Abstract
Vertebrates typically harbor a rich gastrointestinal microbiota, which has coevolved with the host over millennia and is essential for several host physiological functions, in particular maturation of the immune system. Recent studies have highlighted the importance of a single bacterial species, segmented filamentous bacteria (SFB), in inducing a robust T-helper cell type 17 (Th17) population in the small-intestinal lamina propria (SI-LP) of the mouse gut. Consequently, SFB can promote IL-17–dependent immune and autoimmune responses, gut-associated as well as systemic, including inflammatory arthritis and experimental autoimmune encephalomyelitis. Here, we exploit the incomplete penetrance of SFB colonization of NOD mice in our animal facility to explore its impact on the incidence and course of type 1 diabetes in this prototypical, spontaneous model. There was a strong cosegregation of SFB positivity and diabetes protection in females, but not in males, which remained relatively disease-free regardless of the SFB status. In contrast, insulitis did not depend on SFB colonization. SFB-positive, but not SFB-negative, females had a substantial population of Th17 cells in the SI-LP, which was the only significant, repeatable difference in the examined T-cell compartments of the gut, pancreas, or systemic lymphoid tissues. Th17-signature transcripts dominated the very limited SFB-induced molecular changes detected in SI-LP CD4+ T cells. Thus, a single bacterium, and the gut immune system alterations associated with it, can either promote or protect from autoimmunity in predisposed mouse models, probably reflecting their variable dependence on different Th subsets.
Keywords: gender, microbiome, T lymphocyte, autoimmune disease
Colonization of the external and mucosal surfaces of vertebrates by a multitude of microbes is evolutionarily conserved and is essential for many of the host's physiologic processes, including proper maturation of the immune system (1–3). Indeed, animals housed under germ-free (GF) conditions have several dysfunctional or deficient immunological compartments, resulting in abnormal responses to a variety of immune challenges (4).
Recent advances in the molecular discrimination of commensal bacteria colonizing the gastrointestinal tract of mice and in the flow cytometric parsing of murine gut lymphoid compartments have uncovered specific effects of certain bacterial species or genera on particular lymphocyte subsets. For example, the gut-resident commensal, segmented filamentous bacteria (SFB), promotes the development of a robust T-helper cell type 17 (Th17) population in the small-intestinal lamina propria (SI-LP) (5, 6), whereas Clostridia species induce forkhead box P3-positive (Foxp3+) regulatory T cells (Treg) in the colonic lamina propria (7). Relatedly, colonization of the mouse gastrointestinal tract with the human gut commensal Bacteroides fragilis or administration of one of its products, polysaccharide A, can have a substantial impact on both effector and regulatory T-cell subsets, depending on the context (8–10). In addition, bacterial DNA or bacterially produced metabolites, such as ATP or short-chain fatty acids, can exert profound effects on both the innate and adaptive gut immune systems (11–13).
Not unexpectedly, then, gastrointestinal microbiota can alter the incidence and severity of gut autoimmune/inflammatory diseases (14, 15); in fact, inflammatory bowel disease is “transmissible” via gut-resident microbiota under certain experimental conditions (16). More surprising have been recent observations that SFB's influence can be translated systemically to trigger autoimmune arthritis in a predisposed mouse strain (17) or to exacerbate experimental encephalomyelitis (18). Both these diseases depend critically on Th17 cells, a likely explanation for the SFB influence.
Type 1 diabetes (T1D)—especially in the prototypical, spontaneous NOD mouse model (19) —provides an interesting counterpoint. The incidence of T1D is higher in countries with stricter hygiene practices (reviewed in refs. 20, 21), an observation paralleled by the finding that the diabetes incidence in NOD mice often is higher in cleaner colonies, being fully penetrant in GF conditions (22, 23). Furthermore, a null mutation of MyD88, an innate immune system signaling molecule, altered the composition of the gut microbiome and prevented disease development in NOD mice housed in a specific-pathogen-free (SPF) facility (24). The protective effect was attributed to commensals otherwise kept in check by MyD88 signaling, because mice expressing or not expressing this molecule were equally susceptible to disease when housed GF (24). However, the specific commensals responsible for protection from diabetes under routine husbandry conditions remain to be identified.
