Abstract
Background: The TAS1R1 and TAS1R3 G protein–coupled receptors are believed to function in combination as a heteromeric glutamate taste receptor in humans.
Objective: We hypothesized that variations in the umami perception of glutamate would correlate with variations in the sequence of these 2 genes, if they contribute directly to umami taste.
Design: In this study, we first characterized the general sensitivity to glutamate in a sample population of 242 subjects. We performed these experiments by sequencing the coding regions of the genomic TAS1R1 and TAS1R3 genes in a separate set of 87 individuals who were tested repeatedly with monopotassium glutamate (MPG) solutions. Last, we tested the role of the candidate umami taste receptor hTAS1R1-hTAS1R3 in a functional expression assay.
Results: A subset of subjects displays extremes of sensitivity, and a battery of different psychophysical tests validated this observation. Statistical analysis showed that the rare T allele of single nucleotide polymorphism (SNP) R757C in TAS1R3 led to a doubling of umami ratings of 25 mmol MPG/L. Other suggestive SNPs of TAS1R3 include the A allele of A5T and the A allele of R247H, which both resulted in an approximate doubling of umami ratings of 200 mmol MPG/L. We confirmed the potential role of the human TAS1R1-TAS1R3 heteromer receptor in umami taste by recording responses, specifically to l-glutamate and inosine 5′-monophosphate (IMP) mixtures in a heterologous expression assay in HEK (human embryonic kidney) T cells.
Conclusions: There is a reliable and valid variation in human umami taste of l-glutamate. Variations in perception of umami taste correlated with variations in the human TAS1R3 gene. The putative human taste receptor TAS1R1-TAS1R3 responds specifically to l-glutamate mixed with the ribonucleotide IMP. Thus, this receptor likely contributes to human umami taste perception.
INTRODUCTION
Sweet, sour, salty, bitter, and umami constitute the predominant taste qualities that humans perceive. Umami, a Japanese term, was coined by Kikunae Ikeda (1, 2) in 1908 for the taste of a broth created from seaweed (sea tangle), dried fish flakes (skipjack tuna), and mushrooms (shiitake). In English, umami stimuli are often labeled brothy, soupy, meaty, and savory. Compounds that have the umami taste include l-glutamate salts, such as monosodium glutamate (MSG) and monopotassium glutamate (MPG); 5′-ribonucleotides [inosine 5′-monophosphate (IMP) and guanosine 5′-monophosphate (GMP)]; certain other amino acids, such as l-aspartate; and certain peptides (3–5). Glutamate, the prototypical umami stimulus, is found in many protein-rich foods, such as meats, cheeses, wines, and certain fruits and vegetables (green peas, tomatoes, and mushrooms), and sometimes is added directly to selected cuisines as a flavor enhancer (6). A characteristic feature of umami flavor is synergism between l-glutamate and some 5′-ribonucleotides, such as IMP and GMP, which can also elicit a weak umami taste on their own (3, 7, 8).
The idea that umami is a fundamental taste quality, such as sweet and bitter, has long been debated. Psychophysical experiments in humans, neurophysiologic experiments in primates, conditioned taste aversion tests in rats, and genetic studies in mice suggested that umami has unique taste properties (9–17). At the same time, other investigators argued that the umami quality is not unique in rodents because conditioned taste aversions to MSG can generalize to sodium chloride or sucrose in rats (18–21). Importantly, glutamate taste in rodents and in humans may be coded differently, resulting in distinctly different taste qualia for different species.
Some studies suggested an N-methyl-d-aspartate (NMDA) or an NMDA-like ionotropic glutamate receptor may be responsible for detection of glutamate taste (22–24), but more recent work has implicated G protein–coupled receptors (GPCRs) in umami taste transduction (5, 20, 24–30). The first umami GPCR to be discovered was a truncated metabotropic glutamate receptor 4 that was missing most of the N-terminal extracellular domain (ECD), termed taste-mGluR4 (5, 25). The second candidate was a heteromer of receptors TAS1R1 and TAS1R3 that was shown to interact with l-glutamate, showing a response significantly potentiated by 5′-ribonucleotides (28, 30, 31). Recently, 2 variants of metabotropic glutamate receptor 1, mGluR1α and a taste-specific variant of mGluR1, have been suggested as proposed receptors for umami taste perception (32, 33).
The 3 genes of the TAS1R family, TAS1R1, TAS1R2, and TAS1R3, reside in a cluster on human chromosome 1. Proteins coded by these genes function as heteromer taste receptors: TAS1R2 plus TAS1R3 senses sugars and other sweeteners and TAS1R1 plus TAS1R3 senses amino acids in vitro. Although variations in human TAS1R genes have been reported in a multiracial population screen (34), there currently is no known relation of these variations to perceptual phenotypes of human umami taste. Our present study was designed to determine whether the TAS1R genetic variants would be related to umami taste perception. We first conducted a psychophysical investigation of umami sensitivity in 242 subjects who discriminated between sodium chloride and MSG. Ten of those subjects at extremes of sensitivity returned to complete several additional tests of glutamate sensitivity to validate the observation. We next fully sequenced the coding regions of genomic TAS1R1 and TAS1R3 genes for 87 white individuals who were phenotyped for their responses to MPG. We conducted an association analysis to reveal suggestive variants in these genes that are correlated with human umami taste ratings.
SUBJECTS AND METHODS
Subjects: genotyping/phenotyping study
Human genomic DNA was obtained from a population of 87 US subjects, mostly of Dutch ancestry. The youngest subjects in this population were fourth-generation Dutch-American immigrants and the oldest were second generation. All of the subjects were healthy individuals (44% men) recruited from Michigan with a mean age (±SD) of 35 ± 19 y (age range: 14–89 y).
Subjects: psychophysical study
Two hundred forty-two healthy subjects (49% men), from ages 15 to 63 y (mean ± SD age: 30 ± 12 y), were recruited from the Philadelphia area. The 10 subjects who completed the second battery of psychophysical tests had a mean (±SD) age of 27 ± 13 y.
All of the subjects provided informed consent before participating this study on a form approved by the Office of Regulatory Affairs at the University of Pennsylvania and were paid for their participation. All of the subject testing was initiated on or before 12 June 2006.
Stimuli
Concentration-intensity rating
Subjects were trained on the proper use of a general labeled magnitude scale (gLMS) and used it to rate the intensity of the sensation and the taste quality they experienced while tasting a stimulus (35, 36). This gLMS is a semilogarithmic computer-presented scale with the verbal descriptors on a vertical axis: “no sensation,” “barely detectable,” “weak,” “moderate,” “strong,” “very strong,” and “strongest imaginable.” Subjects rated along the axis selecting the point near or between terms that most closely approximated their sensation magnitude, and the computer scored their response as the linear distance along the axis from the origin. Subjects were asked to first determine which descriptor on the scale best describes the intensity of the sensation and then to rate the tastes in the larger context of all sensations rather than in the narrow context of a particular kind of sensation (namely, taste). They were offered a series of prototypical stimuli to show the following qualities: sodium chloride (salty), sucrose (sweet), citric acid (sour), quinine hydrochloric acid (bitter), urea (sour and bitter), and MSG in admixture with IMP (savory). The subjects were instructed that each solution exemplified a quality of taste as its predominant taste quality but not necessarily its only one. They were also instructed in the general meaning of the labels on the scale and the approximate placement of everyday oral stimuli along the scale.
