Abstract
Although CD1 proteins are known to present mycobacterial lipid antigens to T cells, there is little understanding of the in vivo behavior of T cells restricted by CD1a, CD1b and CD1c, and the relative immunogenicity and immunodominance of individual lipids within the total array of lipids that comprise a bacterium. Because bovines express multiple CD1 proteins and are natural hosts of Mycobacterium bovis and Mycobacterium avium paratuberculosis (MAP), we used them as a new animal model of CD1 function. Here, we report the surprisingly divergent responses against lipids produced by these two pathogens during infection. Despite considerable overlap in lipid content, only three out of 69 animals cross-react with M. bovis and MAP total lipid preparations. The unidentified immunodominant compound of M. bovis is a hydrophilic compound, whereas the immunodominant lipid of MAP is presented by CD1b and was identified as glucose monomycolate (GMM). The preferential recognition of GMM antigen by MAP-infected cattle may be explained by the higher expression of GMM by MAP than by M. bovis. The bacterial species-specific nature of the CD1-restricted, adaptive T-cell response affects the approach to development of lipid based immunodiagnostic tests.
Keywords: Animal models, Bacterial infections, CD1 molecules, Lipid antigens, Mycobacteria
Introduction
Mycobacterial diseases are highly prevalent in humans and cattle. Human tuberculosis, caused by M. tuberculosis, and bovine tuberculosis, caused by Mycobacterium bovis, are widespread. Infections with Mycobacterium avium paratuberculosis (MAP), the causative agent of Johne’s disease or bovine paratuberculosis, cause substantial economic losses in the dairy cattle industry [1]. Effective vaccines against mycobacteria are not available for humans and animals despite large efforts in discovery, production, and testing of improved M. bovis BCG strains and mycobacterial protein antigens.
The protective effect of a vaccine is based on priming of the adaptive immune system and the ensuing immunological memory, which is dependent on the development of T-cell memory. Immunization with protein antigens presented by MHC molecules is a very effective way of generating T-cell memory. However, an important disadvantage of immune responses that are dependent on a single or a few proteins is that immune evasion in the form of single mutations may occur. In addition, in outbred populations, vaccine efficacy of a subunit vaccine can be limited at the individual level by the lack of binding capacity of the antigen to the specific set of MHC alleles present in that individual.
The CD1 family of proteins presents lipids, glycolipids and lipopeptides to T cells. The group 1 CD1 isoforms (CD1a, CD1b, CD1c) are largely conserved in mammalian evolution, suggesting that they have an important natural function in the immune response. Targeting the CD1 system for vaccine development has the advantage that the presented antigens are not gene products and are therefore not subject to rapid mutation. Also, the CD1 system shows very limited polymorphism, minimizing interindividual differences within an outbred population in the capacity to present a certain antigen [2]. In addition, CD1-restricted T cells have been shown to be able to express molecules that are important in immune response to mycobacterial infections, including γ-interferon and granulysin [3, 4]. In fact, guinea pigs, immunized with total lipid extract of M. tuberculosis, developed smaller granulomatous lesions upon challenge with M. tuberculosis [5] than control animals, although the bacterial burden was only slightly reduced.
However, a basic unresolved question is whether CD1-restricted T cells, once activated by either natural infection or vaccination, lead to durable adaptive antigen-specific immune responses. Most evidence suggests that CD1d-restricted NKT cells are rapidly but transiently activated such that repeated exposure to antigen can cause anergy, but does not generally lead to priming or durable expansion in ways that lead to protection at time periods long after the initial antigen exposure. Indeed, most models of CD1d-restricted NKT cells emphasize that these cells are innate lymphocytes that, upon stimulation, have a strong, transient effect that is not subject to vaccination. However, unlike CD1d, the three group 1 CD1 isoforms (CD1a, CD1b, CD1c) show inducible expression on dendritic cells that is regulated by Toll-like receptors, and they present more diverse classes of antigens. Accordingly, T cells restricted by group 1 CD1 proteins may have a more diverse pool of precursors that could be primed to become antigen specific memory cells. Thus, answering the question against which antigens CD1-restricted adaptive T-cell responses develop is a fundamental question to the field and of practical importance for vaccine development.
The known CD1-presented mycobacterial lipid antigens are diverse and include mycobacterial mycolates [6-8], diacylglycerols [9], polyisoprenoid lipids [10], sulfotrehalose-containing lipids [11] and lipopeptides [12]. Of note, all these mycobacterial lipids are presented by group 1 CD1 proteins (CD1a, CD1b and CD1c). Because mice do not have orthologs of group 1 CD1 molecules [13] and other non-human models are limited, there have been few studies of the natural functions, targets or frequency of the responding T cells in vivo. Cattle express CD1a, CD1e and multiple CD1b molecules, but lack CD1c and CD1d [14, 15], and this, in combination with a predisposition to mycobacterial infection, allows study of the strength, antigen specificity and duration of mycobacteria-specific T-cell responses to lipids. Because of its close resemblance to human tuberculosis, bovine tuberculosis is an excellent model for human tuberculosis [16].
