Abstract
Several drug delivery designs combine synthetic drug carriers with covalently conjugated targeting moieties. Such modifications of monoclonal antibodies (mAb), or their Fab′ fragments, inevitably result in diminished affinity for their targeted tissue. In an attempt to overcome this limitation, high molecular weight, branched N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers were synthesized and conjugated with Fab′ fragments of the anti-CD20 antibody, 1F5. This produced multivalent conjugates with varying valency (amount of Fab′ per macromolecule) targeted to the B-cell antigen CD20. Evaluation of a multivalent effect was done by determining the apparent KD at low concentrations of conjugates, the Sips heterogeneity factor, a, and the binding enhancement factors of each construct. The results clearly indicated that multivalency could improve the affinity of the HPMA copolymer–Fab′ conjugates to that of unconjugated mAb.
INTRODUCTION
A central tenet of modern drug delivery is to increase the concentration of drug in diseased tissues, while limiting its systemic exposure (1, 2). This translates to increased therapeutic efficiency and reduced side effects. Increasing concentrations in diseased tissues can be challenging and has been addressed through both synthetic (3–5) and biological approaches (6, 7). Several studies have combined high-affinity targeting moieties and high molecular weight drug carriers to confer tissue specificity and enhanced bioactivity to polymer–drug conjugates (8–12). However, covalent modification of targeting moieties resulted in diminished affinity (13). Multivalent conjugate designs were one approach that improved the specificity of the construct (14). Successful studies have demonstrated that multivalent carbohydrates (15), peptides (16), or other small molecules (17) can enhance binding affinity to targeted tissue. Other larger and more specific molecules such as monoclonal antibodies and Fab′ have only been rarely used in such approaches. In this study, we synthesized a high molecular weight soluble polymer that is able to accommodate multiple Fab′ fragments to produce a multivalent drug carrier. Furthermore, we demonstrated the effects of multivalent and multimeric binding of our constructs to the B-cell antigen CD20 in experiments using whole cells.
The soluble polymer poly[N-(2-hydroxypropyl)methacryl-amide] (pHPMA) is inert and biocompatible (18, 19). It has been used extensively as an anticancer drug delivery platform that is able to improve the physicochemical and pharmacokinetic properties of the polymer–drug conjugates by increasing circulating half-life, increasing the solubility of hydrophobic drugs, affecting passive accumulation in solid tumors, and providing the potential to evade multidrug resistance mediated by efflux pumps (18, 20). A number of studies have developed methods of HPMA copolymer synthesis that create high molecular weight, branched copolymers that are soluble in water (21, 22). Similar HPMA copolymers were used to create multivalent conjugates using anti-CD20 Fab′ antibody fragments.
As with any well-designed drug delivery system, the biological target is of critical importance. CD20 is a reliable B-cell marker that is highly expressed on normal and malignant B-cell populations (23, 24) and has been successfully used as a target with the first FDA-approved monoclonal antibody, Rituximab (25, 26). CD20 is a 33–37 kDa, integral membrane receptor that forms dimers and tetramers in the cell membrane (27, 28). In the absence of a known ligand, there is remarkable diversity of epitopes among anti-CD20 mAb clones. The most studied and best characterized clones include Rituximab, B1, 2H7, and 1F5 (29). Our study utilized 1F5, which has a dissociation constant (KD) between 20 and 30 nM (30), while the KD for Rituximab is around 4 nM (31). Rituximab and 1F5 bind distinct epitopes on CD20 (32, 33). Notably, cross-linking certain CD20-ligated mAb, including 1F5, with secondary antibodies (generally goat-antimouse Ab) can induce apoptosis (34, 35). This fact has clinical relevance for Rituximab. Its activity is partially attributed to ligated Rituximab becoming cross-linked by effector cells. Cross-linking, in turn, induces antibody-dependent cell-mediated cytotoxicity (ADCC). However, a full response to Rituximab requires a high-affinity allele of an Fc receptor on effector cells (FcγRIIIa). Patients that are not homozygous for the allele have diminished responses (36). Recent studies have shown that multivalent Rituximab constructs are able to effectively induce apoptosis independent of effector cells (37, 38).
A construct that is able to elicit apoptosis without the need to be internalized by a cell has clinical relevance. Such a construct could be used in a therapeutic regimen such as those used in either Bexxar (39) or Zevalin (40), where anti-CD20 constructs and radioimmunotherapeutics could be given in combination to treat refractory disease. The multivalent construct would induce apoptosis independent of effector cells, and the radioimmunotherapeutic could synergistically eliminate neighboring malignant cell populations which may not express CD20 via a bystander effect.
In this study, several multivalent HPMA copolymer–(anti-CD20) Fab′ conjugates were designed and synthesized. The conjugates varied in the amount of Fab′ per polymer chain to allow studies that could determine the effect of a conjugate’s valence on binding to B-cell antigen CD20. The apparent KD at low concentrations of conjugates, the Sips heterogeneity factor, and the affinity enhancement factors of each construct were determined.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and solvents used were of reagent grade or better unless otherwise stated. N-(2-Hydroxypropyl)methacrylamide (HPMA) was prepared as previously described (41). N-(3-Aminopropyl)methacrylamide hydrochloride (AMA) was purchased from Polysciences (Warrington, PA). Succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) was purchased from Soltec Ventures (Beverly, MA). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and Iodogen tubes were purchased form Thermo Scientific (Waltham, MA). Tetraethylene glycol dimethacrylate (TGD), phthalic dicarboxaldehyde (OPA), and 3-mercaptopropionic acid (MPA) were purchased from Sigma Aldrich (St. Louis, MO). Carrier free radionuclide sodium iodide-125 was purchased from Perkin-Elmer (Waltham, MA).
Cell Lines, Hybridomas, Fab′ Fragment Preparation
Human Burkitt’s B-cell non-Hodgkin’s lymphoma Raji cells (ATCC, Bethesda, MD) were cultured in RPMI-1640 medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 (v/v). All experiments were performed using cells in exponential growth. The anti-CD20 hybridoma clone 1F5 (ATCC, Bethesda, MD) was initially cultured and recloned through limiting dilution in RPMI 1640 supplemented with 10% FBS in a humidified environment with 5% CO2 in air (42). The selected clone was adapted to chemically defined, serum-free media (Invitrogen, Carlsbad, CA). Adapted cells were used to seed a CellMax bioreactor (Spectrum Laboratories, Rancho Dominguez, CA) according to the manufacturer’s instructions. Anti-CD20 mAb was purified on a Protein G Sepharose 4 Fast Flow column (GE Healthcare, Piscataway, NJ) from bioreactor harvest supernatant. Whole IgG2a was digested into F(ab′)2 with lysyl endopeptidase (Wako Chemicals USA, Richmond, VA) following the protocol of Fowers et al. (43). Purity of harvested IgG2a and digested F(ab′)2 were shown to be >95% by SDS-PAGE and SEC (size exclusion chromatography). Immediately prior to using, 5 mg/mL of F(ab′)2 was reduced to Fab′ with 4 mM TCEP in 0.1 M phosphate buffer (pH 6.5) at room temperature for 30 min.
