Abstract
Neuronal development is the result of a multitude of neural migrations, which require extensive cell-cell communication. These processes are modulated by extracellular matrix components, such as heparan sulfate (HS) polysaccharides. HS is molecularly complex as a result of nonrandom modifications of the sugar moieties, including sulfations in specific positions. We report here mutations in HS 6-O-sulfotransferase 1 (HS6ST1) in families with idiopathic hypogonadotropic hypogonadism (IHH). IHH manifests as incomplete or absent puberty and infertility as a result of defects in gonadotropin-releasing hormone neuron development or function. IHH-associated HS6ST1 mutations display reduced activity in vitro and in vivo, suggesting that HS6ST1 and the complex modifications of extracellular sugars are critical for normal development in humans. Genetic experiments in Caenorhabditis elegans reveal that HS cell-specifically regulates neural branching in vivo in concert with other IHH-associated genes, including kal-1, the FGF receptor, and FGF. These findings are consistent with a model in which KAL1 can act as a modulatory coligand with FGF to activate the FGF receptor in an HS-dependent manner.
Keywords: heparan sulfotransferase, Kallmann syndrome
The coordinated assembly of the nervous system in metazoans requires the migration of the large majority of neurons from their place of origin to their final destination in the brain (1). These processes require the complex interplay of many factors, including secreted and transmembrane proteins that mediate communication between cells. The activity of such factors is greatly influenced by the extracellular environment (2). For example, heparan sulfates (HSs), a class of molecularly diverse extracellular glycosaminoglycans, have been shown to be crucial for neural development in mice (3). From work in model organisms, it has become clear that much of the function of HS during neural development is embedded within complex modification patterns of the HS sugar residues (reviewed in refs. 4 and 5). HS modification patterns serve specific and instructive functions during neural development and are believed to regulate ligand-receptor interactions (6–8). These patterns arise as the consequence of nonuniform modifications of the sugar moieties, including sulfations, deacetylations, and epimerizations that are introduced by specific HS-modifying enzymes (9) (Fig. 1A). It is unknown whether the function of HS modifications impinges on normal human development and disease susceptibility.
Fig. 1.
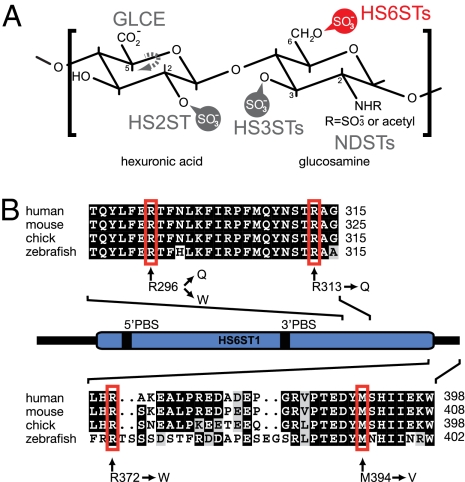
Function and sequence of human HS6ST1 and positions of amino acids mutated in patients with IHH. (A) Characteristic disaccharide of HS consisting of a hexuronic acid and a glucosamine residue. The positions within the HS sugars that can be modified by HS-modifying enzymes are indicated. Vertebrate genomes encode a single HS C-5 glucuronyl epimerase (GLCE) and HS 2-O-sulfotransferase (HS2ST) as well as at least three, four, or seven HS 6-O-sulfotransferases (HS6STs, indicated in red), N-deacetylase/sulfotransferases (NDSTs), or HS 3-O-sulfotransferases (HS3STs), respectively (5). (B) Schematic representation of the human HS6ST1 protein with the conserved sulfotransferase domain indicated in blue. 5′PBS and 3′PBS indicate the phosphoadenosyl-phosphosulfate (PAPS) cofactor binding sites. A multiple sequence alignment of two sections of the C terminus is shown with nonsynonymous changes indicated and amino acid positions denoted on the right. Amino acids shaded in black and gray indicate identical and similar residues, respectively.
Idiopathic hypogonadotropic hypogonadism (IHH) is a clinically and genetically heterogeneous condition that is characterized by lack of sexual maturation and infertility in the absence of other organic etiologies (10). Patients with IHH either have a normal sense of smell [normosmic IHH (nIHH)] or have an impaired sense of smell (anosmia); the combination of IHH and anosmia is termed Kallmann syndrome (KS). The first gene linked to IHH, KAL1, encodes anosmin-1, a neural cell adhesion protein (11, 12) that is required for proper development of the olfactory nerve and for the associated migration of the neurons secreting gonadotropin-releasing hormone (GnRH) (13). GnRH neurons are specified in the olfactory placode during embryonic development and migrate from their place of origin along the olfactory nerves and other olfactory structures into the forebrain. There, they form a neuroendocrine network as part of the hypothalamus, which regulates sexual development (14). A genetic analysis of KAL1 in this process has been hampered by the fact that an obvious KAL1 ortholog cannot be identified in the mouse genome.