Here, we have exploited the particular make-up of our experimental NOD mouse colony to perform a segregation analysis of diabetes development relative to SFB colonization. Cosegregation of SFB positivity and protection from diabetes was subtended by the appearance of a robust Th17 compartment in the SI-LP as well as an increased representation of Th17 signature transcripts, specifically in SI-LP CD4+ T lymphocytes.
Results
Facility and Individual Variation in Colonization of the Gastrointestinal Tract of NOD Mice by SFB.
It was reported more than 3 decades ago that microscopically defined SFB can variably colonize the same mouse strain in different animal facilities (25). More recently, Littman and colleagues, applying a more definitive 16S rRNA quantification method, demonstrated that SFB was present in C57BL/6 (B6) mice housed at Taconic Farms (TAC) but was absent from individuals of the same strain kept at the Jackson Laboratory (JAX) (6). However, it was unclear to what extent all individuals of all strains housed at a particular facility shared SFB status, whether positive or negative. Our primary focus was SFB colonization of the NOD strain, in particular within our experimental colony at the New Research Building (NRB) of Harvard Medical School.
Relative SFB levels in fecal pellets of mice housed at the three sites were determined by quantitative PCR assays that used both specific SFB and conserved eubacterial (EUB) 16S rRNA primers, the latter capable of amplifying the RNA for most bacterial species to permit normalization. [We previously had established that relative SFB levels estimated from fecal pellets reflected well those in the distal small intestine (Fig. S1)]. As anticipated, SFB colonization varied at the different animal facilities, the “cleanest” site being JAX, which showed undetectable levels in the three strains examined (Fig. 1A). Less expected was that TAC harbored both SFB-positive (B6, BALB/c) and SFB-negative (NOD) strains. Interestingly, BALB/c and NOD mice from the NRB facility were variably colonized with SFB; this variation probably reflects the fact that this colony is replenished periodically with new mice from an SFB-negative seed colony that we maintain in Bar Harbor.
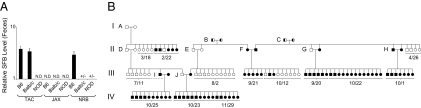
Fig. 1.
SFB status of mice housed in different animal facilities. (A) Variable SFB colonization of different strains at the different sites. PCR-determined relative SFB levels in mice housed at TAC, JAX, or the NRB facility. Each bar represents the average level +/− SEM for six mice (three males and three females). Fecal pellets taken from mice that were shipped to the NRB facility from TAC or JAX were collected and processed or were frozen within 24 h of arrival. Mice were 5–6 wk of age at the time of feces collection. SFB negativity in the NOD strain was confirmed 10 months later with additional sets of mice at 6–8 wk of age. (B) SFB colonization through four generations of NOD mice housed at the NRB facility. Squares represent males; circles indicate females. Black symbols indicate SFB-positive mice; white symbols indicate SFB-negative mice; mixed black/white symbols indicate mice of unknown SFB status. Each litter is delineated by the brackets below, with the date of birth (month/day) within 2010 indicated. SFB status of pups was assessed by PCR of fecal DNA at 4–6 wk of age except for breeding cages A–J. Parental feces were collected and examined for the presence of SFB within 2–3 wk of the litter encompassing the line of descent. The parental age at the time of feces collection varied between 2.5 and 6 mo.
Fig. 1B shows the pattern of SFB transmission within a group of NOD mice housed at the NRB facility, followed across four generations (I–IV) in 2010. Various modes of bacterial transfer are in evidence. (i) Vertical passage by the mother is illustrated by breeding cage J. All offspring from this cage born on either 10/23 or 11/29 were SFB-positive, indicating complete maternal transmission. (ii) Horizontal passage is suggested by breeding cage I. The father in this cage derived from SFB-negative parents and was a littermate of the six SFB-negative offspring of cage D. Likely, he was colonized through cohousing with the unrelated SFB-positive female with which he mated in cage I. Indeed, independent cohousing experiments with mice of the same sex but opposite SFB status have confirmed horizontal transmission within 1–2 wk of cohousing. (iii) Incomplete vertical passage is illustrated by the mixed SFB status of the offspring of breeding cage A, suggesting that this bacterium may be lost over time, leading to incomplete transmission for longer-term cages, resulting in SFB-negative offspring until an SFB-positive breeder is again introduced into the line (as per breeder cage I).
