Abstract
According to theory, gene flow to marginal populations may stall or aid adaptation at range limits by swamping peripheral populations with maladaptive gene flow or by enhancing genetic variability and reducing inbreeding depression, respectively. We tested these contrasting predictions by manipulating patterns of gene flow of the annual plant, Mimulus laciniatus, at its warm range limit. Gene flow was experimentally applied by using crosses within warm-limit populations (selfed and outcrossed), between warm-limit populations, and between warm-limit and central range populations across two elevational transects. We measured the fitness of offspring in a common garden at the warm-edge species range limit. All sources of gene flow increased seedling emergence at the range limit, suggesting local inbreeding depression at both range limit populations; however, lifetime reproductive success only increased significantly when pollen originated from another warm-limit population. Center–to–warm-edge gene flow was maladaptive by delaying time to development at this warm, fast-drying range limit, whereas edge-to-edge gene flow hastened emergence time and time to reproduction. By empirically testing theory on the effects of gene flow on the formation of geographic range limits, we find benefits of gene flow among populations to be greatest when gene flow is between populations occupying the same range limit. Our results emphasize the overlooked importance of gene flow among populations occurring near the same range limit and highlight the potential for prescriptive gene flow as a conservation option for populations at risk from climate change.
Keywords: climate adaptation, ecological gradients, natural selection, phenology, species range limits
Theory on the evolution of range limits predicts that gene flow from large, central populations to edge populations at the range limit could create a flood of maladaptive, nonlocal genes, thereby stalling adaptation and niche expansion (1–3). Alternatively, gene flow from central populations may increase effective population size and genetic variation in edge populations, thereby ultimately increasing fitness at the range limit and perhaps contributing to range expansion (4–6). Overlooked in these models is gene flow among edge populations, which may be especially beneficial because it (i) supplies both favorable alleles or gene combinations that are adaptive at range limits (7) and (ii) enhances genetic variation upon which selection may act. These ideas have not been tested empirically in natural systems at geographic range limits.
With rapid climate change, low genetic variation may constrain the ability of populations to adapt quickly to warming environments (8–10). Globally, climate warming is pushing species ranges upwards in elevation, leaving rear-edge populations to adapt, migrate, or perish (9, 10). Mimulus laciniatus (cut-leaved monkeyflower), an annual plant, inhabits mossy areas on granite seeps between 975 and 3,270 m on the western slope of the California Sierra Nevada Mountains and is representative of species that may experience range contractions under climate change. Snow accumulation is predicted to decrease dramatically in the Sierra Nevada during the 21st century, which could profoundly constrict species geographic ranges (11) and shift vegetation communities (ref. 12; but see ref. 13). Plants from the center of the species range of M. laciniatus mostly occur in middle elevations in mixed coniferous montane woodlands (hereafter referred to as montane habitats) that receive regular snowpack during the winter months (i.e., wet season). The warm, low-elevation, rear-edge range limit in M. laciniatus occurs near the lower edge of the Sierran snow line at approximately 900 m. Near this elevation, precipitation decreases, snow accumulation stops abruptly, and plant communities transition from summer to winter growing seasons within warm, fast-drying, foothill woodland environments (14) (hereafter referred to as foothill environments). M. laciniatus species range limits at low elevations may therefore represent future climates in which a decreased snowpack results in drier or faster-drying environments. Current estimates of Sierran snowpack reduction by the end of this century vary from 25% to more than 90% depending on climate change model and emission scenario (12, 15).
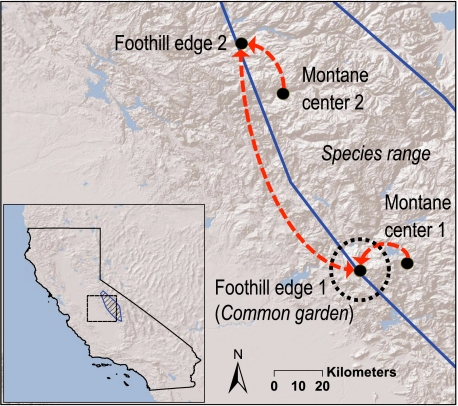
Across the geographic range of M. laciniatus, we tested model predictions of the effects of gene flow on fitness at range limits by creating gene flow (through experimental crosses) between central and range-limit (i.e., warm edge) populations, and between distant populations occupying the same range limit. After a common generation in the greenhouse, crosses were made between plants whose parents were located along two replicate elevational transects occupying different watersheds in the Sierra Nevada. M. laciniatus is primarily a self-fertilizing species and is highly inbred within populations, but does exhibit occasional heterozygosity [fixation index (F) for study populations ranged between 0.867 and 0.953; Table S1]. Despite high rates of selfing, M. laciniatus flowers are showy and can attract pollinators, and we have observed bee visitation to open flowers in the field; thus, outcrossing can happen at low frequency. Given a baseline selfing reproductive mode, we used self-fertilized progeny as a control against which to test the effects of gene flow from local and central populations and the other edge population (i.e., between transects). All crosses were made between 10 sires and 20 dams. Crosses were made as follows: (i) center-to-edge by using montane pollen donors and foothill dams, (ii) edge-to-edge by using foothill sires and dams reciprocally from the two elevational edges, and (iii) local outcrossing within both foothill populations. The seeds created from these crossing treatments from both transects were planted into a common garden at the lower range limit site having the lowest elevation and latitude and the warmest environment (Fig. 1 and Table S1). Lifetime fitness estimates of outcrossed progeny were then compared with local, selfed progeny in each population.
