Abstract
The platelet-tethering function of von Willebrand factor (VWF) is proteolytically regulated by ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13), which cleaves the Tyr1605-Met1606 (P1-P1′) bond in the VWF A2 domain. To date, most of the functional interactions between ADAMTS13 and VWF that have been characterized involve VWF residues that are C terminal to the scissile bond. We now demonstrate that the substrate P3 position in VWF, Leu1603, is a critical determinant of VWF proteolysis. When VWF Leu1603 was substituted with Ser, Ala, Asn, or Lys in a short VWF substrate, VWF115, proteolysis was either greatly reduced or ablated (up to 400-fold reduction in kcat/Km). As Leu1603 must interact with residues proximate to the Zn2+ ion coordinated in the active center of ADAMTS13, we sought the corresponding S3 interacting residues. Substitution of 10 candidate residues in the metalloprotease domain of ADAMTS13 identified two spatially separated clusters centered on Leu198 or Val195 (acting with Leu232 and Leu274, or with Leu151, respectively), as possible subsites interacting with VWF. These experimental findings using the short VWF115 substrate were replicated using full-length VWF. It is hypothesized that VWF Leu1603 interacts with ADAMTS13 Leu198/Leu232/Leu274 and that Val195/Leu151 may form part of a S1 subsite. The recognition of VWF Leu1603 by ADAMTS13, in conjunction with previously reported remote exosites C terminal of the cleavage site, suggests a mechanism whereby the VWF P1-P1′ scissile bond is brought into position over the active site for cleavage. Together with recently characterized remote exosite interactions, these findings provide a general framework for understanding the ADAMTS family substrate interactions.
Keywords: microvascular thrombosis, thrombotic thrombocytopenia purpura
Von Willebrand factor (VWF) is a large multimeric glycoprotein that is essential for normal hemostasis (1). Following vessel injury, VWF binds to exposed subendothelial collagen. Thereafter, in response to the shear forces exerted by the flowing blood, it unfolds from its inactive globular conformation into an active string-like form that can specifically recruit platelets (2–4). VWF multimeric size is a primary determinant of its platelet-tethering function and is proteolytically controlled by the plasma metalloprotease ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) (5–8). The physiological importance of this system is highlighted by the clinical sequelae associated with dysfunction of the VWF/ADAMTS13 axis. Whereas ADAMTS13 deficiency can cause fatal microvascular thrombosis-thrombotic thrombocytopenic purpura (7), excessive VWF proteolysis causes bleeding (i.e., type 2A von Willebrand disease) (1).
ADAMTS13 circulates in plasma as a constitutively active enzyme, which is unusual. Despite this behavior, plasma VWF remains essentially resistant to ADAMTS13 proteolysis in free circulation. This resistance is because shear-dependent unfolding of the VWF A2 domain is first required to expose both ADAMTS13 binding sites and the Tyr1605-Met1606 scissile bond before cleavage can occur. Once the A2 domain has unraveled, however, multiple exosite interactions contribute to the approximation of ADAMTS13 metalloprotease domain with the VWF Tyr1605-Met1606 scissile bond (9–16). The characterized functionally important interactions include the binding of the ADAMTS13 spacer domain with amino acids in the C-terminal region of the VWF A2 domain (9, 12, 14), and the interaction of the ADAMTS13 disintegrin-like domain with Asp1614 in VWF (11). Remarkably, little is known of the molecular interactions between the substrate and the metalloprotease domain of ADAMTS13. A quantitative study of cleavage of VWF short and full-length substrates showed the importance of the P1 and P1′ residues (Tyr1605 and Met1606, respectively), as substitution of these appreciably reduced proteolysis by ADAMTS13 (17). We recently provided further evidence for the importance of the accommodation of the P1′ residue, Met1606, within the S1′ pocket adjacent to the active site (10).
Most attention to date has been paid to regions and residues of VWF that are C terminal to the cleavage site. We hypothesized that a necessary feature of enzymatic cleavage of VWF will be a docking point for ADAMTS13 that is N terminal to the scissile bond. This finding is essential because cleavage must be associated with a reduction in binding affinity of the substrate to enable the protease to recycle, a reduction that will then be provided by bond cleavage. To investigate further the VWF residues important for cleavage by ADAMTS13, we have considered the activities of previously reported short VWF A2 domain fragments that span the cleavage site. One, VWF64 (1605–1668), could not be cleaved by ADAMTS13 (13), but others, such as VWF73 (1596–1668), VWF76 (1593–1668), and VWF115 (1554–1668) are all proteolysed efficiently (14, 18, 19). The difference between the substrates that are and those that are not cleaved resides in the sequence N terminal to the scissile bond. Therefore, we conjectured that the VWF1596–1604 N-terminal sequence must contain a structural determinant that is essential for proteolysis.