In short, different animal facilities, different strains within the same facility, or different individuals within the same strain can show divergent SFB colonization. Given the important impact of SFB on the immune system (5, 6, 17, 26), this variable needs to be taken into account more seriously when considering data from immunological studies.
Protection from Type 1 Diabetes in Female NOD Mice Segregates with SFB Colonization.
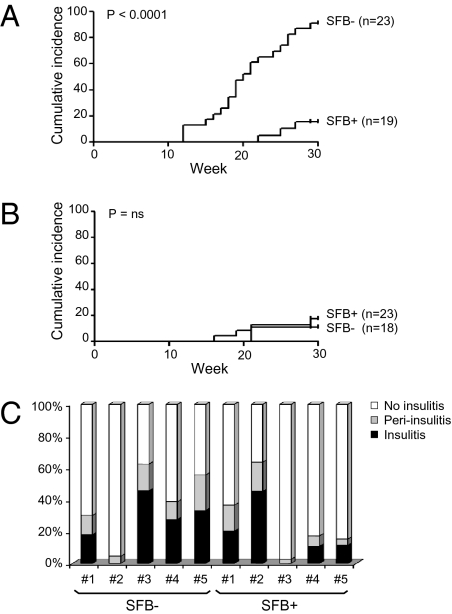
Diabetes incidence in NOD mice housed at the NRB facility is about 50–60% for females and 10–15% for males, in the lower half of the world-wide range (22) and below the ∼80% and 90% incidence currently reported on the websites of the TAC and JAX facilities, respectively. Wondering whether the lower penetrance at our facility might somehow reflect partial colonization by SFB, we cotracked disease development and SFB status in 41 SFB-negative and 42 SFB-positive NRB-housed NOD mice. There was a clear-cut segregation of high diabetes incidence with SFB negativity in the females: only 16% of those colonized by SFB developed disease by 30 wk of age, but 91% of those that were SFB negative did (Fig. 2A). The latter value is similar to the high incidences of diabetes mentioned above for the SFB-negative NOD colonies at TAC and JAX. In contrast, there was no such correlation in males housed at the NRB facility; the incidence of diabetes was similarly low, <20%, whether or not they were colonized with SFB (Fig. 2B).
Fig. 2.
Diabetes incidence and insulitis scores in SFB-negative and SFB-positive NOD mice at the NRB facility. (A and B) Cumulative incidence curves for female (A) and male (B) NOD mice identified as SFB positive or SFB negative at 4–6 wk of age. Diabetes development was monitored weekly starting at 10 wk of age. The difference between the two groups was statistically significant in females (P < 0.0001, log-rank test) but not in males. (C) Insulitis scores from SFB-colonized female NOD mice at 10 wk of age and age-matched SFB-negative controls. Pancreata were prepared and stained with H&E, and islet cell infiltration was scored as described in SI Materials and Methods. Each bar represents an individual mouse.
Insulitis was similar in level and appearance in 10-wk-old SFB-negative and SFB-positive females, most individuals of both types showing at least some degree of frank islet infiltration (Fig. 2C). This finding argues that SFB colonization does not block the disease trigger in NOD mice but instead somehow may modulate diabetes unfolding.
Induction of IL-17–Producing CD4+ T Cells in the SI-LP of SFB-Positive NOD Females.
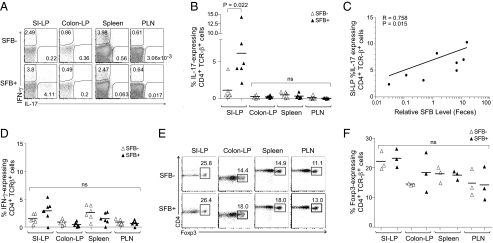
Given reports that SFB rather specifically promotes the development of a robust Th17 population in the SI-LP of B6 mice (6, 17), we used multiparameter flow cytometry to compare the SI-LP and related lymphoid compartments of SFB-positive and SFB-negative individuals of the NOD strain. Indeed, there was a clear induction of IL-17–expressing CD4+TCRβ+ cells in the SI-LP of NOD females colonized with SFB vis-à-vis those not colonized (Fig. 3A). The fraction of IL-17–expressing cells varied widely in individual SFB-positive females (Fig. 3B), but there was a good correlation between this fraction and the fecal pellet SFB titer (Fig. 3C). In contrast, there was no significant induction of Th17 cells in the colonic lamina propria, spleen, or pancreatic lymph nodes (PLN), the site where naive diabetogenic T cells are primed initially by pancreas-derived antigens (27) (Fig. 3 A and B). Nor was there a significant enrichment of the IFN-γ–expressing Th1 population in any of the lymphoid tissues examined (Fig. 3 A and D).
