Abstract
Adeno-associated virus (AAV) vectors are ideal for performing gene repair due to their ability to target multiple different genomic loci, low immunogenicity, capability to achieve targeted and stable expression through integration, and low mutagenic and oncogenic potential. However, many handicaps to gene repair therapy remain. Most notable is the low frequency of correction in vivo. To date, this frequency is too low to be of therapeutic value for any disease. To address this, a point-mutation– based mouse model of the metabolic disease hereditary tyrosinemia type I was used to test whether targeted AAV integration by homologous recombination could achieve high-level stable gene repair in vivo. Both neonatal and adult mice were treated with AAV serotypes 2 and 8 carrying a wild-type genomic sequence for repairing the mutated Fah (fumarylacetoacetate hydrolase) gene. Hepatic gene repair was quantified by immunohistochemistry and supported with reverse transcription polymerase chain reaction and serology for functional correction parameters. Successful gene repair was observed with both serotypes but was more efficient with AAV8. Correction frequencies of up to 10−3 were achieved and highly reproducible within typical dose ranges. In this model, repaired hepatocytes have a selective growth advantage and are thus able to proliferate to efficiently repopulate mutant livers and cure the underlying metabolic disease.
Conclusion
AAV-mediated gene repair is feasible in vivo and can functionally correct an appropriate selection-based metabolic liver disease in both adults and neonates.
Gene therapy is a promising means to cure many monogenic diseases. However, traditional gene therapies are best suited to treat diseases of deficient or absent gene products rather than those diseases caused by aberrantly functioning proteins. Even now, gene therapy efforts remain focused on gene addition strategies using full-length complementary DNA (cDNA) cassettes for the mutated gene of interest, driven by promoter and enhancer sequences.1 Despite many advances, gene addition approaches with adeno-associated virus (AAV) are limited by transient and unregulated expression,2 highly random integrations,3 transgene silencing,4 and increased mutagenic and oncogenic risks.5 Not all protein-coding genes have open reading frames small enough to fit within the low coding capacity of AAV (4.7 kb), thus, this type of gene therapy is not applicable for all disorders.6
Gene repair offers a solution to these drawbacks, where patient genomes are manipulated in vivo using site-specific recombination to actually correct the underlying mutation. Contrary to more widely used gene addition paradigms, gene repair restores gene function within the context of all endogenous regulatory elements, thereby eliminating potential problems with inadequate or inappropriate expression. Different vehicles have been utilized for performing gene repair including single-strand oligonucleotides,7–9 triplex-forming oligonucleotides,10 RNA-DNA hybrids,11,12 small fragment DNA, and AAV.13–15 Of these, AAV has emerged as the most promising. Numerous in vitro studies have shown AAV capable of correcting various types of mutations (insertions, deletions, substitutions) by vector-mediated homologous recombination.16,17 AAV vectors engineered to perform gene repair have the ability to target multiple different genomic loci, show both targeted and stable expression through integration, and have an increased number of applicable human diseases.18 Single-stranded AAV genomes modulate gene repair by integrating site-specifically via homologous recombination and targeting only the disease-causing mutation for replacement with wild-type sequence.19 Gene repair is best suited to correct point-mutation–based diseases that need only one or few nucleotides corrected to restore normal gene expression. This is key, because point mutations are the most frequent genetic abnormality and source of acquired genetic disease.20
To demonstrate targeted hepatic gene repair in vivo for a clinically pertinent disease gene, a hereditary tyrosinemia type I (HTI) mouse model (Fah5981SB) was used. HTI is a fatal genetic disease caused by deficiency of fumarylacetoacetate hydrolase (FAH), the terminal enzyme in the tyrosine catabolic pathway.21 When a FAH deficiency exists, toxic metabolites such as fumarylacetoacetate accumulate in hepatocytes and renal proximal tubules causing death in a cell-autonomous manner.22 Toxic metabolite accumulation can be blocked by 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) administration, a pharmacological inhibitor that blocks the pathway upstream of FAH.23 The Fah5981SB mouse is ideal to study gene repair, because it is point-mutation– based and fully recapitulates the human disease on an accelerated time scale. Strong positive selection for FAH+ cells in the HTI mouse liver has been demonstrated24 and was exploited for in vivo selection of corrected hepatocytes following gene repair. In this system, when AAV vectors containing genomic Fah sequence (hereafter referred to as AAV-Fah) are administered to Fah5981SB mice, only corrected FAH-positive (FAH+) hepatocytes that have undergone integration by homologous recombination can survive and repopulate the liver. The outcome is formation of corrected FAH+ nodules and loss of unintegrated episomal vector genomes. In both neonatal and adult mice treated with AAV-Fah, gene repair restored proper gene and protein expression and cured the underlying HTI phenotype. These results demonstrate proof-of-principle that an appropriate monogenic liver disease can be corrected by AAV-mediated gene repair in vivo.
Materials and Methods
Mouse Strains and Animal Husbandry
The Fah5981SB mouse25 models HTI by bearing a single N-ethyl-N-nitrosourea–induced point mutation in the final nucleotide of exon 8 within the Fah gene.26 This point mutation creates a premature downstream stop codon and exon 8 loss, ultimately leading to formation of truncated, unstable FAH protein that is degraded. Fah5981SB mice die as neonates from acute liver failure if NTBC is not continually administered in the drinking water. NTBC treatment at 4 mg/mL rescues the phenotype and prevents acute hepatocellular and renal injury. Discontinuation of NTBC provides an accurate model of HTI. Mice develop liver and renal disease within 10 days, which progresses to full end-stage liver disease and death within 6–8 weeks.27 The mice have been backcrossed 10 generations onto a C57BL6 background. The Institutional Animal Care and Use Committee of Oregon Health and Science University approved all procedures and mouse experiments.