Preparation of Multivalent Conjugates
The synthesis of copolymers, their fractionation, polymeranalogous modification with SMCC, Fab′ binding, and fractionation of the conjugates are depicted in Scheme 1. The HPMA copolymer containing side chains terminated in NH2 groups (P-NH2) was prepared by radical polymerization using AIBN as the initiator. Briefly, HPMA (658.7 mg, 93 mol %), AMA (53.6 mg, 6 mol %), TGD (16.5 mg, 1 mol %), and AIBN (3.4 mg) were dissolved in 2.9 g of DMSO. After mixing, the polymerization solution was bubbled with argon, sealed in a glass ampule, and then incubated at 50 °C for 24 h. After polymerization, the HPMA copolymer was precipitated and washed in acetone, and then washed with diethyl ether. The precipitate was dried, dissolved in H2O, and dialyzed against distilled water using dialysis tubing with a molecular weight cutoff of 12–14 kDa (Spectrum Laboratories, Rancho Dominguez, CA) to remove any unreacted monomers. The polymer was then lyophilized, dissolved in PBS, and fractionated on a Sephacryl S-500 preparative column (GE Healthcare, Piscataway, NJ) to produce two fractions. The fraction with the very high molecular weight was discarded; the fraction with an upper limit of Mw ~500 kDa was fractionated a second time on a Superose 6 HR16/60 preparative column (GE Healthcare, Piscataway, NJ) to produce a molecular weight “ladder” of ten fractions. Two fractions (P-7NH2: 34 kDa, 5.5 mol % NH2, yield 2.7 wt %; and P-3NH2: 193 kDa, 5.5 mol % NH2, yield 18 wt %) were selected for the synthesis of conjugates. The fractions P-3NH2 and P-7NH2 were dialyzed against water and then lyophilized. The molecular weight averages were estimated using size-exclusion chromatography on an analytical Superose 6 column (GE Healthcare, Piscataway, NJ). All SEC was done using an ÄKTA FPLC system (GE Healthcare, Piscataway, NJ).
Scheme 1.
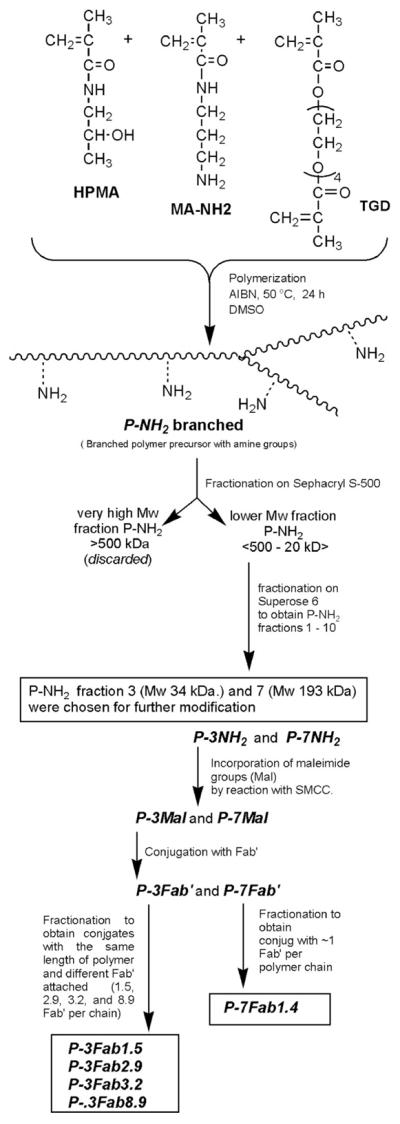
Synthesis of Multivalent HPMA Copolymer–Fab′ Conjugates Targeted to the B-Cell Antigen CD20
Polymeranalogous Conversion of Pendant Amine Groups into Maleimide Groups
The fractions P-3NH2 and P-7NH2 (P-3NH2: 37 mg, 22.5 μmol –NH2; P-7NH2: 6 mg, 3.6 μmol–NH2) were reacted with excess SMCC (14.4 mg, 45.6 μmol; 2.4 mg, 7.2 μmol, respectively) in dry DMF under basic conditions (DIPEA: 26 μL, 72 μmol; 2.1 μL, 12 μmol, respectively) to generate pendant maleimide groups. The activated precursor was precipitated into an excess of acetone, filtered, and washed with ether to remove free SMCC. The conversion of amine groups into maleimide on the polymer proceeded with ~72% to 75% conversion, as determined by modified Ellman’s assay. The products P-3Mal and P-7Mal were then conjugated with reduced 1F5 Fab′ fragments as previously described (13). Briefly, approximately 5 mg of either P-3Mal or P-7Mal (~2.3 μmol maleimide) was dissolved in 50 μL of DMF. Then, 10 mg of Fab′ (0.2 μmol) in 2 mL of PBS (pH 6.5) was added to the reaction, and the solution was mixed overnight at room temperature. The products were fractionated (and unbound Fab′ removed) on a size-exclusion S6 preparative column HR 16/60. HPMA copolymer–Fab′ conjugates containing different amounts of Fab′ per macromolecule, P-3Fab1.5, P-3Fab2.9, P-3Fab3.2, P-3Fab8.9, and P-7Fab1.4, were isolated and characterized. In these abbreviations, the number following Fab′ indicated the amount of Fab′ fragments per macromolecule.
Effective Diameter of HPMA Copolymer–Fab′ Conjugates
The effective diameter for fractions of HPMA copolymer–Fab′ conjugates was evaluated by dynamic light scattering (Brookhaven Scientific Instruments, Holtsville) in PBS at a concentration of at least 1 mg/mL. Prior to analysis, the solution containing conjugates were filtered through a 0.45 μm filter. In addition to the effective diameter, dynamic light scattering (DLS) also calculated the distribution of size, referred to as the polydispersity index (PDI), which differs from molecular weight polydispersity (Mw/Mn). DLS PDI values greater than 0.2 are considered to be heterogeneous, and those below 0.2 are considered homogeneous. The effective diameter of whole Ab served as a positive control with both effective diameters (measured 13.6 nM, literature 14.3) and PDI values (0.06 measured, literature 0.05) compared well with published values (44).
Determination of Fab′ Content per Polymer Chain
The composition of the conjugates, the ratio of protein to polymer, was determined in one step by amino acid analysis. This method enabled measurement of, after acid hydrolysis, both individual amino acids from Fab′ as well as 1-amino-2-propanol released from HPMA monomer units. Calibration was done using free 1F5 Fab′ and HPMA homopolymer. A small amount (>3 mg) of Fab′, homopolymer, or conjugate was dissolved in 6 N HCl in a small ampule, bubbled with nitrogen for 5 min, and then the ampule was sealed. The solution was incubated at 125 °C for 24 h, the ampule was opened, and the hydrolyzed product was dried in vacuo. Hydrolyzed products were dissolved in dH2O, and then derivatized with OPA and MPA prior to being analyzed on a HPLC with an eclipse XDB-C8 column (Agilent, Santa Clara, CA). Fab′ concentration was calibrated with a peak produced by hydrolyzed glutamate, which had an elution time of 9.4 min. Poly(HPMA) concentration was calibrated with a peak produced by 1-amino-2-propanol, which had a retention time of 23.9 min.
Iodination of Fab′ and Multivalent Fab′ Constructs
Reduced anti-CD20 Fab′, anti-CD20 HPMA copolymer–Fab′ conjugates, and whole antiCD20 IgG molecules were labeled with Na125I using Iodo-Gen tubes as previously described (13). Approximately 300 μg of Fab′ or IgG in 0.3 mL PBS was added to a glass vial coated with 4 μg of Iodo-Gen, followed by 0.5 mCi of Na125I. After the solution was incubated (and shaken) at room temperature for 7 min, 125I-labeled protein was separated from free 125I by passing the reaction mixture over a PD-10 desalting column (GE Healthcare, Piscataway, NJ). The labeled molecules were stored at 4 °C in PBS. The specific radioactivity of each conjugate was between 0.7 and 1.5 μCi/μg.
