Abstract
Bipolar spindle formation is pivotal for accurate segregation of mitotic chromosomes during cell division. A growing body of evidence suggests that, in addition to centrosome- and chromatin-based microtubule (MT) nucleation, MT-based MT nucleation plays an important role for proper bipolar spindle formation in various eukaryotic organisms. Although a recently discovered Augmin complex appears to play a central role in this event, how Augmin is regulated remains unknown. Here we provide evidence that a mammalian polo-like kinase 1 (Plk1) localizes to mitotic spindles and promotes MT-based MT nucleation by directly regulating Augmin. Mechanistically, we demonstrated that Cdc2-dependent phosphorylation on a γ-tubulin ring complex (γ-TuRC) recruitment protein, Nedd1/GCP-WD, at the previously uncharacterized S460 residue induces the Nedd1–Plk1 interaction. This step appeared to be critical to allow Plk1 to phosphorylate the Hice1 subunit of the Augmin complex to promote the Augmin–MT interaction and MT-based MT nucleation from within the spindle. Loss of either the Nedd1 S460 function or the Plk1-dependent Hice1 phosphorylation impaired both the Augmin–MT interaction and γ-tubulin recruitment to the spindles, thus resulting in improper bipolar spindle formation that ultimately leads to mitotic arrest and apoptotic cell death. Thus, via the formation of the Nedd1–Plk1 complex and subsequent Augmin phosphorylation, Plk1 regulates spindle MT-based MT nucleation to accomplish normal bipolar spindle formation and mitotic progression.
Keywords: mitosis, spindle assembly, polo-box domain, distributive phosphorylation, Ser/Thr protein kinase
Proper formation of bipolar spindles is a fundamental cellular process centrally required for faithful chromosome segregation and genomic stability. Failure in this process results in immediate biological consequences such as mitotic arrest and apoptosis, while facilitating chromosome missegregation and aneuploidy, a hallmark of cancer. The centrosome is the primary microtubule (MT)-organizing center (MTOC) in most somatic cells surrounded by an electron-dense proteinaceous matrix called pericentriolar material (PCM). PCM contains γ-tubulin ring complexes (γ-TuRC), a large multiprotein complex that promotes MT nucleation (1).
Besides the centrosome-based MT nucleation, a growing body of evidence suggests that noncentrosomal MT nucleation processes also play an important role in bipolar spindle assembly. These include Ran-GTP–driven chromosome-based MT nucleation predominantly observed in acentrosomal divisions such as female meiosis (2, 3) and recently discovered MT-based MT nucleation observed in plant cells, yeast, and more lately in fly and human cells (4–7).
Unlike the much-studied centrosome- and chromosome-based MT nucleation processes, the physiological significance of the MT-based MT nucleation and the components important for this event are only beginning to emerge. Lüders et al. have demonstrated that γ-TuRC localizes not only to centrosomes but also to mitotic spindles in human cultured cells (8). Interestingly, silencing of Nedd1/GCP-WD (for simplicity, Nedd1 hereafter) inhibits recruitment of γ-tubulin to both of these subcellular structures and induces a defect in bipolar spindle assembly (8, 9).
Recent studies in Drosophila and human cells show that a protein complex called Augmin recruits Nedd1 and its associated γ-TuRC to preexisting spindle MTs (7, 10, 11) and promotes MT-based MT nucleation to amplify the MT polymers along the spindles (4, 7, 12–15). Hice1, one of the subunits of the Augmin complex, has been shown to bind to MT directly through its N-terminal domain (16). Thus, investigation on the mechanism underlying the Hice1–MT interaction is likely a key step in understanding the biological processes of how Augmin recruits the Nedd1–γ-TuRC complex to the spindles and mediates MT-based MT nucleation at these sites.
Polo-like kinase 1 (Plk1) is a member of the conserved Polo subfamily of Ser/Thr protein kinases that is essentially required for bipolar spindle formation and mitotic progression (17–20). Loss of the function of Plk1 or its homologs in diverse organisms results in various defects in centrosome maturation and bipolar spindle formation, such as diminished centrosomal MT nucleation, weakened bipolar spindles, and monopolar spindles and consequently induces spindle checkpoint-dependent mitotic arrest and apoptotic cell death. Expression of a dominant-negative polo-box domain (PBD), a phospho-Ser/Thr-binding module (21, 22), also induces a spindle defect similar to that of Plk1 depletion (23, 24), suggesting that PBD-dependent Plk1 function is required for this event.
In this study, we showed that, in addition to the previously reported centrosomes, kinetochores, and midbody localization, Plk1 also localizes along the length of the mitotic spindles. Cdc2 induces the formation of the Nedd1–Plk1 complex by phosphorylating Nedd1 at S460, and this step appears to be central for Plk1-dependent phosphorylation of Hice1, enhancement of the Augmin–MT interaction, and promotion of spindle MT-based MT nucleation.