New insights on anosmin-1 function came from studies in the nematode Caenorhabditis elegans. Cell-specific overexpression of kal-1 (the C. elegans ortholog of KAL1) in a set of C. elegans interneurons resulted in a kal-1–dependent axonal branching phenotype (15). A genetic modifier screen uncovered mutations in the C. elegans HS 6-O-sulfotransferase gene (hst-6) as suppressors of this kal-1 gain-of-function phenotype (15). The enzyme encoded by hst-6 introduces a sulfate specifically in the 6-O-position of the glucosamine sugar moiety within HS (Fig. 1A), indicating that anosmin-1 requires HS with specific 6-O-sulfate modifications to exert its function in vivo (7, 15). Intriguingly, heparan 6-O-sulfation is also required for the function of FGF receptor 1 (FGFR1) and its ligand, FGF8 (16), and loss-of-function mutations in both genes are associated with human GnRH deficiency (17, 18). Experiments in vitro have suggested that KAL1 modulates signaling via FGFR (19), but the genetic relationship between KAL1, FGFR, FGF, and HS remains elusive. In this paper, we investigated HS6ST1, a human homolog of C. elegans hst-6, as a candidate gene for GnRH deficiency and delineated the genetic interactions between these genes using C. elegans as a model.
Results
Mutations in Human HS6ST1 in Individuals with IHH.
Vertebrate genomes contain three genes coding for enzymes with HS 6-O-sulfotransferase activity: HS6ST1, HS6ST2, and HS6ST3 (20). The three genes are expressed dynamically throughout embryonic development in mice, with Hs6st1 being highly expressed in the nervous system, particularly in forebrain and sensory structures (21). Furthermore, immunohistochemical studies in chicken embryos documented high levels of HS in both the olfactory epithelium and the olfactory nerves (22). To test the hypothesis that HS6ST1 is associated with nIHH/KS, we sequenced the coding exons of HS6ST1 and flanking splice sites from genomic DNA of 338 GnRH-deficient patients (271 males and 67 females), including 105 familial cases. We identified 7 subjects with sequence variants: one homozygous [(c.886C > T) + (c.886C > T), p.R296W/R296W] and four heterozygous [(c.887G > A), p.R296Q; (c.938G > A), p.R313Q; (c.1114C > T), p.R372W; and (c.1180A > G), p.M394V] (Fig. 1B and Table S1), indicating that 2% of IHH patients harbor HS6ST1 mutations. The R372W variant was identified in three unrelated probands (nos. 4–6, Table S1). These variants were found neither in the SNP database (Build 132) (http://www.ncbi.nlm.nih.gov/projects/SNP/) nor in a control cohort of 500 ethnically and age-matched unaffected subjects.
Mutations in Human HS6ST1 Display Reduced Activity in Vivo and in Vitro.
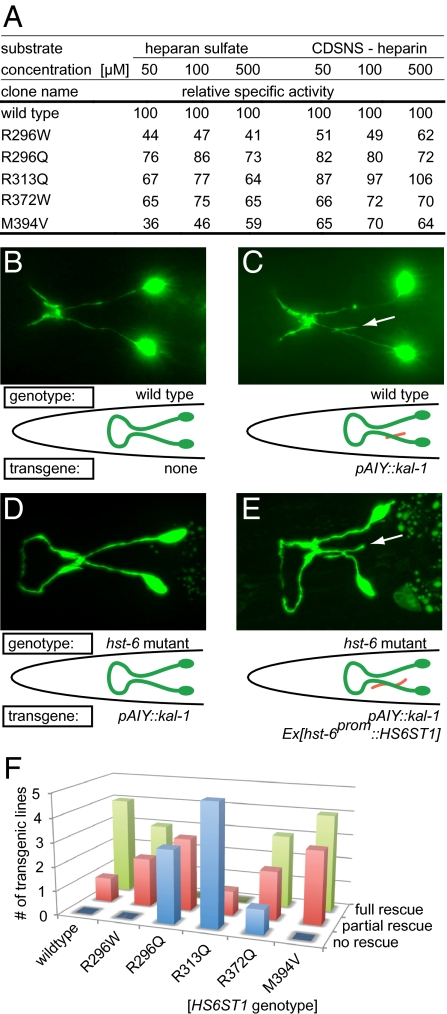
All identified HS6ST1 variants change amino acids that are perfectly conserved in HS6ST1s (Fig. 1B). The amino acids R296 and R313 are located in the sulfotransferase domain and affect basic residues likely to be involved in binding the sulfate donor phosphoadenosyl-phosphosulfate and/or the negatively charged HS substrate (Fig. 1 B and Fig. S1). The R372 and M394 residues map to a region suggested to be critical for the formation of a functional trimer of vertebrate HS sulfotransferases (23). To assess the functional significance of the identified HS6ST1 variants, we determined the HS 6-O-sulfotransferase activity in vitro. The relative specific activity of each purified recombinant mutant enzyme was compared with WT enzyme using two different substrates. All HS6ST1 variants exhibited reduced sulfotransferase activity (36–76% of WT activity at the lowest substrate concentration) with HS as the acceptor substrate, demonstrating that the IHH-associated mutations result in significant loss of enzymatic function (Fig. 2A). Using completely desulfated re–N-sulfated (CDSNS) heparin as a substrate, the activity of the R296W, R296Q, R372W, and M394V mutants was likewise reduced compared with WT (Fig. 2A). In contrast, the enzymatic activity of the R313Q mutant on the CDSNS-heparin substrate was similar to WT (87–106% of WT activity depending on substrate concentration). Because the CDSNS-heparin substrate lacks all O-sulfation (Fig. 1A), the observed discrepancy indicates that O-linked sulfate residues on HS compromise accessibility of the HS substrate to the R313Q mutant enzyme. Therefore, R313 may be involved in substrate recognition or binding, consistent with the position of R313 within the proposed HS6ST1 structure (Fig. S1).