Fig. 1.
The species range (Inset) of the California endemic plant M. laciniatus. Experimental gene flow (red dashed arrows) was applied to populations at the low-elevation foothill woodland range limit, and replicated along two transects from central montane populations. Foothill populations were also mated internally (i.e., local mating) and to each other. All progeny were planted at a common garden at foothill edge 1.
Results and Discussion
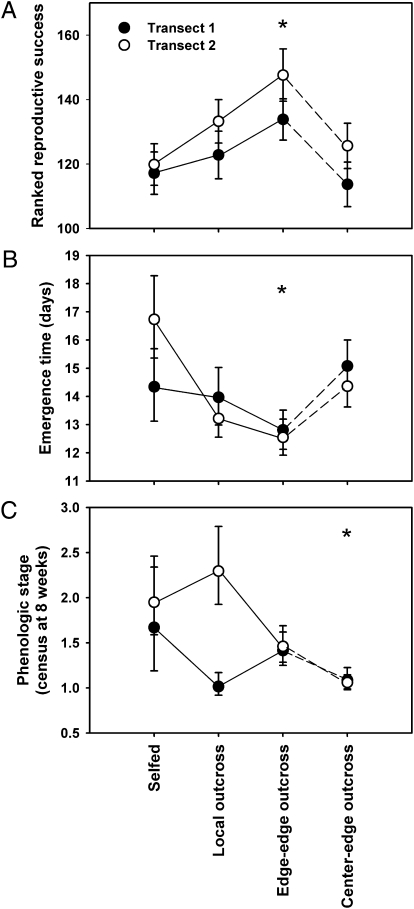
We found evidence supporting the divergent effects of gene flow on range limit populations assumed by two models: although gene flow can increase reproductive success, it does so most when gene flow occurs between populations in similar climatic environments. Edge-to-edge crosses exhibited increased emergence and survival and faster development, and had the highest fitness in the fast-drying edge environment. As a result, the effect of gene flow between warm edge populations that experience similar selective environments was highly beneficial at the warm range limit; progeny produced by selfing in each warm edge population had lower fitness than those from edge-to-edge crosses. In contrast, center-to-edge gene flow increased emergence rates relative to locally selfed plants, but these plants were maladapted to the warm edge, emerging later, and thus achieving lower fitness than edge-to-edge crosses. This result supports range models suggesting that center-to-edge gene flow can be maladaptive (e.g., ref. 1).
Common garden conditions were severe, and (i) outbreeding and (ii) the source of pollen contributing to progeny were very important determinants of plant success at the range limit. Survival of experimental plants to reproduction at the range limit garden was low as a result of desiccation; approximately 22% emerged and approximately 2% of plants survived to produce fruits. Naturally occurring individuals adjacent to experimental blocks also experienced high mortality from desiccation. Gene flow (cross type) significantly affected lifetime reproductive success (P = 0.004; Table 1 and Fig. 2A). The progeny of edge-to-edge crosses had significantly greater lifetime reproductive success than selfed progeny (Dunnett means comparison, P = 0.004; Table 2), resulting from increased survival (χ2 = 12.867; P < 0.005) and increased fruit production (χ2 = 509.51; P < 0.001; SI Materials and Methods and Table S2), whereas progeny from local outcrosses and center-to-edge crosses did not significantly differ from selfed progeny (P = 0.331 and P = 0.996, respectively; Table 2). Plants maternally derived from the transect 2 edge population had greater lifetime reproductive success (P = 0.041), and there was no cross type-by-transect interaction (P = 0.833).
Table 1.
REML analyses for cross-type responses of M. laciniatus at a range-limit common garden for fitness, development, and emergence traits
| Response variable | df | Error df | F statistic | P value |
| Lifetime reproductive success | ||||
| Cross class | 3 | 241 | 4.56 | 0.004* |
| Transect | 1 | 241 | 4.23 | 0.041* |
| Cross class × transect | 3 | 241 | 0.29 | 0.833 |
| Emergence time | ||||
| Cross class | 3 | 347 | 6.1 | 0.001* |
| Transect | 1 | 347 | 0.08 | 0.780 |
| Cross class × transect | 3 | 347 | 0.96 | 0.413 |
| Phenologic stage (at 8 wk) | ||||
| Cross class | 3 | 46 | 4.25 | 0.010* |
| Transect | 1 | 46 | 5.06 | 0.029* |
| Cross class × transect | 3 | 46 | 3.17 | 0.033* |
Table shows a summary of REML analyses for cross-type (i.e., gene flow) responses of M. laciniatus at a range-limit common garden for fitness, development (i.e., phenologic stage), and emergence traits. All traits were rank-transformed.