Five concentrations of MPG (Sigma, St Louis, MO) and one water (Millipore, Billerica, MA) were used in the concentration–intensity rating test: 25, 50, 75, 100, and 200 mmol/L. Five concentrations of sucrose and one water were also offered to 10 of these subjects: 100, 150, 200, 300, and 400 mmol/L. An aliquot of 10 mL of each solution was offered in 40-mL polyethylene medicine cups (Del Val Medical Supply, Pharr, TX) on a numbered tray. Solutions were offered at 21°C. All of the samples were offered in ascending concentration and the series was offered in duplicate. Subjects were asked to sip, rate, and expectorate each solution. On each trial, subjects held 10 mL of solution in their mouth for 5 s and rated the intensity of the solution on a gLMS and were asked to indicate the taste quality of the solution (salty, bitter, sour, sweet, or savory) before expectorating.
Triangle test (3-alternative, forced-choice)
Subjects were offered 24 sets of 3 cups containing 10 mL of 29 mmol MSG/L (Sigma) or 29 mmol NaCl/L [Fluka (subsidiary of Sigma)] solution. In each trial, 2 cups contained one solution and the third contained the other solution. Orders were randomly generated with the random integer generator algorithm at www.random.org. Twelve trials contained 2 cups of sodium chloride and one cup of MSG. Subjects rinsed with filtered deionized water (Millipore) 4 times before the test and 4 times after each sample.
MPG and 5′-ribonucleotide synergy test
The same 10 subjects rated 3 solutions on the gLMS: 1) 10 mL of 20 mmol MPG/L, 2) 20 mmol MPG/L in admixture with 3 mmol IMP/L, and 3) 20 mmol MPG/L in admixture with 3 mmol GMP/L (Sigma). The tasting and rating protocol was the same as described above in Concentration-Intensity Rating.
Two-alternative, forced-choice, intensity test
Ten subjects were offered 10 sets of 2 solutions. Each set contained one medicine cup with 10 mL of 250 mmol sucrose/L (USB, Cleveland, OH) and one with 200 mmol MPG/L. Subjects were asked to select which of the 2 cups in the set was more intense. Orders were randomized.
Modified Harris-Kalmus recognition threshold
Ten subjects were asked to taste MPG samples in an ascending binary dilution series starting at the weakest concentration (bottle 14) and to determine whether they tasted anything. If the answer was “yes,” then they were asked to name the quality. If the answer was “savory,” then they went on to the sorting task; if the answer was another quality, then they went on to the next higher concentration in the series until they reported tasting savory. In the sorting task, subjects were offered 3 cups of the MPG solution at the concentration they reported as savory tasting and 3 cups of water. The solutions were offered in random order and the subjects had to sort them into 3 cups of stimulus and 3 cups of water correctly. If they failed to do so, they were asked to sort the next highest concentration in the series until they could sort correctly.
Genetic sequencing
Single nucleotide polymorphisms (SNPs) were discovered by sequencing genomic DNA for both strands. Each of the exons and adjacent introns of TAS1R1 and TAS1R3 was amplified with the primers designed by software at the Primer3 website (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Polymerase chain reaction (PCR) was performed in a total volume of 20 μL, containing 0.25 mmol/L of each deoxynucleotide (Invitrogen, Carlsbad, CA), 10 pmol of each forward and reverse primer, 1.5 mmol MgCl/L, 10 mmol Tris–hydrochloric acid/L (pH 8.5), 50 mmol KCl/L, 1 U of Hotstar Taq DNA Polymerase (Qiagen, Germantown, MD), and 20 ng of genomic DNA. PCR conditions (MasterCycler, Eppendorf, Hamburg, Germany) were as follows: 40 cycles of denaturation at 95°C for 30 s; annealing at 58°C or 60°C, depending on the primers for 30 s; and extension at 72°C for 1 min. The first step of initial activation and the last step of extension were at 95°C for 10 min and 72°C for 7 min, respectively. Five microliters of the PCR products were separated and visualized in a 1.5% agarose gel. Ten microliters of this PCR product were purified by PureLink PCR Purification Kit (Invitrogen). Sequencing reactions and data analysis were performed by Agencourt Bioscience Company (Beverly, MA).
Functional expression of hTAS1R1-hTAS1R3 in human embryonic kidney 293 (HEK-293) T cells
HEK-293 T cells were transiently transfected with plasmids expressing hTAS1R1 or hTAS1R3. Transfections were performed with Lipofectamine 2000 (Invitrogen). Twenty-two hours after transfection, growth media was removed and cells were fixed with 4% paraformaldehyde. Surface expression of TAS1R1 and TAS1R3 was confirmed by staining with an antibody against an N-terminal epitope tag (FLAG). For functional analysis, HEK-293 T cells were transiently transfected with plasmids expressing hTAS1R1, hTAS1R3, and a chimeric G protein composed of Gα16 containing the last 44 amino acids of Gα-i3. Twenty-two hours after transfection, growth media was removed and cells were washed once with Hank’s balanced salt solution (HBSS) with 20-mmol HEPES/L, then loaded with calcium 4 dye in HBSS with 20-mmol HEPES/L (Molecular Devices, Sunnyvale, CA). Cells were incubated at 37°C for 1 h, then moved to a FlexStation II (Molecular Devices) and set for 30°C. After a 15-min incubation, cell fluorescence was measured for 180 s with an excitation wavelength of 485 nm and an emission wavelength of 525 nm with a 515-nm cutoff. At 30 s, cell cultures were injected with a solution of either 5 mmol l-glutamate/L plus 1 mmol IMP/L or 5 mmol d-glutamate/L plus 1 mmol IMP/L. Data were represented as averaged maximal fluorescence increase (n = 4). Data were analyzed with Prism 5.0 software (GraphPad Software Inc, La Jolla, CA). All of the solutions were pH matched.
Statistical analysis
Psychophysical data were analyzed by analysis of variance (ANOVA) and t tests where indicated. Genotype and allele frequencies in different groups of subjects were compared with the CLUMP program (version 1.9) with 10,000 stimulations (37). The P values reported are 2 tailed and significance was accepted at P < 0.05. A P value of 0.05 was considered significant in tests for Hardy-Weinberg equilibrium. One-factor ANOVA was performed with EXCEL (Microsoft, Redmond, WA). Haplotype frequencies, odds ratio, and 95% CIs were calculated by using the website program http://analysis.bio-x.cn/myAnalysis.php (38).