Fig. 2.
Mutations in HS6ST1 reduce HS6ST1 activity in vitro and in vivo. (A) Relative specific activity of recombinant WT or mutant HS6ST1 variant. Enzymatic activity was determined with two different substrates. Acceptor substrates were used at three different concentrations (50, 250, and 500 μM) previously shown to cover the logarithmic nonsaturated range of HS6ST1 activity in this enzymatic assay (20). All experiments were done in duplicate using equal amounts of protein (Fig. S5). (B–D) Epifluorescent micrographs and schematics of the kal-1–dependent axonal branching phenotype in AIY interneurons. (B) WT morphology of AIY interneurons. (C) Animals overexpressing kal-1 in AIY interneurons display axon branching (indicated in red) (15). (D) Axonal branching is suppressed by a null mutation in the C. elegans hst-6 (15). (E) Transgenic introduction of the human HS6ST1 cDNA in a C. elegans hst-6 null mutant background restores the branches. (F) Quantification of rescue of kal-1–dependent axonal branching in AIY interneurons with human HS6ST1 variants as indicated. Shown are the numbers of transgenic lines for each construct that rescue the phenotype partially, fully, or not at all. Partial rescue was defined as ≥50% of activity and full rescue as ≥95% of activity compared with the mean of human HS6ST1 WT rescuing activity (n = 100–127 per transgenic line). Individual data of transgenic lines are presented in Fig. S2.
To gain insight into the in vivo mechanisms by which HS6ST1 mutations may contribute to IHH pathogenesis, we studied the mutant enzymes in a C. elegans in vivo assay for genetic interactions with kal-1. Transgenic expression of C. elegans kal-1 in AIY interneurons elicits an axon branching phenotype (15) (Fig. 2 B and C), which is suppressed to near-background levels by loss-of-function mutations in hst-6, the single worm HS 6-O-sulfotransferase (7, 15) (Fig. 2D). The suppression of the axonal branching phenotype is rescued (“antisuppressed”) by transgenic expression of a human WT HS6ST1 cDNA (Fig. 2E). In this assay, all mutants display a reduced capacity to rescue the kal-1–dependent branching phenotype (Fig. 2F). Interestingly, the R296W, R372W, and M394V mutant HS6ST1 versions retain some capacity for transgenic rescue of the hst-6 loss-of-function phenotype, with the R296W mutant being the least defective (Fig. 2F). The qualitative differences in activities of different human mutant enzymes in vivo and in vitro are not attributable to different transgenic expression levels (Fig. S2) and might reflect poor conservation of the respective amino acids between the human and C. elegans homologs (Fig. S1). Alternatively, the fact that different mutants result in different levels of loss of function in the KAL1-dependent assay could indicate that HS6ST1 mutations contribute to IHH pathogenesis via both KAL1-dependent and KAL1-independent mechanisms.
kal-1/Anosmin-1 Function Requires the FGFR/egl-15 Receptor and the FGF/egl-17 Ligand in a Context-Dependent Manner.
Several lines of evidence suggest a functional relationship between KAL1, HS6ST1, and possibly FGFR1 and FGF8. First, mutations in FGF/FGFR signaling and in KAL1 are the most frequent among the known genetic lesions associated with nIHH/KS (found in 10% and 5% of patients, respectively) (24). Second, anosmin-1 can directly bind FGFR1 in vitro, and it colocalizes with FGFR1 in cell culture experiments (19). Third, pedigree I segregated mutations in both the FGFR1 and HS6ST1 genes in affected individuals, suggesting possible interactions between these two genes (see below). Because mice do not contain an obvious KAL1 ortholog, we turned to C. elegans to test the genetic relationship between the FGFR, FGF, kal-1, and HS 6-O-sulfotransferase in vivo.