Plasmid Construction
Mus musculus bacterial artificial chromosome (BAC) clone RP23-121N17 from chromosome 7 (Invitrogen) was used as a template for the 4.5-kb long-distance polymerase chain reaction (LD-PCR) amplification of sequence homologous to the region centered on the point mutation in exon 8 of murine Fah (RefSeq NM_010176, chr7:84461356–84481935). Forward primer introducing NotI: 5′-GCGGCCGCTTCCCAGGGTTTTTGTTTGTT-3′; reverse primer: 5′-AGCCCCCACTGACAGCTACAGCT-3′. The PCR resulted in a 4.5-kb product with an introduced 5′-NotI restriction site that allowed cloning into an AAV plasmid backbone as previously described.28
Sequencing
DNA sequencing was performed with an ABI-Prism 3130xl Genetic Analyzer (Applied Biosystems Inc., Foster City, CA) at the Vollum Sequencing Core (Portland, OR). DNA sequences were aligned with MacVector software.
Neonatal Vector Administration
For time course studies, d3 Fah5981SB neonates were injected with 1 × 1011 (AAV2-Fah) or 2 × 1011 (AAV8-Fah) vector genome (vg) in 10 µL volume by intravenous facial vein injection.29 Littermate controls were similarly injected with 1 × 1011 to 2 × 1011 vg of an irrelevant serotype-matched control vector; either AAV2-hAAT,30 or AAV8-GFP.31 All mice were maintained on NTBC throughout. Livers were harvested at 1, 2, or 4 weeks after treatment. For dose-response studies, d3 Fah5981SB neonates were injected with four doses ranging from 3 × 108 to 3 × 1011 vg (in 10 µL volume) of each serotype by intravenous facial vein injection. All mice were maintained on NTBC throughout. Livers were harvested 2 weeks after treatment. For stable integration studies, d3 Fah5981SB neonates were injected with AAV2-Fah at 1 × 1011 vg in 10 µL volume by intravenous facial vein injection. Litter-mate controls were similarly injected with isotonic NaCl solution. Mice were maintained on NTBC until weaning and then withdrawn to select for corrected hepatocytes. Eleven weeks after treatment, a two-thirds partial hepatectomy was performed to induce liver regeneration.32 Livers were harvested >12 weeks after surgery. For random integration studies, d3 Fah5981SB neonates were coinjected with 4 × 1010 vg of both AAV8-Fah and AAV8-hAAT (in 10 µL volume) by intravenous facial vein injection. Mice were maintained on NTBC until weaning and then withdrawn to select for corrected hepatocytes. Serum (for liver function tests) and liver tissue were collected at harvest.
Adult Vector Administration
Adult Fah5981SB mice (age 8–12 weeks) were injected with 1 × 1011 vg of AAV8-Fah (in 100 µL volume) by intravenous tail vein injection. Age-matched littermate controls were similarly injected with isotonic NaCl solution. Mice were placed on NTBC as needed. Serum and liver tissue were harvested >12 weeks after treatment.
Liver Immunohistochemistry
In both adult and neonatal experiments, a minimum of two liver sections were analyzed per mouse and evaluated for the number of FAH+ cell clusters, each representing the clonal expansion of a single corrected hepatocyte. Clonal frequencies, correction factors, hepatocyte counts, fixation, and immunohistochemistry protocols were done as described.33 Quantitation was performed by two separate, blinded investigators.
Statistical Analysis
Experimental results were analyzed for significance by applying a student 2-tailed t-test assuming equal variance. P values <0.05 were considered statistically significant.
Vector Preparation
AAV vector preparation and titering were performed according to standard AAV protocols as described.34
Transplantation
For serial transplantation surgeries, livers were isolated from corrected mice and 3 × 105 to 5 × 105 random hepatocytes were injected intrasplenically at 100 µL volume into Fah5981SB recipient mice as described.35
Fah Quantitative Reverse Transcription PCR
Total RNA was isolated from randomly dissected liver tissue with an RNeasy Mini kit (Qiagen). The cDNA was produced with a Superscript III First-Strand Synthesis kit (Invitrogen). PCR was performed on an iCycler (Bio-Rad Laboratories). Reverse transcription (RT) reaction (100 ng) was subjected to two-step PCR amplification under the following conditions: 1 cycle 95°C × 3 minutes, followed by 45 cycles of 95°C × 15 seconds and 68°C × 50 seconds. Primer sequences: Fah forward: 5′-AGAACTTACTGTCTGCCAGCCAAG-3′; Fah reverse: 5′-GAGGACCATCCCGAAAATGTG-3′; glyceraldehyde 3-phosphate dehydrogenase (Gapdh) forward: 5′-CCACCCCAGCAAGGACACTG-3′; Gapdh reverse 5′-GCTCCCTAGGCCCCTCCTGT-3′. All samples were subjected to ± RT controls and results were normalized to Gapdh expression.
hAAT Copy Number Quantitative PCR
Total DNA was isolated from randomly dissected liver tissue with a MasterPure DNA Purification kit (Epicentre Biotechnologies). PCR was performed on an iQ5 Multicolor Real-Time PCR (Bio-Rad), using the iQ5 Standard Edition Software, version 2.0. Genomic DNA (10 ng) was subjected to a two-step PCR amplification under the following conditions: 1 cycle 95°C × 3 minutes, followed by 45 cycles of 95°C × 15 seconds and 68°C × 40 seconds. Primer sequences: hAAT forward: 5′-TCCTGGGTCAACTGGGCATC-3′; hAAT reverse: 5′-CAGGGGTGCCTCCTCTGTGA-3′; Gapdh forward: 5′-CCACCCCAGCAAGGACACTG-3′; Gapdh reverse 5′-GCTCCCTAGGCCCCTCCTGT-3′. Dilutions of hAAT plasmid into mouse genomic DNA were used to generate copy number standards. Results were normalized to Gapdh expression.