*Significant difference at P < 0.05.
Fig. 2.
Responses of M. laciniatus plants from each gene flow treatment grown at the low-elevation range limit. Center-to-edge responses are distinguished from other treatments by dashed lines. (A) Ranked lifetime reproductive success. Data were ranked in order from zero (no emergence) to plants having the highest total fruit mass. (B) Time to seedling emergence expressed in days. (C) Phenologic stage (measured as 1, vegetative; 2, flower budding; 3, flowering; 4, fruiting) at census period (∼8 wk after planting) when phenologic variation was greatest among plants. All variables were rank-transformed as they could not be transformed to meet parametric assumptions. Means and SEs are back-transformed fitted values from the REML models (Materials and Methods, Tables 1, and 2 show model structure). Error bars are least-square means ± SEs. *Significant differences versus responses of selfed plants, Dunnett mean comparisons within REML models.
Table 2.
Lifetime reproductive success, time to emergence, and phenologic stage results of cross-type treatments versus self-fertilized control progeny of M. laciniatus
| Response variable | df | T statistic | P value |
| Lifetime reproductive success | |||
| Local-edge crosses vs. selfed parentals | 241 | 1.47 | 0.331 |
| Edge–edge vs. selfed parentals | 241 | 3.36 | 0.003* |
| Center–edge vs. selfed parentals | 241 | 0.18 | 0.996 |
| Emergence time | |||
| Local-edge crosses vs. selfed parentals | 347 | −1.91 | 0.118 |
| Edge–edge vs. selfed parentals | 347 | −3.26 | 0.003* |
| Center–edge vs. selfed parentals | 347 | −0.79 | 0.692 |
| Phenologic stage | |||
| Local-edge crosses vs. selfed parentals | 46 | −1.02 | 0.545 |
| Edge–edge vs. selfed parentals | 46 | −0.95 | 0.592 |
| Center–edge vs. selfed parentals | 46 | −2.93 | 0.013* |
Table shows lifetime reproductive success, time to emergence, and phenologic stage (at 8 wk after planting) results for Dunnett mean comparisons of cross-type (i.e., gene flow) treatments to self-fertilized control progeny of Mimulus laciniatus at a range-limit common garden. Dunnett tests were generated within individual REML models for each trait variable and all variables were rank-transformed.
*Significant difference at P < 0.05.
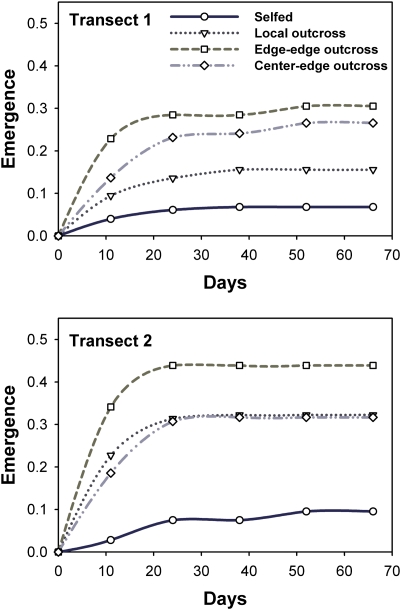
Emergence success and phenologic matching to the environment played a key role in determining lifetime reproductive success. Specifically, earlier development was beneficial in avoiding fitness reductions later in the growing season that were likely caused by soil moisture depletion. Phenotypic selection analysis revealed directional selection against later emergence (β = −1.810; df = 41; F = 10.267; P = 0.003), as well as directional selection for early reproduction (β = 3.233; df = 41; F = 28.141; P < 0.001). The offspring of crosses between foothill edge populations emerged earliest (P < 0.001) and were most reproductively advanced by 8 wk (P = 0.010; Table 2 and Fig. 2B). Although not significant, local outcrossing did appear to affect lifetime reproductive success, emergence time, and phenologic stage in the same qualitative manner as edge–edge outcrossing (Table 2 and Fig. 2); i.e., local outcrossing appeared to be adaptive, but to a lesser degree than edge–edge outcrossing. Locally outcrossed plants had significantly greater emergence rates than selfed plants (df = 3, χ2 = 111.825, P < 0.0001; Fig. 3). In fact, all types of outcrossing improved emergence (Fig. 3), suggesting that these edge populations suffer from inbreeding depression. Plants maternally derived from the transect 2 edge population flowered earlier on average (P = 0.029; Table 1 and Fig. 2C), but this transect effect occurred primarily in selfed and locally outcrossed progeny (interaction of cross type and transect, P = 0.033). Time to emergence did not differ by transect (P = 0.780), and there was no transect by cross-type interaction (P = 0.413; Table 1). However, transect 2 plants had greater emergence (proportional-hazards model, df = 1; χ2 = 12.929; P < 0.001), with no significant interaction effect between transect and cross type (df = 3; χ2 = 4.293; P = 0.232). Performance differences between transects were not likely to have been caused by climate, as transects differed little in temperature and precipitation (Table S1), and there was no evidence for local adaptation between edge populations (edge 1 vs. edge 2). In contrast, plants from the nonlocal transect appeared to have slightly improved emergence rates, phenologic timing, and reproductive success, suggesting how one edge population may benefit another. Edge-to-edge cross types emerged earlier than selfed progeny (Dunnett test, P = 0.003), whereas locally crossed and center-to-edge cross types did not significantly differ from selfed edge plants (P = 0.118 and P = 0.692, respectively; Fig. 2B and Table 2). Thus, the increased lifetime reproductive success of edge-to-edge cross types versus selfed plants resulted from a combination of increased emergence and better phenotype/habitat matching through faster emergence times (Figs. 2B and 3).