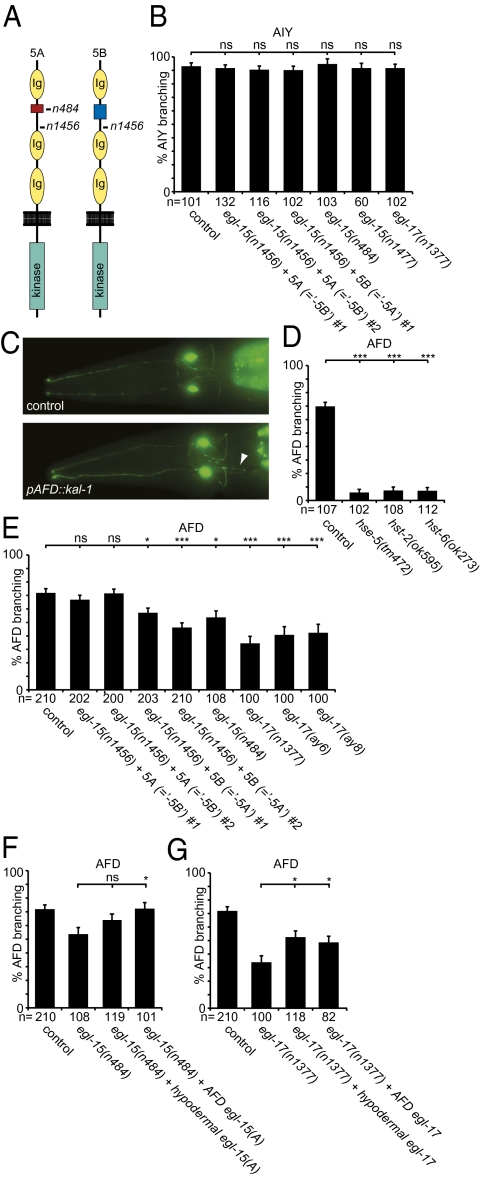
The sole FGFR in C. elegans is encoded by the essential egl-15 gene (25). FGFR/egl-15 is alternatively spliced to produce two splice variants for the extracellular domain, 5A and 5B (Fig. 3A), which have distinct functions during neural maintenance and development (26). We found that the kal-1–dependent branching phenotype in AIY is not dependent on either FGFR/egl-15 splice variant (Fig. 3B). Because complete loss of egl-15 function results in early larval lethality (25), we could not assess whether the 5A and 5B splice variants act redundantly in this cellular context. However, we consider this possibility less likely, because the strong temperature-sensitive egl-15(n1477) allele, which affects all splice variants, does not suppress the kal-1–dependent branching phenotype at the nonpermissive temperature (Fig. 3B).
Fig. 3.
kal-1 function requires hst-6, FGFR/egl-15, and FGF/egl-17. (A) Schematic of the two FGFR/EGL-15 extracellular splice variants 5A and 5B, which differ by a short sequence between Ig domains 1 and 2 indicated in blue and red. The nonsense alleles n484 and n1456 produce premature stop codons in 5A-specific or all splice variants, respectively. The n1456 allele results in complete loss of function. (B) Quantification of kal-1–dependent axonal branching in AIY interneurons in different mutant backgrounds. Indicated are transgenic lines (#1 = otEx1262, #2 = otEx1266 for the “-5B” strains and #1 = otEx1254 for the “-5A” strain) that exclusively express the egl-15(5A) or egl-15(5B) splice variant, respectively, in an egl-15(n1456) null mutant background (26). The egl-15(n484) allele is an egl-15(5A)–specific null allele (43), egl-15(n1477) is a strong temperature-sensitive allele, and egl-17(n1377) is a null allele (27). (C) Ventral view of the pair of AFD sensory interneurons (anterior is to the left). A kal-1–dependent axonal branch (otIs83) (15) is indicated (Lower, arrowhead) that is not observed in WT animals (Upper). (D) kal-1–dependent branching phenotype in AFD sensory neurons (otIs83) is suppressed by loss of the HS C-5 epimerase (hse-5), hst-2, or hst-6. (E) The kal-1–dependent branching phenotype in AFD sensory neurons (otIs83) is suppressed by loss of the FGFR/egl-15(5A) variant or the FGF/egl-17 ligand [using three null alleles: egl-17(n1377), egl-17(ay6), and egl-17(ay8)] (27). Indicated are transgenic lines (#1 = otEx1262, #2 = otEx1266 for the “-5B” strains and #1 = otEx1254, #2 = otEx1255 for the “-5A” strain) that exclusively express the egl-15(5A) or egl-15(5B) splice variant, respectively, in an egl-15(n1456) null mutant background (26). (F) Suppression of the kal-1–dependent branching phenotype in AFD sensory neurons by loss of the FGF receptor egl-15 is rescued by expression of FGFR/egl-15 specifically in AFD neurons (dzEx480, using the gcy-8 promoter) (42) but not in the hypodermis (dzEx484, using the dpy-7 promoter) (41). (G) Suppression of kal-1–dependent branching in AFD sensory neurons by loss of the FGF ligand egl-17 is partially rescued by expression of FGF/egl-17 in the hypodermis (dzEx472, using the dpy-7 promoter) (41) or in AFD neurons (dzEx483, using the gcy-8 promoter) (41). Representative transgenic lines are shown in F and G (for additional transgenic lines see Fig. S3).