Results
Target Vector Design
In Fah5981SB mice, a single point mutation (G→A transversion) at the terminal nucleotide of Fah exon 8 leads to mis-splicing and exon-8 deletion from the messenger RNA (mRNA). Several important criteria derived from the literature18 were considered for the design of the gene repair vector to correct the Fah5981SB point mutation (Fig. 1A). First, the vector should not contain elements needed for driving gene expression such as promoters, enhancers, or cDNA expression cassettes. Second, the fidelity and length of homology should be maximized with the packaging capacity of AAV (4.7 kb) being the limit. Third, the position of the nucleotide targeted for repair should be at the center of the homology. A 4.5-kb PCR product homologous to murine Fah was cloned into an AAV plasmid backbone and verified by DNA sequencing. Recombinant AAV-Fah of serotypes 2 and 8 were produced and administered to Fah5981SB mice as neonates or adults. Correction of the point mutation by homologous recombination (Fig. 1B) leads to normal Fah gene and protein expression.
Fig. 1.
AAV vector design and genomic organization of murine Fah. (A) A 4.5-kb genomic DNA fragment homologous to murine Fah was cloned into an AAV plasmid backbone and verified by DNA sequencing. The position of the nucleotide needed to perform repair was centered within two 2.25-kb homology arms. (B) Mechanism of AAV-mediated homologous recombination. Wild-type Fah has a guanine (G) as the terminal nucleotide of exon 8. In Fah5981SB mice, a G→A transversion occurs at this position leading to mis-splicing and exon-8 deletion. Gene repair of the mutated Fah genomic sequence by AAV-mediated homologous recombination corrects the point mutation and restores proper Fah gene expression.
AAV-Fah Mediates Stable Gene Repair In Vivo
The evaluation of homologous recombination as a strategy for gene repair has traditionally relied on detecting alterations in reporter sequences rather than correcting a disease phenotype. Given the selective advantage of FAH+ hepatocytes in the HTI liver, Fah5981SB mice can be used to study the clinical significance of AAV-mediated gene repair by homologous recombination. Four d3 Fah5981SB neonates were intravenously injected with 1 × 1011 vg of AAV2-Fah and kept on NTBC until weaning, followed by NTBC withdrawal to select for corrected hepatocytes. Two control groups were injected with isotonic NaCl solution. Control group I (n = 3) did not receive a course of NTBC post-weaning, continued to lose weight and died. Control group II (n = 2) did receive one course of NTBC post-weaning but failed to maintain a healthy weight and died. AAV-treated mice began to stabilize in weight at 8 weeks after treatment, suggesting the onset of sufficient liver function. At age 11 weeks, a two-thirds partial hepatectomy was performed to induce liver regeneration and subsequent episomal AAV loss. Continued clinical improvement following partial hepatectomy strongly suggested stable gene repair at the Fah locus. FAH immunohistochemistry showed >50% FAH+ hepatocytes in section overviews (Fig. 2A). The numbers of detectable liver nodules ranged from 21–47 per 50 mm2 section in treated mice and were never detected in controls. Nodules represent the clonal expansion of a single corrected hepatocyte, thus nodule frequency must be corrected for nodule size. For this experiment, the correction factor was estimated to be fourteen. After correction, the initial gene repair frequency ranged from 1/6,300 to 1/11,600 hepatocytes and was within the expected range from previous experiments15 where selection with NTBC did not apply.
Fig. 2.
AAV can mediate stable gene repair in vivo. (A) Liver immunohistology and whole-mount staining for FAH. In the top row from left to right, liver sections are from untreated mice (wild-type [+/+] or Fah5981SB [−/−]); mock-injected Fah5981SB mice; AAV-Fah treated Fah5981SB mice; and Fah5981SB mice after serial transplantation (sTx) with hepatocytes from a previously corrected mouse. Bottom row: Left panel shows wild-type liver as positive control for Fah. The following three panels are low-magnification overviews of liver sections. The second from left panel is from a mock-injected mouse and shows no FAH staining, whereas the two panels at right display multiple brown FAH+ nodules after AAV-Fah injection. Scale bars: solid = 100 µm; dotted = 1 mm. (B) RT-PCR for Fah mRNA. Wild-type mRNA produces a 433–base pair fragment, whereas the exon-8 – deleted mutant mRNA produces a shorter product. The bottom panel shows analysis of nine experimental samples at the left and controls at the right. A “±” indicates whether reverse transcriptase was used. Samples 1–3 are from AAV-treated neonatal mice, samples 4 and 5 are from AAV-treated adults, and samples 6–9 are from serial transplant recipients.