Binding Experiments
Raji cells grown in RPMI containing 10% FBS were washed twice with HBSS. The cells were adjusted to 4 × 106 cells/mL and split into two volumes. To one of the volumes of Raji cells, unlabeled anti-CD20 IgG was added to a final concentration of 500 nM to block CD20 for a negative control. Serial dilutions of 125I-labeled anti-CD20 IgG, Fab′, and multivalent Fab′ conjugates were made from 2000 nM to 1 nM of Fab′ fragment concentration. Millipore 96-well filter plates, coated with a 1% solution of nonfat dry milk in distilled water and incubated at 4 °C overnight, were washed with HBSS. For each of the serial dilutions, 25 μL of negative control cells were added to a single well, and 25 μL of unblocked cells were added to three different wells; then, 25 μL of the respective dilutions were added to each well. The filter-well plates were incubated for 90 min at 120 rpm on a shaker at 25 °C. Following incubation, the Raji cells were washed twice with 200 μL of cold HBSS with 10% FBS, and then collected on filters. The filters were allowed to air-dry, removed from the plate, and counted. Negative control wells were used to correct for nonspecific binding.
The equilibrium binding was evaluated using either single-site binding parameters (eq 1) or the Sips equation (eq 2) using Sigma Plot (Systat Software, San Jose, CA). Data was also drafted in Scatchard plots.
| (1) |
| (2) |
where B is the concentration of bound ligand, F is the concentration of free ligand, Bmax is the theoretical limit of bound ligand, KD is the dissociation constant, and a is the Sips heterogeneity factor. When a is equal to 1, the Sips equation simplifies to the single-site binding equation.
RESULTS
Synthesis of Multivalent Conjugates
Copolymerization of HPMA with a small amount of the cross-linker TGD, at conditions below the gel point, yielded an HPMA copolymer with broadly distributed molecular weight that ranged from 20 kDa to greater than 1000 kDa (Scheme 1). The yield of this polymerization reaction was 69%, with a reproducible molecular weight profile after three separate polymerization experiments. Macromolecules with a molecular weight higher than 500 kDa were removed by fractionation on a SEC column. The fraction containing macromolecules with M < 500 kDa (45% by weight of original copolymer) was used to build a molecular weight “ladder” with narrowly disperse molecular weights by a second fractionation. The amine content of these fractions was 5.5 mol %. Two of the fractions, P-3NH2 (Mw =193 kDa) and P-7NH2 (Mw = 34 kDa), were selected for the use in conjugate synthesis and were activated by a reaction with SMCC. The maleimide content in polymer precursors P-3Mal and P-7Mal was 4.1 mol %.
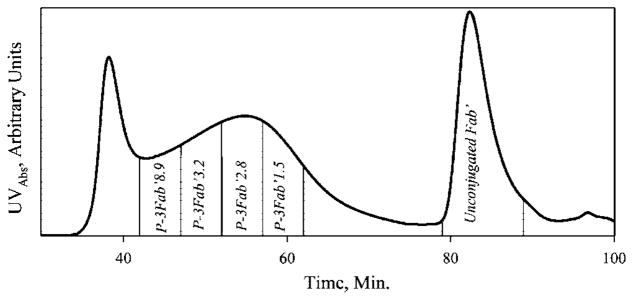
Reduced Fab′ fragments of the anti-CD20 mAb were conjugated to polymer precursors P-3Mal and P-7Mal via thioether bonds in an overnight reaction. The P-3Fab′ conjugate was fractionated on a SEC column to provide fractions with different amounts of bound Fab′ fragments per macromolecule. The free (unbound) Fab′ could be easily separated from the conjugate fractions (Figure 1). However, only a single fraction was isolated from the P-7Fab′ conjugate (data not shown). The fractions are referred to by the fraction number (e.g. P-3 or P-7) and by the number of Fab′ fragments per polymer chain as determined by modified amino acid analysis (e.g., P-3Fab′8.9).
Figure 1.
Fractionation of the P-3Fab′ conjugate by size exclusion chromatography. Elution profile of reaction mixture on a Superose S6 preparative column (HR 10/30, FPLC system); buffer PBS pH 6.5; flow rate 1 mL/min; amount of sample applied 10 mg in 2 mL; detection absorbance at 280 nm. The numerical suffix at the fraction designation is the average valence as determined by amino acid analysis.
Determination of Valence
A modified amino acid analysis protocol was used to determine the Fab′ per polymer chain or valence of each fraction. The method cleaved 1-amino-2-propanol from the HPMA monomer units. Following derivatization with OPA and MPA, the 1-amino-2-propanol related peak was well-separated from Fab′-associated amino acid peaks. It is important to note that the analysis determined the average valence, which is dependent on the polydispersity of the conjugate fraction. The polydispersity within each conjugate fraction was not determined in this study, but the elution volumes for each fraction were sufficiently small to guarantee a low polydispersity (1.2–1.3).
Amino acid analysis demonstrated that Fab′ “loading” was responsible for the broadened FPLC profile of the conjugates as seen in Figure 1. The fractions of P-3 conjugate with the highest valence of 8.9 Fab′/chain eluted first. The P-3 fraction with the lowest valence P-3Fab1.5 eluted last. Determination of valence also confirmed that Fab′ loading was directly related to the size of the polymeric precursor. After conjugation, the larger polymer fraction P-3 had a broad distribution of Fab′ per chain with a median valence of ~3 Fab′/chain, while the fraction P-7 (Mw 34 kDa) had an average valence of 1.4 Fab′ per polymer chain (Table 1).
Table 1.
Chemical and Physical Analysis of Polymeric Precursors and Conjugates
| conjugate | Mwa | Pdb | valencec | effective diameterd | PDIe |
|---|---|---|---|---|---|
| P-7Fab′1.4 | 34 | 1.16 | 1.4 | 10.8 | 0.070 |
| P-3Fab′1.5 | 193 | 1.25 | 1.5 | 17.8 | 0.203 |
| P-3Fab′2.9 | 193 | 1.25 | 2.9 | 21.3 | 0.19 |
| P-3Fab′3.2 | 193 | 1.25 | 3.2 | 30.2 | 0.168 |
| P-3Fab′8.9 | 193 | 1.25 | 8.6 | 38.7 | 0.174 |
Molecular weight average of polymeric precursor as determined by size exclusion chromatography.
Polydiversity, ratio of molecular weight averages given by weight average over number average.
Valence of conjugate or Fab′ per polymer chain as determined by modified amino acid analysis (see Experimental Procedures).
Effective diameter of conjugates as determined by dynamic light scattering (DLS).
Value of polydispersity index of the diameter of the conjugate determined by DLS.