Fig. 3.
Time-to-emergence plots (displayed by transect) showing the proportion of M. laciniatus plants emerged from gene flow treatments when offspring of experimental crosses (n = 1,640) were grown at the low-elevation range limit. Time periods shown occurred before most drought-induced mortality.
Seasonal timing is an important factor for climate adaptation (16), and the observed later flowering in higher elevation genotypes (center-to-edge crosses) may be adaptive when developing plants encounter slower growing degree day accumulation, longer photoperiods, and greater precipitation at higher elevations (14). For example, Waser et al. (17) found the date of snowmelt to be the strongest predictor of differentiation in a study of morphological variation and mating success among populations of Mimulus guttatus occupying different elevations of the Rocky Mountains. Clausen and Hiesey (18) observed that later flowering was adaptive at higher elevations in the Sierran plant, Potentilla glandulosa (19) [However, see Angert et al. (20).] Our findings for this highly selfing species show that center-to-edge crosses did express maladaptive traits (i.e., late flowering); however, benefits of gene flow probably also arose from overcoming local inbreeding depression (4), and allowed those plants to achieve equivalent lifetime reproductive success to selfed edge genotypes with better adapted emergence phenotypes, but slower development times. Lifetime reproductive success of center-to-edge plants was similar to that of selfed progeny (Fig. 2A and Table 2); although mean performance was lower, the top performers (i.e., upper 95th percentile) from center-to-edge crosses had a mean total fruit mass of 1.159 mg, compared with 0.653 mg, 1.488 mg, and 4.450 mg for selfed, locally crossed, and edge-to-edge groups, respectively. Thus, center-to-edge gene flow may create adaptive opportunities within a subset of individuals, but comes at a cost of introducing maladaptive alleles overall in the M. laciniatus system.
By using modeling approaches, Holt and Gomulkiewicz (4) demonstrated that, even in small sink populations, maladaptive gene flow may increase effective population size and genetic variation as long as offspring have absolute fitnesses greater than one. In contrast, gene flow from similar edge habitats may provide the benefits of increased variation without such costs because of similar selective regimes at range edges. Isolation across the elevation gradient (e.g., within watersheds as a result of elevation-induced phenology variation) may help expand the ecological niche by preventing maladaptive gene flow (sensu ref. 1). In contrast, isolation between similar elevations (e.g., between watersheds) may stall the adaptive process by preventing beneficial admixture. Edge populations are more genetically isolated from each other than they are to their respective center populations along the elevation gradient; pairwise codominant genetic distance estimates are 0.175, 0.067, and 0.049 for edge-to-edge, center-to-edge 1, and center-to-edge 2, respectively (Table S3); isolation may be a result of distance between populations as well as opportunities for gene flow that might differ within versus across watersheds. All populations were significantly differentiated from each other (P < 0.05) after accounting for multiple comparisons (21).
Gene flow between populations adapted to different environments can have a variety of outcomes, as illustrated by comparing the results from this study to those of similar studies in the literature. If some “hybrids” exhibit higher fitness than parental genotypes, gene flow can be beneficial (22). Raabová et al. (23) showed outbreeding to be advantageous in populations of the rare perennial herb Aster amellus when gene flow originated from distant, but ecologically similar, environments. Willi et al. (24) demonstrated fitness benefits from outbreeding that lasted through the second generation in the rare plant, Ranunculus reptans, particularly when populations were small and inbred. On the contrary, outcrossing may not always be beneficial, and outbreeding depression, resulting from effects of locally maladaptive alleles or gene combinations, can develop in successive generations (25). Little to no benefit of gene flow was shown in an experimentally subdivided population of the selfing species Triticum aestivum (26) and in matings among remnant prairie populations in the outcrossing species Echinacea angustifolia (27). In crosses between nonnative, diverged populations of the selfing plant, Avena barbata, Johansen-Morris and Latta (28) demonstrated that, although early-generation hybrids did not differ from the parental lineages in field and novel greenhouse conditions, late-generation hybrids had lower fitness. Nevertheless, several hybrid genotypes outperformed parental genotypes, suggesting some adaptive opportunities via novel genetic combinations. In sum, these contrasting results suggest that many factors influence the outcome of gene flow, including population size, ecological and spatial distance between target and source populations, and selective regime. Frankham et al. (29) recently provided evidence that outbreeding depression is more likely to develop between populations that meet at least one of the following conditions: have different karyotypes, have been isolated for many generations, or occupy different environments. Edge-to-edge crosses in M. laciniatus showed immediate benefits over parental genotypes; this increased fitness in early-generation hybrids creates opportunities to colonize novel (22, 28) or, in this case, stressful environments to which both maternal and paternal populations are adapted.