Results
We introduced a panel of single and multiple amino acid substitutions into the short A2 domain substrate, VWF115, which spans residues Glu1554-Arg1668 (Fig. S1). The synthetic peptide designated N-terminal peptide (NTP) that spans Asp1596-Tyr1605 is also shown. VWF115 and its variants were expressed, purified, and quantified as previously described, before functional analyses (11, 18).
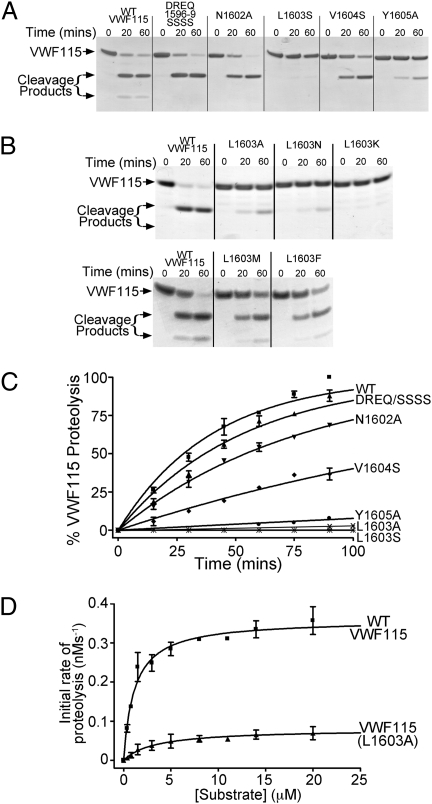
Cleavage of WT VWF115 by ADAMTS13 was rapid when examined by SDS/PAGE to visualize the cleavage products (Fig. 1A). The VWF115 variants in which the P10-P7 residues, VWF115 Asp1596-Gln1599, were all replaced by Ser (DREQ1596-9SSSS), or the P4 residue, VWF115 Asn1602, was substituted for Ala, were both cleaved rapidly, very similar to WT VWF115 (Fig. 1A). Point substitution of VWF115 Val1604 (the P2 residue) to Ser resulted in a modest reduction in proteolysis, whereas substitution of the P1 residue, VWF115 Tyr1605, to Ala predictably reduced appreciably the rate of cleavage (Fig. 1A). However, substitution of VWF115 Leu1603 (at the P3 position) with Ser abolished proteolysis (Fig. 1A). In additional experiments, VWF115 Leu1603 was also substituted with Ala, Asn, and Lys: all of these variants had severely reduced (Leu1603Ala) or undetectable (Leu1603Asn and Leu1603Lys) cleavage (Fig. 1B, Upper). In contrast, substitution of VWF Leu1603 with large hydrophobic residues Met and Phe had minimal influence on cleavage (Fig. 1B, Lower). Cleavage of selected variants was studied quantitatively by HPLC under conditions in which the catalytic efficiency, kcat/Km, could be determined (Fig. 1C). Minimal cleavage of VWF115 Leu1603Ser was observed (Fig. 1C). Table 1 summarizes the specificity constants derived from replicate experiments. The proteolysis of the VWF115 Leu1603Ser mutant was essentially ablated (>400 fold reduction in catalytic efficiency). Although this reduction in kcat/Km was appreciably larger even than the 38-fold reduction of the VWF115 Tyr1605Ala variant with the substituted P1 residue, the P1 residue substitution to Ser in a VWF115 Tyr1605Ser variant also had barely detectable cleavage when examined by SDS/PAGE (Fig. S2). In additional quantitative experiments, the cleavage of VWF115 Leu1603Ala was studied by HPLC. Time-course experiments with 1 nM ADAMTS13 indicated that even this conservative residue change produced ∼100-fold reduction in cleavage efficiency (Fig. 1C); however, quantitation of such low level of proteolysis is imprecise. Using higher concentrations (10 nM) of ADAMTS13 to induce sufficient cleavage of the VWF115 Leu1603Ala variant, substrate titration of the initial rate of proteolysis (Fig. 1D) of the variant demonstrated reduced (n = 3) kcat of 0.11 ± 0.07 (compare with WT ADAMTS13, 0.33 ± 0.18) s−1 and increased Km of 6.8 ± 3.6 (compare with WT ADAMTS13, 0.91 ± 0.51) μM.
Fig. 1.
Cleavage of VWF115 and its variants by ADAMTS13. (A) Composite and point substitution variants of VWF115 (6μM), N terminal to and including the scissile bond, are incubated with 3.5 nM ADAMTS13 and reaction products visualized by SDS/PAGE. (B) Proteolysis of different point substitution variants of VWF115 Leu1603 by ADAMTS13 was analyzed as in A. (Upper) Cleavage of VWF115 Leu1603Ala, -Asn, and -Lys; (Lower) Cleavage of VWF115 Leu1603Met and -Phe. (C) Graph showing the time course of cleavage of 2.5 μM VWF115 point and composite substitution variants by 1nM ADAMTS13. VWF115 proteolysis was quantified by HPLC. (D) Determination of kcat and Km for WT VWF115 and for the VWF115 Leu1603Ala variant. High concentrations of ADAMTS13 (10 rather than 1 nM) were used to induce proteolysis of VWF Leu1603Ala, the initial rates of which were determined by HPLC, corrected for protease concentration and plotted as a function of substrate concentration.