The in vitro generation of T-cell lines has been extremely useful for identifying CD1-presented antigens but has real limitations for understanding the role of natural antigens during infection in vivo and the phenotype and functions of the T cells that recognize these antigens in vivo. For example, many lipid-reactive T cells have been enriched by selection for cells that lack CD4 and CD8 or by culturing them with CD1-expressing antigen-presenting cells derived from MHC mismatched donors. Such manipulations may cause the observations to represent relatively rare T cells or identify lipid antigens of subdominant antigenicity. Also, many long-term T-cell clones show a drift toward a Th1 phenotype and so may not accurately reflect functions that normally occur in vivo. Data obtained from non-manipulated material obtained from in vivo models of mycobacterial infections can provide valuable additional insights into antigen specificity and phenotype of T cells in vivo. Because non-human models that express orthologs of CD1a, CD1b or CD1c are currently limited to guinea pigs [5, 17], most ex vivo data are obtained in humans. A limited number of studies describe lipid-reactive T cells expanded in the acute phase of human infection, using total lipid extracts or antigens that had previously been identified using T-cell clones [10, 11, 18]. From among the various mycobacterial non-protein antigens known to date, the particular antigens that dominate the response against a certain mycobacterial species or strain are unknown. However, identifying the antigens and T cells that generate the strongest adaptive responses are the key pieces of information necessary to use CD1 technology to develop diagnostic tests and vaccines.
in vitro, CD1-restricted T cells have been shown to precisely discriminate detailed structures of mycobacterial lipids such as mycolic acids, glucose monomycolates (GMM), sulfotrehaloses, dideoxymycobactins and mycoketides. Because these lipids vary naturally among pathogenic and environmental strains of mycobacteria, these results suggest that lipid-reactive T cells might respond differently to individual species of mycobacteria. For example, GMM is made using glucose derived from the host and is not known to be made by mycobacteria growing in the environment, leading to the hypothesis that this antigen is a marker that distinguishes productive infections from environmental growth [8]. An example of a mycobacterial species-specific lipopeptide is the MAP-specific lipopentapeptide L5P [19]. Differing species of mycobacteria also share many classes of lipids with a common basic design and even though some of these have species-specific molecular characteristics, cross-reactive T cells are known to exist [20] These considerations suggest that mixtures of mycobacterial lipids could be used to distinguish mycobacterial infections from infection by non-mycobacterial species, but species-specific immunodiagnosis requires further work to define the immunodominant antigens within a given species.
To provide evidence for priming of adaptive, CD1-restricted T cells during natural infection and to identify immunodominant antigens, we describe the ex vivo T-cell responses against lipids during bovine tuberculosis, caused by acute infection with M. bovis, and paratuberculosis, caused by chronic infection with MAP. We found significantly elevated immune responses against lipids among animals naturally infected with either M. bovis or MAP as compared with controls, providing evidence for antigen specific T-cell responses, primed by in vivo infection. Further, we found that these two mycobacterial species cause mycobacterial species-specific T-cell responses against lipids, and showed that it is possible to determine immunodominance of lipids in freshly isolated T cells.
Results
M. bovis and MAP infection generate strong antilipid T-cell responses in vivo
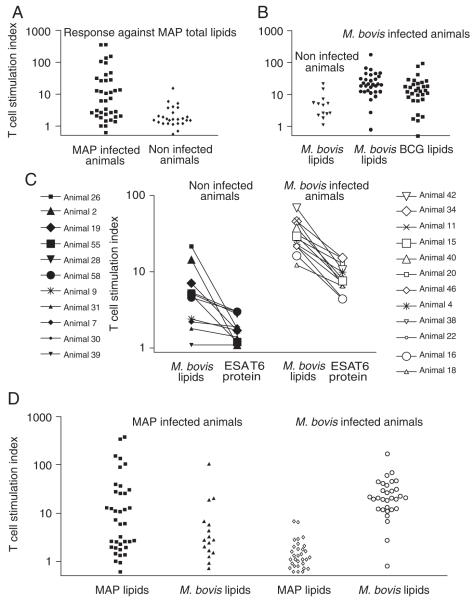
Adult animals suffering from a fecal culture-confirmed infection with MAP, resulting from natural oral exposure (n = 38) and MAP-culture-negative controls or animals obtained from farms that have not resulted in any positive MAP cultures during the last 5 years (called “MAP negative controls”, n = 26) were tested for T-cell reactivity against MAP total lipid extract from the strain MAP 280105. In MAP-infected animals, the stimulation index, defined as the proliferative response obtained with antigenic stimulation divided by the response obtained with incubation with medium alone, was significantly higher (median 6.9) against total extracts from MAP compared with MAP-negative controls (median 1.6; p = 0.01) (Fig. 1A). Similarly, M. bovis-infected animals (n = 31) show significantly higher responses to M. bovis 4913C total lipid extract (median 20.3) than bovine purified protein derivative (PPD) skin test-negative controls (n = 13; median 4.6; p = 0.001) (Fig. 1B). To compare the strength of the antilipid response with an antiprotein response, we also performed stimulations with ESAT6 protein, an immunogenic protein expressed by members of the M. tuberculosis complex, including M. bovis. M. bovis-infected animals (n = 12) show a significantly higher response to ESAT6 (median 7.9) than bovine PPD skin test-negative controls (n = 11; median 1.8; p = 0.00003) (Fig. 1C). The strength of the anti-ESAT6 response was in all cases lower than the antilipid response. Of note, two skin test-negative animals with a relatively high response against M. bovis total lipid extract (animal 25 and animal 2) do not respond to ESAT6. We conclude that natural infection of cattle stimulates a robust and durable immune response to lipid extracts, which is stronger but less specific than the response against the M. bovis protein ESAT6.
Figure 1.
T-cell responses of MAP- and M. bovis-infected and non-infected animals to total lipid extracts. (A) Freshly isolated PBMC of MAP-infected animals or age-matched controls from MAP-free farms were stimulated with MAP total lipid extract and with M. bovis total lipid extract. (B) M. bovis-infected cattle and buffalo and non-infected controls from the same herd were stimulated with M. bovis total lipid extract. (C) M. bovis-infected buffalo and non-infected controls from the same herd were stimulated with M. bovis total lipid extract and recombinant, pure ESAT6 protein. Each symbol represents one individual animal. (D) Cross reactivity between lipid extracts was determined by stimulating PBMC of singly infected animals with total lipid extracts of MAP and M. bovis. The T-cell stimulation index was calculated as described in the Materials and methods section.