Theoretical models predicting the formation of range limits via failure to adapt across ecological gradients make assumptions about the effects of gene flow on fitness (7). In the Kirkpatrick and Barton model (1), range limits arise as a result of steep environmental gradients, asymmetrical gene flow from large central populations to smaller peripheral populations, and low adaptive potential in peripheral populations. The relative magnitude of these forces, and the demographic impacts of initially maladaptive gene flow from center-to-edge populations, may determine the ultimate impact of gene flow on the persistence of range edge populations (5, 6, 30). Across species, peripheral populations generally do not differ in abundance from central populations (7, 31), but they do appear to be more genetically isolated and differentiated (32). It is still unknown whether peripheral populations tend to be more or less adapted to selective pressures at range limits than their interior counterparts; our study found that traits of center-to-edge crosses were indeed less adaptive at the range edge, but any loss in fitness caused by maladaptation (e.g., slower development time) was counterbalanced by increased seedling emergence that offset these disadvantages. Other advantages or disadvantages to gene flow from the center of the range might have been found had additional generations (F2 and beyond) been tested; e.g., the mean fitness of hybrids from different environments often decreases in subsequent generations (22, 25, 28). Clearly, adaptive tradeoffs can exist across ecological gradients (see ref. 20) and may lead to adaptation to novel environments at range limits and, ultimately, niche evolution (33). Local mating did not significantly increase lifetime reproductive success, whereas nonlocal mating did. Thus, if the warm-edge range limit in M. laciniatus is caused by a failure to adapt to the drier conditions found at lower elevations in the Sierran foothills (14), our results support the hypothesis that stalled adaptation results from insufficient genetic variation (34).
This work demonstrates the capacity of gene flow to increase the fitness of populations at range limits, and that this capacity also depends on the source of gene flow. We provide experimental support for models in which gene flow, per se, can increase absolute fitness and population size in a marginal environment (4), as well as the intuitive prediction that gene flow from similar environments can be more beneficial than gene flow from dissimilar ones. We view these results as having several important implications. Generally, gene flow among previously isolated populations may be an important mechanism that expands the realized niche of a species if novel genetic combinations increase the evolutionary potential of populations to respond to strong selection. For example, Lavergne and Molofsky (35) have shown that novel genotypes and increased within-population genetic variation are linked to increased colonization ability in the invaded range of reed canary grass (Phalaris arundinacea). Conversely, bottleneck events and increased isolation among range-edge populations (32, 36) may reduce genetic variation upon which selection can act.
Clausen and Hiesey (18) considered ecological races to be reservoirs of genetic variation. Here we show that mixing of previously isolated populations from similar climates can increase lifetime reproductive success in relatively stressful habitats that may represent future environments predicted by climate-warming scenarios (12). Populations inhabiting novel environments, such as at range limits, should be considered collectively for their potential to produce novel, adaptive genetic combinations. In the race to facilitate species tracking of rapidly changing climates, prescriptive gene flow among range limit populations may represent a form of genetic rescue (24, 37) that increases the evolutionary capacity of range limit populations to respond to rapidly changing selective regimes. Populations at range edges, and the unique properties they possess, should be highly prioritized in conservation planning efforts that seek to decrease biodiversity losses caused by human-caused climate warming (9, 38).
Materials and Methods
Sample Populations.
M. laciniatus was sampled along two widely spaced geographic transects (Fig. 1 and Table S1) across the elevation gradient where climate varies in several ways to affect plant species distributions, including growing season length, temperature, and water availability (14, 39, 40). These transects were chosen to capture replicate gradients, and were part of different watersheds occupied by this creek- or seep-dependent species. Range limits were defined as areas supporting the lowest elevation individuals along the elevation gradient. The range limit for each transect was verified by an exhaustive search within potentially suitable habitats at lower elevations. Temperature and precipitation estimates were derived from Bioclim model averages (41) for each locality based on climate data between the years 1950–2000 (Table S1).
Experimental Gene Flow Crosses.
All crosses were conducted between plants that were the offspring of an intervening greenhouse generation used to minimize maternal effects. Within each transect, 20 gene-flow recipients (dams) from the lower range limit (foothill) population received pollen originating from one of four sources: (i) 10 randomly selected sires from the central (montane) site on that same transect, (ii) 10 randomly selected local sires from the same foothill population, (iii) 10 randomly selected sires at the lower range limit (foothill) population of the other transect (i.e., edge-to-edge cross; Fig. 1), and (iv) pollen from the same individual via automatic selfing, as a natural control for outcrossing. The same mating design was then repeated for populations in the second transect. In edge-to-edge crosses, plants received nonlocal pollen from foothill edge population plants of the other transect (Table S1). In center-to-edge treatments, pollen sources were plants whose parents occupied higher, montane elevations with cooler, wetter climatic selection regimes than edge populations.
Common Garden Experiment at the Lower Range Limit.