RESULTS
Individual variation in MSG sensitivity
MSG compared with sodium chloride triangle test
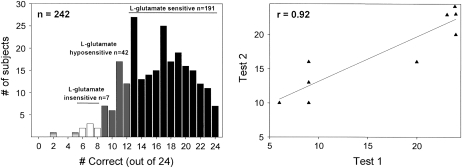
In a series of 24 triangle test trials using the method described by Lugaz et al (39), 242 subjects were tested for sensitivity to MSG by comparing 29 mmol MSG/L with 29 mmol NaCl/L. A histogram of their results is shown in Figure 1 (left plot). Random chance performance results in ≈8 of 24 responses correct. Responses of ≥13 correct out of 24 are significantly above chance in a 3-alternative forced-choice task (P < 0.05; 40). We regarded responses of 6 to 8 out of 24 correct as random guesses and labeled subjects who had those scores as insensitive to l-glutamate taste (Figure 1, white bars; n = 7). Subjects who had ≤5 responses correct were not categorized because these responses deviate from random performance (n = 2) and are marked as the shaded open bars. Subjects who had responses of 9 to 12 out of 24 correct were regarded as hyposensitive subjects to glutamate taste (Figure 1, gray bars; n = 42). Subjects who scored 13 to 24 out of 24 correct were labeled as sensitive to l-glutamate taste (Figure 1, black bars; n = 191).
FIGURE 1.
Variation in l-glutamate taste sensitivity. The left panel depicts a histogram of 242 subjects’ performances in a discrimination task: 29 mmol monosodium l-glutamate/L was tested against 29 mmol NaCl/L in 24 trials of a 3-alternative, forced-choice, triangle test. White bars depict chance performance and indicate monosodium l-glutamate–insensitive subjects. Dark gray bars indicate monosodium l-glutamate–hyposensitive subjects’ performances. Black bars indicate the performance of subjects who can significantly distinguish l-glutamate from sodium chloride. Two subjects depicted as light gray bars performed below chance. The right panel shows the test-retest correlation of 5 insensitive subjects and 5 sensitive subjects. The performances of 2 insensitive subjects were the same and thus are superimposed in the figure.
Reliability test
We asked 10 subjects, 5 insensitive (scoring between 6 and 8 correct) and 5 sensitive (scoring between 20 and 24 correct), to take the 24 trial triangle tests again at the completion of the experiment 3 to 6 mo later. The test-retest correlation was r = 0.92 (Figure 1, right panel).
Validity tests
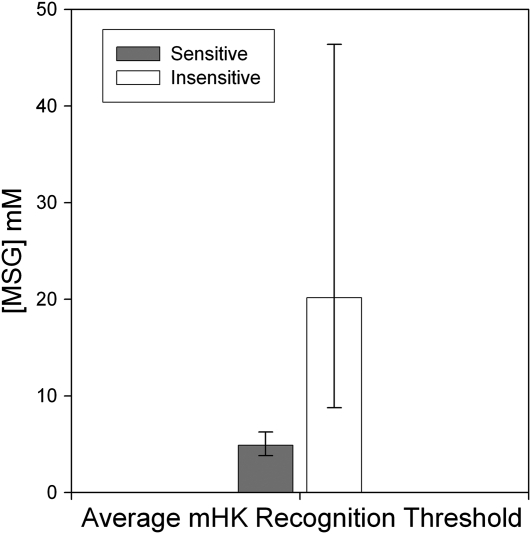
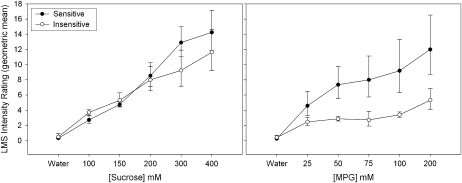
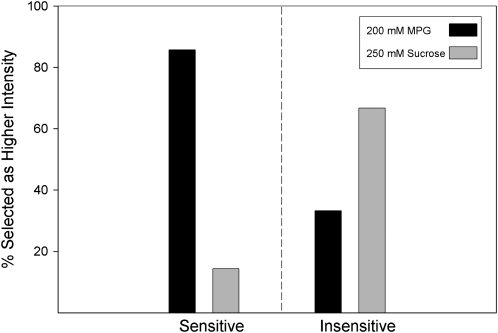
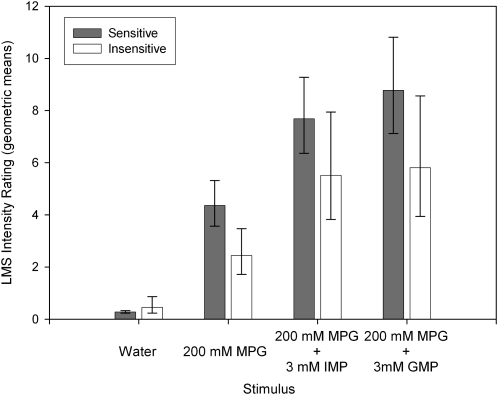
The same 10 subjects who were retested ( also participated in 1) an umami taste quality recognition threshold test, a modified Harris-Kalmus recognition threshold; 2) MPG and sucrose concentration intensity comparisons; 3) 2-alternative, forced-choice, intensity tests between sucrose and MPG; and 4) umami synergy tests with l-glutamate mixed with 5′-ribonucleotides IMP and GMP. The modified Harris-Kalmus recognition threshold test revealed that the sensitive subjects detected umami taste from MSG at a 4-fold lower concentration than did the insensitive subjects (Figure 2). Concentration-intensity functions for sucrose (Figure 3, left panel) and MPG (Figure 3, right panel) show the specificity of the differences in taste for these 2 groups. The 2 umami sensitivity groups did not differ in perception of sucrose but did for MPG across the concentration function range at 50, 75, 100, and 200 mmol/L (ANOVA, P < 0.05). The concentrations used in the forced-choice intensity function (200 mmol MPG/L compared with 250 mmol sucrose/L) were selected with the goal of showing a reversal of perceived intensity for the 2 groups of subjects. For subjects who are sensitive to umami taste, MPG was selected as more intense than sucrose in 43 of 50 trials (86%), and for the insensitive subjects, MPG was selected as more intense in only 16 of 50 trials (32%), thus showing the reversal of which stimulus was perceived as more intense (P < 0.05; Figure 4). The synergy test confirmed that insensitive subjects remained less sensitive to umami taste than the sensitive subjects even when the 5′-ribonucleotides IMP and GMP were added (Figure 5). This test also showed that the umami-insensitive subjects were not umami blind; they showed an enhanced umami taste when IMP and GMP were added, albeit weaker in intensity than for the sensitive subjects.
FIGURE 2.
Mean (±SEM) modified Harris-Kalmus (mHK) recognition thresholds for umami taste. The same 10 subjects as were tested in Figure 1 (right panel) had their umami taste quality recognition thresholds measured by testing with monosodium l-glutamate (MSG). The dark bar represents the average response of umami-sensitive subjects, as categorized in Figure 1, and the light bar represents insensitive subjects. The y axis represents the concentration of MSG in mmol/L that was correctly identified as umami tasting; P < 0.05 (t test).
FIGURE 3.
Mean (±SEM) concentration-intensity functions for sucrose and l-glutamic acid potassium salt (MPG). The left panel depicts the taste intensity rating functions on a general labeled magnitude scale (LMS) for sucrose and for MPG (right panel) for the same 10 subjects as were tested in Figures 1 and 2. Five concentrations of each stimulus and water were rated. The filled symbols represent the umami-sensitive subjects, and the open symbols represent the insensitive subjects as categorized in Figure 1. MPG was significantly different between the groups by using ANOVA at 50, 75, 100, and 200 mmol/L (P < 0.05).
FIGURE 4.