Results
Plk1 Localizes to Mitotic Spindles and Binds to a Previously Uncharacterized Nedd1 p-S460 Motif in a Cdc2-Dependent Manner.
In an effort to identify components critically required for mediating Plk1-dependent bipolar spindle formation, we have investigated whether any of the previously characterized centrosomal proteins directly bind to the PBD of human Plk1. This screening led to the identification of Nedd1 as a strong PBD-binding protein. Nedd1 localizes to the centrosomes and spindles and recruits γ-TuRC to these subcellular structures (8, 9). To examine whether Plk1 also localizes to these locations, we immunostained asynchronously growing HeLa cells. In addition to centrosomes, kinetochores, and midbody, a significant level of endogenous Plk1 was clearly detected along the mitotic spindles (Fig. 1A, Left and Fig. S1A). Consistent with a previous report (25, 26), EGFP-fused Plk1 signals were also evident along the spindle, when expressed at the physiological level (Fig. 1A, Right and Fig. S1B).
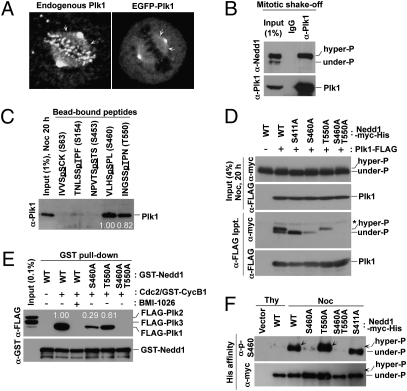
Fig. 1.
Plk1 localizes to mitotic spindles and binds to both Nedd1 p-S460 and p-T550 epitopes generated by Cdc2. (A) Asynchronously growing HeLa cells either untreated (Left) or infected with lentivirus expressing EGFP–Plk1 at the physiological level (Right) were immunostained for confocal microscopy. Arrows indicate Plk1 signals along the mitotic spindles. (B) Plk1 immunoprecipitates were prepared from mitotic HeLa cells and subjected to immunoblotting analyses. (C) Indicated Nedd1 peptides cross-linked to the beads were incubated with mitotic HeLa cell lysates and precipitated. Phosphorylated residues are underlined. Numbers, relative efficiencies of Plk1 precipitation. (D) Mitotic 293T cells transfected with the indicated constructs were subjected to coimmunoprecipitation analyses. Note that, unlike the endogenous Nedd1, the slow-migrating forms are less manifest in the input because of less efficient phosphorylation of the transfected constructs. (E) Indicated GST–Nedd1 proteins were reacted with Cdc2/GST–Cyclin B1 and then incubated with total lysates containing FLAG–Plk1 to 3. The bead-bound GST–Nedd1 ligands were precipitated and analyzed. Numbers represent relative efficiencies of Plk1 precipitation. (F) The indicated Nedd1 proteins were affinity purified from transfected 293T cells and then immunoblotted. Arrows indicate hyperphosphorylated, slow-migrating Nedd1 WT and T550A mutant.
To understand the nature of the Nedd1–Plk1 interaction in vivo, we first carried out coimmunoprecipitation and PBD pull-down assays. Results showed that Plk1 PBD interacts specifically with a slow-migrating, hyperphosphorylated form of Nedd1 during the early M phase of the cell cycle. (Fig. 1B and Fig. S1C). Visual scanning of the primary sequence of Nedd1 isoform b (hereafter referred to as Nedd1 for simplicity) led to the identification of five potential PBD-binding sequences (22). Testing of the phosphorylated peptides derived from these sequences revealed that a phospho-S460 (p-S460) peptide efficiently interacted with Plk1, whereas a phospho-T550 (p-T550) peptide interacted with Plk1 at a somewhat lesser level (Fig. 1C). Provision of either one of the PBD-binding phosphopeptides disrupted the Nedd1–Plk1 interaction (Fig. S1D).
Subsequent analyses revealed that the S460 residue is phosphorylated both in vivo and in vitro in a manner that requires the Cdc2 kinase activity (Fig. S1 E and F and Fig. S2A). Interestingly, mutation of S460 to A (S460A) eliminated the PBD interaction with the hyperphosphorylated, slow-migrating form of Nedd1, whereas mutation of T550 to A (T550A) abolished the interaction with the underphosphorylated form of Nedd1 (Fig. 1D). The S460A T550A double mutations annihilated all of the interactions. Notably, the S411A mutation eliminated the slow-migrating form but failed to diminish the Plk1–Nedd1 interaction (Fig. 1D). We also observed that Cdc2/Cyclin B1-phosphorylated Nedd1 specifically interacted with Plk1, but not with Plk2 or Plk3, and this interaction required both S460 and, to a lesser degree, T550 (Fig. 1E). Taken together, our results in part confirm the previous observation that the p-T550 motif serves as a Plk1 PBD binding site (26) and suggest that the newly identified p-S460 motif is likely critical for PBD-dependent mitotic functions.