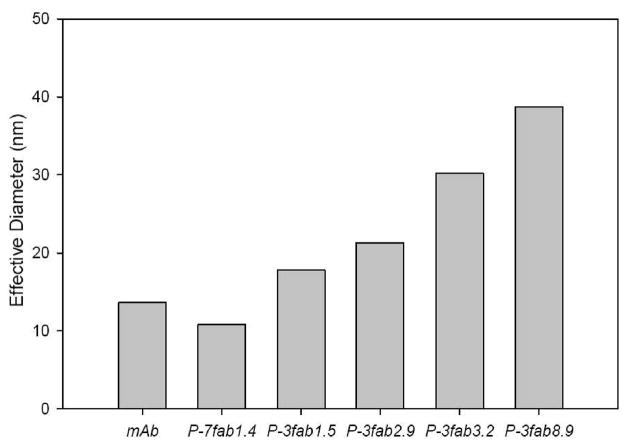
Analysis of the effective diameters of HPMA copolymer–Fab′ conjugates by DLS demonstrated that Fab′ loading directly corresponded with the size of the conjugate. Loading of Fab′ extended HPMA copolymers from their native random coil structure. As the valence of the polymeric precursor increased, the effective diameter also increased (Figure 2). In the case of P-3 fractions, the fraction with the most Fab′ per chain, P-3fab8.9, had an effective diameter of 38.7 nm, while the fraction with the least Fab′ per chain, P-3fab1.5, had an effective diameter of 17.8 nm. Also, fractionation of the conjugate produced low polydispersity index values near or below 0.2 as shown in Table 1. To ensure that the fractions were not aggregates, a Zimm plot was also used to calculate the molecular weight of the conjugate P-3fab′8.9. The calculated molecular weight was 611 ± 43 kDa, a value that corresponded well with the molecular weight estimates made by adding the molecular weight of polymer and the total molecular mass of Fab′ attached to the conjugate.
Figure 2.
Effective diameters of the multivalent HPMA copolymer–Fab′ conjugates as determined by dynamic light scattering (DLS). The effective diameter corresponded to the valence of each conjugate.
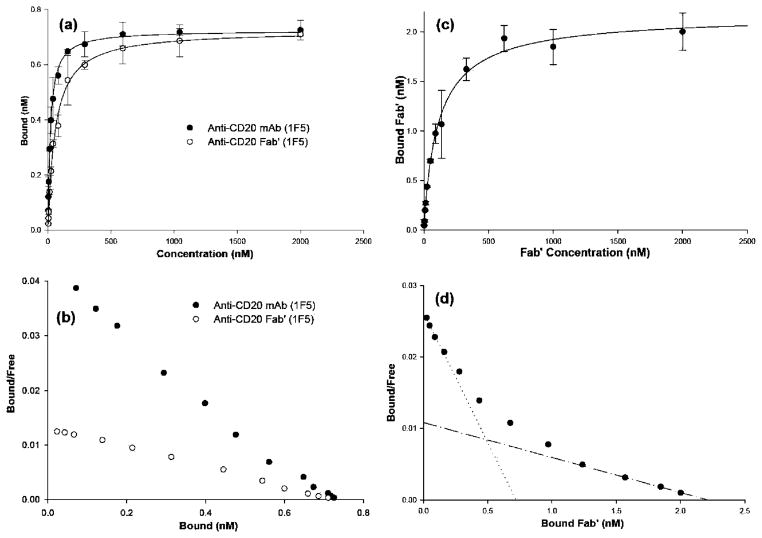
Binding Isotherms and Scatchard Plots
The single-site binding equation produced a good fit to experimental data for the analysis of CD20 binding with whole Ab or with Fab′. The binding constants (KD) were 19 nM and 58 nM, respectively. The average number of CD20 receptors per cells was 217 000 ± 27 000 and 217 000 ± 39 000 as calculated from binding experiments with whole Ab and Fab′ fragments, respectively. Both KD and receptors/cell were close to other published values (30, 31). However, the single-site binding equation failed to adequately model the binding of multivalent conjugates to the CD20 receptor, with the most significant deviation occurring at low concentrations of Fab′. A better fit was produced by the Sips equation; typical data for conjugate P-3Fab′3.2 are shown on Figure 3c,d. The Sips equation uses the same KD and Bmax fitting parameters as the single-site binding equation and adds a heterogeneity constant, a, as a fitting parameter (eq 1) (45). If the heterogeneity constant is set to 1, the Sips equation simplifies to the single-site binding equation. For that reason, KD and Bmax values listed in Table 1 for mAb and Fab′ are listed as KD,Sips and Bmax,Sips. Figure 3 provides a representative example of how the Sips equation was used to fit the binding isotherm data and of the curved Scatchard plot. For the conjugates evaluated, the Sips heterogeneity constant was within the interval 〈0.78; 0.99〉 (Table 2). The isotherms for conjugates P-3Fab′1.5, P-3Fab′2.9, P-3Fab′8.9, and P-7Fab′1.4 are presented in Supporting Information.
Figure 3.
Representative examples of binding of 125I-labeled anti-CD20 mAb (1F5), Fab′, and multivalent conjugate P-3Fab′3.2 to CD20(+) Raji cells. (a) Isotherms of mAb or Fab′ fit with the single-site binding equation using a least-squares method. (b) The Scatchard plot for the binding of mAb and Fab′. (c) Binding isotherm of P-3Fab′3.2 fitted with the Sips equation. (d) Binding data for P-3Fab′3.2 evaluated using a Scatchard plot. The slope of the dotted line in is equal to 1/KD,low[Fab′], while the slope of the dash–dot line is equal to 1/KD,sat[Fab′]. Other multivalent conjugates had similar isotherms and Scatchard plots as P-3Fab′3.2 (see Supporting Information). Each concentration was corrected for nonspecific binding as described in the Experimental Procedures.
Table 2.
Results of Binding Analysis
| targeting molecule | KD,Sips (nM)a | KD,low[Fab′] (nM) b | KD,Sat[Fab′] (nM) c | Bmax,Sips (nM) d | Bmax,scat (nM) e | af | valence g | valence h |
|---|---|---|---|---|---|---|---|---|
| mAb | 19 ± 1 | NA | NA | 0.72 ± 0.13 | 0.72 | 1 | - | - |
| Fab′ | 58 ± 4 | NA | NA | 0.72 ± 0.09 | 0.72 | 1 | - | - |
| P-7Fab′1.4 | 85 ± 11 | 47 | 109 | 0.98 ± 0.04 | 1.01 | 0.99 ± 0.08 | 1.4 | 1.4 |
| P-3Fab′1.5 | 86 ± 14 | 32 | 190 | 1.09 ± 0.05 | 1.14 | 0.78 ± 0.06 | 1.6 | 1.5 |
| P-3Fab′2.9 | 182 ± 35 | 34 | 226 | 2.04 ± 0.01 | 2.01 | 0.81 ± 0.07 | 2.8 | 2.9 |
| P-3Fab′3.2 | 115 ± 25 | 27 | 204 | 2.18 ± 0.10 | 2.21 | 0.93 ± 0.09 | 3.1 | 3.2 |
| P-3Fab′8.9 | 439 ± 165 | 15 | 257 | 7.5 ± 1.10 | 6 | 0.78 ± 0.16 | 8.2 | 8.9 |
Affinity binding constant determined by a least-squares fit of binding isotherm data.
Affinity binding constant derived from the inverse slope of Scatchard curves at low concentrations of Fab′.
Affinity binding constant derived from the inverse slope of Scatchard curves at saturating concentrations of Fab′.
Theoretical limit of bound ligand determined by a least-squares fit of the single-site binding equation.
Theoretical limit of bound ligand determined by graphical extrapolation of Scatchard plots to the x-axis.
Heterogeneity factor determined by the Sips equation.
Valence of conjugates determined with modified amino acid analysis.