We next characterized an alternate kal-1–dependent branching phenotype. The AFD sensory neurons that are presynaptic to AIY interneurons display a kal-1–dependent axon branching phenotype that is similar in appearance to the branching phenotype in AIY neurons (15) (Fig. 3C). The phenotype is both dependent on misexpression of kal-1 and sensitive to dosage of the kal-1 transgene (15) (Fig. S3 A and B). Surprisingly, the branching phenotype in AFD neurons requires HS not only with HS 6-O-sulfation and C-5 epimerization but with 2-O-sulfation (Fig. 4D), in contrast to the phenotype in AIY interneurons, which is less dependent on HS 2-O-sulfation (15). Moreover, AFD neurons require FGFR/egl-15 for normal development (26). These data suggest that (i) the molecular mechanisms for both normal development and kal-1–dependent branching in AIY and AFD neurons are distinct and (ii) the branching phenotype of AFD neurons requires HS with different sulfation patterns (including 2-O- and 6-O-sulfation).
Fig. 4.
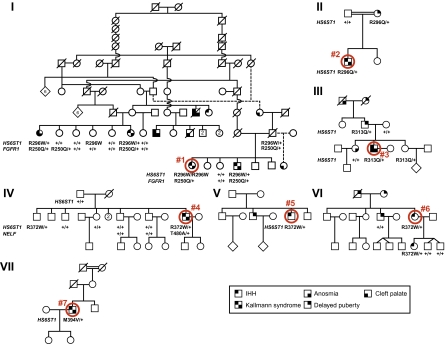
Complex inheritance of KS/nIHH in families with HS6ST1 mutations. Pedigrees of seven families with IHH (nIHH/KS). Note that 11 of 11 genotyped individuals with IHH (nIHH/KS) carry one of the loss-of-function mutations in HS6ST1 described here. Phenotypic symbols are listed in the key, and probands are indicated by a red circle and a unique number (compare Table S1 and SI Text for phenotypic details). Available genotypes are indicated below each individual. + denotes a WT allele. Numbers within symbols denote the number of additional siblings.
We found that kal-1–dependent branching in AFD neurons is significantly suppressed by loss of egl-15(5A) but not egl-15(5B) (Fig. 3E). Similarly, loss of FGF/egl-17, the canonical FGF ligand for egl-15(5A) (27), significantly suppresses the branching phenotype (Fig. 3E). Interestingly, suppression by loss of the FGF/egl-17 ligand is stronger than by loss of FGFR/egl-15(5A), albeit not complete (Fig. 3E). This suggests that FGF/egl-17 may act through alternate pathways [e.g., FGFR/egl-15(5B)– or FGFR/egl-15–independent mechanisms]. In addition, HS-dependent yet FGF/egl-17–independent mechanisms must exist that allow the formation of kal-1–dependent branches in AFD neurons. One possible explanation is redundancy of FGF/egl-17 with let-756, the second FGF ligand encoded in the C. elegans genome (28).
To determine in which tissue FGFR/egl-15 and FGF/egl-17 act to elicit kal-1–dependent branching in AFD neurons, we conducted cell-specific rescue experiments. We found that expression of egl-15 in AFD neurons but not in the hypodermis can rescue the suppression of branching in AFD neurons as a result of loss of egl-15(5A) function (Fig. 3F). This suggests that FGFR/egl-15 is required in AFD neurons for kal-1–dependent branching. In contrast, either hypodermal or AFD neuron-specific expression of egl-17 partially restores the branching in egl-17 null mutants, suggesting that the FGF/egl-17 ligand can act cell-nonautonomously to elicit kal-1–dependent branches in AFD neurons (Fig. 3G).
Taken together, these studies provide the first in vivo evidence that the anosmin-1 ortholog kal-1 can function in different neuronal contexts in a manner that is dependent on distinct HS modifications (including 6-O-sulfation, 2-O-sulfation, and C-5 epimerization) and may or may not depend on the FGFR and its ligand(s). Moreover, these in vivo findings are consistent with a model in which KAL1 may serve a modulatory role as a coligand with FGF in an HS-dependent manner as part of a ternary complex with FGFR, as previously suggested by cell culture studies (19). Oligogenic mutations in humans that directly or indirectly affect the genetic network comprising anosmin-1, the FGF-FGFR signaling module, and HS-modifying enzymes may thus synergize to cause the clinical phenotype of IHH (see below).
IHH Associated with HS6ST1 Mutations Displays Clinical Heterogeneity and Complex Inheritance.