A 2-alternative, forced-choice, intensity test of l-glutamic acid potassium salt (MPG) compared with sucrose. The same 10 subjects as were tested in Figures 1–3 each received 10 trials of 2 solutions in which they had to determine whether the 200 mmol MPG/L or 250 mmol sucrose/L tasted more intense. The solid bars represent the overall percentage of trials that MPG was selected as more intense, and the gray bars represent the overall percentage of trials that sucrose was selected as more intense. The left panel depicts data for 5 umami-sensitive subjects and the right panel for the 5 umami-insensitive subjects as categorized in Figure 1. P < 0.05 for the reversal (chi-square test).
FIGURE 5.
Mean (±SEM) umami synergy tests with 200 mmol l-glutamic acid potassium salt (MPG)/L mixed with 3 mmol 5′-inosine monophosphate (IMP) or guanosine 5′-monophosphate (GMP)/L. The same 10 subjects as were tested in Figures 1–4 were tested. The dark bars represent the taste intensity ratings of solutions on a general labeled magnitude scale (LMS) for umami-sensitive subjects, and the white bars represent the insensitive subjects, as categorized in Figure 1. The x axis shows the stimuli: water, MPG, MPG mixed with IMP, and MPG mixed with GMP. Synergy was significant for both groups but was greater in intensity for the sensitive group; P < 0.05 (ANOVA).
Genetic analysis
Phenotyping
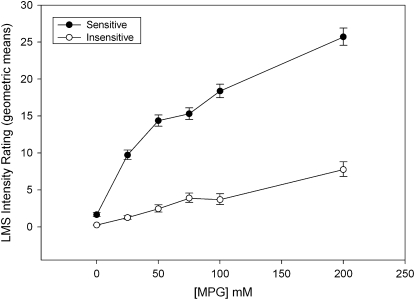
A total of 87 subjects from the extremes of sensitivity who generated MPG concentration-intensity curves were categorized and divided into sensitive or insensitive groups on the basis of the ratings categorization criteria for the 242-subject population (Figure 3, right panel). The results seen in Figure 6 are similar to those obtained with our other population. We separated subjects approximately evenly into sensitive and insensitive categories to enable further statistical genetic analysis correlating genetic variations with perceptual variations. The sensitive group rated MPG as twice a strong as the insensitive group (Figure 6).
FIGURE 6.
Mean (±SEM) concentration-intensity curves for l-glutamic acid potassium salt (MPG) from a total of 87 subjects with extreme phenotypes whose TAS1R genes were sequenced. The figure depicts the overall average MPG concentration-taste intensity ratings on a general labeled magnitude scale (LMS). The x axis represents MPG concentration, and the y axis the average taste intensity ratings. The 87 subjects were divided into 2 approximately even groups of either umami-sensitive or -insensitive subjects. Differences in umami taste were evident between the 2 groups at 25, 50, 75, 100, and 200 mmol/L; P < 0.05 (ANOVA).
Sequencing and genotyping
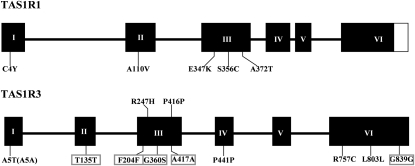
We identified variations in these subjects’ TAS1R1 and TAS1R3 genes by sequencing genomic DNA from all of the individuals. SNPs observed in these genes are shown in Table 1. Comparisons of aligned exonic sequences revealed 5 SNPs in TAS1R1 (all nonsynonymous) and 12 SNPs in TAS1R3 (4 nonsynonymous and 8 synonymous). Most of these SNPs are in the third and sixth exons of TAS1R genes (Figure 7). In total, 9 of 17 polymorphisms led to changes in the amino acids encoded at that position. Five SNPs in TAS1R3 gene have never been reported before (Figure 7, boxes surrounding SNP identities).
TABLE 1.
Details of single nucleotide polymorphisms (SNPs) in TAS1R genes1
| Gene | dbSNP2 | Exon | cSNP3 | Allele | Position of cSNP | Amino acid encoded | Amino acid position | Allele frequency4 |
| TAS1R1 | rs35375392 | 1 | G11→A | G | 11 | Cys | 4 | Singleton |
| Chromosome location: 1p36.23 | A | Tyr | ||||||
| Size: 2526 bp5 | rs41278020 | 2 | C329→T | C | 329 | Ala | 110 | 0.90 |
| Amino acid: 8416 | T | Val | 0.10 | |||||
| rs10864628 | 3 | G1039→A | G | 1039 | Glu | 347 | 0.99 | |
| A | Lys | 0.01 | ||||||
| rs41307749 | 3 | C1067→G | C | 1067 | Ser | 356 | Singleton | |
| G | Cys | |||||||
| rs34160967 | 3 | G1114→A | G | 1114 | Ala | 372 | 0.71 | |
| A | Thr | 0.29 | ||||||
| TAS1R3 | — | 1 | G13→A | G | 13 | Ala | 5 | 0.87 |
| Chromosome location: 1p36.33 | A | Thr | 0.13 | |||||
| Size: 2559 bp5 | — | 1 | T15→A | T | 15 | Ala | 5 | 0.87 |
| Amino acid: 8526 | A | Ala | 0.13 | |||||
| — | 2 | C459→T | C | 459 | Thr | 153 | 0.97 | |
| T | Thr | 0.03 | ||||||
| — | 3 | C612→T | C | 612 | Phe | 204 | Singleton | |
| T | Phe | |||||||
| — | 3 | G740→A | G | 740 | Arg | 247 | 0.93 | |
| A | His | 0.07 | ||||||
| — | 3 | G1078→A | G | 1078 | Gly | 360 | Singleton | |
| A | Ser | |||||||
| rs3813210 | 3 | C1248→T | C | 1248 | Pro | 416 | 0.92 | |
| T | Pro | 0.08 | ||||||
| — | 3 | G1251→A | G | 1251 | Ala | 417 | Singleton | |
| A | Ala | |||||||
| — | 4 | G1323→A | G | 1323 | Pro | 441 | 0.99 | |
| A | Pro | 0.01 | ||||||
| rs307377 | 6 | C2269→T | C | 2269 | Arg | 757 | 0.98 | |
| T | Cys | 0.02 | ||||||
| — | 6 | C2407→T | C | 2407 | Leu | 803 | Singleton | |
| T | Leu | |||||||
| T | Leu | |||||||
| — | 6 | C2517→T | C | 2517 | Gly | 839 | Singleton | |
| T | Gly |
dbSNP, Single Nucleotide Polymorphism database (http://www.ncbi.nlm.nih.gov/); cSNP, coding region single nucleotide polymorphism; bp, base pair.
Published identifiers of each known SNP.
SNP and its position.
Frequency of the polymorphism within all 174 chromosomes sequenced.
Total number of base pairs in each gene.
Number of coded amino acids in the gene.
FIGURE 7.
The open reading frame structures of the human TAS1R1 and TAS1R3 genes and their observed single nucleotide polymorphisms. The black boxes represent exons and the connecting spanners introns. The letter-number codes depict the locations and amino acid positions of identified polymorphisms. The first letter depicts the common amino acid and the second letter the substituted amino acid. The same letter indicates that the nucleotide change did not affect the amino acid code (synonymous change). The boxes indicate previously unreported polymorphisms.