Valence of conjugates determined by the ratio of Bmax of the conjugate over Bmax of the Fab′
The Scatchard plots of the multivalent conjugates were concave curves, whereas the Scatchard plots for whole Ab and Fab′ were linear. The curvature of the Scatchard plots was relevant in describing the multimeric nature of binding; the latter was also described by the heterogeneity constant, a, of the Sips equation. Curvature of Scatchard plots may suggest a number of phenomena, including heterogeneity of binding sites, and nonspecific binding. In the analysis of experimental data, nonspecific binding was subtracted from total binding. Also, the heterogeneity of binding sites was considered to be minimal, since the thioether bond would be distant from the binding site of the Fab′ (13). In this instance, a concave Scatchard plot suggests negative cooperativity. Consequently, the strongest binding will occur at the lowest concentration of multivalent conjugate. At low concentrations, a multivalent conjugate can bind multiple receptors. As the concentration of the conjugate increases, the probability of a single multivalent conjugate binding multiple receptors is reduced to the point that each multivalent molecule only binds a single receptor (all of the receptors are occupied by different molecules). As a result, the ratio of the conjugate’s Bmax divided by the Bmax of the Fab′ is indicative of the total valence of the conjugate or Fab′ per polymer chain. The valence of conjugates (Fab′ per macromolecule) determined with binding isotherms was in good agreement with the values determined by chemical analysis (Table 2).
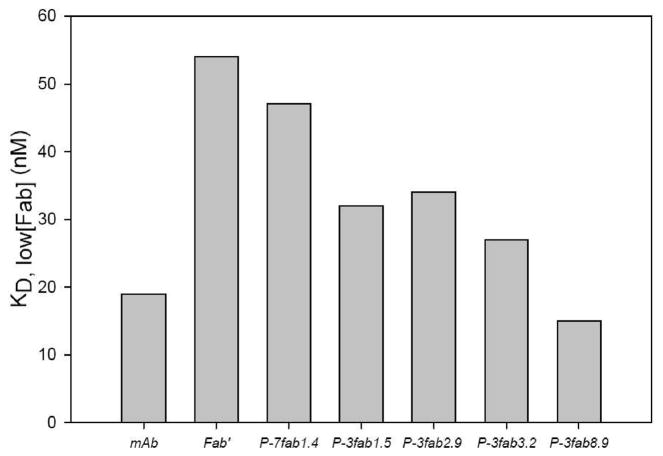
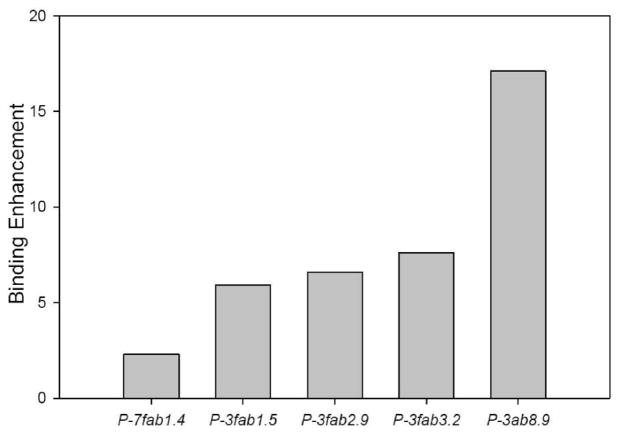
The multivalent or multimeric binding effect may be quantified by the enhancement factor, the ratio of the apparent binding constant, KD,Sat[Fab′], at saturating concentrations of Fab′ and the apparent binding constant, KD,low[Fab′], at low concentrations of Fab′ (46). The apparent binding constants were extrapolated from relevant sections of Scatchard plots. The KD,low[Fab′] are shown in Figure 4, and the enhancement factors in Figure 5. The largest enhancement factor, 17.7, of the investigated conjugates was found for P-3Fab′8.9. The KD,low[Fab′] for P-3Fab′8.9 was 15 nM; the KD,Sat[Fab′] was 256.5 nM. The smallest enhancement factor was for conjugate P-7Fab′1.4, which had a KD,low[Fab′] of 47 nM, a KD,Sat[Fab′] of 108.1 nM, and an enhancement factor of 2.3.
Figure 4.
Apparent binding constants, KD,low[Fab′], for anti-CD20 Ab, Fab′ fragment, and HPMA copolymer–Fab′ conjugates. The KD,low[Fab′] is indicative of the slope of the Scatchard plot at low concentrations of Fab′ (see Figure 3d). Generally, the KD,low[Fab′] decreased (affinity increased) as the valence of the construct increased.
Figure 5.
Binding enhancement for anti-CD20 Ab, Fab′ fragment, and HPMA copolymer–Fab′ conjugates. Enhancement is derived from increase in affinity due to multiple binding sites per construct. It was calculated by dividing the KD,sat[Fab′] by KD,low[Fab′].
The KD,low[Fab′] values provide good evidence of a multivalent, multimeric binding enhancement as shown in Figure 4. The value observed for P-3Fab′8.9 (15 nM) was better than those for free Ab (19 nM) and free Fab′ (58nM).
DISCUSSION
The aim of this study was to demonstrate that water-soluble polymers have potential as multivalent drug carriers. To this end, high molecular weight branched HPMA copolymers were prepared by copolymerization of HPMA with a cross-linking agent (TGD) at conditions below the gel point. Fractions of different molecular weight were used in the synthesis of multivalent HPMA copolymer–Fab′ conjugates. The Fab′ fragment (of anti-CD20 1F5 antibodies) was attached to the copolymers via a thioether bond. The maximum enhancement of binding affinity for the target Raji cells achieved here (17.7 times) bodes well for the development of novel, more effective long-circulating polymer–drug conjugates. To ensure biocompatibility, degradable sequences have to be incorporated into the HPMA copolymer carrier structure, using primary chains with molecular weight below the renal threshold. This can be achieved by connecting primary HPMA copolymer chains via enzymatically degradable cross-links (bridges). Previously, two approaches have been used for the synthesis of degradable, water-soluble, cross-linked (branched) copolymers short of gel point. The first approach consisted of cross-linking reactive HPMA copolymer precursors, containing side chains terminated in reactive ester groups, with degradable α,ω-diaminooligopeptides (47–49). The second approach was to use biodegradable cross-linkers and prepare the conjugates by copolymerization. One cross-linker, N2,N5-bis(N-methacryloylglycylphenylananylleucylglycyl)ornithine methyl ester, has been used to reproducibly synthesize high molecular weight branched HPMA copolymers that can be lysosomally degraded to primary chains to facilitate elimination from the organism by renal filtration (21). It is interesting to note that the efficacy of doxorubicin conjugates with branched HPMA copolymers, in an ovarian carcinoma xenograft model, increased with increasing molecular weight of the conjugate (22).
The enlargement of the effective diameter of HPMA copolymer with conjugation Fab′ was demonstrated qualitatively by SEC and quantitatively with DLS. This result is not unexpected. HPMA copolymers, in an aqueous environment, have a dynamic, random coil conformation. Dispersive forces would exist between intrachain Fab′ that would result in stretching out the polymer chain. This report demonstrated that enlargement of the HPMA copolymer was directly related to the amount of Fab′ that had been loaded onto the polymer. Due to the nature of HPMA copolymers, the conjugates would maintain a dynamic and flexible structure, a property that may increase the effective valence of the conjugates, and the ability of intrachain Fab′ to gain access and bind to CD20.
The valence of the conjugates was analyzed chemically with a modified amino acid analysis method (50). The valence was confirmed by comparing the Bmax of each conjugate with the Bmax of unconjugated Fab′ (Table 2). The two determinations corresponded well and infer that the polydispersity among the fractions was narrow (1.2–1.3). However, the effective valence, or the maximum amount of Fab′ per conjugate that could bind CD20, was not determined. It can be hypothesized that most intrachain Fab′ could bind with CD20 simultaneously at low concentrations of conjugate. The formation of dynamic oligomers of CD20 in lipid rafts (29, 51) would create several local binding sites for the targeted conjugate. After the first Fab′ of a conjugate bound to a CD20 molecule, the dynamic and flexible nature of the HPMA copolymer backbone (52) would allow a conformational response within the polymer, so that intrachain Fab′ could bind proximal CD20 molecules. Also, the size of the conjugates appears to favor multivalent interactions, since conjugates with higher orders of valency had effective diameters in a saline solution greater than whole mAb, 14.3 nm (44), and in the theoretical range of cross-linked Abs.