To demonstrate that FAH staining was not artifactual and that proper Fah gene expression had indeed been restored, Fah RT-PCR was performed on RNA from treated livers. The presence of correctly spliced mRNA was demonstrated in all treated mice (Fig. 2B). To further demonstrate the stability of correction, 3 × 105 random hepatocytes from a corrected mouse were serially transplanted into four secondary adult Fah5981SB recipients. Serial transplantation is another means to induce hepatocyte turnover and eliminate episomal AAV genomes.35 Serial transplant recipients had successful engraftment and displayed clinical improvement, whereas untransplanted controls showed continuous weight loss and died. FAH immunohistochemistry from livers of serial transplant recipients had extensive hepatocellular FAH staining, further demonstrating stability of the gene repair (Fig. 2A).
Time course comparison of AAV8-Fah and AAV2-Fah
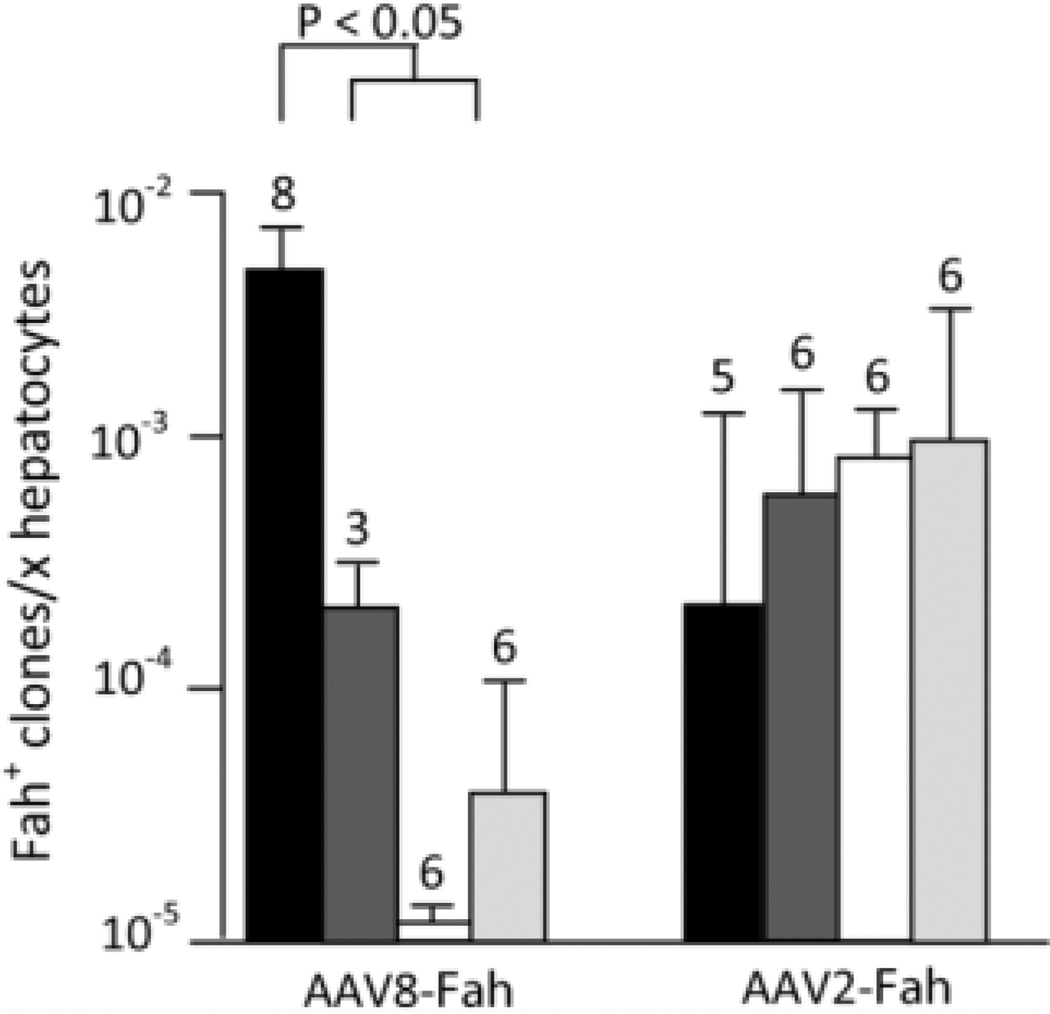
AAV8 is the preferred serotype for liver transduction because of its strong hepatic tropism, rapid capsid disassembly and genome release.36 In contrast, although AAV2 has been shown to transduce liver, it is characterized by slow capsid disassembly and genome release. To address the question whether AAV serotypes 8 and 2 have different gene repair dynamics in vivo, d3 Fah5981SB neonates were treated with 2 × 1011 vg of AAV8-Fah or 1 × 1011 vg of AAV2-Fah and analyzed after 1, 2, or 4 weeks post-treatment for the presence of FAH+ hepatocytes (Fig. 3). In AAV8-Fah treated mice, the highest number of FAH+ hepatocytes seen (up to 1/180 hepatocytes) were detected within the first week post-treatment. Correction frequencies declined with time and stabilized after 4 weeks. In contrast, AAV2-treated mice had little detectable Fah expression within the first seven days, supporting the fact that AAV2 uncoats more slowly than AAV8. Week two showed an increase in Fah expression that remained stable until week four. No FAH+ hepatocytes were detected at any time point in control mice injected with serotype-matched irrelevant control vectors AAV8-GFP or AAV2-hAAT at equivalent doses. These results conclusively demonstrate that emergence of FAH+ hepatocytes were neither due to spontaneous reversion, nor gene repair stimulated non-specifically by mere AAV transduction.
Fig. 3.