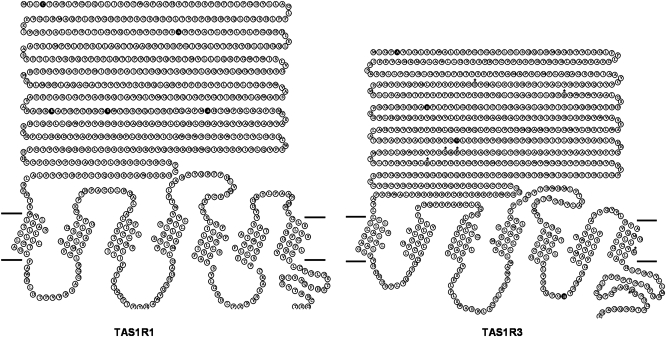
After placing the polymorphisms into the ribbon diagrams of the TAS1R1 and TAS1R3 proteins (Figure 8), we showed that the majority (14 of 17) of the variant amino acid positions resided in the large N-terminal ECD of these receptors, which comprises the putative “venus fly trap” ligand binding domain of the receptors. The other 3 variants of TAS1R3 are located in the third intracellular loop, the seventh transmembrane domain (TMD), and C-terminal intracellular domains (Figure 8).
FIGURE 8.
Amino acid “ribbon” plots of the protein sequences for hTAS1R1 and hTAS1R3 with polymorphisms. TAS1R1 is represented on the left and TAS1R3 on the right. Note that TAS1R1 is a slightly larger protein than TAS1R3. The left side of each protein is the amino terminal, and the right side is the carboxy terminal. The dark horizontal lines depict the cellular membrane. Above these lines is extracellular space, and below these lines is intracellular space. Circles represent amino acids, and the single letter codes within each circle represent the amino acid identity. Black amino acids indicate the protein locations of nonsynonymous single nucleotide polymorphisms (SNPs), and amino acids with an asterisk above indicate the position of synonymous SNPs.
SNP analysis
Some of the SNPs we detected are singletons (a variant allele was observed only once in all of the chromosomes) and so were not analyzed. We analyzed 2 SNPs of TAS1R1 (A110V and A372T) and 6 SNPs of TAS1R3 (A5T, A5A, T153T, R247H, P416P, and R757C) because of their relatively high allele frequency (>0.01). ANOVA revealed 3 suggestive polymorphisms, all in TAS1R3. Two were in the long amino terminal of this projected protein (Figure 8, black circles in plot). A5T (nucleotide G13→A) resulted in a doubling of umami taste intensity ratings for the rare allele at 100 and 200 mmol MPG/L (P = 0.052 and P = 0.054, respectively). R247H (nucleotide G740→A) resulted in a doubling of umami taste intensity ratings for the rare allele at 200 mmol MPG/L (P = 0.054). The third suggestive SNP resided in the third intracellular loop (Figure 8, black circle). Unlike the other 2 SNPs, which had effects only at high concentrations, this SNP R757C (nucleotide C2269→T) seemed only to affect the umami intensity ratings of 25 mmol MPG/L (P = 0.03). Heterozygous subjects who have the rarer CT genotype rated 25 mmol MPG/L twice as umami as did those who have the common CC genotype.
Group analyses
We performed chi-square tests as a qualitative analysis on 2 subject groups (sensitive and insensitive) separated by their ratings of MPG, but no differences in genotype distribution of any single SNP between groups were shown (data not shown). In addition, we observed 3 haplotypes in TAS1R1 and 6 haplotypes in TAS1R3. Haplotype analysis results did not show any difference between these 2 groups in frequencies of haplotypes constructed by the SNPs.
Functional receptor expression
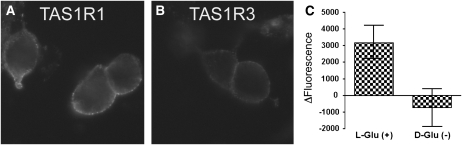
HEK-293 T cells were transiently transfected with plasmids expressing TAS1R1 or TAS1R3. Proteins were successfully expressed on the cell surface, as depicted in Figure 9A (TAS1R1) and Figure 9B (TAS1R3), and confirmed by staining with an antibody against an N-terminal epitope tag (FLAG). HEK-293 T cells that were transiently transfected with plasmids expressing TAS1R1, TAS1R3, and a chimeric G protein comprised of Ga16 containing the last 44 amino acids of Ga-i3 together showed enhanced calcium fluorescence responses to 5 mmol l-glutamate/L mixed with 1 mmol IMP/L but not to 5 mmol d-glutamate/L mixed with 1 mmol IMP/L ( Figure 9C). Experiments were repeated in quadruplicate (n = 4), and bar heights represent mean fluorescence change. Data were analyzed with Prism 5.0 software (GraphPad Software Inc). Thus, this candidate human umami taste receptor responded specifically to l-glutamate when mixed with IMP but not to d-glutamate mixed with IMP.
FIGURE 9.
Expression and calcium mobilization by TAS1R1/TAS1R3. A, B: Human embryonic kidney 293 (HEK293) T cells were transiently transfected with plasmids that express TAS1R1 or TAS1R3. Surface expression of TAS1R1 (A) and TAS1R3 (B) was confirmed by staining with an antibody against an N-terminal epitope tag (FLAG). C: HEK-293 T cells were transiently transfected with plasmids that express TAS1R1, TAS1R3, and a chimeric G protein composed of Ga16 containing the last 44 amino acids of Ga-i3. Cell cultures were injected with a solution of either 5 mmol l-glutamate (L-Glu)/L and 1 mmol inosine 5′-monophosphate (IMP)/L or 5 mmol d-glutamate (D-Glu)/L and 1 mmol IMP/L. Data are represented as an average maximal fluorescence increase (n = 4; bars represent mean ± SEM). Only solutions containing l-glutamate induced calcium mobilization.
DISCUSSION
Although umami taste is associated most closely with the taste of MSG, it has been argued that MSG should not be used as a chemical synonym for umami taste (41). Indeed, some subjects do not seem to taste MSG as umami. A specific ageusia (loss of taste) for MSG has been reported in human subjects (39). In that study, 8 of 109 subjects had no ability to distinguish 29 mmol MSG/L from 29 mmol NaCl/L and showed markedly decreasing MSG taste sensitivity, which suggested a severe dysfunction of sensing umami. These hypotasters probably perceived mostly the sodium cation in the l-glutamate salt. To avoid the possibly confusing Na+ salty taste that may be introduced by MSG, we used MPG as the umami stimulus in several of our experiments, as other researchers have done (42–44).
We have confirmed that a small percentage of subjects [7% (or possibly 9%) of 242 subjects] were unable to distinguish 29 mmol MSG/L from 29 mmol NaCl/L in a series of 24 triangle tests. We showed that subject performance was reliable and valid. Umami-insensitive subjects had higher umami recognition thresholds and rated the intensity of a MPG concentration series as weaker in taste than did their sensitive counterparts, whereas the 2 groups did not differ in how they rated the sweetness of a sucrose concentration series and reported that 250 mmol sucrose/L was stronger tasting than 200 mmol MPG/L; umami-insensitive subjects reported the opposite and showed depressed umami ratings to mixtures of MPG mixed with IMP and GMP relative to their umami-sensitive counterparts. This last umami synergy test also revealed that umami-insensitive subjects are not umami ageusic or umami blind. They are much less sensitive to umami taste specifically but nevertheless are able to perceive weak umami taste from MPG mixed with ribonucleotides.