Multivalency is important for the clinical efficacy of Rituximab. Ghetie and co-workers demonstrated enhanced efficacy of tetravalent homodimers prepared by cross-linking of Rituximab with heterobifunctional reagents (37). Homodimers and trimers possessed similar binding affinity to lymphoma B cells in vitro but a slower rate of dissociation when compared to the monomer. Zhang et al. (38) cross-linked Rituximab with oxidized dextran (Mw = 6000) into high molecular weight conjugates (no data on molecular weight provided) possessing similar binding avidity as Rituximab monomer. In both cases (37, 38), multivalent conjugates induced apoptosis in CD20(+) cells.
It is notable that the single-site binding equation accurately modeled isotherms for both mAb and unconjugated Fab′ bound to CD20, but failed with multivalent constructs. In this case, the Sips equation provided a good model. This is likely due to the thermodynamic assumptions of the single-site binding equation. It assumes that there is a single energy of adsorption, regardless of ligand concentration (53, 54). This assumption is not valid for a multivalent conjugate that can bind multimerically to a cell surface (55). In the case of a multivalent Fab′ conjugate such as P-3Fab′8.9, a single conjugate molecule could bind multiple CD20 receptors at low concentrations. As the concentration of P-3Fab′8.9 is increased, it would become less likely that the conjugate could bind multiple receptors to the point that each multivalent conjugate could only bind to a single receptor (56). Just as the probability of a conjugate binding multiple receptors is reduced, so is the energy of adsorption reduced with an increase of conjugate concentration. This is reflected in a concave Scatchard plot and a heterogeneity factor, a, that is less than 1. In this context, the factor, a, represents the distribution of adsorption energies at all concentrations of conjugate and the binding constant KD represents the average affinity of binding (45, 57). However, in an in vivo setting, the concentration of the conjugate at the target site is expected to be low. Consequently, the binding enhancement for the multivalent conjugates (Figure 5) should be fully operative.
Evidence of multivalent interactions involving the conjugates and CD20 include a general increase of affinity of the conjugates toward CD20 (Figure 4) and an increase of the enhancement factor (Figure 5) with increase of a conjugate’s valence. The theoretical basis of a multivalent interaction is that the first univalent interaction creates a complex in which the remaining free binding sites on both species are at greatly enhanced local concentrations. Such “cooperative” interactions are also responsible for two strands of DNA annealing, viruses binding to cells, and the folding of proteins into stable structures. Several studies have demonstrated that multivalent enhancement can improve the binding affinity of low-affinity targeting moieties, such as saccharides binding to concavanalin A, by several orders of magnitude (58–60). Depending on the adsorption kinetics, these studies have displayed similar binding isotherms (61). However, the multivalent effect dramatically increased the average binding constant KD of the multivalent ligand, rather then merely recovering the binding constant associated with the monomeric ligand, as we were able to demonstrate. The best explanation for this is the increased effective diameter of the construct, which would not be observed in studies that involve small, low-affinity multivalent molecules. The increased diameter of the multivalent construct directly impacts the Kon rate of the construct by limiting the construct diffusivity (62).
Several studies have evaluated the stability of the thioether bond made by maleimide and cysteine under saline and serum conditions. Studies of HPMA copolymer–Fab′ conjugates have demonstrated stability under saline conditions (13). In another study that measured the half-life of antibody–drug conjugates, it was shown that the thioether bond has a half-life of seven days in serum (63). Interestingly, this did not affect the overall effectivity of drug, and the half-life could be extend by substituting the thioether bond with a bromoacetamide linker, a strategy that could also be utilized in a future design. Other HPMA copolymer constructs conjugated to Fab′ fragments by thioether bonds have shown stability in vivo (64).
In summary, methods to produce multivalent HPMA copolymer–anti-CD20 Fab′ conjugates and to analyze their multivalent binding to the B-cell antigen CD20 were examined. Binding of the multivalent constructs was characterized by the binding constant, KD, the Sips heterogeneity factor, a, and the binding enhancement factor. The multivalent constructs bound the B-cell antigen CD20 in a manner that was consistent with negative cooperative models. It was revealed that using Fab′ conjugated to soluble branched HPMA copolymer can result in binding enhancement that can surpass binding affinities of whole mAb. Furthermore, the multivalent binding of these conjugates may be useful in exploring cell-surface dynamics involved with induction of apoptosis, initiated by cross-linking the B-cell antigen CD20.
Supplementary Material
Acknowledgments
The research was supported in part by the NIH grant CA51578 from the National Cancer Institute.
Footnotes
Supporting Information Available: Binding isotherms and Scatchard plots for P-3Fab1.5, P-3Fab2.9, P-3Fab8.9, and P-7Fab1.4. This material is available free of charge via the Internet at http://pubs.acs.org.
LITERATURE CITED
- 1.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 2.Couvreur P, Vauthier C. Nanotechnology: intelligent design to treat complex disease. Pharm Res. 2006;23:1417–1450. doi: 10.1007/s11095-006-0284-8. [DOI] [PubMed] [Google Scholar]
- 3.Duncan R, Kopečková-Rejmanová P, Strohalm J, Hume I, Cable HC, Pohl J, Lloyd JB, Kopeček J. Anticancer agents coupled to N-(2-hydroxypropyl)methacrylamide copolymers. I. Evaluation of daunomycin and puromycin conjugates in vitro. Br J Cancer. 1987;55:165–174. doi: 10.1038/bjc.1987.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan R, Spreafico F. Polymer conjugates. Pharmacokinetic considerations for design and development. Clin Pharmacokinet. 1994;27:290–306. doi: 10.2165/00003088-199427040-00004. [DOI] [PubMed] [Google Scholar]
- 5.Putnam D, Kopeček J. Polymer conjugates with anticancer activity. Adv Polym Sci. 1995;122:55–123. [Google Scholar]
- 6.Schuster JM, Bigner DD. Immunotherapy and monoclonal antibody therapies. Curr Opin Oncol. 1992;4:547–552. doi: 10.1097/00001622-199206000-00020. [DOI] [PubMed] [Google Scholar]
- 7.von Mehren M, Adams GP, Weiner LM. Monoclonal antibody therapy for cancer. Annu Rev Med. 2003;54:343–369. doi: 10.1146/annurev.med.54.101601.152442. [DOI] [PubMed] [Google Scholar]