Fig. 3.
Induction of Th17 cells in the SI-LP of SFB-positive NOD females. (A) Lymphocytes were isolated from the indicated lymphoid tissues of SFB-positive or SFB-negative female NOD mice at 6–8 wk of age. Intracellular cytokine expression was enhanced by a 4-h culture in the presence of phorbol ester and ionomycin. Cells were gated by side-scatter and as CD45+, CD19−, CD8−, TCRβ+, and CD4+. Shown is a representative plot for IL-17 and IFN-γ staining from six mice analyzed. (B) Frequencies of IL-17–producing CD4+TCRβ+ cells in various lymphoid organs of SFB-positive and SFB-negative NOD mice. Gating was done as per A. Statistical analysis was performed using the nonparametric Mann–Whitney test. (C) Correlation of relative SFB levels in fecal pellets (determined as per Fig. 1) and percentage of IL-17–expressing CD4+TCRβ+ cells (as per A and B). Statistical analysis was performed using the Pearson correlation. (D) Frequencies of IFN-γ–producing CD4+TCRβ+ cells in the same lymphoid organs of the NOD mice depicted in B. Gating was done as per A. (E and F) Representative flow cytometry plots of three mice analyzed and frequencies of Foxp3-expressing CD4+TCRβ+ cells for various lymphoid tissues of age-matched SFB-positive and SFB-negative NOD mice. Cells were gated by side-scatter and as CD45+, CD19−, CD8−, TCRβ+, and CD4+. Ns, not significant.
Because it has been argued that Foxp3+CD4+ Treg cells may have a reciprocal relationship with Th17 cells (28, 29), we paid special attention to this subset. However, SFB-positive and SFB-negative NOD females housed at the NRB facility had very similar Treg fractions in each of the four lymphoid tissues examined, notably in the colonic lamina propria (Fig. 3 E and F).
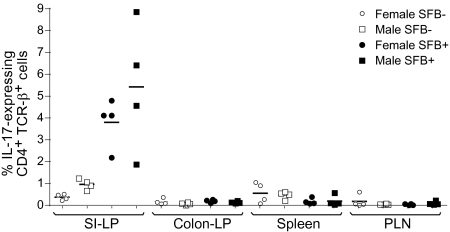
More broadly, we saw no substantial, repeatable differences in the total αβ, total CD4+, CD8+, or γδ T-cell compartments of SFB-colonized and SFB-free female NOD mice. For all of the populations examined to date, males and females were indistinguishable; most importantly, SFB-negative NOD males, just like their female counterparts, had few IL-17–producing CD4+TCRβ+ cells in the SI-LP and colon lamina propria, and, vice versa, the SFB-positive male and female pairs examined concomitantly had similarly high numbers of SI-LP Th17 cells (Fig. 4).
Fig. 4.
Comparison of Th17 cell numbers in the SI-LP of female versus male NOD mice classed by SFB status. Frequencies of IL-17–producing CD4+TCRβ+ cells in various lymphoid organs of SFB-positive and SFB-negative female and male NOD mice. Lymphocytes were isolated and stained and were gated as per Fig. 3A. IL-17 expression levels from four SFB-positive and four SFB-negative female NOD mice from Fig. 3 were paired with SFB-positive and SFB-negative male mice processed on the same day. Statistical analysis was performed using the paired t test. No statistically significant differences between SFB-positive and SFB-negative NOD mice were found in any compartment except the SI-LP. The P values for SI-LP IL-17 levels from SFB-positive versus SFB-negative NOD mice are 0.0097 for females and 0.0459 for males.