Genetic analysis of mouse strains showed differences in preference for sweeteners and glutamate (16, 45–47), which suggests that nonsynonymous polymorphisms in TAS1R genes are associated with preference and ligand sensitivity. In agreement with a previous report in humans (34), our investigation confirmed significant nucleotide and protein sequence diversity in the candidate human taste receptors, TAS1R1 and TAS1R3. The identification of DNA variant sites within the whole coding region of TAS1R1 and TAS1R3 genes detected 17 SNPs, 9 of which cause amino acid substitutions. The third exon, which is the second largest of the 6 exons, carries more polymorphisms than any other exon in both TAS1R1 (3 of 5) and TAS1R3 (5 of 12).
We observed the majority of amino acid sequence variations in the large ECD of TAS1Rs, which is in contrast with the TAS2R taste receptor family that does not possess large amino terminal domains and carries most of their polymorphisms in the TMDs. These data parallel Kim et al’s findings (48). Interestingly, we detected more polymorphisms in TAS1R3 than in TAS1R1 (34). This difference from previous reports likely stems from the composition of the populations used in the studies. Within the multiracial sample, Kim et al showed many more variations in a Cameroonian population than in the other 7 populations. In white populations, however, only 2 SNPs in TAS1R1 (A110V and A372T) and 4 in TAS1R3 (A5T; P441P; R757C and L803L) were detected, which is consistent with our observations.
Like other members of class C GPCRs, each TAS1R possesses a large N-terminal ECD, 7 transmembrane spanning segments (TM1–TM7) separated by alternating intracellular and extracellular loops (i1–i3 and e1–e3, respectively), and an intracellular C-terminal domain. Previous studies suggested that the ECD of TAS1R1 contains binding sites of umami stimuli (49) and the ECD of TAS1R3 also is required for the receptor-ligand binding activity (50). Although the binding role of TAS1R ECDs for human taste perception has not been determined, our findings support the hypothesis that the majority of polymorphisms occur in the ECDs of TAS1Rs, thereby affecting sensitivity to umami stimuli. The TMDs of class C GPCRs are involved in G protein coupling and contain binding sites for allosteric modulators (51, 52). Protein interaction experiments show that the TMD of human TAS1R1 displays robust ligand-independent constitutive activity and efficiently activates G proteins, whereas TAS1R3 couples poorly to G proteins (53). One surprising observation is that by interacting with the TMDs of TAS1R3, the sweet taste inhibitor lactisole also can suppress umami taste of l-glutamate (49, 54).
We detected a genetic variation in the third intracellular loop of TAS1R3 (R757C), which may be involved in the activation of the G protein or binding to an allosteric modulator. Interestingly, we showed that the rare allele of this variant resulted in greater umami ratings of low concentrations (25 mmol/L) of MPG. Subjects who possessed a T allele at this locus rated MPG as twice as strong as those who possessed only the C allele, which codes for l-arginine. In view of the predicted role of TMD of TAS1R3 protein as the binding site to allosteric modulators, R757C in this region may be an influential factor in the receptor pathway of umami taste. It is worth noting that the C allele is the common allele of R757C in our sample population, which may not reflect frequencies in other populations (34). The discrepancy in allele frequencies may be explained by small sample size and lack of racial diversity in the present report.
We have also identified 2 other suggestive SNPs in the amino terminal domain of TAS1R3. Unlike the R757C variant, which affected only the ratings of low MPG concentrations, the A5T and the R247H variants resulted in greater taste ratings of high concentrations of MPG in those possessing the less common alleles. The position of these SNPs suggests that they may influence binding with l-glutamate resulting in stronger activation of the taste system. Most of the variations identified are in the large ECDs of TAS1R proteins, suggesting that the amino acid diversity in this TAS1R region reflects the receptor’s recognition and binding functions for umami taste. We also detected more polymorphisms in TAS1R3 than in TAS1R1. This difference in the number of variations may be due to the common role of TAS1R3 in both sweet and umami taste in humans.
Because there have been few examples of the human TAS1R1-TAS1R3 heteromer responding to l-glutamate, we also sought to independently confirm this observation to further strengthen the argument that this receptor contributes to human umami taste perception. We expressed the native human TAS1R1-TAS1R3 proteins together with a Gα protein in HEK-293 T cells and showed that this receptor enabled the cells to respond specifically to l-glutamate in mixture with IMP but not to d-glutamate in mixture with IMP. This observation together with our findings that sequence variants in TAS1R3 correlate with umami taste perception from MPG support the hypothesis that a TAS1R1-TAS1R3 heteromer contributes directly to the detection and perception of umami taste from l-glutamate. (Other articles in this supplement to the Journal include references 55–83.)
Acknowledgments
We thank Noriatsu Shigemura for his invaluable advice regarding functional expression of human TAS1R1 and TAS1R3 heteromers in HEK cells.
The authors’ responsibilities were as follows—PASB: oversaw all aspects of this study and writing of the manuscript; SM: assisted with the design, data collection, analysis of psychophysical data, and writing of the manuscript; AT, OMA, and NLE: assisted with the psychophysical data collection and analysis; QYC: designed genetic studies, collected sequence data, analyzed the TAS1R genetic data, and was principal author of the manuscript; and TAG and JR: were consultants in the design of the study and performed all functional expression experiments and analysis of the functional data. The presenting author’s (PASB) expenses associated with participation in the symposium were paid by the conference sponsor, the International Glutamate Technical Committee, a nongovernmental organization funded by industrial producers and users of glutamate in food. None of the authors had financial conflicts of interest with this research.
REFERENCES
- 1.Ikeda K. Japanese patent 4805. 1908.