- 8.Hongrapipat J, Kopečková P, Liu J, Prakongpan S, Kopeček J. Combination chemotherapy and photodynamic therapy with Fab′ fragment targeted HPMA copolymer conjugates in human ovarian carcinoma cells. Mol Pharmaceutics. 2008;5:696–709. doi: 10.1021/mp800006e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan R. Polymer conjugates for drug targeting. From inspired to inspiration. J Drug Targeting. 2006;14:333–335. doi: 10.1080/10611860600833880. [DOI] [PubMed] [Google Scholar]
- 10.Merdan T, Callahan J, Petersen H, Kunath K, Bakowsky U, Kopečková P, Kissel T, Kopeček J. Pegylated polyethylenimine-Fab′ antibody fragment conjugates for targeted gene delivery to human ovarian carcinoma cells. Bioconjugate Chem. 2003;14:989–996. doi: 10.1021/bc0340767. [DOI] [PubMed] [Google Scholar]
- 11.Říhová B, Jelínková M, Strohalm J, Št’astný M, Hovorka O, Plocová D, Kovář M, Dráberová L, Ulbrich K. Antiproliferative effect of a lectin- and anti-Thy-1.2 antibody-targeted HPMA copolymer-bound doxorubicin on primary and metastatic human colorectal carcinoma and on human colorectal carcinoma transfected with the mouse Thy-1.2 gene. Bioconjugate Chem. 2000;11:664–673. doi: 10.1021/bc9901696. [DOI] [PubMed] [Google Scholar]
- 12.Říhová B, Kopečková P, Strohalm J, Rossmann P, Větvička V, Kopeček J. Antibody-directed affinity therapy applied to the immune system: in vivo effectiveness and limited toxicity of daunomycin conjugated to HPMA copolymers and targeting antibody. Clin Immunol Immunopathol. 1988;46:100–114. doi: 10.1016/0090-1229(88)90010-4. [DOI] [PubMed] [Google Scholar]
- 13.Omelyanenko V, Kopečková P, Gentry C, Shiah JG, Kopeček J. HPMA copolymer-anticancer drug-OV-TL16 antibody conjugates. 1. Influence of the method of synthesis on the binding affinity to OVCAR-3 ovarian carcinoma cells in vitro. J Drug Targeting. 1996;3:357–373. doi: 10.3109/10611869608996827. [DOI] [PubMed] [Google Scholar]
- 14.Mammen M, Choi S, Whitesides GM. Polyvalent interaction in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem, Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor-ligand binding mechanisms with multivalent ligand architecture. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 16.Ding H, Prodinger WM, Kopeček J. Identification of CD21-binding peptides with phage display and investigation of binding properties of HPMA copolymer-peptide conjugates. Bioconjugate Chem. 2006;17:514–523. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 17.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chem Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Kopeček J, Kopečková P, Minko T, Lu Z. HPMA copolymer-anticancer drug conjugates: design, activity, and mechanism of action. Eur J Pharm Biopharm. 2000;50:61–81. doi: 10.1016/s0939-6411(00)00075-8. [DOI] [PubMed] [Google Scholar]
- 19.Říhová B, Bilej M, Větvička V, Ulbrich K, Strohalm J, Kopeček J, Duncan R. Biocompatibility of N-(2-hydroxypropyl) methacrylamide copolymers containing adriamycin. Immunogenicity, and effect on haematopoietic stem cells in bone marrow in vivo and mouse splenocytes and human peripheral blood lymphocytes in vitro. Biomaterials. 1989;10:335–342. doi: 10.1016/0142-9612(89)90075-6. [DOI] [PubMed] [Google Scholar]
- 20.Minko T, Kopečková P, Kopeček J. Efficacy of chemotherapeutic action of HPMA copolymer-bound doxorubicin in a solid tumor model of ovarian carcinoma. Int J Cancer. 2000;86:108–117. doi: 10.1002/(sici)1097-0215(20000401)86:1<108::aid-ijc17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Dvořák M, Kopečková P, Kopeček J. High-molecular weight HPMA copolymer-adriamycin conjugates. J Controlled Release. 1999;60:321–332. doi: 10.1016/s0168-3659(99)00087-5. [DOI] [PubMed] [Google Scholar]
- 22.Shiah JG, Dvořák M, Kopečková P, Sun Y, Peterson CM, Kopeček J. Biodistribution and antitumour efficacy of long-circulating N-(2-hydroxypropyl)methacrylamide copolymer-doxorubicin conjugates in nude mice. Eur J Cancer. 2001;37:131–139. doi: 10.1016/s0959-8049(00)00374-9. [DOI] [PubMed] [Google Scholar]
- 23.Anderson KC, Bates MP, Slaughenhoupt BL, Pinkus GS, Schlossman SF, Nadler LM. Expression of human B cell-associated antigens on leukemias and lymphomas: a model of human B cell differentiation. Blood. 1984;63:1424–1433. [PubMed] [Google Scholar]
- 24.Stashenko P, Nadler LM, Hardy R, Schlossman SF. Expression of cell surface markers after human B lymphocyte activation. Proc Natl Acad Sci USA. 1981;78:3848–3852. doi: 10.1073/pnas.78.6.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boye J, Elter T, Engert A. An overview of the current clinical use of the anti-CD20 monoclonal antibody rituximab. Ann Oncol. 2003;14:520–535. doi: 10.1093/annonc/mdg175. [DOI] [PubMed] [Google Scholar]
- 26.Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83:435–445. [PubMed] [Google Scholar]
- 27.Tedder TF, McIntyre G, Schlossman SF. Heterogeneity in the B1 (CD20) cell surface molecule expressed by human B-lymphocytes. Mol Immunol. 1988;25:1321–1330. doi: 10.1016/0161-5890(88)90047-8. [DOI] [PubMed] [Google Scholar]
- 28.Tedder TF, Schlossman SF. Phosphorylation of the B1 (CD20) molecule by normal and malignant human B lymphocytes. J Biol Chem. 1988;263:10009–10015. [PubMed] [Google Scholar]
- 29.Deans JP, Li H, Polyak MJ. CD20-mediated apoptosis: signalling through lipid rafts. Immunology. 2002;107:176–182. doi: 10.1046/j.1365-2567.2002.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel RB, Mattes MJ. Intracellular accumulation of the anti-CD20 antibody 1F5 in B-lymphoma cells. Clin Cancer Res. 2002;8:2701–2713. [PubMed] [Google Scholar]
- 31.Cardarelli PM, Quinn M, Buckman D, Fang Y, Colcher D, King DJ, Bebbington C, Yarranton G. Binding to CD20 by anti-B1 antibody or F(ab′)2 is sufficient for induction of apoptosis in B-cell lines. Cancer Immunol Immunother. 2002;51:15–24. doi: 10.1007/s00262-001-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polyak MJ, Deans JP. Alanine-170 and proline-172 are critical determinants for extracellular CD20 epitopes; heterogeneity in the fine specificity of CD20 monoclonal antibodies is defined by additional requirements imposed by both amino acid sequence and quaternary structure. Blood. 2002;99:3256–3262. doi: 10.1182/blood.v99.9.3256. [DOI] [PubMed] [Google Scholar]
- 33.Binder M, Otto F, Mertelsmann R, Veelken H, Trepel M. The epitope recognized by rituximab. Blood. 2006;108:1975–1978. doi: 10.1182/blood-2006-04-014639. [DOI] [PubMed] [Google Scholar]
- 34.Hofmeister JK, Cooney D, Coggeshall KM. Clustered CD20 induced apoptosis: src-family kinase, the proximal regulator of tyrosine phosphorylation, calcium influx, and caspase 3-dependent apoptosis. Blood Cells Mol Dis. 2000;26:133–143. doi: 10.1006/bcmd.2000.0287. [DOI] [PubMed] [Google Scholar]
- 35.Shan D, Ledbetter JA, Press OW. Apoptosis of malignant human B cells by ligation of CD20 with monoclonal antibodies. Blood. 1998;91:1644–1652. [PubMed] [Google Scholar]