Low cross-reactivity of T-cell responses against lipids from M. bovis and MAP
It is known that M. tuberculosis and M. leprae have lipids in common that generate cross-reactive T-cell responses in humans [20]. We studied whether the same is true for M. bovis and MAP in cattle, or whether mycobacterial species-specific lipids are targets of the T-cell response. M. bovis-infected animals were tested for reactivity against MAP lipids, and MAP-infected animals were tested for reactivity against M. bovis lipids (Fig. 1D). Of the 16 MAP-infected animals tested, only three (7.8%) responded with a stimulation index of 10 or higher to M. bovis lipids. None of the 31 M. bovis-infected animals responded with a stimulation index of greater than 10 to MAP lipids. The low cross-reactivity prompted a search for specific lipid antigens of MAP and M. bovis.
Along with the tests for cross-species reactivity, we determined whether the response was specific for the mycobacterial strain used and for any possible effects of the medium in which it was grown. First, we tested whether animals with reactivity against M. bovis 4913C lipids also responded to lipids from the related vaccine strain, M. bovis BCG. M. bovis-infected animals showed an increased response to M. bovis BCG lipids (p<0.001) with response rates similar to that of WT M. bovis (Fig. 1B), although the stimulation index against M. bovis lipids was higher (median 20.3) than against BCG lipids (median 13.2). Stimulation with lipid extracts of M. bovis 4913C grown in 7H9 supplemented with oliec acid dextrose complex (OADC) or grown in 7H9 supplemented with 1% d-glucose or M. bovis ATCC 19210 grown in 7H9 supplemented with OADC or grown in 7H9 supplemented with 1% d-glucose resulted in highly comparable T-cell responses in M. bovis-infected animals (data not shown). In addition, we compared lipid extracts of MAP strain ATCC 19698 grown in 7H9 medium supplemented with OADC and mycobactin J (MycJ) and MAP strain 280105 grown in Watson–Reid medium supplemented with MycJ, which resulted in highly comparable T-cell responses in MAP-infected animals (data not shown).
To determine the value of lipid-based T-cell assays as a diagnostic tool, we compared our results with the reference diagnostics, consisting of skin testing for M. bovis infections and culture-based protocols for MAP infections, as described in the Materials and methods. If a stimulation index of 10 or higher was considered a positive response to lipids, the sensitivity and specificity of this were calculated to be 87 and 85% for M. bovis-infected animals tested for reactivity against M. bovis lipids, and 47 and 96% for MAP-infected animals tested for reactivity against MAP lipids. The sensitivity of the T-cell assay was calculated as the percentage of animals with a T-cell stimulation index of 10 or higher of the total number of animals with a positive test result, using the reference diagnostics. Specificity refers to the percentage of animals with a T-cell stimulation index lower than 10 of the total number of negative animals using the reference diagnostics.
Differing immunodominant substances in MAP and M. bovis total lipid extracts
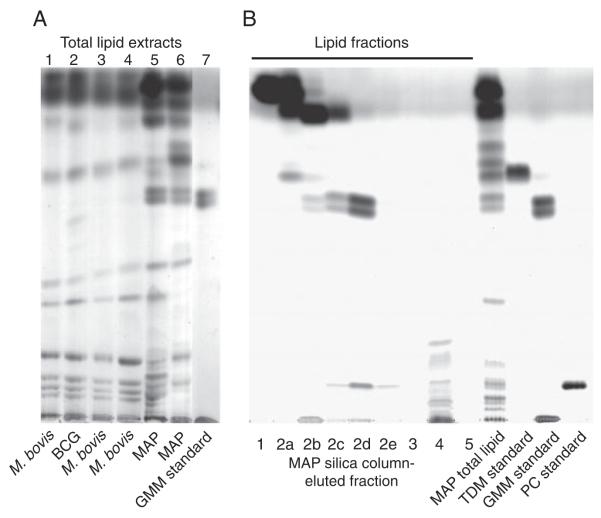
Thin layer chromatography (TLC) analyses performed on lipid extracts of several MAP, BCG and M. bovis strains cultured on independent occasions and in different media revealed that the overall composition of the total lipid extracts of each bacterial species was similar and reproducible (Fig. 2A). Of note, much more GMM, resolving as a doublet, was present in MAP. It is known that for the production of GMM, glucose needs to be present in the culture medium. d-glucose was present in all media that we used for mycobacterial culture in concentrations between 0.2% (7H9 supplemented with OADC) and 1.2% w/v (Watson-Reid supplemented with OADC). Lipid extracts of MAP consistently contained more GMM than M. bovis and M. bovis BCG, independently from the type of medium and the glucose concentration present in the medium (Fig. 2A).
Figure 2.
TLC analysis of lipid extracts. (A) An aliquot of 10 μg of total lipids, extracted from bacterial cultures by chloroform/methanol extraction, was applied to a silica TLC plate and developed as described in the Materials and methods section. Lane 1: M. bovis ATCC 19210 grown in 7H9/OADC; lane 2: BCG Pasteur grown in 7H9/OADC; lane 3: M. bovis 4913C grown in 7H9/OADC; lane 4: M. bovis ATCC 19210 grown in 7H9/D-glucose; lane 5: MAP ATCC 19698 grown in 7H9/OADC/MycJ; lane 6: MAP 280105, grown in Watson–Reid medium/MycJ; lane 7: purified M. phlei-derived GMM. Additional lanes between lanes 6 and 7 that were present on the original TLC plate but not relevant for this figure were electronically removed. (B) An aliquot of 4 mg of MAP total lipids was loaded on a silica column and eluted with increasingly hydrophilic eluents (fraction 1: chloroform; fraction 2a–e: chloroform:acetone mixtures in ratios of 9:1, 8:2, 7:3, 6:4 and 5:5 v:v; fraction 3: acetone; fraction 4: methanol; fraction 5: water). A fixed percentage of each fraction was loaded on a TLC plate and developed and visualized. Trehalose dimycolate (TDM), GMM and phosphatidylcholine (PC) were used as standards.