Because phosphorylation of the T550 residue has shown to be important for proper centrosome-based MT nucleation (26), we then focused on characterizing the p-S460–dependent Nedd1–Plk1 interaction. We observed that the p-S460 epitope was detected on mitotic Nedd1 wild-type (WT), T550A, and S411A proteins at similar levels (Fig. 1F), suggesting that the p-S460 epitope is generated independently of T550 or S411 phosphorylation. Close examination of the cells releasing from a G1/S block revealed that phosphorylation at S460 occurs concurrently with Cdc27 phosphorylation, but persists until after Cyclin B1 degradation (Fig. S2B). Notably, the p-S460 epitope was found associated only with the slow-migrating Nedd1 form (Fig. S2B), suggesting that S460 phosphorylation occurs after S411 phosphorylation-dependent migration shift.
p-S460–Dependent Nedd1–Plk1 Interaction Is Important for Targeting Nedd1 to the Spindles.
To investigate the physiological significance of the p-S460–dependent Nedd1–Plk1 interaction in comparison with that of p-T550–dependent interaction, we generated HeLa cells stably expressing sh-Nedd1–insensitive WT or mutant forms of Nedd1, which were then depleted of endogenous Nedd1 before analyses (Fig. 2A). Examination of asynchronously growing cultures revealed that ∼18% of the S460A cells exhibited a mitotic chromosome morphology and ∼3% of them displayed apoptotic cell death (Fig. 2B). On the other hand, the T550A cells showed only a modest level (∼8%) of mitotic cells. The S460A T550A double mutant produced a mitotic index (∼20%) slightly greater than the S460A mutant, but lower than the cells expressing the spindle-localization–defective S411A mutant or the control vector (Fig. 2B). The S460A and S411A mutants exhibited a high level of multipolar spindle morphologies, whereas the T550A mutant displayed a significant level of monopolar spindles. The S460A T550A double mutant appeared to possess an additive spindle defect, although it was less defective than the cells expressing the control vector (Fig. 2C). An increase in cells with chromosome segregation defect was commonly observed in all of the mutants (Fig. 2C). As a consequence of these defects, both S460A and T550A mutants exhibited a significantly longer preanaphase than the WT (Fig. S3 A and B and Movies S1, S2, S3, and S4).
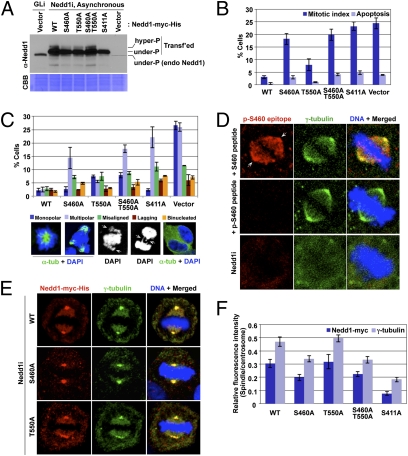
Fig. 2.
Role of S460 and T550 phosphorylation in the subcellular localization and mitotic functions of Nedd1. (A–C) HeLa cells expressing various RNAi-insensitive Nedd1 constructs were depleted of endogenous Nedd1 by sh-Nedd1 (Nedd1i). The resulting cells were immunoblotted (A) or immunostained to quantify mitotic defects (n > 600 cells). (B and C) Arrows in C, misaligned and lagging chromosomes. Error bars, SD. (D) HeLa cells were subjected to immunostaining analyses after preincubating the antibody with 10 μg/mL of the indicated peptide or depleting the endogenous Nedd1 by sh-Nedd1 (Nedd1i). Arrows indicate centrosomes. (E and F) The cells generated in A were immunostained (E) and relative fluorescence intensities were quantified (n > 20 cells per sample) (F). Error bars, SD.
In immunostaining analyses, we observed that the Nedd1 p-S460–specific signal was detected both at the centrosomes and along the spindles (Fig. 2D). Remarkably, mutation of S460 to A was sufficient to impair the ability of Nedd1 to localize to the spindles, whereas mutation of T550 to A did not significantly alter this event (Fig. 2 E and F; also see Fig. S3 C and D for the actual numbers of signal intensities). In good agreement with the previous finding (8), the S411A mutant also failed to localize to the spindles. The subcellular localization of γ-tubulin closely mirrored that of Nedd1 in all cases. These observations suggest that the defect associated with the S460A or S411A mutation can be attributable to the impaired Nedd1 localization to the spindles.