Specific Induction of Th17 Signature Genes in CD4+ T Cells from the SI-LP of SFB-Positive NOD Mice.
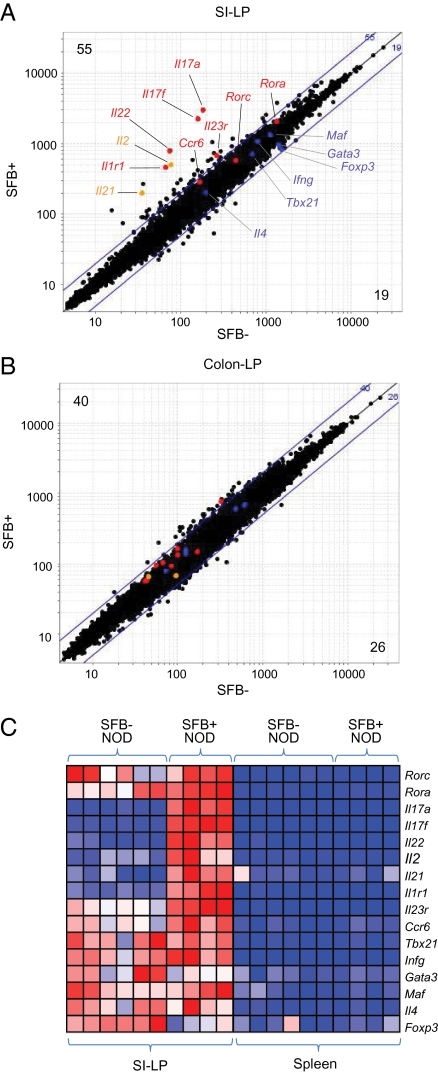
To explore further SFB-induced changes in the CD4+ T-cell compartment of the SI-LP in an unbiased manner, we performed gene-expression profiling using Affymetrix microarrays. The profiles of CD4+ SI-LP T cells from 6- to 10-wk-old SFB-positive (n = 4) and SFB-negative (n = 6) NOD mice were quite similar overall: In individuals colonized by SFB, only 55 and 19 transcripts were up- and down-regulated, respectively, at an arbitrary fold-change threshold of 2 [see Fig. 5A, and Table S1 for raw data and false discovery rates (FDR) and Fig. S2 for individual gene plots], compared with 16 and 15 transcripts over- or underrepresented, respectively, in randomized control datasets. Consistently up-regulated in SFB-positive NOD mice was a set of Th17 signature genes, including Il17a, Il17f, Il22, Il1r1, and Il23r (indicated in red). In contrast, the levels of transcripts characteristic of Th1, Th2, and Treg cells (shown in blue) were not influenced by SFB status. SFB colonization also increased the representation of Il2 and Il21 transcripts in CD4+ SI-LP T cells, of interest because the corresponding genes fall within one of the more potent diabetes susceptibility intervals, idd3 (30). In line with the flow cytometric data (Fig. 3 A and B), Th17 signature genes were not induced in CD4+ T cells residing in the colonic lamina propria (Fig. 5B and Table S2).
Fig. 5.
Specific induction of Th17 transcripts in SI-LP CD4+ T cells of SFB-positive NOD mice. (A) Affymetrix microarray analysis of SI-LP CD4+ T cells from SFB-negative (x axis) versus SFB-positive (y axis) NOD mice 6–10 wk of age. Highlighted are several genes characteristic of Th17 cells (red type), other CD4+ T-cell subsets (blue type), or genes located within the idd3 diabetes susceptibility interval (orange type). Values in the upper left and lower right corners refer to the numbers of loci up- or down-regulated, respectively, by more than twofold. (B) As in A, except that values for colonic lamina propria transcripts are plotted. (C) An expression heatmap for transcription factors and cytokines diagnostic of Th17 cells and other CD4+ T-cell subsets. Data are from SI-LP or splenic CD4+ T cells from NOD mice that were positive or negative for SFB. All mice were 6–10 wk of age. Data are row-normalized.