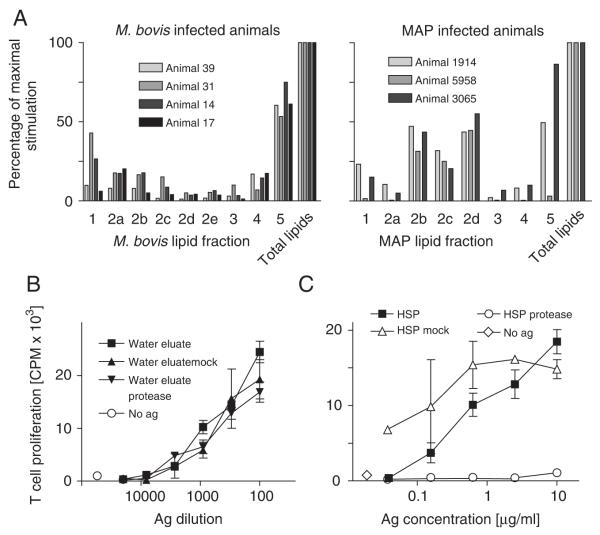
To determine which constituents of the MAP and M. bovis total lipid extract were responsible for the observed T-cell stimulation, total lipid extracts were fractionated on open silica columns using chloroform, mixtures of chloroform and acetone, acetone, methanol and water as eluents. A TLC analysis of the MAP fractions confirmed the expected separation of compounds into distinct classes (Fig. 2B). Infected animals were tested for T-cell reactivity against each fraction. Of the M. bovis lipid fractions, we noted that each of four animals tested showed similar profiles of reactivity and that the water eluate was the only fraction to which T cells consistently reacted at high levels (Fig. 3A, left panel). Each of three cattle infected with MAP tested showed a similar pattern of reactivity to fractions, and this pattern was different from that observed in M. bovis-infected cattle. The immunodominant fractions of MAP were more hydrophobic compounds eluting from the silica column in chloroform:acetone mixtures in ratios of 8:2, 7:3, and 6:4 v:v (Fig. 3A, right panel). Because TLC analysis had shown that each of these stimulatory fractions was highly enriched for the doublet corresponding to GMM (Fig. 2B), animals that responded to them were subsequently tested for reactivity against pure GMM standard purified from M. phlei (Fig. 4A, left panel). The purity and identity of this GMM standard was confirmed by mass spectrometry (Supporting Information Fig. 1b, lower panel). Cross reactivity of T cells against GMM from different sources is expected because when mycolic acid derivatives are presented by CD1 to T cells, the length variation, as well as the variable meromycolate modifications, are buried in the CD1 molecule and not directly recognized by the T-cell [7, 8, 21]. T cells from each of the animals that recognized GMM-containing MAP fractions also recognized the GMM standard, and the GMM standard is as potent as the MAP fraction that contains GMM. From this we conclude that GMM is an immunodominant lipid of MAP. M. phlei GMM was not recognized by M. bovis-infected animals, indicating that the development of this response in vivo is specific for the species of the pathogen (Fig. 4A, right panel).
Figure 3.
T-cell responses against subfractions of MAP and M. bovis lipid extracts. (A) M. bovis (left panel) and MAP (right panel) infected animals that respond to total lipid extracts were tested for reactivity against fractions eluted from a silica column loaded with total lipid extract. The following eluents were used: fraction 1: chloroform; fraction 2a–e: chloroform:acetone mixtures in ratios of 9:1, 8:2, 7:3, 6:4 and 5:5 v:v; fraction 3: acetone; fraction 4: methanol; fraction 5: water. [3H]thymidine incorporation by freshly isolated PBMC was measured to determine T-cell proliferation. (B) Protease treatment of MAP water eluate did not diminish its antigenicity. (C) A control protein antigen, HSP70, that was protease-treated in parallel with the water eluate, was effectively destroyed as shown by its greatly diminished T-cell stimulatory effect on T cells from healthy animals that had been immunized with this protein.
Figure 4.
Pure GMM is recognized by MAP-infected animals and presented by CD1b. (A) The GMM-containing mixture of compounds eluting from a MAP lipid-loaded silica column with a 7:3 v:v chloroform:acetone mixture, and pure GMM from M. phlei were used to stimulate freshly isolated PBMC of MAP (left panel) and M. bovis (right panel) infected animals. [3H]thymidine incorporation was measured after 3 days of incubation to determine T-cell proliferation. Representative data of two animals are shown. (B) Reactivity against the 7:3 v:v chloroform:acetone mixture-eluted compounds could be blocked with the BCD1b3.1 antibody that recognizes bovine CD1b, but not with and isotype control antibody.
Although GMM-containing fractions produced the most potent response, two of the three MAP-infected animals also responded to compounds present in the water eluent from the silica column.