HS6ST1 mutations were found in patients who had IHH with either normal olfaction (nIHH) or variable degrees of olfactory dysfunction (KS) as well as with either normal or abnormal olfactory structures (Fig. 4, Table S1, and SI Text). The same wide spectrum of severity and timing of onset of GnRH deficiency described for other IHH genes was observed (29). Two patients had microphallus, one of whom also had unilateral cryptorchidism, both hallmarks of GnRH deficiency during the neonatal period (30). Three patients presented with absent puberty (severe GnRH deficiency), whereas three male patients had partial puberty as evidenced by some spontaneous testicular development (Table S1 and Fig. S4). After discontinuing his testosterone therapy of several years, proband 7 experienced sustained reversal of his hypogonadism (31) (Fig. S4) suggesting that a hormonal and/or environmental component (possibly operating via epigenetic mechanisms) can modify the clinical course of HS6ST1 mutation-associated IHH. Reminiscent of the midline defects observed in patients harboring mutations in FGFR1 or FGF8 (17, 18), probands 1 and 3 exhibited a high arched palate and cleft palate, respectively. Clinical variability is evident both within and across families carrying the same genetic variation. For instance, heterozygosity for R313Q is associated with a severe phenotype (KS and cleft palate) in proband 3; delayed puberty in his father; and normal smell, puberty, and reproductive function in his sister (Fig. 4 and Table S1). In pedigrees V and VI, heterozygosity for R372W is associated with nIHH, whereas in pedigree IV, the same defect is associated with KS in proband 4 and with infertility of unknown etiology with normal puberty and smell in his brother. Thus, IHH-associated HS6ST1 mutations display incomplete penetrance and variable expressivity of the disease phenotype.
Though HS6ST1 is an autosomal gene, the associated GnRH deficiency shows complex inheritance patterns (Fig. 4) not readily conforming to Mendelian definitions of autosomal dominant or recessive transmission. Three pedigrees are consanguineous (I, II, and VII), a feature that usually suggests an autosomal recessive mode of disease inheritance, yet only one patient is homozygous (proband 1); all other patients with HS6ST1 mutations are heterozygous, even within the homozygous subject's family (pedigree I). On the other hand, two nonconsanguineous pedigrees (III and VI) show transgenerational inheritance of GnRH deficiency, suggesting an autosomal dominant mode (Fig. 4). However, the fact that both, rather than just one, of the IHH proband's unrelated parents (VI) or grandparents (III) are affected with attenuated phenotypes (delayed puberty or anosmia) argues against a dominant defect. Finally, some subjects with HS6ST1 mutations had delayed puberty and not IHH (Fig. 4, pedigrees II and III), strongly indicating that these mutations can contribute to but are not sufficient for the full-blown GnRH deficiency. Thus, additional genetic and/or epigenetic mechanisms likely cooperate with HS6ST1 mutations to determine the clinical presentation of the patients.
To test if additional genetic factors contribute to the observed variability, we sequenced the coding regions of eight additional IHH genes: KAL1, GNRHR, NELF, GPR54, PROK2, PROKR2, FGFR1, and FGF8 (SI Text). Six subjects in pedigree I had a heterozygous variant in FGFR1 (p.R250Q) and one subject in pedigree IV had a heterozygous variant in NELF (p.T480A) (Fig. 4 and Table S1). The combined presence of mutations in HS6ST1 and FGFR1 (digenicity) in pedigree I is predictive of the KS phenotype: all four genotyped KS patients carry both the HS6ST1 R296W mutation and the FGFR1 loss-of-function variant R250Q, whereas either gene defect alone does not cause KS (Fig. 4). FGFR1 R250 is involved in binding FGF8 (32), and was independently associated with another case of GnRH deficiency (33). Because FGFR1 requires HS 6-O-sulfation for interaction with FGF8 (32), the HS6ST1 R296W and FGFR1 R250Q mutations may synergize to compromise FGF signaling, consistent with prior reports of digenicity (17, 24, 34, 35). The fact that carriers of HS6ST1 mutations do not always manifest IHH (pedigrees I, III, and IV) is consistent with the presence of monoallelic gene defects in IHH-associated genes in as many as 10% of unaffected controls (24). In two of three such pedigrees (I and IV), we found mutations in other IHH-associated genes (FGFR1 and NELF). Thus, in addition to HS6ST1 mutations, other genetic lesions are likely involved in this digenic or oligogenic disease, with the paralogous HS6ST2 or HS6ST3 gene being possible candidates. In conclusion, we propose that hypomorphic HS6ST1 alleles contribute to IHH pathogenesis by interacting with mutant alleles in other disease-associated genes.
Discussion
This work implicates mutations in HS modification enzymes in human development and disease. Several lines of evidence support a role for HS6ST1 as a gene associated with the pathogenesis of nIHH/KS. First, all identified mutations affect amino acid residues that are highly conserved in HS6ST1 (Fig. 1B) and are absent from 500 healthy control subjects as well as from the SNP database. Second, the HS6ST1 mutations identified in patients with IHH reduce enzyme activity in vitro and in a C. elegans in vivo assay for hst-6 function (Fig. 2). Lastly, the C. elegans ortholog hst-6 displays genetic interactions with kal-1, the FGFR egl-15, and the FGF ligand egl-17 in cell context-dependent manner.