The heatmap of Fig. 5C reemphasizes the specific induction of transcripts characteristic of Th17 cells in the CD4+ SI-LP T cells of SFB-positive versus SFB-negative NOD mice housed at the NRB facility. It also illustrates the striking difference vis-à-vis splenic CD4+ T cells, which show no signs of induction, again consistent with the flow cytometry data (Fig. 3 A, B, and D).
Discussion
That autoimmune diabetes is exacerbated in NOD mice housed under GF conditions was first reported almost 25 y ago (23), prompting widespread speculation that commensal microbiota normally keep islet-directed autoimmunity in check. An influence of gut-resident microbes would be consistent with several indications of a privileged link between the gut and pancreas/PLN, notably a preferential antigen-trafficking route (31) and shared lymphocyte homing addressins (32, 33). However, more recent reports that GF and SPF NOD mice show very similar disease profiles (24, 34, 35) have clouded the issue. This report offers an explanation for at least some of these discrepancies. Extending recent observations that B6 mice housed in the TAC and JAX animal facilities were differentially colonized by SFB (6), we showed that NOD mice kept at different sites also carried different loads of this gut commensal. In our variably infected NRB colony, SFB-negative females had a high diabetes incidence, approaching complete penetrance, whereas SFB-positive females were protected from diabetes. Obviously, then, disease exacerbation when comparing GF and SPF mice would be most evident in a colony infected with SFB and would be only minimal in an SFB-negative colony.
When considering a possible mechanism for SFB-mediated protection from NOD diabetes, a role for Th17 cells immediately comes to mind. De novo introduction of SFB into mice in several experimental contexts quite specifically promoted the development of a robust Th17 compartment in the SI-LP (6, 17) [although one group did observe a broader influence on Th subsets (5)], and the impact of these cells was translated distally to promote nongut autoimmune diseases (17, 18). The function of IL-17–producing cells in NOD diabetes has been a topic of much debate. Some investigators have argued for a critical role, in particular in later, effector processes (36–38), although others have cautioned about possible misinterpretations of findings from Th17 transfer experiments because of the conversion to or expansion of Th1 cells (39, 40). In clear contradiction, IL-17–producing cells were found either to inhibit diabetes in NOD mice, as well as in BioBreeding rats, in several experimental contexts (41–43) or to have no effect (44). These divergent conclusions likely reflect multiple complexities: that different Th subsets may play variable roles according to the particular disease stage; that multiple cell types can produce IL-17; and that Th17 cells produce cytokines other than IL-17. Nonetheless, an attractive hypothesis to pursue is that SFB colonization of NOD mice induces the emergence of a robust Th17 population in the gut, and that this population can impact diabetogenesis by inhibiting the islet-directed Th1 response, a notion consistent with a recent report that Th17 cells inhibit Th1 effector cells in a colitis model (45). Because insulitis appeared to be typical in 10-wk-old SFB-positive females, Th17 cells would not influence disease triggering but rather would dictate the “flavor” of the insulitis that sets in and/or its evolution to overt diabetes.
It also is possible that Th17-produced mediators other than the signature cytokine IL-17A are responsible for SFB-promoted protection from NOD diabetes. For example, IL-22, a member of the IL-10 cytokine family, is thought to confer transepithelial resistance to injury (as reviewed in refs. 46, 47). Breach of the gastrointestinal barrier with subsequent leakage of potentially immunogenic dietary components has been proposed to promote T1D in NOD mice (48) and is a well-established trigger of celiac disease, an anti–gliadin-directed autoimmune disorder that frequently accompanies T1D in humans (49, 50). Moreover, we previously demonstrated that gut irritation in NOD mice alters the priming of diabetogenic islet-derived self-antigens in the PLNs (31). It is possible that, in the absence of SFB, other gut microbes provoke intestinal stress in female NOD mice; SFB-induced Th17 cells might produce mediators, such as IL-22, that are beneficial for epithelial barrier integrity. The unusually intimate interactions of SFB with the gut epithelium might be another element impacting the gut barrier (51).