Bovine T-cell responses to GMM are CD1b-restricted
The only known mechanism by which T cells respond to GMM is via presentation by CD1 proteins. In humans, GMM is presented to T cells by CD1b, and only this isoform is known to have a groove large enough to accommodate the large (C80) mycolic acids. Cattle express three CD1b molecules: CD1b1, CD1b3 and CD1b5. Therefore, we hypothesized that this response might be dependent on one or more of the bovine CD1b orthologs. The BCD1b3.1 monoclonal antibody developed against human CD1b and is considered to be a broadly cross-reactive anti-CD1b antibody because it recognizes guinea pig CD1b [17] and bovine CD1b3 [14]. This antibody but not an isotype control antibody blocked the response to GMM, implicating bovine CD1b as necessary for the response (Fig. 4B). Similarly, reactivity against the MAP fractions eluting in chloroform:acetone 8:2, 7:3 and 6:4v:v from the silica column could be blocked with the BCD1b3.1 antibody.
Partial analysis of the silica column water eluates
TLC analysis of the water eluents from silica columns of M. bovis and MAP lipid extracts revealed no spots other than some material at the origin. Weak staining at the origin was obtained with the general cupric acetate staining and with ninhydrin staining for amide groups, but not with the phosphate-specific Dittmer reagent. Because the water eluate represents the most hydrophilic fraction of the total lipid extract we considered the possibility that these fractions contain contaminating bacterial proteins. To test this, a bicinchoninic acid protein assay was performed on the samples. The standard curve was generated using serial dilutions of BSA and the detection limit of the assay was determined to be 3.5 μg/mL. Using this method, we measured absorbances corresponding with 13, 60, 86 and 40 μg/mL protein in the undiluted water eluates of MAP 280105, MAP ATCC 19698, M. bovis AN5 and M. bovis 4913C, respectively. Because this type of protein assay is not equally sensitive for all amino acids, and because lipids, biogenic amines and other substances are known to produce positive signal as well, we also performed an HPLC-based analysis of the total amino acid content after acid hydrolysis (Table 1). These data show that the maximum protein content was 27, 20 and 11 μg/mL in the undiluted water eluates of MAP ATCC 19698, M. bovis AN5 and M. bovis 4913C lipids, respectively. Of note, the two different M. bovis water eluates contained only a partly over-lapping amino acid content.
Table 1.
Amino acid content water eluates of total lipid extract-loaded silica columns
| Silica column water eluate from lipids from: | |||
|---|---|---|---|
| Amino acid | |||
| MAP ATCC 19698 |
M. bovis AN5 |
M. bovis 4913C |
|
| Amino acid concentration (μM) | |||
| Asn+Asp | 35 | 22 | 7 |
| Glu+Gln | 77 | 28 | 14 |
| Cys | 2 | 4 | 8 |
| Ser | 12 | 20 | 8 |
| His | 4 | 3 | 3 |
| Gly | 24 | 40 | 15 |
| Thr | 6 | 0 | 0 |
| Arg | 2 | 0 | 2 |
| Ala | 14 | 23 | 6 |
| Tau | 4 | 3 | 37 |
| Tyr | 1 | 1 | 0 |
| Val | 5 | 15 | 0 |
| Phe | 3 | 6 | 1 |
| Ile | 3 | 7 | 4 |
| Leu | 8 | 18 | 4 |
| Orn | 4 | 0 | 0 |
| Lys | 37 | 17 | 2 |
| Protein (μg/mL)a) | 27 | 20 | 11 |
Protein content was calculated based on the MW of the residuals of the determined set of amino acids, excluding Tau and Orn, on a μg/mL basis.
Positive-mode and negative-mode mass spectra were collected using nanoelectrospray ionization mass spectrometry (Supporting Information Fig. 1). In all T-cell assays performed on M. bovis-infected animals, the response to the water-eluted compounds of the lipid extracts of two different M. bovis strains was alwayscomparable and the same was true for the water-eluted compounds of total lipid preparations of two different MAP strains when used to stimulate T cells from MAP-infected animals. Therefore, we hypothesized that the chemical nature of the antigenic substance in the water-eluted fractions derived from different strains of one mycobacterial species is likely to be identical. For this reason we scanned the mass spectra for peaks shared between preparations derived from one mycobacterial species, but we could not identify any shared ions.
To test whether the strong T-cell responses against the silica column water eluates were dependent on proteins, we performed experiments using proteases. When the water-eluting compounds from M. bovis or MAP were pretreated with either of two broad spectrum proteases, proteinase K and pronase, they retained their ability to stimulate T cells as the same level as mock-treated samples (Fig. 3B). In contrast, pronase and proteinase K greatly reduced or abolished recognition of the protein antigen HSP70 by T cells of animals that had been vaccinated with this antigen (Fig. 3C). Thus, the antigenicity of the water eluates from silica columns loaded with M. bovis and MAP lipids was not dependent on proteins.
Discussion
Almost all of the reported ex vivo, polyclonal T-cell responses are directed against proteins or peptides. Here, we show that mycobacterial lipids are strongly recognized by ex vivo T cells of bovines that suffer from infections with MAP or M. bovis. Even though mycobacterial lipid extracts of different species share many lipid compounds and are known to be able to induce cross-reactive T-cell responses when responses against M. tuberculosis and M. leprae are compared [20], we found little cross reactivity between the lipid extracts of MAP and M. bovis in infected cattle. Reactivity to the lipid preparations was exclusively detected in infected animals, showing that the observed results reflected adaptive, in vivo primed responses, and not some general immunostimulatory effect of the lipids leading to an innate-like immune response. Although slightly weaker, reactivity against BCG lipids was detected in the same animals in which reactivity against M. bovis lipids was detected. Similar responses were expected because of the close genetic relationship between M. bovis and BCG and this suggests that future efforts to generate species-specific immunodiagnostic reactions for such closely related organisms will rely on identifying species specific lipids, such as trehalose sulfoglycolipids, rather than administering more complex mixtures of lipids.