Time-course study of AAV-mediated gene repair frequencies in neonates. Vectors are noted below each set and were administered at 1 × 1011 to 2 × 1011 vg. Frequencies were quantified by counting single FAH+ clones per × hepatocytes (1/x) from neonates harvested 1, 2, or 4 weeks after treatment. Means ± standard deviation are shown, with the number of animals analyzed indicated above each bar. Black bar = 1 week after treatment; gray bar = 2 weeks after treatment; white bar = 4 weeks after treatment.
Gene Repair in Response to Different Vector Doses
To examine dose responses, d3 Fah5981SB neonates were injected with four AAV-Fah concentrations ranging from 3 × 108 to 3 × 1011 vg for each serotype and kept on NTBC until harvest at weaning to prevent metabolic selection of FAH+ cells. In general, AAV8-Fah displayed a linear dose response over the range of doses administered (Fig. 4) where the highest doses administered produced the greatest gene repair. The difference in repair frequencies between the highest dose and all other doses administered was significant. In contrast, AAV2-Fah had no significant change in repair frequency over the entire range of doses administered. Overall, results indicate that AAV8-mediated gene repair is superior to that with AAV2.
Fig. 4.
Dose response study of AAV-mediated gene repair frequencies in neonates. Vectors administered are noted below each data set and were administered at 3 × 1011 to 3 × 108 vg. Frequencies were quantified by counting single FAH+ clones per × hepatocytes (1/x) from neonates harvested 3 weeks after treatment. Means ± standard deviation are shown, with the number of animals analyzed indicated above each bar. Black bar = 3 × 1011 vg; dark gray bar = 3 × 1010 vg; white bar = 3 × 109 vg; light gray bar = 3 × 108 vg.
AAV-Mediated Gene Repair Is Feasible in Quiescent Liver
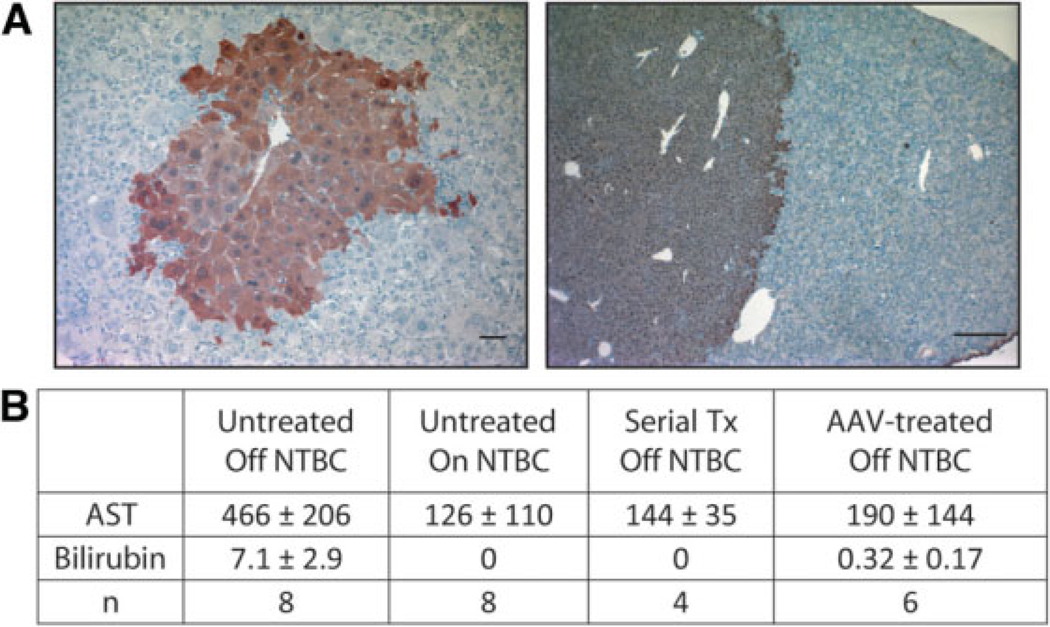
The adult liver has considerably less cellular turnover than neonatal liver undergoing rapid growth and proliferation. Thus, gene repair frequencies are predicted to be lower in adults as homologous recombination is most prevalent during mitotic S-phase.37 AAV8 was chosen to test the feasibility of gene repair in the nearly quiescent adult liver as it had now been demonstrated to be both faster and more efficient at gene repair than AAV2. Adult Fah5981SB mice (8–12 weeks old) were injected with 1 × 1011 vg of AAV8-Fah (n = 6), whereas age-matched littermate controls were injected with isotonic NaCl (n = 8). Mice were withdrawn from NTBC to allow selection of corrected hepatocytes. Serum for liver function tests and liver tissue were harvested >12 weeks after treatment. Mice treated with AAV8-Fah showed clinical improvement and repopulation with FAH+ hepatocytes (Fig. 5A), whereas all mice in the control group had to be euthanized and showed no hepatic repopulation. Surprisingly, the initial correction frequency of FAH+ nodules was comparable to that seen with neonatal administration. The clonal expansion of corrected hepatocytes was able to reverse the tyrosinemic phenotype and was highly reproducible. Liver function tests for AST and bilirubin demonstrated near complete correction when compared to controls (Fig. 5B).
Fig. 5.