Table 1.
Quantitative analysis of scissile bond cleavage by ADAMTS13 for WT VWF and its variants
| VWF115 | kcat/Km (×105 M−1s−1) | Fold reduction |
| WT | 9.3 ± 2.3 | — |
| DREQ/SSSS | 6.4 ± 0.7 | 1.4 |
| N1602A | 4.3 ± 0.4 | 2.2 |
| L1603S | <0.02 | >400 |
| L1603A | 0.1 ± 0.04 | 100 |
| V1604S | 1.8 ± 0.1 | 5.1 |
| Y1605A | 0.2 ± 0.05 | 38 |
| M1606A | * | 18 |
| D1614A | ** | 6.5 |
| Δ1660–8 | *** | 12.5–15 |
Specificity constants, kcat/Km, (±SD) were determined by HPLC quantitation of VWF115 proteolysis by ADAMTS13 (n ≥ 3). Values for VWF115 Leu1603 variants must be regarded as approximate because of very low cleavage rates. Results indicated by asterisks are from the literature: *, ref. 10; **, ref. 11; ***, refs. 9 and 13.
Results from the literature are summarized in the lower part of Table 1. The reduction in cleavage constant for the VWF115 Leu1603Ser variant is appreciably greater than the 18-fold reduction for the VWF115 Met1606Ala variant in which the P1′ residue was substituted (10), the 6.5-fold reduction for the VWF115 Asp1614Ala that disrupts the disintegrin-like domain binding exosite (11), and the 12.5- to 15-fold reduction for the Δ1660–1668 C-terminal deletion variant that abolishes the spacer domain binding exosite (9, 13). Together, these results clearly demonstrate an essential role for VWF residue Leu1603, which is N terminal to the scissile bond, acting with VWF P1 residue, Tyr1605, in the cleavage process.
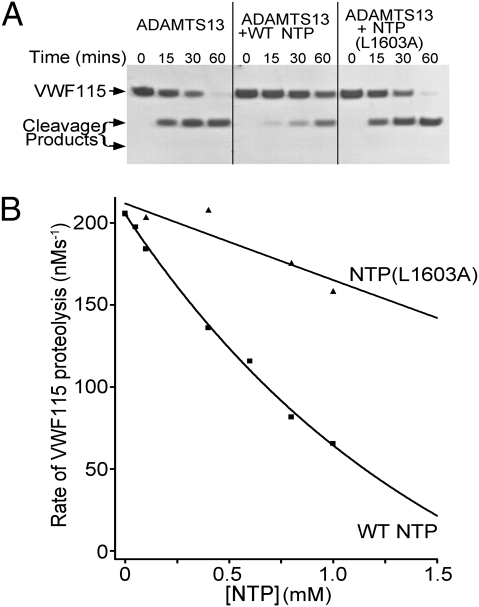
Further evidence for an important docking site within VWF Asp1596-Tyr1605 was obtained using an NTP containing this sequence to inhibit ADAMTS13 cleavage of VWF115. Addition of 1 mM WT NTP clearly reduced cleavage of VWF115 by ADAMTS13 when visualized by SDS/PAGE (Fig. 2A). In contrast, an NTP variant containing the Leu1603Ala substitution was without apparent effect, confirming Leu1603 as the key residue in this sequence. The influence of the NTP upon the initial rate of VWF115 proteolysis by ADAMTS13 was quantified by HPLC (Fig. 2B). Although WT NTP produced a large concentration dependent reduction in cleavage rate, the effect of NTP Leu1603Ala was very modest.
Fig. 2.
Inhibition of cleavage of VWF115 by the NTP. (A) Proteolysis of 6 μM VWF115 by 3.5 nM ADAMTS13 in the presence and absence of 1 mM NTP and NTP(Leu1603Ala). VWF115 cleavage is visualized by SDS/PAGE. (B) Inhibition of the initial rate of cleavage of 2.5 μM VWF115 by 1 nM ADAMTS13 in the presence of 0 to 1 mM NTP or NTP(Leu1603Ala). The initial rate of proteolysis was quantified using HPLC.