Requirement of the Nedd1 p-S460–Plk1 Interaction for Proper Hice1 Phosphorylation and Augmin–MT Interaction.
In an effort to understand the function of p-S460–dependent Nedd1–Plk1 interaction, we have searched for potential Plk1 substrates downstream of the Nedd1–Plk1 interaction. Nedd1 has been shown to interact with a spindle-associating complex known as Augmin (7, 11). Thus, we examined whether Plk1 phosphorylates any of the Augmin subunits. In vitro Plk1 kinase assays with 6 of the 8 Augmin subunits that we expressed revealed that Plk1 significantly phosphorylates Hice1 and also weakly phosphorylates CCDC5 (Fig. S4A). Notably, whereas the kinase activity of Plk1 has been shown to be required for the Plk1–FAM29A interaction (11), Plk1 does not appear to phosphorylate FAM29A (Fig. S4 A–C).
We next examined whether Nedd1 associates with Hice1 and, if so, whether the Nedd1-bound Plk1 phosphorylates Hice1 in vivo to regulate the Hice1–MT interaction. We observed that affinity purification of Nedd1 WT, S460A, T550A, or S460A T550A coprecipitated Hice1 efficiently, whereas S411A did not (Fig. 3A). Thus, Nedd1 S460A associates with Augmin without localizing to the spindle, and S411A may have a structural alteration that prevents its interaction with the Hice1-containing Augmin complex.
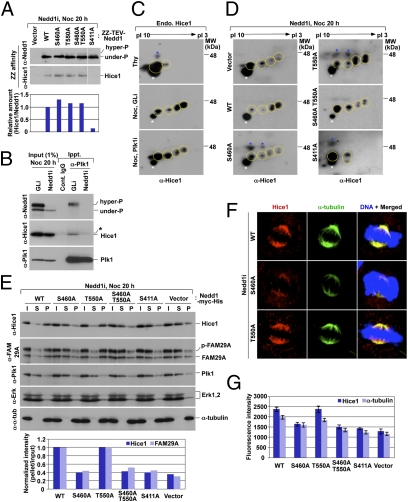
Fig. 3.
Requirement of the Nedd1 p-S460–Plk1 interaction for both Plk1-dependent Hice1 phosphorylation and Hice1-mediated Augmin–MT interaction. (A) ZZ-affinity purification was carried out using transfected 293T cells arrested with nocodazole (Noc). After TEV digestion, eluted Nedd1 and its associated proteins were analyzed by immunoblotting. (B) Mitotic HeLa cells silenced for either control luciferase (GLi) or Nedd1 (Nedd1i) were subjected to coimmunoprecipitation analyses. Asterisk, a cross-reacting protein. (C) Total lysates were prepared from HeLa cells treated with either thymidine (Thy) for 20 h or first infected with lentivirus expressing control sh-luciferase (GLi) or sh-Plk1 (Plk1i) and then treated with nocodazole (Noc) for 20 h. Yellow dotted circle, various forms of Hice1; white asterisks, a reproducibly appearing, nonspecific dot signal that serves as a convenient marker to position the Hice1 protein; blue asterisks, randomly detected cross-reacting proteins. (D and E) Mitotic total lysates prepared from the cells in Fig. 2A were subjected to 2D gel electrophoresis (D) or to MT cosedimentation assay (E). (F and G) The same cells used in D and E were immunostained to examine the effect of the indicated Nedd1 mutations on the localization of Hice1 and γ-tubulin along the spindles. Fluorescence intensities for the cells in F were quantified (n > 20 cells per sample) (G). Error bars, SD.
In a second experiment, we observed that Plk1 interacted with phosphorylated Nedd1 and this complex coprecipitated Hice1. Depletion of Nedd1 abolished the Plk1–Hice1 interaction (Fig. 3B), suggesting that Nedd1 mediates the Plk1–Hice1 interaction.