Seeds from the experimental pollination treatments (N = 1,680) were sown randomly into 20 trays (experimental blocks) filled with a moss-based planting medium that emulates natural moss substrates and soil depths. When the wet season had begun, trays were placed in the field in areas supporting natural populations along seeps on capillary matting (WA-cpm; Greenhouse Megastore). This matting absorbs water in a manner similar to mossy tufts where plants grow naturally. The growing season ends as the seeps dry out. Plants were monitored biweekly throughout the growing season from early March until mid-June 2008. During each monitoring period, plants were recorded as being present (emerged) and alive or dead, and received a phenologic index score (1, vegetative; 2, flower buds present; 3, open flowers present; 4, developing fruit present) referred to here as a “phenologic stage.” Plants were harvested when fruits were mature, and whole-plant total fruit mass was used as a lifetime fitness proxy (total seed mass correlation with fruit mass, r = 0.929; df = 139; P < 0.0001).
Analysis.
Fitness analysis was restricted to blocks in which at least some plants survived to produce fruits. Plants that never emerged and emerged plants that did not survive to produce fruits were assigned zero fitness. Our fitness measure, referred to here as “lifetime reproductive success,” thus combines survival and reproductive output. Plants differed most from each other in phenology at approximately 8 wk, and this census period was used to test for phenologic stage differences among populations and cross types. We used REML (procedure MIXED in SAS statistical package, version 9.1) to test for differences in lifetime reproductive success, time to emergence, and phenologic stage at 8 wk among gene flow treatments by using block as a random effect and cross type and transect as fixed effects. These data were rank-transformed because standard transformations failed to meet distributional assumptions of parametric analyses (42). Within REML models, we used Dunnett means comparison tests to test for significant variation among progeny from each cross type and self-fertilized seeds (pooled across transects) for the aforementioned traits. Self-fertilized progeny were considered the control condition in the Dunnett tests because selfing is the primary mode of fertilization in M. laciniatus.
We used two-component mixture regression models (43, 44) to assess whether differences in lifetime reproductive success arose because of differential survival, seed production, or both (SI Materials and Methods and Table S2). Blocks were treated as fixed effects because these models do not accommodate random effects. To determine the effect of cross type on timing and proportion of seedling emergence, we used a proportional-hazards model (45) whereby data were right-censored after 9 wk (by which time drought stress had essentially halted further reproduction) or at an earlier census if a plant's soil was found to be completely dry during monitoring.
Finally, to understand how fitness variation among individuals can be explained by traits expressed in this warm range-edge environment, phenotypic selection analysis (46) was performed for emergence time and phenologic stage at 8 wk. An individual's relative fitness (i.e., lifetime reproductive success) could be calculated based on grand means or block means. We found that both methods gave the same results qualitatively because the same effects were significant in each analysis. Here, we report the analysis based on fitness expressed relative to the grand mean. Phenotypic selection analysis was performed on individuals pooled across all cross types.
Genetic Estimates of Inbreeding and Distance Between Populations.
We extracted genomic DNA from plant tissue raised from seed by using a modified cetyltrimethyl ammonium bromide protocol (47). To conduct population genetic analyses, we used 11 codominant markers (Table S4); these included three markers that vary in single-copy nuclear gene intron lengths (48–50) and eight microsatellites (51). PCR products were analyzed with an ABI 3730 DNA analyzer. Amplified fragment sizes were scored in GeneMarker (SoftGenetics). All markers were located on different linkage groups and therefore represent genetically independent loci. For each population (n = 31, n = 45, n = 33, and n = 40 for edge 1, edge 2, center 1, and center 2, respectively), we calculated F by using GenAlEx version 6 (52) and the codominant genetic distance between population pairs (53) by using FSTAT version 2.9.3.2 (54) (Table S1 and S3). All populations were tested for significant differentiation after accounting for multiple comparisons by using FSTAT.
Supplementary Material
Acknowledgments
We thank Amy Angert, Jean Burns, Ivalú Cacho, Joanna Clines, Nancy Emery, Jon Haloin, Monique Kolster, Rick Lankau, Andrew Latimer, Bob Latta, Patrick McIntyre, John McKay, Stephanie Porter, Meghan Skaer, Jens Stevens, and Michael Turelli for helpful discussions. Bernie May, Maureen Stanton, and two anonymous reviewers provided comments that greatly improved the manuscript. We thank the following individuals for field and laboratory assistance: Clare Aslan, Arish Aziz, Ashley Bateman, Brooke Baythavong, Lily Cai, Alexa Carleton, Annie Chang, Donna Chen, Dana Chou, Ruthie Chow, Zacharia Costa, Megan DeMarche, Tomas Gepts, Oscar Gonzalez, Nikhil Gopal, Bryant Gross, Dena Grossenbacher, Yasmine Hernandez, Jayden Hoekstra, Jazzmyn Hoekstra, Luke Hoekstra, Carrie Huynh, Christina Islas, Marta Hura, Tina Lam, Tihua Lee, Jerell Maneja, Drew Maraglia, Lauren McGeoch, Joshua Mopas, Cassandra Morales-de-Silvestore, Leesa Mullins, Thuy Nguyan, Jessenia Perez, Tam Tran, Randeep Uppal, Marit Wilkerson, and Jennifer Wolf. Matthew Hufford, Jim Kami, David Lowry, Harry Meimberg, and John Willis provided assistance in selecting genetic markers and optimizing PCR. Neil Willits provided statistical advice. Yosemite National Park, Devils Postpile National Monument, the US Forest Service, and Bonnie Bladen and Raymond Laclergue provided land and plant resources. This work was funded by grants and fellowships (to J.P.S.) from the California Native Plant Society; US Forest Service Native Plant Materials Program NFN3; National Science Foundation (NSF) Doctoral Dissertation Improvement Grant NSF-DEB 0808607; NSF Division of Graduate Education Biological Invasions and Responding to Rapid Environmental Change (REACH) Integrative Graduate Education and Research Traineeship Awards (0114432 and 0801430, respectively) at University of California, Davis; University of California, Davis, Plant Sciences Department; University of California, Davis, Jastro Shields research program; and University of California, Davis, Center for Population Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100404108/-/DCSupplemental.