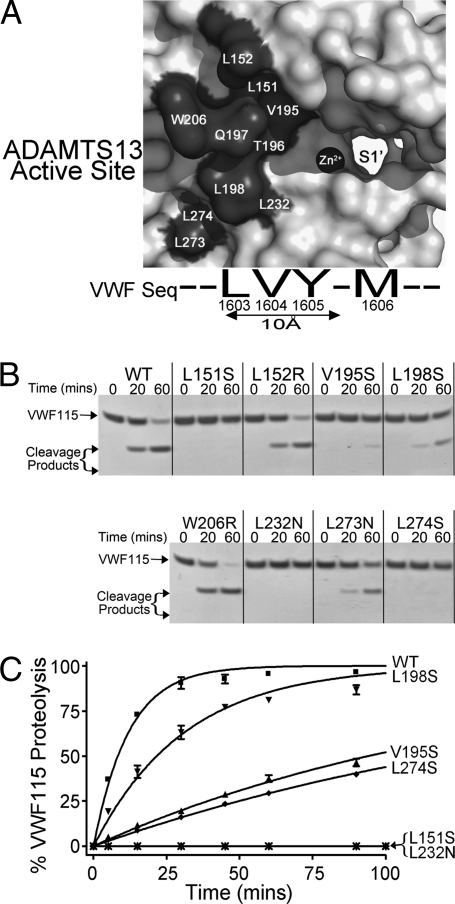
The VWF P3 residue, Leu1603, is located within ∼10 Å of the scissile bond. It must therefore interact with residues on the surface of the ADAMTS13 metalloprotease domain in close proximity to the catalytic Zn2+ ion coordinated within the active site. Using a model of ADAMTS13, we identified 10 candidate S3 residues in ADAMTS13 (Leu151, Leu152, Val195, Thr196, Gln197, Leu198, Trp206, Leu232, L273, and L274) that might interact with VWF Leu1603 because of their proximity to the active site (Fig. 3A). ADAMTS13 residues Thr196 and Gln197 were substituted in a recent report and the variants found to have normal proteolytic activity against VWF115 (10), and therefore were excluded from further investigation. When the activities of ADAMTS13 variants Leu151Ser, Leu152Arg, Val195Ser, Leu198Ser, Trp206Arg, Leu232Asn, Leu273Asn, and Leu274Ser were examined by SDS/PAGE using WT VWF115 (Fig. 3B), we found that ADAMTS13 variants Leu151Ser, Val195Ser, Leu198Ser, Leu232Asn, and Leu274Ser exhibited reduced ability to cleave VWF115. The activities of these selected variants were therefore examined quantitatively by HPLC (Fig. 3C). This experiment revealed an approximately fivefold reduction in kcat/Km for ADAMTS13 Leu198Ser compared with WT ADAMTS13 (9.3 ± 2.3 × 105 M-1·s−1, n = 3) and a 13- to 16-fold, reduction in kcat/Km for ADAMTS13 variants Val195Ser and Leu274Ser. No proteolytic cleavage fragments could be detected for the ADAMTS13 Leu232Asn and Leu151Ser variants, which had poor secretion. The ADAMTS13 residues which, when substituted resulted in reduced protease activity, fall into two clusters: Leu198/Leu232/Leu274 and Val195/Leu151.
Fig. 3.
Determinants of VWF cleavage by ADAMTS13 and its variants. (A) Model of the metalloprotease domain of ADAMTS13 showing 10 candidate S3 residues: the catalytic Zn2+ ion and proposed S1′ pocket are also labeled. The VWF sequence around the scissile bond is indicated below the model. (B) Proteolysis of 6 μM VWF115 by 1 nM WT ADAMTS13 and ADAMTS13 variants Leu151Ser, Leu152Arg, Val195Ser, Leu198Ser, Trp206Arg, Leu232Asn, Leu273Asn, and Leu274Ser. VWF proteolysis is visualized by SDS/PAGE. (C) HPLC quantitation of 2.5 μM VWF115 proteolysis by 3 nM WT ADAMTS13 and by ADAMTS13 variants Leu151Ser, Val195Ser, Leu198Ser, Leu232Asn, and Leu274Ser.
The approximately fivefold reduction in kcat/Km for proteolysis of VWF115 by the ADAMTS13 Leu198Ser variant is very similar to that for the cleavage of VWF115 Val1604Ser by WT ADAMTS13 (Table 1). This finding raised the possibility that ADAMTS13 Leu198 might interact with VWF Val1604 (P2 residue) rather than VWF Leu1603 (P3 residue). However, this contention was excluded by experiments in which WT VWF115 and VWF115 Val1604Ser were proteolysed by catalytic amounts (1–3 nM) of WT ADAMTS13 or ADAMTS13 Leu198Ser (Fig. S3). These results clearly suggest that the effects of both of these substitutions were independent of each other, making the interaction between the residues unlikely.
In a similar manner, we next examined whether the ADAMTS13 variants Leu198Ser, Val195Ser, and Leu274Ser induce cleavage of the VWF115 Leu1603Ser variant, which is of decreased magnitude compared with that of WT ADAMTS13. For these experiments, it was necessary to use high concentrations (30 nM) of the ADAMTS13 variants, as the usual catalytic concentrations (1–3 nM) failed to induce proteolysis of VWF115 Leu1603Ser. For the ADAMTS13 Leu198Ser variant, similar, minimal cleavage of the VWF115 Leu1603Ser occurred to that observed for WT ADAMTS13 (Fig. S4). For the ADAMTS13 Val195Ser and Leu274Ser variants, there was a suggestion of less cleavage of the VWF115 Leu1603Ser variant.