Next, we investigated whether Plk1 directly regulates Hice1 through the interaction with Nedd1. Comparative 2D gel electrophoresis analyses for control sh-luciferase and sh-Hice1 cells enabled us to identify multiply phosphorylated Hice1-specific signals (Fig. S4D). Although Hice1 did not appear to be significantly phosphorylated under thymidine-treated (S phase) conditions, it displayed multiply phosphorylated forms under nocodazole-treated (M phase) conditions (Fig. 3C; note the appearance of multiple, slow-migrating Hice1 spots with sizable shifts toward the acidic side). Interestingly, depletion of Plk1 greatly diminished the levels of multiply phosphorylated forms (Fig. 3C). As expected if a specific Nedd1–Plk1 interaction were required for proper phosphorylation of Hice1, cells expressing the S460A mutant, but not the T550A mutant, greatly reduced the level of Hice1 phosphorylation (Fig. 3D). The S460A T550A double mutant exhibited an impaired Hice1 phosphorylation at a level similar to that of the S460A cells. The S411A mutant, defective in both Augmin interaction and spindle localization, was severely crippled in Hice1 phosphorylation (Fig. 3D). These observations suggest that the p-S460–dependent Nedd1–Plk1 interaction is critical for proper Plk1-dependent Hice1 phosphorylation and the p-S411–dependent Nedd1–Augmin interaction is a prerequisite for this event.
To investigate the physiological significance of the Plk1-dependent Hice1 phosphorylation, MT cosedimentation assays were performed in the presence of taxol using nocodazole-arrested lysates prepared from WT and various Nedd1 mutants. In the cells expressing WT Nedd1, Hice1 efficiently interacted with the polymerized MT pellet fraction (Fig. 3E). FAM29A also associated with the MT fraction, thus corroborating the Hice1–MT interaction. Under these conditions, both S460A and S460A T550A cells exhibited much diminished levels of the Hice1–MT and FAM29A–MT interactions, whereas the T550A mutant displayed largely unaltered levels of the interactions (Fig. 3E). Closely correlating with the drastically diminished Hice1 phosphorylation in Fig. 3D, the levels of the Hice1 and FAM29A interactions with the MTs were severely impaired in both S411A and control vector cells (Fig. 3E). Consistent with these observations, Hice1 localization to the spindle was normal in the WT Nedd1 or T550A cells, whereas it was greatly diminished in the cells expressing S460A, S460A T550A double, S411A, or the control vector (Fig. 3 F and G).
Plk1-Dependent Hice1 Phosphorylation Promotes the Hice1–MT Interaction and Hice1 Localization to the Spindle.
To directly examine the role of Plk1-dependent Hice1 phosphorylation, we first determined Plk1-dependent Hice1 phosphorylation sites in vitro by mass spectrometry. Among them, 17 sites were also found to be phosphorylated in vivo. We then generated various Hice1 mutants by converting phosphorylated residues to Ala, and tested their ability to bind to MTs. Multiple mutations on the clustered S129, T130, S131, and S133 residues and single mutations on the S143A and S151A residues significantly diminished the ability of Hice1 to interact with MTs (Fig. S5). Thus, we combined all these mutations and generated a 6A mutant (S129A, T130A, S131A, S133A, S143A, and S151A). We also generated the corresponding 6D mutant by mutating these sites to negatively charged Asp residues. Expression of the 6A or 6D mutant in Hice1 RNAi cells did not influence the level of FAM29A, whereas expression of control vector diminished it to an undetectable level (Fig. 4A). Because depletion of any one of the Augmin subunits leads to destabilization of other subunits in the complex (4, 14), these findings suggest that both the 6A and 6D mutants are capable of forming a proper Augmin complex. The 6A mutations greatly diminished multiply phosphorylated Hice1 forms to a degree similar to that of the Plk1i cells in Fig. 3C (Fig. 4B). Remarkably, although the 6D mutant did not appear to exhibit an augmented MT binding, it bound to MTs as efficiently as the mitotic Hice1 WT (Fig. 4C). In contrast, the 6A mutant was severely impaired in MT binding. Confirming these results, the levels of FAM29A bound to MTs were similar to those of Hice1 (Fig. 4C).
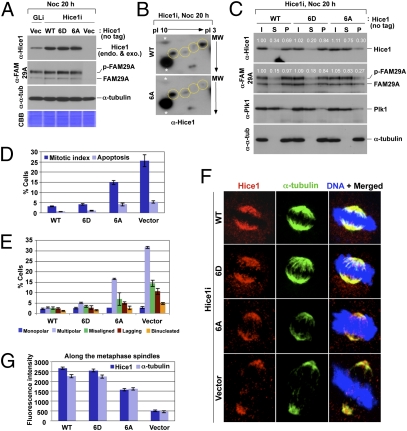
Fig. 4.