References
- 1.Kirkpatrick M, Barton NH. Evolution of a species’ range. Am Nat. 1997;150:1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- 2.Antonovics J. The nature of limits to natural selection. Ann Mo Bot Gard. 1976;63:224–247. [Google Scholar]
- 3.Haldane JBS. The relation between density regulation and natural selection. Proc R Soc Lond B Biol Sci. 1956;145:306–308. doi: 10.1098/rspb.1956.0039. [DOI] [PubMed] [Google Scholar]
- 4.Holt RD, Gomulkiewicz R. How does immigration influence local adaptation? A reexamination of a familiar paradigm. Am Nat. 1997;149:563–572. [Google Scholar]
- 5.Alleaume-Benharira M, Pen IR, Ronce O. Geographical patterns of adaptation within a species’ range: Interactions between drift and gene flow. J Evol Biol. 2006;19:203–215. doi: 10.1111/j.1420-9101.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 6.Barton N. Adaptation at the edge of a species’ range. In: Silvertown J, Antonovics J, editors. Integrating Ecology and Evolution in a Spatial Context. London: Blackwell; 2001. pp. 365–392. [Google Scholar]
- 7.Sexton JP, Mcintyre PJ, Angert AL, Rice KJ. Evolution and ecology of species range limits. Annu Rev Ecol Evol Syst. 2009;40:415–436. [Google Scholar]
- 8.Kellermann V, van Heerwaarden B, Sgrò CM, Hoffmann AA. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325:1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- 9.Hampe A, Petit RJ. Conserving biodiversity under climate change: The rear edge matters. Ecol Lett. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- 10.Bridle JR, Vines TH. Limits to evolution at range margins: When and why does adaptation fail? Trends Ecol Evol. 2007;22:140–147. doi: 10.1016/j.tree.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Loarie SR, et al. Climate change and the future of California's endemic flora. PLoS ONE. 2008;3:e2502. doi: 10.1371/journal.pone.0002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayhoe K, et al. Emissions pathways, climate change, and impacts on California. Proc Natl Acad Sci USA. 2004;101:12422–12427. doi: 10.1073/pnas.0404500101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crimmins SM, Dobrowski SZ, Greenberg JA, Abatzoglou JT, Mynsberge AR. Changes in climatic water balance drive downhill shifts in plant species’ optimum elevations. Science. 2011;331:324–327. doi: 10.1126/science.1199040. [DOI] [PubMed] [Google Scholar]
- 14.Barbour M, Keeler-Wolf T, Schoenherr A. Terrestrial Vegetation of California. Berkeley, CA: Univ California Press; 2007. [Google Scholar]
- 15.Cayan DR, Maurer EP, Dettinger MD, Tyree M, Hayhoe K. Climate change scenarios for the California region. Clim Change. 2008;87:21–42. [Google Scholar]
- 16.Bradshaw WE, Holzapfel CM. Genetic response to rapid climate change: It's seasonal timing that matters. Mol Ecol. 2008;17:157–166. doi: 10.1111/j.1365-294X.2007.03509.x. [DOI] [PubMed] [Google Scholar]
- 17.Waser NM, Vickery RKJ, Price MV. Patterns of seed dispersal and population differentiation in Mimulus guttatus. Evolution. 1982;36:753–761. doi: 10.1111/j.1558-5646.1982.tb05441.x. [DOI] [PubMed] [Google Scholar]
- 18.Clausen J, Hiesey WM. Experimental Studies on the Nature of Species. IV. Genetic Structure of Ecological Races. Publication #615. Washington, DC: Carnegie Institution; 1958. [Google Scholar]
- 19.Nunez-Farfan J, Schlichting CD. Natural selection in Potentilia glandulosa revisited. Evol Ecol Res. 2005;7:105–119. [Google Scholar]
- 20.Angert AL, Bradshaw HD, Jr, Schemske DW. Using experimental evolution to investigate geographic range limits in monkeyflowers. Evolution. 2008;62:2660–2675. doi: 10.1111/j.1558-5646.2008.00471.x. [DOI] [PubMed] [Google Scholar]
- 21.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 22.Barton NH. The role of hybridization in evolution. Mol Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- 23.Raabová J, Münzbergová Z, Fischer M. Consequences of near and far between-population crosses for offspring fitness in a rare herb. Plant Biol (Stuttg) 2009;11:829–836. doi: 10.1111/j.1438-8677.2008.00186.x. [DOI] [PubMed] [Google Scholar]
- 24.Willi Y, van Kleunen M, Dietrich S, Fischer M. Genetic rescue persists beyond first-generation outbreeding in small populations of a rare plant. Proc Biol Sci. 2007;274:2357–2364. doi: 10.1098/rspb.2007.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1768. doi: 10.1111/j.1558-5646.1999.tb04560.x. [DOI] [PubMed] [Google Scholar]