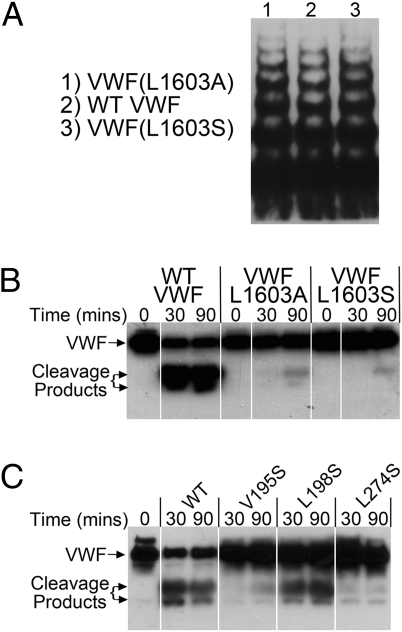
To verify our findings of the importance of the P3 residue in VWF proteolysis by ADAMTS13, we introduced both the Leu1603Ala and Leu1603Ser substitutions into full-length recombinant VWF. These variants were expressed normally and had normal multimer distribution (Fig. 4A). Full-length WT VWF was rapidly and effectively cleaved by ADAMTS13 (Fig. 4B, visualized on SDS/PAGE and Western blotting following reduction of the protein), whereas minimal cleavage was observed with the VWF Leu1603Ala and Leu1603Ser variants (Fig. 4B). Additional experiments examining the ability of ADAMTS13 Leu198Ser, Val195Ser, and Leu274Ser variants to cleave full-length WT VWF were also performed. These experiments showed a small reduction in VWF cleavage by ADAMTS13 Leu198Ser, but large reductions for the other two ADAMTS13 variants (Fig. 4C).
Fig. 4.
Proteolysis of full-length VWF by ADAMTS13. (A) Recombinant WT VWF and VWF Leu1603Ala and Leu1603Ser were examined for their multimer composition using SDS-agarose gel electrophoresis and Western blotting for VWF. (B) Proteolysis of recombinant full-length WT VWF, VWF Leu1603Ala, and Leu1603Ser (1.0 μg/mL) by 20 nM ADAMTS13. Changes in VWF were examined by SDS/PAGE under reducing conditions with Western blotting for VWF. (C) Proteolysis of full-length WT VWF (1.0 μg/mL) by 15 nM WT ADAMTS13 and ADAMTS13 Val195Ser, Leu198Ser, and Leu274Ser variants examined by SDS/PAGE with Western blotting, as in B.
Further support for an essential role of VWF Leu1603 and ADAMTS13 Leu198 and Val195 residues in the cleavage reaction is provided by species alignments showing their structural conservation. VWF Leu1603 is perfectly conserved in all species (Fig. S5). Interestingly, complete conservation of the functionally important P9′ residue in VWF, Asp1614, can also be noted. Similar regional alignment of the ADAMTS13 metalloprotease domains also shows strong conservation of Leu151, Val195, Leu198, Leu232, and Leu274 (Fig. S6).
Discussion
Cleavage of VWF by ADAMTS13 requires approximation of the substrate and protease. As ADAMTS13 circulates as a constitutively active enzyme, VWF cleavage is not driven by changes in the protease, such as an activation step that characterizes most zymogen to protease transitions in blood coagulation factors. Rather, proteolysis is uniquely influenced by changes in the conformation of VWF, which must first unfold. A critical feature of VWF unfolding has been shown to be the unfolding of the A2 domain itself. This domain is stabilized by a hydrophobic plug comprised of vicinal Cys residues located at the C terminus of the domain, which is inserted in the center of the domain near to the scissile bond (3, 20). This hydrophobic plug must first be removed for the A2 domain. Only once unfolded does VWF reveal the Tyr1605-Met1606 scissile bond and enable access of ADAMTS13. As with many reactions in hemostasis, remote exosites assist the cleavage process by bringing the active center of the protease into position over the scissile bond (9–11, 13, 14, 21). These exosite interactions which are C terminal to the scissile bond, either directly or indirectly facilitate the P1-P1′ bond positioning and cleavage. We recently provided evidence that P1′ residue, VWF Met1606, interacts with ADAMTS13 metalloprotease domain residues Asp252-Pro256 that form the S1′ pocket, and that specificity of ADAMTS13 can be altered by substituting these residues (10).