- 2.Ikeda K. On a new seasoning. J Tokyo Chem Soc 1909;30:820–36 [Google Scholar]
- 3.Yamaguchi S. Basic properties of umami and effects on humans. Physiol Behav 1991;49:833–41 [DOI] [PubMed] [Google Scholar]
- 4.Halpern BP. Glutamate and the flavor of foods. J Nutr 2000;130:910S–4S [DOI] [PubMed] [Google Scholar]
- 5.Chaudhari N, Landin AM, Roper SD. A metabotropic glutamate receptor variant functions as a taste receptor. Nat Neurosci 2000;3:113–9 [DOI] [PubMed] [Google Scholar]
- 6.Kuninaka A. Taste and flavor enhancers. New York, NY: Marcel Dekker, 1981 [Google Scholar]
- 7.Yamaguchi S. The synergistic taste effect of monosodium glutamate and disodium 5′-inosinate. J Food Sci 1967;32:473–8 [Google Scholar]
- 8.Zhang F, Klebansky B, Fine RM, et al. Molecular mechanism for the umami taste synergism. Proc Natl Acad Sci USA 2008;105:20930–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohara I, Tanaka Y, Otsuka SI. Discrimination of monosodium glutamate and sodium chloride solutions by rats. Physiol Behav 1979;22:877–82 [DOI] [PubMed] [Google Scholar]
- 10.Kawamura Y, Kare MR, Umami: a basic taste. New York, NY: Marcel Dekker, 1987 [Google Scholar]
- 11.Ninomiya Y, Funakoshi M. Behavioral discrimination between glutamate and the four basic taste substances in mice. Comp Biochem Physiol A Comp Physiol 1989;92:365–70 [DOI] [PubMed] [Google Scholar]
- 12.Baylis LL, Rolls ET. Responses of neurons in the primate taste cortex to glutamate. Physiol Behav 1991;49:973–9 [DOI] [PubMed] [Google Scholar]
- 13.Kawamura Y, Kurihara K, Nicolaidis S, Oomura Y, Wayner MJ. Umami: proceedings of the Second International Symposium on Umami. Physiol Behav 1991;49:833–1030 [PubMed] [Google Scholar]
- 14.Rolls ET, Critchley HD, Wakeman EA, Mason R. Responses of neurons in the primate taste cortex to the glutamate ion and to inosine 5′-monophosphate. Physiol Behav 1996;59:991–1000 [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi S. Umami: aspecial issue. Food Rev Int 1998;14:123–337 [Google Scholar]
- 16.Bachmanov AA, Tordoff MG, Beauchamp GK. Intake of umami-tasting solutions by mice: a genetic analysis. J Nutr 2000;130:935S–41S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolls ET. The representation of umami taste in the taste cortex. J Nutr 2000;130:960S–5S [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto T, Yuyama N, Kato T, Kawamura Y. Gustatory responses of cortical neurons in rats. III. Neural and behavioral measures compared. J Neurophysiol 1985;53:1370–86 [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto T, Matsuo R, Fujimoto Y, Fukunaga I, Miyasaka A, Imoto T. Electrophysiological and behavioral studies on the taste of umami substances in the rat. Physiol Behav 1991;49:919–25 [DOI] [PubMed] [Google Scholar]
- 20.Stapleton JR, Roper SD, Delay ER. The taste of monosodium glutamate (MSG), L-aspartic acid, and N-methyl-D-aspartate (NMDA) in rats: are NMDA receptors involved in MSG taste? Chem Senses 1999;24:449–57 [DOI] [PubMed] [Google Scholar]
- 21.Heyer BR, Taylor-Burds CC, Tran LH, Delay ER. Monosodium glutamate and sweet taste: generalization of conditioned taste aversion between glutamate and sweet stimuli in rats. Chem Senses 2003;28:631–41 [DOI] [PubMed] [Google Scholar]
- 22.Faurion A. Are umami taste receptor sites structurally related to glutamate CNS receptor sites? Physiol Behav 1991;49:905–12 [DOI] [PubMed] [Google Scholar]
- 23.Brand JG, Teeter JH, Kumazawa T, Huque T, Bayley DL. Transduction mechanisms for the taste of amino acids. Physiol Behav 1991;49:899–904 [DOI] [PubMed] [Google Scholar]
- 24.Lin W, Kinnamon SC. Physiological evidence for ionotropic and metabotropic glutamate receptors in rat taste cells. J Neurophysiol 1999;82:2061–9 [DOI] [PubMed] [Google Scholar]
- 25.Chaudhari N, Yang H, Lamp C, et al. The taste of monosodium glutamate: membrane receptors in taste buds. J Neurosci 1996;16:3817–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bigiani A, Delay RJ, Chaudhari N, Kinnamon SC, Roper SD. Responses to glutamate in rat taste cells. J Neurophysiol 1997;77:3048–59 [DOI] [PubMed] [Google Scholar]
- 27.Nakashima K, Katsukawa H, Sasamoto K, Ninomiya Y. Behavioral taste similarities and differences among monosodium L-glutamate and glutamate receptor agonists in C57BL mice. J Nutr Sci Vitaminol (Tokyo) 2001;47:161–6 [DOI] [PubMed] [Google Scholar]
- 28.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA 2002;99:4692–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damak S, Rong M, Yasumatsu K, et al. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science 2003;301:850–3 [DOI] [PubMed] [Google Scholar]
- 30.Zhao GQ, Zhang Y, Hoon MA, et al. The receptors for mammalian sweet and umami taste. Cell 2003;115:255–66 [DOI] [PubMed] [Google Scholar]
- 31.Nelson G, Chandrashekar J, Hoon MA, et al. An amino-acid taste receptor. Nature 2002;416:199–202 [DOI] [PubMed] [Google Scholar]
- 32.Toyono T, Seta Y, Kataoka S, Kawano S, Shigemoto R, Toyoshima K. Expression of metabotropic glutamate receptor group I in rat gustatory papillae. Cell Tissue Res 2003;313:29–35 [DOI] [PubMed] [Google Scholar]
- 33.San Gabriel A, Uneyama H, Yoshie S, Torii K. Cloning and characterization of a novel mGluR1 variant from vallate papillae that functions as a receptor for L-glutamate stimuli. Chem Senses 2005;30(suppl 1):i25–6 [DOI] [PubMed] [Google Scholar]
- 34.Kim UK, Wooding S, Riaz N, Jorde LB, Drayna D. Variation in the human TAS1R taste receptor genes. Chem Senses 2006;31:599–611 [DOI] [PubMed] [Google Scholar]
- 35.Bartoshuk LM, Duffy VB, Green BG, et al. Valid across-group comparisons with labeled scales: the gLMS versus magnitude matching. Physiol Behav 2004;82:109–14 [DOI] [PubMed] [Google Scholar]
- 36.Green BG, Dalton P, Cowart B, Shaffer G, Rankin K, Higgins J. Evaluating the 'Labeled Magnitude Scale' for measuring sensations of taste and smell. Chem Senses 1996;21:323–34 [DOI] [PubMed] [Google Scholar]
- 37.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 1995;59:97–105 [DOI] [PubMed] [Google Scholar]
- 38.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005;15:97–8 [DOI] [PubMed] [Google Scholar]
- 39.Lugaz O, Pillias AM, Faurion A. A new specific ageusia: some humans cannot taste L-glutamate. Chem Senses 2002;27:105–15 [DOI] [PubMed] [Google Scholar]
- 40.Bi J. Sensory discrimination tests and measurements: statistical principles, procedures, and tables. Hoboken, NJ: Blackwell Publishing, 2006 [Google Scholar]
- 41.Halpern BP. What's in a name? Are MSG and umami the same? Chem Senses 2002;27:845–6 [DOI] [PubMed] [Google Scholar]
- 42.Sako N, Yamamoto T. Analyses of taste nerve responses with special reference to possible receptor mechanisms of umami taste in the rat. Neurosci Lett 1999;261:109–12 [DOI] [PubMed] [Google Scholar]
- 43.He W, Yasumatsu K, Varadarajan V, et al. Umami taste responses are mediated by alpha-transducin and alpha-gustducin. J Neurosci 2004;24:7674–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruyama Y, Pereira E, Margolskee RF, Chaudhari N, Roper SD. Umami responses in mouse taste cells indicate more than one receptor. J Neurosci 2006;26:2227–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Max M, Shanker YG, Huang L, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet 2001;28:58–63 [DOI] [PubMed] [Google Scholar]
- 46.Reed DR, Li S, Li X, et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci 2004;24:938–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winnig M, Bufe B, Meyerhof W. Valine 738 and lysine 735 in the fifth transmembrane domain of rTas1r3 mediate insensitivity towards lactisole of the rat sweet taste receptor. BMC Neurosci 2005;6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat 2005;26:199–204 [DOI] [PubMed] [Google Scholar]
- 49.Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA 2004;101:14258–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Zhang F, Xu H, Li Q.2007. PCT/US patent 2006/041158.