AAV-mediated gene repair is feasible in adult liver. (A) Liver sections stained for FAH from adult Fah5981SB mice more than 12 weeks after treatment with AAV8-Fah. Scale bars = 100 µm. (B) Liver function test results. Aspartate aminotransferase (AST [U/I]); bilirubin (mg/dL). Values (± standard deviation) represent adult untreated Fah5981SB mice off or on NTBC, serially transplanted (Tx) Fah5981SB mice off NTBC, and adult AAV-treated Fah5981SB mice off NTBC.
Frequency of Random Integration
Although phenotypic reversion of Fah5981SB mice indicates successful site-specific gene repair, random integration could also occur.38 To assess random integration frequencies, d3 Fah5981SB neonates were coinjected with 4 × 1010 vg of AAV8-Fah and an irrelevant serotype-matched control vector AAV8-hAAT. Post-weaning, mice were subjected to NTBC withdrawal to select for corrected hepatocytes. To ensure no episomes remained, 5 × 105 random hepatocytes were then serially transplanted into eight secondary Fah5981SB recipients. After >12 weeks off NTBC, serum and liver tissue were collected at harvest. qPCR was used to determine Fah and hAAT copy numbers in each mouse (Table 1). The frequency of randomly integrated hAAT ranged from 0 (undetectable) to 0.06/dGE and averaged 0.005/dGE. Only half the hepatocytes in repopulated livers were donor-derived, thus frequencies were corrected by a factor of two, resulting in an average random integration frequency of 0.01/dGE (1%) in corrected hepatocytes. This number is similar to multiple estimates of random integration of AAV8 from the literature.35 Liver function tests in serially transplanted mice demonstrated near complete reconstitution by normalization of AST and bilirubin levels (Fig. 5B). Differences in serum AST and bilirubin levels were significant (P < 0.05). In addition, in neonatal follow-ups 16 weeks post-treatment, no tumors were observed (n = 15, data not shown).
Table 1.
Frequency of Random Integration
| hAAT Copy Number in Recipient Mice | |||
|---|---|---|---|
| Donor | sTx Recipient |
Number of Copies/dGE |
Average |
| 1 | 1a | 0.009 | 0.028 ± 0.023 |
| 1b | 0.026 | ||
| 1c | 0.002 | ||
| 1d | 0.042 | ||
| 1e | 0.058 | ||
| 2 | 2a | 0.002 | 0 |
| 2b | 0.001 | ||
| 2c | 0 | ||
| 2d | 0 | ||
| 2e | 0 | ||
| Total | 0.005 | ||
| Correction factor | 2 | ||
| Random integration | 0.01 = 1% | ||
After neonatal coinjection of 4 × 1010 vg of both AAV8-Fah and AAV8-hAAT, FAH+ hepatocytes were selected for and then serially transplanted to remove any episomes. Data represent the number of copies of hAAT per diploid genome equivalent (dGE) in serial transplant (sTx) recipients. Because only 50% of the hepatocytes in these repopulated livers were derived from the original AAV-Fah treated donor liver, the frequency was corrected by a factor of 2 to give an estimated random integration frequency of 1%.
Discussion
AAV has emerged as the vector of choice for gene repair as its single-stranded nature facilitates correction by homologous recombination. Numerous studies have demonstrated successful AAV-mediated gene repair to correct different mutation types in vitro.16,17 In doing so, these studies provided the essential validation and framework for all AAV-mediated gene repair studies in vivo. Few publications exist demonstrating repair in vivo,39,40 and they are hindered by the fact that they target clinically irrelevant marker mutations in exogenously provided transgenes like green fluorescent protein (GFP) or LacZ. One report has shown limited efficacy in vivo using a neonatal mouse model of the disease mucopolysaccharidosis type VII.15 In that study, a single point mutation in the β-glucuronidase gene was corrected at frequencies of 10−4 to 10−5 using AAV2 and AAV6 at 2 × 1011 to 6 × 1011 vg doses. Nonetheless, the low correction frequencies were not therapeutic in treated mice, because no selective advantage exists for corrected hepatocytes in that model. This study differs from our own in several ways. Our study is the first to demonstrate the stability of gene correction in both adult and neonatal mice. In addition to AAV2, our study demonstrated greater correction using AAV8, the most hepatotropic of all the naturally occurring AAV serotypes. Furthermore, correction frequencies of up to 10−3 as early as 3 weeks after treatment in adult mice were shown; rather than 10−4 in 12–24 weeks after treatment in the previous study. Finally, our work tested a range of AAV doses from 1011 to 108 vg and was able to demonstrate correction at all doses administered.
Numerous handicaps to AAV-mediated gene repair remain. Most notable is the low frequency of correction in vivo. To date, this frequency is too low to be of therapeutic value for many diseases. However, our work demonstrates that AAV-mediated gene repair has the capacity to be a real therapeutic alternative in a suitable selection-based disease. In hereditary tyrosinemia type I, corrected hepatocytes have a selective growth advantage and can clonally expand to restore liver function, even if the initial gene repair efficiency is low. While this situation is an exception, there are several disorders in which selection has been shown, including Fanconi’s anemia,41 the copper storage disorder Wilson’s disease,42 many bile-acid transporter defects,43 and junctional epidermolysis bullosa44 to name a few. If correction frequencies were increased even 10-fold, they would become clinically relevant for an even broader range of diseases. For example, it has been predicted that gene repair frequencies of 10−2 would have therapeutic benefit in patients with hemophilia A.45
Our study establishes the utility of two different AAV serotypes (AAV8 and AAV2) for hepatic gene targeting of both adult and neonatal mice in vivo. Interestingly, the biology of these two serotypes differed considerably in terms of gene targeting. Different kinetics for the two serotypes have been described previously with gene addition approaches wherein higher doses of AAV correlated with higher levels of gene expression.36 Here, we observed a similar phenomenon where the highest doses administered produced the greatest gene repair frequencies in vivo. Targeting was confirmed by immunohistochemistry, RT-PCR, and functional measures of liver correction using serum liver function tests.