A small number of residues N terminal to the VWF scissile bond have previously been examined for their influence on cleavage by ADAMTS13, although this has been done in the context of polymorphisms that might influence VWF function rather than exploring specific exosite interactions (17). Interestingly, the report containing these results also demonstrated the requirement for the P1 residue to have an aromatic side chain, which is a determinant of its accommodation in the S1 pocket. This finding very likely explains the 38-fold reduction in proteolysis of the VWF115 Tyr1605Ala variant and minimal proteolysis of the VWF115 Tyr1605Ser variant. We reasoned that an N-terminal docking site other than the P1 residue must also be essential for scissile bond cleavage. Our systematic investigation reported here of the influence of residues N terminal to the scissile bond, prompted by examination of the reported activities of small VWF substrates, enabled us to identify the functional importance of VWF Leu1603 at the P3 substrate position. Remarkably, substitution of this single residue has the largest effect of any prior reported residue change in VWF. Indeed, the reduction in cleavage specificity of the variant VWF Leu1603Ser is far greater (>400-fold) than those of composite substitutions or deletions of prior identified exosites within the VWF sequence (Table 1). This finding is further supported by strict sequence conservation of VWF Leu1603 between species, which is greater even than the cleavage site residues (Fig. S5). The functional importance of VWF Leu1603 points to an important role in the cleavage mechanism, acting together with the P1 residue, Tyr1605, to orient and anchor the scissile bond. To identify potential interacting residues for the P3 residue on the ADAMTS13 protease domain, we substituted potential hydrophobic interacting partners close to the active center and examined VWF cleaving activity of these variants, which suggested that two separate clusters of ADAMTS13 residues involving residues Leu198 and Val195 may provide interaction sites for VWF. Although mutation of these residues did not induce a comparable reduction in cleaving activity as replacement of VWF Leu1603, mutation of adjacent residues Leu274, Leu232, and Leu151 in ADAMTS13 either reduced or abolished cleavage. The very low expression levels of these two latter ADAMTS13 mutants suggests that mutation-induced misfolding of the ADAMTS13 metalloprotease domain cannot be excluded as a contributor to reduced activity.
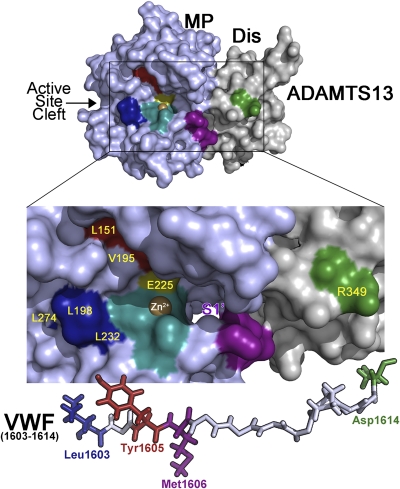
We now propose a provisional mechanism for cleavage of VWF using a model of the ADAMTS13 metalloprotease and disintegrin-like domains (Fig. 5). This mechanism shows ADAMTS13 Arg349 (green) that provides a docking point for VWF Asp1614 C terminal to the catalytic center (orange with Zn2+ shown). Based on our results, two potential VWF interaction sites, involving Leu198 and Val195, are shown. We postulate that ADAMTS13 Val195 and Leu151 may be positioned to form part of the S1 rather than S3 subsite and might therefore interact with the aromatic ring of VWF Tyr1605, rather than with VWF Leu1603. If so, we propose that ADAMTS13 Leu198, Leu232, and Leu274 contribute to the S3 subsite for VWF Leu1603. Based on a prior report, the likely position of the S1′ pocket flanked by ADAMTS13 Asp252-Pro256 (purple) is shown: this subsite is predicted to accommodate Met1606 (10). Underneath is shown the VWF1603-1614 polypeptide with potentially complementary interacting residues color coordinated. It is proposed, therefore, that VWF Leu1603, Tyr1605, and Asp1614 contact ADAMTS13 Leu198, Val195, and Arg349 in ADAMTS13, respectively, ensuring that the scissile bond is brought into position over the active center for cleavage to occur. Our results suggest that VWF Leu1603 interaction with the metalloprotease is absolutely essential for proteolysis to occur. The interactions of ADAMTS family members with their substrates are generally not well understood and the detailed understanding of how VWF is positioned and cleaved by ADAMTS13 may provide a useful general model for this multidomain metalloprotease family.
Fig. 5.
Model of the ADAMTS13 active center aligned with VWF. ADAMTS13 metalloprotease (MP, light blue) and disintegrin-like (Dis, gray) domains is shown with the area in the box enlarged to illustrate specific positioning of the scissile bond and potential docking points for the VWF peptide around the scissile bond. ADAMTS13 residues implicated in VWF proteolysis derived from the present and other reports are highlighted; Leu198, Leu232, Leu274 (dark blue), Val195, Leu151 (dark brown), Arg349 (green), active center (light blue, with Zn2+ shown in brown) with catalytic Glu225 and residues flanking the S1′ pocket (purple). The S1 pocket that harbors the P1 residue Tyr1605 is predicted to lie around the Val195/Leu151 cluster in this model. We propose that the cluster of Leu198, Leu232, and Leu274 provides the S3 subsite for VWF Leu1603. Beneath the figure is the VWF1603-1614 polypeptide (shown to scale), with identified interacting residues (P3, P1, P1′, and P9′) highlighted in complementary colors.