Interestingly, SFB was not primarily responsible for the low incidence of diabetes in male NOD mice in our colony: Both SFB-positive and SFB-negative males had a low incidence of <20%. Because a substantially higher penetrance is achievable in other colonies (in particular GF colonies) (e.g., ref. 24), we suspect that another microbe (or other microbes) might be responsible. A different downstream mechanism probably is involved, because SFB-negative males and females had similarly low levels Th17 cells in the SI-LP (and elsewhere), and Th17 cells and signature transcripts were similarly induced in SFB-positive mice of both sexes. A comparison of the male and female gastrointestinal microbiomes via high-throughput sequencing should prove informative in this regard.
In conclusion, we have demonstrated that cosegregation of diabetes protection with a single gut-resident commensal can explain variable penetrance of an organ-specific autoimmune disease in different mouse colonies. It is likely that patterns of colonization by immunomodulatory commensals also are at play in other immunologic disease models with varying penetrance. Finally, we speculate that the development of autoimmune disease in genetically predisposed humans also might be strongly influenced by commensal colonization patterns, especially considering that microbiota represent a rich set of “environmental modifiers,” with more than 100 trillion bacteria persistently colonizing our mucosal surfaces (52). SFB itself has not yet been identified in humans, but the possibility that another microbe fills its gut niche remains unexplored.
Materials and Methods
Mice.
B6, BALB/c, and NOD mice were purchased from TAC or JAX or were housed in our animal facility at the NRB. Details of strains and their handling are given in SI Materials and Methods.
Tissue Collection and DNA Isolation.
Procedures entailed in feces collection, dissection of intestinal segments, lymphoid tissue preparation, and genomic DNA isolation are detailed in SI Materials and Methods.
SFB Quantification.
Genomic DNA from fecal pellets or tissues was amplified with SFB-specific [SFB736F and SFB884R (53)] and conserved EUB 1114F and 1221R (54) 16S ribosomal DNA primers. Primer sequences are listed in SI Materials and Methods. The ratio of the EUB/SFB values, calculated as in ref. 6, yielded relative SFB levels.
Diabetes and Insulitis Assessments.
Mice were screened weekly for diabetes development as described in SI Materials and Methods. Insulitis was assessed at 10 wk of age as detailed in SI Materials and Methods.
Preparation of Intestinal Cell Suspensions for Immunologic Analyses.
Cell suspensions were prepared from the SI-LP and colon lamina propria of 6- to 10-wk-old NOD mice as recently described (17) and as detailed in SI Materials and Methods. Cell preparations from the spleen and PLN were by standard procedures, as described in ref. 31.
Flow Cytometry.
Flow cytometric analyses of lymphocytes from the small intestine, colon, spleen, and PLN were performed as described previously (17, 55) and are explained in more detail in SI Materials and Methods.
Microarray Analysis.
Cells were sorted, RNA was isolated and hybridized to GeneChip Mouse Gene 1.0 ST arrays (Affymetrix), and data were analyzed as described in SI Materials and Methods. Datasets are available at the National Center for Biotechnology Information (accession no. 29806).
Statistical Analyses.
Data are presented as mean ± SEM. Statistical significance was assessed using the student's t test, Mann–Whitney test, or Pearson correlation, as indicated in the figure legends. The log-rank test was used for the cumulative incidence analyses. P values <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank A. Wilcox, K. Hattori, and A. Ortiz-Lopez for assistance with mice; K. Leatherbee and J. Ericson for help with microarrays; and C. Laplace for figure preparation. This work was supported by Grant 5R01 DK59658 from the National Institutes of Health and by a generous gift from the Howalt family to D.M. and C.B. J.A.H. was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research and the Canadian Diabetes Association. H.-J.W. was supported by a grant from the Arthritis National Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The datasets reported in this paper are available at the National Center for Biotechnology Information (accession no. 29806).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108924108/-/DCSupplemental.