Plk1-dependent phosphorylation of Hice1 in the N-terminal region is critical for proper Hice1–MT interaction and mitotic spindle localization. (A) HeLa cells expressing the indicated RNAi-insensitive Hice1 constructs were depleted of either control luciferase (GLi) or endogenous Hice1 (Hice1i) and analyzed by immunoblottings. (B) Cells in A were treated with nocodazole for 20 h, and total lysates were prepared and subjected to 2D gel electrophoresis. Yellow dotted circle, various forms of Hice1; white asterisks, nonspecific dot signals that serve as convenient markers to position the Hice1 protein. (C) The cells in A were arrested in mitosis and then subjected to MT cosedimentation assay. Numbers, relative signal intensities. (D–G) The cells in A cultured under subconfluent conditions were immunostained to quantify mitotic defects (n > 600 cells per sample) (D and E) or subcellular localization patterns (n > 22 cells per sample) (F and G) of various forms of Hice1. Error bars, SD.
Analyses of asynchronously growing cells generated in Fig. 4A showed that cells expressing either 6A or control vector, but not WT or 6D, exhibited significantly higher rates of mitotic index and apoptosis with substantially increased levels of multipolar spindles and missegregating chromosomes (Fig. 4 D and E). As expected if Plk1-dependent Hice1 phosphorylation is critical for the recruitment of Hice1 to the mitotic spindles, the 6A mutant, but not the WT and 6D mutant, was impaired in spindle localization (Fig. 4 F and G).
Plk1-Mediated Hice1 Phosphorylation Is Required for Proper MT-Based MT Nucleation.
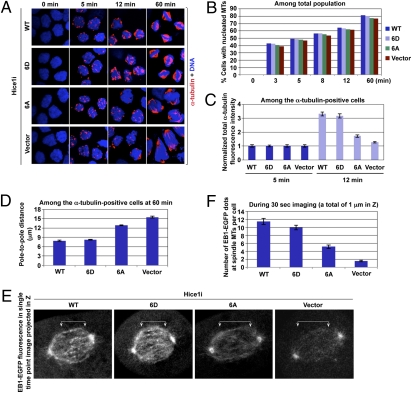
We then carried out a microtubule regrowth assay, using HeLa cells expressing either WT or mutant forms of Hice1 where endogenous Hice1 was depleted. Results showed that the cells expressing WT Hice1 effectively generated a detectable level of nucleating MTs as early as 3–5 min after release (Fig. 5 A and B). The presence of multiple MT nucleation sites in a single cell is likely due to multiple MT nucleating activities arising from both centrosomes and mitotic chromatin, as previously reported (8). Because of the dominant role of centrosomes in this process, these cells established normal bipolar spindles in ∼80% of the population 60 min after release (Fig. 5 A and B). Interestingly, the Hice1 6D-expressing cells generated MTs as effectively as the WT Hice1 cells, suggesting that the 6D mutant is fully capable of rescuing the Hice1 RNAi defect. However, although the Hice1 6A mutant nucleated MTs as efficiently as WT Hice1 at early time points (3 min and 5 min), it soon failed to continue to increase the α-tubulin fluorescence intensities at later time points (see the 12-min image in Fig. 5 A and C). As a result, these cells eventually exhibited much weakened bipolar spindle morphologies at the 60-min time point, although the total fraction (∼77%) of cells with detectable MTs was similar to that of WT cells (Fig. 5 A and B). The control vector-expressing cells exhibited a similar level of MT nucleation at an early stage, but displayed greatly impaired bipolar spindle morphologies at later time points (Fig. 5 A–C). Consistent with these results, the pole-to-pole distance for 6A- or control vector-expressing cells was significantly longer than that for WT or 6D cells (Fig. 5D).
Fig. 5.
Plk1-dependent Hice1 phosphorylation is important for MT-based MT nucleation. (A–D) The cells generated in Fig. 4A were subjected to MT regrowth assay (A). The efficiency of MT nucleation (n > 500 cells per sample) (B), total α-tubulin fluorescence intensity (n > 23 cells per sample) (C), and pole-to-pole distance (n > 21 cells per sample) (D) were quantified. Error bars, SD. (E and F) The cells described in Fig. 4A were infected with lentivirus expressing EB1-EGFP and then depleted of the endogenous Hice1 by sh-RNA (Hice1i). Cells were imaged using Zeiss VivaTome time-lapse microscopy. The EB1-EGFP fluorescences were detected as dotted signals in the single time-point image in E. Arrowed brackets, EB1-EGFP signals along the spindles. The number of EB1-EGFP dot signals per cell (n > 21 cells per sample) (F) was quantified from the cells imaged in E. Error bars, SD.
To closely examine the defect associated with the 6A mutation during mitosis, we then performed time-lapse microscopy in metaphase cells expressing an MT plus end tracking protein, EB1, C-terminally fused with EGFP. The results showed that Hice1 WT- or 6D-expressing cells exhibited an average of ∼10–11 EB1-EGFP dots within 1 μm in Z (which is approximately one-tenth of cell depth) of a cell in a period of 30 s (Fig. 5 E and F and Movies S5, S6, S7, and S8). Under these conditions, Hice1 6A-expressing cells exhibited an average of 5 EB1-EGFP dots with significantly weakened EB1-EGFP fluorescence along the spindle, whereas control vector-expressing cells displayed an average of less than 2 EB1-EGFP dots along the spindle. Because the abundance of EB1 could serve as an indirect indicator for MT nucleation and polymerization activity, these observations suggest that Plk1-dependent Hice1 phosphorylation is required for proper MT-based MT nucleation and spindle stability.