We also evaluated the frequency of random integration in cells with proper gene repair using coinjection with a second, nonselectable AAV vector. The average copy number of the irrelevant vector corrected for repopulation efficiency indicated that 0.5%–1% of targeted cells also had a random integration. This number is similar to multiple estimates of random integration of AAV8 from the literature.35 Therefore, it can be concluded that gene repair does not result in a higher random integration frequency.
In summary, our experiments demonstrated stable hepatic gene repair in both adult and neonatal mice with AAV-Fah serotypes 2 and 8. Serial transplantation was possible without difficulty and serially reconstituted animals had normal hepatic function. Most importantly, this work was the first to show functional metabolic correction of a disease model using AAV-mediated gene repair and can be envisioned as a therapeutic strategy for disorders with a selective advantage in corrected cells. Although these experiments focused on correcting the metabolic disease HTI, the novel approach described herein can serve as a model for gene repair in any monogenic disease caused by point mutations.
Acknowledgment
We thank Angela Major for histology support and Terry Storm for AAV preparations.
This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases to M.G. (RO1-DK48252) and the National Cancer Institute to N.P. (F31CA130116). The funding organizations played no role in experimental design, data analysis, or manuscript preparation.
Abbreviations
- AAV
adeno-associated virus
- AST
aspartate aminotransferase
- dGE
diploid genome equivalent
- FAH
fumarylacetoacetate hydrolase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- hAAT
human alpha-1 antitrypsin
- HTI
hereditary tyrosinemia type I
- LD-PCR
long-distance polymerase chain reaction
- NTBC
2-(2-nitro-4-trifluoro-methylbenzol)-1,3-cyclohexanedione
- RT-PCR
reverse transcription polymerase chain reaction
- vg
vector genome
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Mueller C, Flotte TR. Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther. 2008;15:858–863. doi: 10.1038/gt.2008.68. [DOI] [PubMed] [Google Scholar]
- 2.Russell DW, Kay MA. Adeno-associated virus vectors and hematology. Blood. 1999;94:864–874. [PMC free article] [PubMed] [Google Scholar]
- 3.Hendrie PC, Hirata RK, Russell DW. Chromosomal integration and homologous gene targeting by replication-incompetent vectors based on the autonomous parvovirus minute virus of mice. J Virol. 2003;77:13136–13145. doi: 10.1128/JVI.77.24.13136-13145.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 5.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 6.Verma IM, Somia N. Gene therapy–promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 7.Andrieu-Soler C, Halhal M, Boatright JH, Padove SA, Nickerson JM, Stodulkova E, et al. Single-stranded oligonucleotide-mediated in vivo gene repair in the rd1 retina. Mol Vis. 2007;13:692–706. [PMC free article] [PubMed] [Google Scholar]
- 8.Huen MS, Lu LY, Liu DP, Huang JD. Active transcription promotes single-stranded oligonucleotide mediated gene repair. Biochem Biophys Res Commun. 2007;353:33–39. doi: 10.1016/j.bbrc.2006.11.146. [DOI] [PubMed] [Google Scholar]
- 9.Wu XS, Xin L, Yin WX, Shang XY, Lu L, Watt RM, et al. Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression. Proc Natl Acad Sci U S A. 2005;102:2508–2513. doi: 10.1073/pnas.0406991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 11.Kren BT, Parashar B, Bandyopadhyay P, Chowdhury NR, Chowdhury JR, Steer CJ. Correction of the UDP-glucuronosyltransferase gene defect in the gunn rat model of crigler-najjar syndrome type I with a chimeric oligonucleotide. Proc Natl Acad Sci U S A. 1999;96:10349–10354. doi: 10.1073/pnas.96.18.10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kren BT, Bandyopadhyay P, Steer CJ. In vivo site-directed mutagenesis of the factor IX gene by chimeric RNA/DNA oligonucleotides. Nat Med. 1998;4:285–290. doi: 10.1038/nm0398-285. [DOI] [PubMed] [Google Scholar]
- 13.Vasileva A, Jessberger R. Precise hit: adeno-associated virus in gene targeting. Nat Rev Microbiol. 2005;3:837–847. doi: 10.1038/nrmicro1266. [DOI] [PubMed] [Google Scholar]
- 14.Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS, Russell DW. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- 16.Inoue N, Dong R, Hirata RK, Russell DW. Introduction of single base substitutions at homologous chromosomal sequences by adeno-associated virus vectors. Mol Ther. 2001;3:526–530. doi: 10.1006/mthe.2001.0283. [DOI] [PubMed] [Google Scholar]
- 17.Inoue N, Hirata RK, Russell DW. High-fidelity correction of mutations at multiple chromosomal positions by adeno-associated virus vectors. J Virol. 1999;73:7376–7380. doi: 10.1128/jvi.73.9.7376-7380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrie PC, Russell DW. Gene targeting with viral vectors. Mol Ther. 2005;12:9–17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Parekh-Olmedo H, Ferrara L, Brachman E, Kmiec EB. Gene therapy progress and prospects: targeted gene repair. Gene Ther. 2005;12:639–646. doi: 10.1038/sj.gt.3302511. [DOI] [PubMed] [Google Scholar]