References
- 1.Finke D. Induction of intestinal lymphoid tissue formation by intrinsic and extrinsic signals. Semin Immunopathol. 2009;31:151–169. doi: 10.1007/s00281-009-0163-6. [DOI] [PubMed] [Google Scholar]
- 2.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 10.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 12.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 16.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2010;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson MS, Bluestone JA. The NOD mouse: A model of immune dysregulation. Annu Rev Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 20.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 21.Zipris D. Epidemiology of type 1 diabetes and what animal models teach us about the role of viruses in disease mechanisms. Clin Immunol. 2009;131:11–23. doi: 10.1016/j.clim.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Pozzilli P, Signore A, Williams AJK, Beales PE. NOD mouse colonies around the world—recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki T, et al. In: Immune-Deficient Animals in Biomedical Research. Rygaard J, Brunner N, Groem N, Spang-Thomsen M, editors. New York: S. Karger Publishers Inc; 1987. pp. 112–116. [Google Scholar]
- 24.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2009;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Höglund P, et al. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J Exp Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridgway WM, et al. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol. 2008;100:151–175. doi: 10.1016/S0065-2776(08)00806-7. [DOI] [PubMed] [Google Scholar]
- 31.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci USA. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hänninen A, Salmi M, Simell O, Jalkanen S. Mucosa-associated (beta 7-integrinhigh) lymphocytes accumulate early in the pancreas of NOD mice and show aberrant recirculation behavior. Diabetes. 1996;45:1173–1180. doi: 10.2337/diab.45.9.1173. [DOI] [PubMed] [Google Scholar]
- 33.Yang XD, Sytwu HK, McDevitt HO, Michie SA. Involvement of beta 7 integrin and mucosal addressin cell adhesion molecule-1 (MAdCAM-1) in the development of diabetes in obese diabetic mice. Diabetes. 1997;46:1542–1547. doi: 10.2337/diacare.46.10.1542. [DOI] [PubMed] [Google Scholar]
- 34.Alam C, et al. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia. 2011;54:1398–1406. doi: 10.1007/s00125-011-2097-5. [DOI] [PubMed] [Google Scholar]
- 35.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS ONE. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, et al. Adjuvant treatment suppresses IL-17 production by T cell-independent myeloid sources in nonobese diabetic mice. Mol Immunol. 2010;47:2397–2404. doi: 10.1016/j.molimm.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Jain R, et al. Innocuous IFNgamma induced by adjuvant-free antigen restores normoglycemia in NOD mice through inhibition of IL-17 production. J Exp Med. 2008;205:207–218. doi: 10.1084/jem.20071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emamaullee JA, et al. Inhibition of Th17 cells regulates autoimmune diabetes in NOD mice. Diabetes. 2009;58:1302–1311. doi: 10.2337/db08-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bending D, et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin-Orozco N, Chung Y, Chang SH, Wang YH, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol. 2009;39:216–224. doi: 10.1002/eji.200838475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikoopour E, et al. Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J Immunol. 2010;184:4779–4788. doi: 10.4049/jimmunol.0902822. [DOI] [PubMed] [Google Scholar]
- 42.Tse HM, et al. NADPH oxidase deficiency regulates Th lineage commitment and modulates autoimmunity. J Immunol. 2010;185:5247–5258. doi: 10.4049/jimmunol.1001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau K, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 44.Komiyama Y, et al. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J Immunol. 2006;177:566–573. doi: 10.4049/jimmunol.177.1.566. [DOI] [PubMed] [Google Scholar]
- 45.O'Connor W, Jr, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connor W, Jr, Zenewicz LA, Flavell RA. The dual nature of T(H)17 cells: Shifting the focus to function. Nat Immunol. 2010;11:471–476. doi: 10.1038/ni.1882. [DOI] [PubMed] [Google Scholar]
- 47.Sonnenberg GF, Fouser LA, Artis D. Functional biology of the IL-22-IL-22R pathway in regulating immunity and inflammation at barrier surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 48.Lee AS, et al. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia. 2010;53:741–748. doi: 10.1007/s00125-009-1626-y. [DOI] [PubMed] [Google Scholar]
- 49.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: The complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visser J, Rozing J, Sapone A, Lammers K, Fasano A. Tight junctions, intestinal permeability, and autoimmunity: Celiac disease and type 1 diabetes paradigms. Ann N Y Acad Sci. 2009;1165:195–205. doi: 10.1111/j.1749-6632.2009.04037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chase DG, Erlandsen SL. Evidence for a complex life cycle and endospore formation in the attached, filamentous, segmented bacterium from murine ileum. J Bacteriol. 1976;127:572–583. doi: 10.1128/jb.127.1.572-583.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 53.Barman M, et al. \ salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 55.Feuerer M, et al. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci USA. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.