Cleavage of the VWF scissile bond by ADAMTS13 is essential for normal hemostasis. An absence of cleavage, caused by inherited or acquired deficiencies of ADAMTS13, predisposes to thrombotic thrombocytopenic purpura. Numerous point mutations in the ADAMTS13 gene have now been identified in patients with this disorder. Mostly, these mutations reduce the synthesis/secretion of ADAMTS13 and it is believed that accumulation of ultra large VWF multimer contributes to the acute microvascular thrombotic occlusions by forming platelet clumps. The present results suggest that residue substitutions within VWF could also potentially play a role in microvascular thrombosis. It will be of interest to screen families and individuals suffering from microvascular thrombosis for mutations in the VWF gene around the cleavage site, particularly any cases of thrombosis known to be without gene mutations in ADAMTS13.
Materials and Methods
Expression and Purification of VWF115 and VWF115 Variants.
An expression vector for the VWF A2 domain cleavage fragment, VWF115 (spanning VWF residues 1554–1668), its mutagenesis, and expression have been previously described (18).
Expression of Recombinant Full-Length VWF and the VWF(Leu1603Ser and Leu1603Ala) Mutants.
An expression vector for full-length human VWF and its mutagenesis have been previously described (22). Recombinant full-length WT VWF and these variants were expressed transiently in HEK293T cells (23). The concentration of VWF was determined by a specific ELISA (24), and VWF multimer analysis was assessed as previously described (24).
Expression of Recombinant ADAMTS13 and ADAMTS13 Variants.
The expression vector for WT ADAMTS13 and its mutagenesis have been described elsewhere (25). WT ADAMTS13 and ADAMTS13 variants were transiently expressed in HEK293T cells (23). Expression and secretion of ADAMTS13 was confirmed by Western blotting using a polyclonal rabbit anti-ADAMTS13 antibody (26). After 3 to 4 d, conditioned medium was harvested, cleared, and concentrated using 50-kDa molecular weight centrifugal filter devices (Millipore). ADAMTS13 concentration was determined using a specific in-house ADAMTS13 ELISA, as previously described (26, 27).
VWF115 Cleavage by ADAMTS13.
WT ADAMTS13 or ADAMTS13 variant (1 or 3.5 nM, unless otherwise indicated) in 20 mM Tris, 150 mM NaCl (pH 7.8) was incubated for 1 h with 5 mM CaCl2. Next, 6 μM VWF115, or VWF115 variant was added and at regular intervals aliquots of 15 μL were removed and stopped with 5 μL 30 mM EDTA. Thereafter, proteolysis of VWF115 was visualized by SDS/PAGE with Coomassie staining to detect cleavage products. Quantitative analysis of VWF115 proteolysis was performed by reactions that were set up similarly, except 2.5 μM VWF115 were used and samples were analyzed by HPLC from which the catalytic efficiencies for proteolysis (kcat/Km) were derived, as previously described (18). Determination of individual catalytic constants, kcat and Km, were also performed as described (18), using 1 nM ADAMTS13 with WT VWF115 and 10 nM ADAMTS13 with VWF Leu1603Ala. The protease was titrated with increasing concentrations of the substrates, initial rates of proteolysis determined by HPLC, and these were corrected for differences in protease concentrations. All functional assays were repeated a minimum of three times and with different preparations of VWF115 and ADAMTS13.
Cleavage of Full-Length Multimeric VWF by ADAMTS13.
To compare the cleavage of full-length WT VWF and VWFLeu1603Ser and Leu1603Ala variants, ADAMTS13 (20 nM) was preincubated for 30 min in 20 mM Tris, 0.5% BSA, 5 mM CaCl2. Recombinant WT VWF or VWF variant, 1.0 μg/mL, were preincubated in the presence of 3.0 M urea (final concentration 1.0 M) at 37 °C. For the analysis of the ADAMTS13 Val195Ser, Leu198Ser, and Leu274Ser variants, WT VWF (1.0 μg/mL) and 15 nM of the variants were used. Subsamples were removed reactions were stopped with EDTA. Changes in VWF resulting from ADAMTS13 proteolysis were analyzed by SDS/PAGE under reducing conditions and Western blotting for VWF, as previously described (14).
Peptide Inhibition Assay.
For peptide inhibition assay, 1 mM synthetic peptide containing VWF Asp1596-Tyr1605 sequence (termed NTP) (Alta Bioscience) or NTP(Leu1603Ala) was incubated with 3.5 nM ADAMTS13 and 5 mM CaCl2 at 37 °C for 30 min, and 6 μM VWF115 was then added. The reaction was stopped (from 0–60 min) with EDTA and analyzed by SDS/PAGE with Coomassie staining. To quantify the effect of peptides on VWF115 proteolysis by ADAMTS13, 0 to 1 mM NTP or NTP(Leu1603Ala) were individually incubated with 1 nM ADAMTS13 and CaCl2 at 37 °C for 30 min, and 2.5 μM VWF115 was then added. After 15 min, the reaction was stopped with EDTA. Thereafter, VWF115 proteolysis was quantified by HPLC.