- 26.Rousselle Y, Thomas M, Galic N, Bonnin I, Goldringer I. Inbreeding depression and low between-population heterosis in recently diverged experimental populations of a selfing species. Heredity. 2011;106:289–299. doi: 10.1038/hdy.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagenius S, Hangelbroek HH, Ridley CE, Shaw RG. Biparental inbreeding and interremnant mating in a perennial prairie plant: Fitness consequences for progeny in their first eight years. Evolution. 2010;64:761–771. doi: 10.1111/j.1558-5646.2009.00860.x. [DOI] [PubMed] [Google Scholar]
- 28.Johansen-Morris AD, Latta RG. Fitness consequences of hybridization between ecotypes of Avena barbata: Hybrid breakdown, hybrid vigor, and transgressive segregation. Evolution. 2006;60:1585–1595. [PubMed] [Google Scholar]
- 29.Frankham R, et al. Predicting the probability of outbreeding depression. Conserv Biol. 2011;25:465–475. doi: 10.1111/j.1523-1739.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- 30.Kawecki TJ. Adaptation to marginal habitats. Annu Rev Ecol Evol Syst. 2008;39:321–342. [Google Scholar]
- 31.Sagarin RD, Gaines SD. The ‘abundant centre’ distribution: To what extent is it a biogeographical rule? Ecol Lett. 2002;5:137–147. [Google Scholar]
- 32.Eckert CG, Samis KE, Lougheed SC. Genetic variation across species’ geographical ranges: The central-marginal hypothesis and beyond. Mol Ecol. 2008;17:1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- 33.Holt RD. On the evolutionary ecology of species’ ranges. Evol Ecol Res. 2003;5:159–178. [Google Scholar]
- 34.Hoffmann AA, Blows MW. Species borders: Ecological and evolutionary perspectives. Trends Ecol Evol. 1994;9:223–227. doi: 10.1016/0169-5347(94)90248-8. [DOI] [PubMed] [Google Scholar]
- 35.Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pujol B, Pannell JR. Reduced responses to selection after species range expansion. Science. 2008;321:96. doi: 10.1126/science.1157570. [DOI] [PubMed] [Google Scholar]
- 37.Edmands S. Between a rock and a hard place: Evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- 38.Aitken SN, Yeaman S, Holliday JA, Wang TL, Curtis-McLane S. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evolutionary Applications. 2008;1:95–111. doi: 10.1111/j.1752-4571.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clausen J, Keck DD, Hiesey WM. Experimental Studies on the Nature of Species. III. Environmental Responses of Climatic Races of Achillea. Publication #581. Washington, DC: Carnegie Institution; 1948. [Google Scholar]
- 40.Angert AL, Schemske DW. The evolution of species’ distributions: Reciprocal transplants across the elevation ranges of Mimulus cardinalis and M. lewisii. Evolution. 2005;59:1671–1684. [PubMed] [Google Scholar]
- 41.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 42.Conover W, Iman R. Rank transformations as a bridge between parametric and nonparametric statistics. Am Stat. 1981;35:124–129. [Google Scholar]
- 43.Zeileis A, Kleiber C, Jackman S. Regression models for count data in R. J Stat Softw. 2008;27:1–25. [Google Scholar]
- 44.Lambert D. Zero-inflated Poisson regression, with an application to defects in manufacturing. Technometrics. 1992;34:1–14. [Google Scholar]
- 45.Cox D. Regression models and life-tables. J R Stat Soc, B. 1972;34:187–220. [Google Scholar]
- 46.Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 47.Lin J-Z, Ritland K. Flower petals allow simpler and better isolation of DNA for plant RAPD analyses. Plant Mol Biol Rep. 1995;13:210–213. [Google Scholar]
- 48.Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sweigart AL, Fishman L, Willis JH. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics. 2006;172:2465–2479. doi: 10.1534/genetics.105.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowry DB, Rockwood RC, Willis JH. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008;62:2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly AJ, Willis JH. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol Ecol. 1998;7:769–774. [Google Scholar]
- 52.Peakall R, Smouse PE. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 54.Goudet J. FSTAT: A program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001. Available at www2.unil.ch/popgen/softwares/fstat.htm. Accessed on November 1, 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.