The chemical nature of the immunostimulatory compound(s) present in the highly immunogenic silica column water eluates of the total lipids could not be determined. Mass spectrometric analysis did not lead us to clear candidate antigenic compounds in the water eluates. It is possible that the antigenic compounds are difficult to detect by TLC and mass spectrometry and have therefore escaped detection. It is unlikely that proteins are responsible for the observed responses against the water eluates because protease digestion did not diminish the immunogenicity of the water eluates. Also, based on direct measurement of the protein and amino acid content of the undiluted water eluates, we calculated that the dilutions of the water eluates that effectively stimulated T cells in our in vitro assays contained 10–50 ng/mL protein, which is about 100-fold below the usually reported optimal concentration of a protein antigen. Last, the methods used for the extraction of total lipids, of which the water eluate is a subfraction, is known to lead to low protein contamination [5].
In animals infected with MAP we identified GMM as an immunodominant lipid. Even though M. bovis is known to produce GMM as well, animals infected with M. bovis did not recognize GMM; so it seems that in vivo, M. bovis has less GMM response-priming capacity than MAP. TLC analysis of the total lipid extracts of our bacterial cultures showed that in vitro MAP produces much more GMM than M. bovis, and that these differences could not be related to the culture medium used. It is likely that similar quantitative differences in GMM content in vivo play a role in the differential capacities of these two mycobacterial species to prime an anti-GMM T-cell response in vivo. Recent studies show that GMM is produced by the action of a mycolate transferase known as antigen 85A, suggesting that this gene and other potentially redundant mycolyl transferases are now candidates for controlling immunogenicity in vivo [22].
The immunogenicity of GMM in humans has been previously demonstrated in a T-cell line derived from a skin biopsy of a leprosy patient after extensive in vitro culture. GMM has been shown to be recognized weakly by non-manipulated ex vivo T cells of tuberculosis patients [18], also in a CD1-restricted fashion. The data presented here show that also in naturally infected cattle, GMM is immunogenic and presented by CD1b. From this we conclude that the previously reported presentation of GMM to the LDN5 cell line is not a rare event, but rather a phenomenon that takes place in several mycobacterial infections in at least two different host species. However, the quantity of GMM produced by the infecting mycobacterium may be crucial for effective CD1 presentation of GMM and CD1-restricted T-cell activation, as suggested by the lack of GMM reactivity by M. bovis-infected animals.
Our work shows that the identification of potential lipid or other non-protein subunit vaccine candidates in bovines is possible by in vitro stimulation of freshly isolated PBMC. The in vitro use of T cells that have been primed in vivo by a natural infection provides insights into immunogenicity and immunodominance of substances that have not yet received as much attention as protein antigens.
Materials and methods
Bacteria
MAP clinical isolate 280105, isolated in The Netherlands, and ATCC strain 19698 were grown in open liquid cultures using Watson–Reid medium (containing 1% d-glucose) supplemented with 0.5 μg/mL MycJ (Allied Monitor, Fayette, MO, USA), or Middlebrook 7H9 medium (Difco) supplemented with Middleb-rook OADC (BD BBL) and 0.5 μg/mL MycJ. M. bovis clinical isolate 4913C (isolated from cattle in South Africa), ATCC strain 19210, the strain AN5 and M. bovis BCG Pasteur were grown in 7H9 medium with OADC or in 7H9 medium with 1% d-glucose. M. phlei was grown in 7H9 medium supplemented with 0.5 mg/mL Tween-80 and 1% d-glucose. 7H9 medium does not contain glucose, but supplementation with OADC as we did leads to a final concentration of 0.2% d-glucose; thus, all bacteria used had been grown in the presence of glucose. The lipid content of the different strains grown in the different media was compared to assess the variation between strains and the effect of different media (Fig. 2A).
Antigen preparation
Bacterial cultures were subjected to centrifugation for 20 min at 4000 g and the pellet was washed once with distilled water. The wet pellets were extracted for 2 h at room temperature in chloroform:methanol 1:2 v:v and 2:1 v:v consecutively. The total lipid extracts were dried in a rotating evaporator at room temperature and re-dissolved in chloroform and quantification and analysis was done by TLC. Fractionation of the MAP total lipid extracts was performed by loading on a silica solid phase extraction column with a bed weight of 2 g (Supelco) and consecutive elution with three bed volumes (12 mL) of each of the following eluents: chloroform, 9:1, 8:2, 7:3, 6:4, 5:5 v:v mixtures of chloroform and acetone, acetone, methanol and then water. The GMM standard was isolated from M. phlei by preparative TLC. Preparative TLC plates (200 μm) were resolved in 60:30:6 v:v chloroform:methanol:water, dried and sprayed with water to visualize bands. Individual bands were scraped and silica dust was extracted in 2:1 v:v chloroform:methanol. The identity and purity of GMM was confirmed as described in the “lipid analysis” section below.
HSP70 was prepared as previously described [23]. Pronase is a mixture of endopeptidases and exopeptidases (carboxy-peptidases and aminopeptidases) that cleave denatured and native proteins down into individual amino acids. Proteinase K is an endopeptidase that has a preference for cleavage between an aliphatic, aromatic or hydrophobic and any other amino acid; however, it will digest any peptidic bond if added in excess and over long incubation periods. Pronase and proteinase K were used to digest water eluates from silica columns loaded with MAP and M. bovis lipids, and HSP70 in protease buffer (10 mM CaCl, 10 mM HEPES buffer, 25 mM ammoniumbicarbonate) for 4 h at 40°C, followed by 10 min of inactivation at 851C. Mock treatment of antigens was performed in the same buffer and at the same temperatures, but without addition of the proteases.