- 36.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcγRIIIa gene. Blood. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 37.Ghetie MA, Bright H, Vitetta ES. Homodimers but not monomers of Rituxan (chimeric anti-CD20) induce apoptosis in human B-lymphoma cells and synergize with a chemotherapeutic agent and an immunotoxin. Blood. 2001;97:1392–1398. doi: 10.1182/blood.v97.5.1392. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Khawli LA, Hu P, Epstein AL. Generation of rituximab polymer may cause hyper-cross-linking-induced apoptosis in non-Hodgkin’s lymphomas. Clin Cancer Res. 2005;11:5971–5980. doi: 10.1158/1078-0432.CCR-05-0554. [DOI] [PubMed] [Google Scholar]
- 39.Horning SJ, Younes A, Jain V, Kroll S, Lucas J, Podoloff D, Goris M. Efficacy and safety of tositumomab and iodine-131 tositumomab (Bexxar) in B-cell lymphoma, progressive after rituximab. J Clin Oncol. 2005;23:712–719. doi: 10.1200/JCO.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 40.Wiseman GA, White CA, Witzig TE, Gordon LI, Emmanouilides C, Raubitschek A, Janakiraman N, Gutheil J, Schilder RJ, Spies S, Silverman DH, Grillo-Lopez AJ. Radioimmunotherapy of relapsed non-Hodgkin’s lymphoma with zevalin, a 90Y-labeled anti-CD20 monoclonal antibody. Clin Cancer Res. 1999;5:3281s–3286s. [PubMed] [Google Scholar]
- 41.Kopeček J, Bažilová H. Poly[N-(2-hydroxypropyl)methacrylamide]: 1. Radical polymerization and copolymerization. Eur Polym J. 1973;9:7–14. [Google Scholar]
- 42.Ong GL, Elsamra SE, Goldenberg DM, Mattes MJ. Single-cell cytotoxicity with radiolabeled antibodies. Clin Cancer Res. 2001;7:192–201. [PubMed] [Google Scholar]
- 43.Fowers KD, Callahan J, Byron P, Kopeček J. Preparation of Fab′ from murine IgG2a for thiol reactive conjugation. J Drug Targeting. 2001;9:281–294. doi: 10.3109/10611860108997936. [DOI] [PubMed] [Google Scholar]
- 44.Allen TM, Brandeis E, Hansen CB, Kao GY, Zalipsky S. A new strategy for attachment of antibodies to sterically stabilized liposomes resulting in efficient targeting to cancer cells. Biochim Biophys Acta. 1995;1237:99–108. doi: 10.1016/0005-2736(95)00085-h. [DOI] [PubMed] [Google Scholar]
- 45.Nisonoff A, Pressman D. Heterogeneity and average combining constants of antibodies from individual rabbits. J Immunol. 1958;80:417–428. [PubMed] [Google Scholar]
- 46.Crespo-Biel O, Lim CW, Ravoo BJ, Reinhoudt DN, Huskens J. Expression of a supramolecular complex at a multivalent interface. J Am Chem Soc. 2006;128:17024–17032. doi: 10.1021/ja0637705. [DOI] [PubMed] [Google Scholar]
- 47.Cartlidge SA, Duncan R, Lloyd JB, Kopečková-Rejmanová P, Kopeček J. Soluble, crosslinked N-(2-hydroxypropyl)methacrylamide copolymers as potential drug carriers: 3. Targeting by incorporation of galactosamine residues Effect of route of administration. J Controlled Release. 1987;4:265–278. [Google Scholar]
- 48.Cartlidge SA, Duncan R, Lloyd JB, Kopečková-Rejmanová P, Kopeček J. Soluble crosslinked N-(2-hydroxypropyl)methacrylamide copolymers as potential drug carriers: 2. Effect of molecular weight on blood clearance and body distribution in the rat after intravenous administration Distribution of unfractionated copolymer after intraperitoneal, subcutaneous or oral administration. J Controlled Release. 1987;4:253–264. [Google Scholar]
- 49.Cartlidge SA, Duncan R, Lloyd JB, Rejmanová P, Kopeček J. Soluble crosslinked N-(2-hydroxypropyl)methacrylamide copolymers as potential drug carriers: 1. Pinocytosis by rat visceral yolk sacs and rat intestine cultured in vitro Effect of molecular weight on uptake and intracellular degradation. J Controlled Release. 1986;3:55–56. [Google Scholar]
- 50.Irvine GB. Amino acid analysis. Precolumn derivatization methods. Methods Mol Biol. 1997;64:131–138. doi: 10.1385/0-89603-353-8:131. [DOI] [PubMed] [Google Scholar]
- 51.DeLisi C. The effect of cell size and receptor density on ligand--receptor reaction rate constants. Mol Immunol. 1981;18:507–511. doi: 10.1016/0161-5890(81)90128-0. [DOI] [PubMed] [Google Scholar]
- 52.Bohdanecký M, Bažilová H, Kopeček J. Poly[N-(2-hydroxypropyl)methacrylamide] - II Hydrodynamic properties of dilute solutions. Eur Polym J. 1974;10:405–410. [Google Scholar]
- 53.Frankel ME, Gerhard W. The rapid determination of binding constants for antiviral antibodies by a radioimmunoassay. An analysis of the interaction between hybridoma proteins and influenza virus. Mol Immunol. 1979;16:101–106. doi: 10.1016/0161-5890(79)90051-8. [DOI] [PubMed] [Google Scholar]
- 54.Scatchard G. Equilibrium thermodynamics and biological chemistry. Science. 1942;95:27–32. doi: 10.1126/science.95.2454.27. [DOI] [PubMed] [Google Scholar]
- 55.Dam TK, Roy R, Das SK, Oscarson S, Brewer CF. Binding of multivalent carbohydrates to concanavalin A and Dioclea grandiflora lectin. Thermodynamic analysis of the “multivalency effect”. J Biol Chem. 2000;275:14223–14230. doi: 10.1074/jbc.275.19.14223. [DOI] [PubMed] [Google Scholar]
- 56.Dower SK, Titus JA, Segal DM. Cell surface dynamics: concepts and models. In: Perelson AS, DeLisi C, Wiegel FW, editors. Receptors and ligands in intercellular communication. Vol. 3. Dekker; New York: 1984. p. xvii. [Google Scholar]
- 57.Sips R. On the structure of a catalyst surface. J Chem Phys. 1948;16:490–495. [Google Scholar]
- 58.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. Control of multivalent interactions by binding epitope density. J Am Chem Soc. 2002;124:1615–1619. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
- 59.Mammen M, Dahmann G, Whitesides GM. Effective inhibitors of hemagglutination by influenza virus synthesized from polymers having active ester groups. Insight into mechanism of inhibition. J Med Chem. 1995;38:4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 60.Roy R, Page D, Perez SF, Bencomo VV. Effect of shape, size, and valency of multivalent mannosides on their binding properties to phytohemagglutinins. Glycoconj J. 1998;15:251–263. doi: 10.1023/a:1006945028547. [DOI] [PubMed] [Google Scholar]
- 61.Dam TK, Roy R, Page D, Brewer CF. Negative cooperativity associated with binding of multivalent carbohydrates to lectins. Thermodynamic analysis of the “multivalency effect”. Biochemistry. 2002;41:1351–1358. doi: 10.1021/bi015830j. [DOI] [PubMed] [Google Scholar]
- 62.Shoup D, Szabo A. Role of diffusion in ligand binding to macromolecules and cell-bound receptors. Biophys J. 1982;40:33–39. doi: 10.1016/S0006-3495(82)84455-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alley SC, Benjamin DR, Jeffrey SC, Okeley NM, Meyer DL, Sanderson RJ, Senter PD. Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjugate Chem. 2008;19:759–765. doi: 10.1021/bc7004329. [DOI] [PubMed] [Google Scholar]
- 64.Lu ZR, Shiah JG, Kopečková P, Kopeček J. Polymerizable Fab’ antibody fragment targeted photodynamic cancer therapy in nude mice. STP Pharma Sci. 2003;13:69–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.