SNPs on SCCHN risk is crucial to elucidate the possible synergistic effect of gene-gene and gene-environment interactions in cancer susceptibility.
Acknowledgments
The first two authors (AKM and SVS) are recipients of Research Fellowship from Council of Scientific and Industrial Research, India. The work was supported by CSIR network project NWP0034.
Footnotes
Conflict of Interest: None declared.
References
- 1.Lindahl T, Karran P, Wood RD. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–69. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 2.Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H, et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–25. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- 3.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 4.Mohrenweiser HW, Wilson DM, 3rd, Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res. 2003;526:93–125. doi: 10.1016/s0027-5107(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 5.Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597–609. doi: 10.1089/15230860152542952. [DOI] [PubMed] [Google Scholar]
- 6.Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog. 2005;42:127–41. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 7.Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23:1229–34. doi: 10.1093/carcin/23.7.1229. [DOI] [PubMed] [Google Scholar]
- 8.Sugimura H, Kohno T, Wakai K, Nagura K, Genka K, Igarashi H, et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1999;8:669–74. [PubMed] [Google Scholar]
- 9.Xing DY, Tan W, Song N, Lin DX. Ser326Cys polymorphism in hOGG1 gene and risk of esophageal cancer in a Chinese population. Int J Cancer. 2001;95:140–3. doi: 10.1002/1097-0215(20010520)95:3<140::aid-ijc1024>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–7. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen K, Schlink K, Gotte W, Hippler B, Kaina B, Oesch F. DNA repair activity of 8-oxoguanine DNA glycosylase 1 (OGG1) in human lymphocytes is not dependent on genetic polymorphism Ser326/Cys326. Mutat Res. 2001;486:207–16. doi: 10.1016/s0921-8777(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 12.Arcand SL, Provencher D, Mes-Masson AM, Tonin PN. OGG1 Cys326 variant, allelic imbalance of chromosome band 3p25.3 and TP53 mutations in ovarian cancer. Int J Oncol. 2005;27:1315–20. [PubMed] [Google Scholar]
- 13.Hashimoto T, Uchida K, Okayama N, Imate Y, Suehiro Y, Hamanaka Y, et al. Interaction of OGG1 Ser326Cys polymorphism with cigarette smoking in head and neck squamous cell carcinoma. Mol Carcinog. 2006;45:344–8. doi: 10.1002/mc.20140. [DOI] [PubMed] [Google Scholar]
- 14.Monteiro E, Varzim G, Silva R, da Costa B, Lopes C. Polymorphisms of the human OGG1 gene in laryngeal cancer: implications in radiotherapy response and survival. Rev Laryngol Otol Rhinol (Bord) 2005;126:135–40. [PubMed] [Google Scholar]
- 15.Nohmi T, Kim SR, Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Takezaki T, Gao CM, Wu JZ, Li ZY, Wang JD, Ding JH, et al. hOGG1 Ser(326)Cys polymorphim and modification by environmental factors of stomach cancer risk in Chinese. Int J Cancer. 2002;99:624–7. doi: 10.1002/ijc.10400. [DOI] [PubMed] [Google Scholar]
- 17.Tsukino H, Hanaoka T, Otani T, Iwasaki M, Kobayashi M, Hara M, et al. hOGG1 Ser326Cys polymorphism, interaction with environmental exposures, and gastric cancer risk in Japanese populations. Cancer Sci. 2004;95:977–83. doi: 10.1111/j.1349-7006.2004.tb03186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel U, Olsen A, Wallin H, Overvad K, Tjonneland A, Nexo BA. No association between OGG1 Ser326Cys and risk of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:1680–1. [PubMed] [Google Scholar]
- 19.Xu J, Zheng SL, Turner A, Isaacs SD, Wiley KE, Hawkins GA, et al. Associations between hOGG1 sequence variants and prostate cancer susceptibility. Cancer Res. 2002;62:2253–7. [PubMed] [Google Scholar]
- 20.Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, et al. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder PP. Ethnic populations of India as seen from an evolutionary perspective. J Biosci. 2001;26(4 Suppl):533–45. doi: 10.1007/BF02704750. [DOI] [PubMed] [Google Scholar]
- 22.Malhotra KC. Morphological composition of the people of India. J Hum Evol. 1978;7:45–63. [Google Scholar]
- 23.Gadgil M, Joshi NV, Prasad UV, Manoharan S, Patil S. Hyderabad: Universities Press; 1998. In the Indian human heritage; pp. 100–29. [Google Scholar]
- 24.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.Packer BR, Yeager M, Burdett L, Welch R, Beerman M, Qi L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D21. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Packer BR, Yeager M, Staats B, Welch R, Crenshaw A, Kiley M, et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:D528–D32. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 29.Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 30.Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- 31.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and head and neck cancer risk (Review) Int J Oncol. 2008;32:945–73. [PubMed] [Google Scholar]
- 32.Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF, et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol Biomarkers Prev. 2003;12:1100–4. [PubMed] [Google Scholar]
- 33.Gorgens H, Muller A, Kruger S, Kuhlisch E, Konig IR, Ziegler A, et al. Analysis of the base excision repair genes MTH1, OGG1 and MUTYH in patients with squamous oral carcinomas. Oral Oncol. 2007;43:791–5. doi: 10.1016/j.oraloncology.2006.10.004. [DOI] [PubMed] [Google Scholar]