Recombinant ESAT6 protein was described previously [24].
Lipid analysis
For analytical TLC, approximately 25 μg of lipids were loaded on a 20×20 cm Silica 60 TLC plate (Merck) and developed in 60:16:2 v:v:v chloroform:methanol:water. Plates were sprayed with 3% w/v cupric acetate in 8% v/v phosphoric acid, dried and baked for 1 h at 140°C. Alternatively, plates were sprayed with 0.2% w/v ninhydrin and 3% v/v acetic acid in water-saturated butanol for detection of free amino groups, or with Dittmer–Lester reagent for the detection of phosphates. Total amino acid content was analyzed by HPLC (Jasco) after acid hydrolysis in 6 M HCl for 16 h and pre-column ortho-phthalaldehyde derivatization. Nanoelectrospray ionization mass spectrometry (ThermoFinnigan LCQ Advantage) was performed on fractions dissolved in a 1:1 v:v water:methanol mixture and on purified GMM in a 1:2 v:v chloroform:methanol mixture using borosilicate glass pipettes pulled to a final orifice of 1–2 μm and an internal stainless steel electrode.
Animals
Adult Holstein–Frisian cattle infected with MAP were obtained from dairy cattle farms in The Netherlands with a known history of MAP infections. Diagnosis of MAP infection was performed using a fecal culture system [25] at the National Veterinary Health Service, Deventer, The Netherlands. Samples were checked for bacterial growth every 4 wk and considered negative if after a culture period of 16 wk no bacterial growth was observed. Bacteria were confirmed to be MAP based on mycobactin dependence of the culture and the confirmation of the presence of the MAP-specific IS900 insertion sequence by PCR [26]. This culture-based protocol is not highly sensitive at the level of an individual animal, but highly specific, allowing confirmed MAP culture-positive animals into the study. Because all animals included in our study were more than 2 years of age, and because infection with MAP takes place in very young calves [1], all MAP-infected animals used can be considered chronically infected. Control animals originated from farms with unknown MAP status if they were confirmed to be MAP culture-negative, or from Dutch dairy farms, which have participated in a national paratuberculosis control program for at least 5 years during which no test-positive animals were found. On these farms all animals older than 2 years of age are checked for paratuberculosis infection on a yearly basis by the National Veterinary Health Service using an ELISA (Pourquier) and pooled fecal culture.
All animals from The Netherlands were considered tuberculosis-free as bovine tuberculosis is not endemic in The Netherlands. Tuberculosis-infected dairy cattle were collected during natural outbreaks in Belgium and in South Africa (Afrikaner, Jersey, Nguni). In addition, two bovine tuberculosis-infected free-ranging African buffalo herds were sampled in the Hluhluwe-iMfolozi Park in South Africa. The diagnosis of bovine tuberculosis was determined at the level of the individual animal by single or comparative skin testing using bovine and avian PPD. Animals were considered positive when the induration caused by bovine PPD was at least 2 mm bigger than the induration caused by avian PPD, or at least 6 mm if the single tuberculin test was used. This skin test, based on the same principles as the Mantoux test for human tuberculosis, depends on adaptive, mycobacterial protein-specific T-cell responses, and turns positive in bovines 3 or 4 wk after infection with M. bovis [27, 28]. In addition, the diagnosis of bovine tuberculosis was confirmed at herd level by post-mortem examination and M. bovis culture from visible lesions from at least one animal from the herd.
For some control experiments, we used healthy animals that had been immunized with recombinant HSP70 protein in dimethyl dioctadecyl ammoniumbnomide (DDA), as described earlier [29]. In all cases 20–100 mL of heparinized blood was drawn from the jugular vein. Experiments were conducted according to the regulations of the Veterinary and Agrochemical Research Centre CODA CERVA, KwaZulu Natal parks, and approved by the Animal Ethical Committee of the University of Utrecht, The Netherlands (protocol number 2007.II.06.152/Vervolg1).
T-cell assays
PBMC were isolated by standard Ficoll–Hypaque gradient centrifugation of heparinized blood that was drawn from the jugular vein. For biological testing lipids were evaporated to dryness under nitrogen and sonicated with T-cell medium. T-cell medium was made by supplementing 500 mL of RPMI medium with 50 mL fetal calf serum (Hyclone), penicillin (Gibco), streptomycin (Gibco), 20 mM HEPES (Gibco) and 4 mL 1 N NaOH solution. T-cell activation was measured by incubating 2×105 PBMC with antigens in series of dilutions. Proliferation was measured after coculture for 3 days with antigen, followed by a 7 h pulse of 1 μCi of [3H]thymidine before harvesting and counting β emissions. Stimulation indices were calculated by dividing the number of counts per minute obtained with the optimal antigen dilution by the number obtained with incubation with medium alone. Anti-human CD1b (BCD1b3.1), which is known to cross react with bovine CD1b3 [14], and an IgG1 isotype control (P3), were used for blocking studies at 20 μg/mL.
Statistical analyses
Logarithmic transformation of stimulation indices of T-cell proliferation assays was performed to achieve homogeneity of variance and to better approximate a normal distribution. To test the differences in T-cell reactivity to total lipid extracts between infected and non-infected groups of animals, a two-sided, non-paired t-test was used.
Supplementary Material
Acknowledgements
This work was supported by the Wellcome Trust (GR078283 to I.V.R.), the NIH (R01 AI071155 and AI049313 to D.B.M.) and a Burroughs Wellcome Fund Translational Investigator Award to D.B.M. The authors would like to thank Claus Aagaard, Statens Serum Institut, Copenhagen, Denmark, for providing ESAT6, Karl Walravens, Kristin Kremer, and Tiny Hlokwe for providing mycobacterial cultures, Paul van Haard for amino acid analysis, and the Belgian Food Safety Agency to facilitate blood sampling.