- 51.Gasparini F, Kuhn R, Pin JP. Allosteric modulators of group I metabotropic glutamate receptors: novel subtype-selective ligands and therapeutic perspectives. Curr Opin Pharmacol 2002;2:43–9 [DOI] [PubMed] [Google Scholar]
- 52.Pin JP, Kniazeff J, Liu J, et al. Allosteric functioning of dimeric class C G-protein-coupled receptors. FEBS J 2005;272:2947–55 [DOI] [PubMed] [Google Scholar]
- 53.Sainz E, Cavenagh MM, Lopez Jimenez ND, et al. The G-protein coupling properties of the human sweet and amino acid taste receptors. Dev Neurobiol 2007;67:948–59 [DOI] [PubMed] [Google Scholar]
- 54.Galindo-Cuspinera V, Breslin PA. The liaison of sweet and savory. Chem Senses 2006;31:221–5 [DOI] [PubMed] [Google Scholar]
- 55.Fernstrom JD. Introduction to the symposium. Am J Clin Nutr 2009;90(suppl):705S–6S [DOI] [PubMed] [Google Scholar]
- 56.Krebs JR. The gourmet ape: evolution and human food preferences. Am J Clin Nutr 2009;90(suppl):707S–11S [DOI] [PubMed] [Google Scholar]
- 57.Curtis RI. Umami and the foods of classical antiquity. Am J Clin Nutr 2009;90(suppl):712S–8S [DOI] [PubMed] [Google Scholar]
- 58.Kurihara K. Glutamate: from discovery as a food flavor to role as a basic taste (umami). Am J Clin Nutr 2009;90(suppl):719S–22S [DOI] [PubMed] [Google Scholar]
- 59.Beauchamp GK. Sensory and receptor responses to umami: an overview of pioneering work. Am J Clin Nutr 2009;90(suppl):723S–7S [DOI] [PubMed] [Google Scholar]
- 60.Sano C. History of glutamate production. Am J Clin Nutr 2009;90(suppl):728S–32S [DOI] [PubMed] [Google Scholar]
- 61.Li X. T1R receptors mediate mammalian sweet and umami taste. Am J Clin Nutr 2009;90(suppl):733S–7S [DOI] [PubMed] [Google Scholar]
- 62.Chaudhari N, Pereira E, Roper SD. Taste receptors for umami: the case for multiple receptors. Am J Clin Nutr 2009;90(suppl):738S–42S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.San Gabriel A, Maekawa T, Uneyama H, Torii K. Metabotropic glutamate receptor type 1 in taste tissue. Am J Clin Nutr 2009;90(suppl):743S–6S [DOI] [PubMed] [Google Scholar]
- 64.Yasumatsu K, Horio N, Murata Y, et al. Multiple receptors underlie glutamate taste responses in mice. Am J Clin Nutr 2009;90(suppl):747S–52S [DOI] [PubMed] [Google Scholar]
- 65.Kinnamon SC. Umami taste transduction mechanisms. Am J Clin Nutr 2009;90(suppl):753S–5S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachmanov AA, Inoue M, Ji H, Murata Y, Tordoff MG, Beauchamp GK. Glutamate taste and appetite in laboratory mice: physiologic and genetic analyses. Am J Clin Nutr 2009;90(suppl):756S–63S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shigemura N, Shirosaki S, Ohkuri T, et al. Variation in umami perception and in candidate genes for the umami receptor in mice and humans. Am J Clin Nutr 2009;90(suppl):764S–9S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mennella JA, Forestell CA, Morgan LK, Beauchamp GK. Early milk feeding influences taste acceptance and liking during infancy. Am J Clin Nutr 2009;90(suppl):780S–8S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raliou M, Wiencis A, Pillias A-M, et al. Nonsynonymous single nucleotide polymorphisms in human tas1r1, tas1r3, and mGluR1 and individual taste sensitivity to glutamate. Am J Clin Nutr 2009;90(suppl):789S–99S [DOI] [PubMed] [Google Scholar]
- 70.Donaldson LF, Bennett L, Baic S, Melichar JK. Taste and weight: is there a link? Am J Clin Nutr 2009;90(suppl):800S–3S [DOI] [PubMed] [Google Scholar]
- 71.Rolls ET. Functional neuroimaging of umami taste: what makes umami pleasant? Am J Clin Nutr 2009;90(suppl):804S–13S [DOI] [PubMed] [Google Scholar]
- 72.Blachier F, Boutry C, Bos C, Tomé D. Metabolism and functions of l-glutamate in the epithelial cells of the small and large intestines. Am J Clin Nutr 2009;90(suppl):814S–21S [DOI] [PubMed] [Google Scholar]
- 73.Kokrashvili Z, Mosinger B, Margolskee RF. Taste signaling elements expressed in gut enteroendocrine cells regulate nutrient-responsive secretion of gut hormones. Am J Clin Nutr 2009;90(suppl):822S–5S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akiba Y, Kaunitz JD. Luminal chemosensing and upper gastrointestinal mucosal defenses. Am J Clin Nutr 2009;90(suppl):826S–31S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kondoh T, Mallick HN, Torii K. Activation of the gut-brain axis by dietary glutamate and physiologic significance in energy homeostasis. Am J Clin Nutr 2009;90(suppl):832S–7S [DOI] [PubMed] [Google Scholar]
- 76.Tomé D, Schwarz J, Darcel N, Fromentin G. Protein, amino acids, vagus nerve signaling, and the brain. Am J Clin Nutr 2009;90(suppl):838S–43S [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto S, Tomoe M, Toyama K, Kawai M, Uneyama H. Can dietary supplementation of monosodium glutamate improve the health of the elderly? Am J Clin Nutr 2009;90(suppl):844S–9S [DOI] [PubMed] [Google Scholar]
- 78.Burrin DG, Stoll B. Metabolic fate and function of dietary glutamate in the gut. Am J Clin Nutr 2009;90(suppl):850S–6S [DOI] [PubMed] [Google Scholar]
- 79.Brosnan ME, Brosnan JT. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr 2009;90(suppl):857S–61S [DOI] [PubMed] [Google Scholar]
- 80.Stanley CA. Regulation of glutamate metabolism and insulin secretion by glutamate dehydrogenase in hypoglycemic children. Am J Clin Nutr 2009;90(suppl):862S–6S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hawkins RA. The blood-brain barrier and glutamate. Am J Clin Nutr 2009;90(suppl):867S–74S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Magistretti PJ. Role of glutamate in neuron-glia metabolic coupling. Am J Clin Nutr 2009;90(suppl):875S–80S [DOI] [PubMed] [Google Scholar]
- 83.Fernstrom JD. Symposium summary. Am J Clin Nutr 2009;90(suppl):881S–5S [DOI] [PubMed] [Google Scholar]