Sequence Alignments and Molecular Modeling.
Amino acid sequences of either VWF or ADAMTS13 from different species retrieved from PubMed were aligned using AlignX software (Informax).
The ADAMTS13 metalloprotease domain was initially modeled using the HHPred server based on its sequence homology to ADAMTS1, -4, and -5, for which the crystal structures are available (28–30). Models were manipulated with Pymol software (Delano Scientific LLC).
Supplementary Material
Acknowledgments
This work was supported by grants from the British Heart Foundation (RG/06/007 and PG/09/038).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018559108/-/DCSupplemental.
References
- 1.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Zhang CZ, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466:992–995. doi: 10.1038/nature09295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q, et al. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci USA. 2009;106:9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furlan M, Robles R, Lämmle B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87:4223–4234. [PubMed] [Google Scholar]
- 6.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 7.Levy GG, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, et al. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 9.Pos W, et al. An autoantibody epitope comprising residues R660, Y661, and Y665 in the ADAMTS13 spacer domain identifies a binding site for the A2 domain of VWF. Blood. 2010;115:1640–1649. doi: 10.1182/blood-2009-06-229203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot R, Lane DA, Crawley JT. The ADAMTS13 metalloprotease domain: Roles of subsites in enzyme activity and specificity. Blood. 2010;116:3064–3072. doi: 10.1182/blood-2009-12-258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot R, Bardhan A, Ramroop N, Lane DA, Crawley JT. Essential role of the disintegrin-like domain in ADAMTS13 function. Blood. 2009;113:5609–5616. doi: 10.1182/blood-2008-11-187914. [DOI] [PubMed] [Google Scholar]
- 12.Wu JJ, Fujikawa K, McMullen BA, Chung DW. Characterization of a core binding site for ADAMTS-13 in the A2 domain of von Willebrand factor. Proc Natl Acad Sci USA. 2006;103:18470–18474. doi: 10.1073/pnas.0609190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112:1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci USA. 2006;103:19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanardelli S, et al. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114:2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7:2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruss CM, Notley CR, Hegadorn CA, O'Brien LA, Lillicrap D. ADAMTS13 cleavage efficiency is altered by mutagenic and, to a lesser extent, polymorphic sequence changes in the A1 and A2 domains of von Willebrand factor. Br J Haematol. 2008;143:552–558. doi: 10.1111/j.1365-2141.2008.07266.x. [DOI] [PubMed] [Google Scholar]
- 18.Zanardelli S, et al. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281:1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 19.Kokame K, Matsumoto M, Fujimura Y, Miyata T. VWF73, a region from D1596 to R1668 of von Willebrand factor, provides a minimal substrate for ADAMTS-13. Blood. 2004;103:607–612. doi: 10.1182/blood-2003-08-2861. [DOI] [PubMed] [Google Scholar]
- 20.Luken BM, Winn LY, Emsley J, Lane DA, Crawley JT. The importance of vicinal cysteines, C1669 and C1670, for von Willebrand factor A2 domain function. Blood. 2010;115:4910–4913. doi: 10.1182/blood-2009-12-257949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci USA. 2009;106:19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKinnon TA, Chion AC, Millington AJ, Lane DA, Laffan MA. N-linked glycosylation of VWF modulates its interaction with ADAMTS13. Blood. 2008;111:3042–3049. doi: 10.1182/blood-2007-06-095042. [DOI] [PubMed] [Google Scholar]
- 23.Gardner MD, et al. A functional calcium-binding site in the metalloprotease domain of ADAMTS13. Blood. 2009;113:1149–1157. doi: 10.1182/blood-2008-03-144683. [DOI] [PubMed] [Google Scholar]
- 24.O'Donnell JS, McKinnon TA, Crawley JT, Lane DA, Laffan MA. Bombay phenotype is associated with reduced plasma-VWF levels and an increased susceptibility to ADAMTS13 proteolysis. Blood. 2005;106:1988–1991. doi: 10.1182/blood-2005-02-0792. [DOI] [PubMed] [Google Scholar]
- 25.Crawley JT, et al. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005;105:1085–1093. doi: 10.1182/blood-2004-03-1101. [DOI] [PubMed] [Google Scholar]
- 26.Chion CK, Doggen CJ, Crawley JT, Lane DA, Rosendaal FR. ADAMTS13 and von Willebrand factor and the risk of myocardial infarction in men. Blood. 2007;109:1998–2000. doi: 10.1182/blood-2006-07-038166. [DOI] [PubMed] [Google Scholar]
- 27.Crawley JT, Lane DA, Woodward M, Rumley A, Lowe GD. Evidence that high von Willebrand factor and low ADAMTS-13 levels independently increase the risk of a non-fatal heart attack. J Thromb Haemost. 2008;6:583–588. doi: 10.1111/j.1538-7836.2008.02902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerhardt S, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 29.Mosyak L, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shieh HS, et al. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.