The human mutations identified in HS6ST1 have distinct effects in a KAL1-dependent assay, implying that HS6ST1 may be required in vivo for KAL1-dependent and -independent pathways. Known genes involved in human GnRH deficiency comprise several ligand-receptor systems, including GNRH1 (MIM ID 152760) and GNRHR (MIM ID 138850) (36, 37), FGF8 (MIM ID 600483) and FGFR1 (MIM ID 136350) (17, 18), PROK2 (MIM ID 607002) and PROKR2 (MIM ID 607123) (34, 38), and TAC3 (MIM ID 162330) and TACR3 (MIM ID 162332) (39). Given the role of HS fine structures in regulating diverse ligand-receptor interactions (32, 40), it seems plausible that human HS6ST1 mutations can contribute to GnRH biology by compromising signaling through one or more of these pathways. Different mutations could result in substantially different defects in the fine structure of HS. Mechanistically, this could be the result of defects in (i) catalytic activity, (ii) substrate recognition, or (iii) protein functions that are independent of catalytic activity (e.g., protein-protein interactions). The observed qualitative differences between in vivo and in vitro activity of different mutations in HS6ST1 (Fig. 2) could be the result of such different mechanisms. The R313Q mutation that modestly affects in vitro activity on one but not another substrate (Fig. 2A), yet has profound defects in vivo (Fig. 2 B–F), provides credence to such hypotheses.
HS fine structure is important for the interaction of ligands and receptors, such as FGF and FGFR (8, 32). Thus, certain HS6ST1 mutations could specifically compromise defined molecular pathways. Consistent with this notion, we identified additional mutations in FGFR1 in the family that segregates the R296W mutation in HS6ST1, the mutation that is most defective in vitro (Fig. 2A) but least defective in the kal-1–dependent AIY branching phenotype in C. elegans (Fig. 2 B–F). One could envision that the R296W mutation alters HS6ST1 enzyme function in a way that results in HS fine structure defects that primarily impinge on the KAL-1–independent functions rather than the KAL-1–dependent functions of FGFR1. Such a scenario is in accord with our genetic analyses of kal-1, FGFR/egl-15, FGF/egl-17, and hst-6 interactions in C. elegans, which show that kal-1 function can be mediated by both FGF-dependent and -independent mechanisms, depending on the cellular context and likely the HS composition (AFD and AIY neurons, respectively) (Fig. 3). Importantly, HS with distinct modifications is required for kal-1–dependent branching in both AFD and AIY interneurons, underscoring the significance of HS as a crucial modifier of kal-1–dependent signaling in vivo.
IHH associated with HS6ST1 mutations segregates as a complex trait in families (Fig. 4). Such inheritance patterns likely result from oligogenic interactions (24) between HS6ST1 and other known (e.g., FGFR1, NELF; Fig. 4) as well as yet unknown disease-associated genes. Thus, the identified HS6ST1 missense mutations may not be sufficient to cause disease. Rather, HS6ST1 is an important gene that contributes pathogenic alleles to the genetic network responsible for the neuroendocrine control of human reproduction. Additional genetic and/or epigenetic mechanisms likely contribute to the pathogenesis of GnRH deficiency in a genetic background that is sensitized by reducing HS6ST1 function. Identifying these factors will be an important goal in the future to understand the interaction between HS modifications and cell-cell signaling. Because HS has been implicated in many other signaling pathways mediating cell-cell communication during development, it should be explored whether mutations in HS-modifying enzymes could also contribute to other human diseases by interacting with different genetic networks. Conversely, genes mediating extracellular sugar modifications other than 6-O-sulfation (e.g., 2-O-sulfation, C-5 epimerization) are plausible candidate genes for IHH.
Materials and Methods
Phenotyping and Genotyping of Human Subjects.
IHH was defined as absent or incomplete puberty by the age of 18 y in the absence of other causes. KS was defined as IHH with anosmia or hyposmia. Detailed histories and physical examinations are presented in SI Text, as are details on genotyping. The Human Research Committee of Massachusetts General Hospital approved this study, and all subjects provided written informed consent before participation.
In Vitro and in Vivo Assays.
Sulfotransferase activities in vitro were determined as described using CDSNS-heparin or HS (from pig aorta) as the acceptor substrate (20) (SI Text). HS6ST1 activity in vivo was determined with a transgenic rescue assay, using cell-specific heterologous promoters (SI Text). Neuroanatomy was scored as described (7). Statistical significance was calculated using the z test, and values were subjected to the Bonferroni correction where applicable. Unless noted otherwise, statistical significance is indicated as follows: ns, not significant, *P < 0.05; **P < 0.005; and ***P < 0.0005.
Supplementary Material
Acknowledgments
We are indebted to patients, family members, and the Massachusetts General Hospital General Clinical Research Center nursing staff. We thank R. Townley for protein sequence alignments, molecular modeling studies, and discussions and M. Stern for egl-17 alleles. We are grateful to J. Brandler and P. Weinberg for technical assistance and to N. Baker, T. Boulin, S. Emmons, O. Hobert, B. Morrow, and members of the Pitteloud and Bülow laboratories for comments. This work was supported by National Institutes of Health Grants R01HD055380 and R01HD055380-03S1 (to H.E.B.) and Grants 5U54HD028138, 1R01HD056264, 1 UL1 RR025758-01, and M01-RR-01066 (to N.P.) and by a postdoctoral fellowship of the Academy of Finland (to J.T.). H.E.B. is an Alfred P. Sloan Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102284108/-/DCSupplemental.