Discussion
The Nedd1 p-S460–Plk1 Complex-Dependent Augmin Regulation.
Augmin is a recently identified protein complex that is critically required for spindle MT-based MT nucleation, as observed in Drosophila and human cells (4, 7). The Hice1 subunit of the complex is thought to directly bind to MTs to promote this process (16). However, how Hice1 interacts with MT and recruits MT-nucleating activities to the spindles remains unknown. Hice1 and the other Augmin subunits localize to the interphase centrosomes even in the presence of cytoplasmic MTs (7, 13, 16), suggesting that mitotic-specific posttranslational modifications such as phosphorylation may target Augmin to the spindles. In this regard, our finding that Cdc2-dependent formation of the Nedd1 p-S460–Plk1 complex is central for Plk1-dependent Hice1 phosphorylation and spindle MT-based MT nucleation is intriguing. Moreover, the unique mechanism of phosphorylating Hice1 by Nedd1 p-S460–bound Plk1 is the first example of the Plk1-dependent ‘‘distributive phosphorylation'' model proposed by Lowery et al., in which a PBD-dependent interaction with its binding target enables the catalytic activity of Plk1 to phosphorylate a third protein, associating either directly or indirectly with the PBD-binding target (27).
We observed that, in addition to Hice1, Plk1 phosphorylates another Augmin subunit, CCDC5, in vitro (Fig. S4A), raising the possibility that Plk1-dependent Augmin regulation occurs via additional biochemical pathways. Furthermore, loss of Hice1 function is more detrimental than loss of the Plk1-dependent Hice1 phosphorylation sites, hinting that other kinases may also regulate Hice1 function. Alternatively, unphosphorylated Hice1 may have a basal level of MT-binding activity.
A recent report suggested that Plk1 recruits FAM29A, another subunit of the Augmin complex, to spindles, and promotes MT-based MT nucleation (11), implying that Plk1 may regulate FAM29A directly. However, Plk1 failed to significantly phosphorylate FAM29A in vitro or alter the level of phosphorylation in vivo (Fig. S4 A–C). Our finding that Plk1 directly regulates the recruitment of Hice1, and therefore the Augmin complex, to the spindle may help explain why Plk1 activity is required for FAM29A recruitment to the spindle and subsequent MT-based MT nucleation from within the spindle.
Two-Step Mechanism Underlying Augmin Recruitment to the Spindle and Significance of Plk1-Dependent MT-Based MT Nucleation.
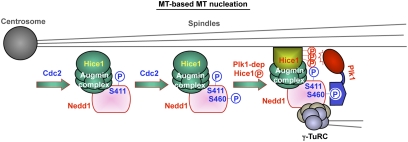
The molecular mechanism of how Augmin is targeted to mitotic spindles has been elusive. It has been shown that mutation of S411 to Ala abolishes Nedd1 localization to the spindle (8). Our results demonstrate that the S411A mutant, but not the S460A or T550A mutant, is defective in interacting with the Augmin complex (Fig. 3A), suggesting that phosphorylation at S411 is specifically required for Nedd1 to interact with Augmin. Interestingly, however, the S460A mutant bearing phosphorylated S411 residue is still significantly defective in spindle localization, albeit at a lesser degree than the S411A mutant. Hence, phosphorylation at S411 is necessary but not sufficient for spindle localization. These observations led us to propose that the recruitment of the Nedd1-bound Augmin to the spindle is regulated at two sequential steps. First, phosphorylation at S411 induces a structural change in Nedd1 in a way that enables the Nedd1–γ-TuRC complex to associate with Augmin. This event, however, is not sufficient for the complex to translocate to the spindle MTs. Second, subsequent phosphorylation at S460 recruits Plk1, thereby allowing the latter to phosphorylate Hice1 and promote the Augmin-spindle MT interaction (Fig. 6). This two-step model predicts that the Nedd1-bound Plk1 recruits itself to the Augmin-associated spindles through the Nedd1 p-S411–dependent Augmin interaction and the Nedd1 p-S460–dependent Hice1 phosphorylation. This model also helps explain why the S411A mutant, which is crippled at the first step, is more severely impaired in Hice1 phosphorylation and subsequent Hice1–MT interaction than is the Hice1 binding-competent, but phosphorylation-incompetent, S460A mutant (Fig. 3 D–G).