Abbreviations
- GMM
glucose monomycolate
- MAP
Mycobacterium avium paratuberculosis
- MycJ
Mycobactin J
- TLC
thin layer chromatography
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Supporting Information for this article is available at www.wiley-vch.de/contents/jc_2040/2009/39619_s.pdf
References
- 1.Harris NB, Barletta RG. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin. Microbiol. Rev. 2001;14:489–512. doi: 10.1128/CMR.14.3.489-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han M, Hannick LI, DiBrino M, Robinson MA. Polymorphism of human CD1 genes. Tissue Antigens. 1999;54:122–127. doi: 10.1034/j.1399-0039.1999.540202.x. [DOI] [PubMed] [Google Scholar]
- 3.Stenger S, Mazzaccaro RJ, Uyemura K, Cho S, Barnes PF, Rosat JP, Sette A, et al. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 4.Stenger S, Hanson DA, Teitelbaum R, Dewan P, Niazi KR, Froelich CJ, Ganz T, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 5.Dascher CC, Hiromatsu K, Xiong X, Morehouse C, Watts G, Liu G, McMurray DN, et al. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int. Immunol. 2003;15:915–925. doi: 10.1093/intimm/dxg091. [DOI] [PubMed] [Google Scholar]
- 6.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 7.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 8.Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, Porcelli SA. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J. Exp. Med. 2000;192:965–976. doi: 10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 10.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 11.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted t cells during infection with Mycobacterium tuberculosis. J. Exp. Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 13.Dascher CC, Brenner MB. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 2003;24:412–418. doi: 10.1016/s1471-4906(03)00179-0. [DOI] [PubMed] [Google Scholar]
- 14.Van Rhijn I, Koets AP, Im JS, Piebes D, Reddington F, Besra GS, Porcelli SA, et al. The bovine CD1 family contains group 1 CD1 proteins, but no functional CD1d. J. Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 15.Looringh van Beeck FA, Reinink P, Hermsen R, Zajonc DM, Laven MJ, Fun A, Troskie M, Schoemaker NJ, et al. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Mol. Immunol. 2009;46:1424–1431. doi: 10.1016/j.molimm.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Rhijn I, Godfroid J, Michel A, Rutten V. Bovine tuberculosis as a model for human tuberculosis: advantages over small animal models. Microbes Infect. 2008;10:711–715. doi: 10.1016/j.micinf.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Dascher CC, Hiromatsu K, Naylor JW, Brauer PP, Brown KA, Storey JR, Behar SM, et al. Conservation of a CD1 multigene family in the guinea pig. J. Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- 18.Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA. T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect. Immun. 2003;71:3076–3087. doi: 10.1128/IAI.71.6.3076-3087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biet F, Bay S, Thibault VC, Euphrasie D, Grayon M, Ganneau C, Lanotte P, et al. Lipopentapeptide induces a strong host humoral response and distinguishes Mycobacterium avium subsp. paratuberculosis from M. avium subsp. avium. Vaccine. 2008;26:257–268. doi: 10.1016/j.vaccine.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 20.Sieling PA, Torrelles JB, Stenger S, Chung W, Burdick AE, Rea TH, Brennan PJ, et al. The human CD1-restricted T cell repertoire is limited to cross-reactive antigens: implications for host responses against immunologically related pathogens. J. Immunol. 2005;174:2637–2644. doi: 10.4049/jimmunol.174.5.2637. [DOI] [PubMed] [Google Scholar]
- 21.Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat. Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 22.Matsunaga I, Naka T, Talekar RS, McConnell MJ, Katoh K, Nakao H, Otsuka A, et al. Mycolyltransferase-mediated glycolipid exchange in Mycobacteria. J. Biol. Chem. 2008;283:28835–28841. doi: 10.1074/jbc.M805776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koets AP, Rutten VP, de Boer M, Bakker D, Valentin-Weigand P, van Eden W. Differential changes in heat shock protein-, lipoarabinomannan-, and purified protein derivative-specific immunoglobulin G1 and G2 isotype responses during bovine Mycobacterium avium subsp. paratuberculosis infection. Infect. Immun. 2001;69:1492–1498. doi: 10.1128/IAI.69.3.1492-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aggerbeck H, Madsen SM. Safety of ESAT-6. Tuberculosis (Edinb) 2006;86:363–373. doi: 10.1016/j.tube.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen JB. An improved medium for culture of Mycobacterium paratuberculosis from bovine faeces. Acta Vet. Scand. 1982;23:325–335. doi: 10.1186/BF03546784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vary PH, Andersen PR, Green E, Hermon-Taylor J, McFadden JJ. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne’s disease. J. Clin. Microbiol. 1990;28:933–937. doi: 10.1128/jcm.28.5.933-937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thom ML, Hope JC, McAulay M, Villarreal-Ramos B, Coffey TJ, Stephens S, Vordermeier HM, Howard CJ. The effect of tuberculin testing on the development of cell-mediated immune responses during Mycobacterium bovis infection. Vet. Immunol. Immunopathol. 2006;114:25–36. doi: 10.1016/j.vetimm.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Francis J. Bovine Tuberculosis. Staples Press Limited; London: 1947. [Google Scholar]
- 29.Koets A, Hoek A, Langelaar M, Overdijk M, Santema W, Franken P, Eden W, Rutten V. Mycobacterial 70 kD heat-shock protein is an effective subunit vaccine against bovine paratuberculosis. Vaccine. 2006;24:2550–2559. doi: 10.1016/j.vaccine.2005.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.