References
- 1.Jessell TM, Sanes JR. Development. The decade of the developing brain. Curr Opin Neurobiol. 2000;10:599–611. doi: 10.1016/s0959-4388(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez F, Rifkin DB. Cell signaling events: A view from the matrix. Matrix Biol. 2003;22:101–107. doi: 10.1016/s0945-053x(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 3.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 4.Holt CE, Dickson BJ. Sugar codes for axons? Neuron. 2005;46:169–172. doi: 10.1016/j.neuron.2005.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bülow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 6.Bülow HE, et al. Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Curr Biol. 2008;18:1978–1985. doi: 10.1016/j.cub.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bülow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 8.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 10.Seminara SB, Hayes FJ, Crowley WF., Jr Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): Pathophysiological and genetic considerations. Endocr Rev. 1998;19:521–539. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 11.Franco B, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 12.Legouis R, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 13.Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 14.Wierman ME, et al. Molecular mechanisms of gonadotropin-releasing hormone neuronal migration. Trends Endocrinol Metab. 2004;15:96–102. doi: 10.1016/j.tem.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Bülow HE, Berry KL, Topper LH, Peles E, Hobert O. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc Natl Acad Sci USA. 2002;99:6346–6351. doi: 10.1073/pnas.092128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo BM, Salmivirta M. Heparin/Heparan sulfate domains in binding and signaling of fibroblast growth factor 8b. J Biol Chem. 2002;277:32616–32623. doi: 10.1074/jbc.M204961200. [DOI] [PubMed] [Google Scholar]
- 17.Falardeau J, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. doi: 10.1172/JCI34538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodé C, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, et al. Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. J Biol Chem. 2009;284:29905–29920. doi: 10.1074/jbc.M109.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habuchi H, et al. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 21.Sedita J, Izvolsky K, Cardoso WV. Differential expression of heparan sulfate 6-O-sulfotransferase isoforms in the mouse embryo suggests distinctive roles during organogenesis. Dev Dyn. 2004;231:782–794. doi: 10.1002/dvdy.20173. [DOI] [PubMed] [Google Scholar]
- 22.Nishizuka M, Arai Y. Glycosaminoglycans in the olfactory epithelium and nerve of chick embryos: An immunocytochemical study. Neurosci Res. 1996;24:165–173. doi: 10.1016/0168-0102(95)00990-6. [DOI] [PubMed] [Google Scholar]
- 23.Bethea HN, Xu D, Liu J, Pedersen LC. Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis. Proc Natl Acad Sci USA. 2008;105:18724–18729. doi: 10.1073/pnas.0806975105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sykiotis GP, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci USA. 2010;107:15140–15144. doi: 10.1073/pnas.1009622107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeVore DL, Horvitz HR, Stern MJ. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell. 1995;83:611–620. doi: 10.1016/0092-8674(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 26.Bülow HE, Boulin T, Hobert O. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron. 2004;42:367–374. doi: 10.1016/s0896-6273(04)00246-6. [DOI] [PubMed] [Google Scholar]
- 27.Burdine RD, Chen EB, Kwok SF, Stern MJ. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:2433–2437. doi: 10.1073/pnas.94.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roubin R, et al. let-756, a C. elegans fgf essential for worm development. Oncogene. 1999;18:6741–6747. doi: 10.1038/sj.onc.1203074. [DOI] [PubMed] [Google Scholar]
- 29.Hardelin JP, Dodé C. The complex genetics of Kallmann syndrome: KAL1, FGFR1, FGF8, PROKR2, PROK2, et al. Sex Dev. 2008;2:181–193. doi: 10.1159/000152034. [DOI] [PubMed] [Google Scholar]
- 30.Grumbach MM. A window of opportunity: the diagnosis of gonadotropin deficiency in the male infant. J Clin Endocrinol Metab. 2005;90:3122–3127. doi: 10.1210/jc.2004-2465. [DOI] [PubMed] [Google Scholar]
- 31.Raivio T, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357:863–873. doi: 10.1056/NEJMoa066494. [DOI] [PubMed] [Google Scholar]
- 32.Mohammadi M, Olsen SK, Goetz R. A protein canyon in the FGF-FGF receptor dimer selects from an à la carte menu of heparan sulfate motifs. Curr Opin Struct Biol. 2005;15:506–516. doi: 10.1016/j.sbi.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Raivio T, et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–4390. doi: 10.1210/jc.2009-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodé C, et al. Kallmann syndrome: Mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitteloud N, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Roux N, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 37.Bouligand J, et al. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med. 2009;360:2742–2748. doi: 10.1056/NEJMoa0900136. [DOI] [PubMed] [Google Scholar]
- 38.Pitteloud N, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104:17447–17452. doi: 10.1073/pnas.0707173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topaloglu AK, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeCouter J, et al. The endocrine-gland-derived VEGF homologue Bv8 promotes angiogenesis in the testis: Localization of Bv8 receptors to endothelial cells. Proc Natl Acad Sci USA. 2003;100:2685–2690. doi: 10.1073/pnas.0337667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilleard JS, Barry JD, Johnstone IL. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol Cell Biol. 1997;17:2301–2311. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci USA. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130:3757–3766. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.