- 20.Bergeron B. Case Studies in Genes and Disease. 1st ed. Philadelphia: American College of Physicians Press; 2004. [Google Scholar]
- 21.Mitchell GA, Grompe M, Lambert M, Tanguay R. The Metabolic and Molecular Basis of Inherited Disease. 8th ed. New York: McGraw-Hill; 2001. Hypertyrosinemia; pp. 1777–1805. [Google Scholar]
- 22.Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis. 2001;21:563–571. doi: 10.1055/s-2001-19035. [DOI] [PubMed] [Google Scholar]
- 23.Al-Dhalimy M, Overturf K, Finegold M, Grompe M. Long-term therapy with NTBC and tyrosine-restricted diet in a murine model of hereditary tyrosinemia type I. Mol Genet Metab. 2002;75:38–45. doi: 10.1006/mgme.2001.3266. [DOI] [PubMed] [Google Scholar]
- 24.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 25.Aponte JL, Sega GA, Hauser LJ, Dhar MS, Withrow CM, Carpenter DA, et al. Point mutations in the murine fumarylacetoacetate hydrolase gene: Animal models for the human genetic disorder hereditary tyrosinemia type 1. Proc Natl Acad Sci U S A. 2001;98:641–645. doi: 10.1073/pnas.98.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Culiat CT, Klebig ML, Liu Z, Monroe H, Stanford B, Desai J, et al. Identification of mutations from phenotype-driven ENU mutagenesis in mouse chromosome 7. Mamm Genome. 2005;16:555–566. doi: 10.1007/s00335-005-0032-0. [DOI] [PubMed] [Google Scholar]
- 27.Grompe M, Lindstedt S, al-Dhalimy M, Kennaway NG, Papaconstantinou J, Torres-Ramos CA, et al. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1995;10:453–460. doi: 10.1038/ng0895-453. [DOI] [PubMed] [Google Scholar]
- 28.Nakai H, Montini E, Fuess S, Storm TA, Meuse L, Finegold M, et al. Helper-independent and AAV-ITR-independent chromosomal integration of double-stranded linear DNA vectors in mice. Mol Ther. 2003;7:101–111. doi: 10.1016/s1525-0016(02)00023-0. [DOI] [PubMed] [Google Scholar]
- 29.Sands MS, Barker JE. Percutaneous intravenous injection in neonatal mice. Comp Med. 2000;50:107. [PubMed] [Google Scholar]
- 30.Chen ZY, Yant SR, He CY, Meuse L, Shen S, Kay MA. Linear DNAs concatemerize in vivo and result in sustained transgene expression in mouse liver. Mol Ther. 2001;3:403–410. doi: 10.1006/mthe.2001.0278. [DOI] [PubMed] [Google Scholar]
- 31.Grimm D, Pandey K, Nakai H, Storm TA, Kay MA. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Montini E, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. Kinetics of liver repopulation after bone marrow transplantation. Am J Pathol. 2002;161:565–574. doi: 10.1016/S0002-9440(10)64212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 35.Nakai H, Montini E, Fuess S, Storm TA, Grompe M, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- 36.Thomas CE, Storm TA, Huang Z, Kay MA. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 38.Nakai H, Wu X, Fuess S, Storm TA, Munroe D, Montini E, et al. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J Virol. 2005;79:3606–3614. doi: 10.1128/JVI.79.6.3606-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vijg J, Dolle ME, Martus HJ, Boerrigter ME. Transgenic mouse models for studying mutations in vivo: applications in aging research. Mech Ageing Dev. 1997;98:189–202. doi: 10.1016/s0047-6374(97)00107-3. [DOI] [PubMed] [Google Scholar]
- 40.Nickerson HD, Colledge WH. A LacZ-based transgenic mouse for detection of somatic gene repair events in vivo. Gene Ther. 2004;11:1351–1357. doi: 10.1038/sj.gt.3302311. [DOI] [PubMed] [Google Scholar]
- 41.Battaile KP, Bateman RL, Mortimer D, Mulcahy J, Rathbun RK, Bagby G, et al. In vivo selection of wild-type hematopoietic stem cells in a murine model of Fanconi anemia. Blood. 1999;94:2151–2158. [PubMed] [Google Scholar]
- 42.Allen KJ, Cheah DM, Wright PF, Gazeas S, Pettigrew-Buck NE, Deal YH, et al. Liver cell transplantation leads to repopulation and functional correction in a mouse model of Wilson's disease. J Gastroenterol Hepatol. 2004;19:1283–1290. doi: 10.1111/j.1440-1746.2004.03451.x. [DOI] [PubMed] [Google Scholar]
- 43.De Vree JM, Ottenhoff R, Bosma PJ, Smith AJ, Aten J, Oude Elferink RP. Correction of liver disease by hepatocyte transplantation in a mouse model of progressive familial intrahepatic cholestasis. Gastroenterology. 2000;119:1720–1730. doi: 10.1053/gast.2000.20222. [DOI] [PubMed] [Google Scholar]
- 44.Ortiz-Urda S, Lin Q, Yant SR, Keene D, Kay MA, Khavari PA. Sustainable correction of junctional epidermolysis bullosa via transposon-mediated nonviral gene transfer. Gene Ther. 2003;10:1099–1104. doi: 10.1038/sj.gt.3301978. [DOI] [PubMed] [Google Scholar]
- 45.Kay MA, High K. Gene therapy for the hemophilias. Proc Natl Acad Sci U S A. 1999;96:9973–9975. doi: 10.1073/pnas.96.18.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]