Fig. 6.
Model proposing the mechanism of Plk1-dependent Augmin recruitment to the spindle and spindle MT-based MT nucleation. See text for details.
Here, we provide evidence that Plk1 promotes bipolar spindle formation by forming the Nedd1–Plk1 complex and directly regulating the function of one of the Augmin subunits, Hice1. Defects in these processes result in abnormal bipolar spindle formation that leads to mitotic arrest, chromosome missegregation, and apoptosis. Plk1 has been shown to promote centrosome-based MT nucleation. Thus, although how this event is coordinated with the Plk1-mediated MT-based MT nucleation described here remains to be further investigated, our data suggest that Plk1 plays a key role in the formation of bipolar spindles.
Materials and Methods
Plasmid Construction, Cell Culture, Virus Generation and Infection, and Antibodies.
Detailed information is provided in SI Materials and Methods. All of the shRNA target sequences used in this study are listed in Table S1. Primary antibodies used in this study are listed in Table S2.
Microtubule Cosedimentation and Regrowth Assays.
MT cosedimentation assays were performed in the presence of taxol using nocodazole-arrested lysates. Further details are described in SI Material and Methods. For microtubule regrowth assay, HeLa cells were arrested in S phase by the addition of 2.5 mM thymidine for 20 h. Cells were released for 7 h and treated with 200 nM of nocodazole for 4 h. After washing the cells with cold medium, cells were placed on ice for 30 min. MT regrowth was induced by replacing the cold medium with prewarmed medium at 37 °C. At the indicated time points after the medium replacement, the cells were fixed and immunostained with anti–α-tubulin antibody and a Texas Red-conjugated secondary antibody.
Supplementary Material
Acknowledgments
We are grateful to Mary Dasso, Dan L. Sackett, and Yasuhiko Terada for critical reading of the manuscript; Gohta Goshima, Jens Lüders, Andreas Merdes, Dan L. Sackett, Tim Stearns, Ryota Uehara, and Michael B. Yaffe for reagents and technical assistance; and Susan Garfield, Valarie Barr, Brian Svedberg, and Elise Shumsky for their assistance in confocal and time-lapse microscopies. This work was supported in part by National Cancer Institute intramural grants (to K.S.L. and T.D.V.), a Food and Drug Administration (FDA) grant (to L.-R.Y.), Korea Basic Science Institute's high field NMR research program Grant T3022B (To J.K.B.), and the World Class Institute research program of the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology of Korea (B.Y.K.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 11301.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106223108/-/DCSupplemental.
References
- 1.Wiese C, Zheng Y. Microtubule nucleation: Gamma-tubulin and beyond. J Cell Sci. 2006;119:4143–4153. doi: 10.1242/jcs.03226. [DOI] [PubMed] [Google Scholar]
- 2.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Karsenti E, Vernos I. The mitotic spindle: A self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- 4.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata T, et al. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 7.Uehara R, et al. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci USA. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lüders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 9.Haren L, et al. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol. 2006;172:505–515. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Coppinger JA, Jang CY, Yates, JR, III, Fang G. FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol. 2008;183:835–848. doi: 10.1083/jcb.200807046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Fang K, Fang G. FAM29A, a target of Plk1 regulation, controls the partitioning of NEDD1 between the mitotic spindle and the centrosomes. J Cell Sci. 2009;122:2750–2759. doi: 10.1242/jcs.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawo S, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Meireles AM, Fisher KH, Colombié N, Wakefield JG, Ohkura H. Wac: A new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol. 2009;184:777–784. doi: 10.1083/jcb.200811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wainman A, et al. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 2009;23:1876–1881. doi: 10.1101/gad.532209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G, et al. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol Cell Biol. 2008;28:3652–3662. doi: 10.1128/MCB.01923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 18.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 19.Petronczki M, Lénárt P, Peters JM. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 20.van de Weerdt BC, Medema RH. Polo-like kinases: A team in control of the division. Cell Cycle. 2006;5:853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- 21.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 23.Hanisch A, Wehner A, Nigg EA, Silljé HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell. 2006;17:448–459. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seong YS, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–32293. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- 25.Kishi K, van Vugt MA, Okamoto K, Hayashi Y, Yaffe MB. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J Cell Sci. 2009;122:2240–2251. doi: 10.1242/jcs.042747. [DOI] [PubMed] [Google Scholar]
- 27.Lowery DM, Mohammad DH, Elia AE, Yaffe MB. The Polo-box domain: A molecular integrator of mitotic kinase cascades and Polo-like kinase function. Cell Cycle. 2004;3:128–131. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.