Abstract
Ig class-switch recombination (CSR) is directed by the long and repetitive switch regions and requires activation-induced cytidine deaminase (AID). One of the conserved switch-region sequence motifs (AGCT) is a preferred site for AID-mediated DNA-cytosine deamination. By using somatic gene targeting and recombinase-mediated cassette exchange, we established a cell line-based CSR assay that allows manipulation of switch sequences at the endogenous locus. We show that AGCT is only one of a family of four WGCW motifs in the switch region that can facilitate CSR. We go on to show that it is the overlap of AID hotspots at WGCW sites on the top and bottom strands that is critical. This finding leads to a much clearer model for the difference between CSR and somatic hypermutation.
Keywords: DNA recombination and repair, immunoglobulin gene rearrangement
Immunoglobulins can be categorized into several different classes (or isotypes) depending on the identity of the constant (C) region of its heavy chain. This portion of the molecule is responsible for the effector function that is critical for the clearance of invading pathogens during a humoral response. The heavy-chain germ-line chromosome contains a tandem array of C “genes” (μ, δ, γ3, γ1, γ2b, γ2a, ε, and α in mouse; μ, δ, γ3, γ1, α1, γ2, γ4, ε, and α2 in human). In naive B cells, Cμ/δ is used by default, as it is the first C region downstream of the variable region. In antigen-stimulated B cells, an intrachromosomal deletion known as class-switch recombination (CSR) can replace Cμ/δ with one of the downstream C regions, resulting in a “switch” of Ig isotype from IgM/IgD to IgG (γ), IgE (ε), or IgA (α).
CSR occurs through a cut-and-paste mechanism that involves the generation and repair of DNA double-strand breaks (DSBs) in a large zone of repetitive DNA upstream of the C region, known as the switch (S) region (1, 2). S regions are characterized by a large number of tandem repeats that are heterogeneous and lack a clear boundary. Only several short sequence motifs are common to all S regions, including G-rich pentamers GAGCT and GGG(G/C)T (3–5). Mouse and human have two distinct families of S regions. The repeats in Sμ, Sε, and Sα are composed almost entirely of pentamers. The Sγ repeats (Sγ3, Sγ1, Sγ2b, Sγ2a, etc.), although also containing these pentamers, have a unique 49- to 52-bp unit. S regions harbor most recombination breakpoints during CSR and are therefore considered important for this process. Although the unique structural features of S regions were well appreciated, it wasn't until 2003, when elegant studies from Alt and colleagues provided direct evidence that not any sequence could function as a switch region. For example, Xenopus Sμ, but not a size-matched irrelevant intronic sequence from the XPV gene, could functionally substitute for mouse Sγ1 in CSR to IgG1 (6). Therefore, the S region is not just a sequence-occupying space to absorb DSBs, but rather contains critical sequence motifs required for CSR. Unfortunately, because of the length and heterogeneity of S-region sequences and the lack of association of CSR breakpoints with any specific DNA sequence, it remains unclear what distinguishes S regions from other DNA sequences in the genome.
CSR requires a B-cell–specific factor called activation-induced cytidine deaminase (AID) that is also required for somatic hypermutation (SHM) of the Ig variable region (7, 8). AID is a cytosine deaminase that converts cytosines to uracils on DNA (9). AID-generated uracils are thought to be the source of DNA lesions required for the initiation of CSR and SHM. Uracils in DNA are usually repaired by the base excision repair (BER) and mismatch repair (MMR) pathways. In BER, uracils are removed by uracil-N-glycosylase (UNG). The resulting abasic sites are then cleaved by apurinic/apyrimidinic endonuclease (APE). Alternatively, uracils in DNA can be recognized as U:G mismatches by MSH2/6 and repaired via MMR. Indeed, CSR is inhibited in the absence of UNG or MSH2, and completely abolished in the absence of both (10–12).
In addition to S-region sequences and AID, CSR also requires germ-line transcription (GLT) through the S region (1, 2). At the onset of CSR, GLT initiates from a so-called I exon upstream of the S region and terminates after the C region. The GLT is spliced and polyadenylated, and the S region looped-out as an intron. The requirement for transcription may be partially explained by the fact that AID is a single-stranded DNA-specific cytosine deaminase (13–16). Transcription would allow transient separation of the two DNA strands to allow AID access to its single-stranded substrate, and in this regard, chromosomal R-loops have been described in switch regions of activated primary B cells (17–19).
In vitro experiments have shown that AID deamination efficiency on any given cytosine is influenced by its surrounding nucleotides (15, 20). AID prefers sequences that conform to WRC (W = A,T; R = A,G) (21), matching the SHM hotspot motif previously identified in vivo. WRC motifs are highly enriched in S regions, mostly in the form of AGCT that is conserved in all S regions (22). More than half of the cytosines on the nontemplate DNA strand of the S region are in a WRC motif. However, whether this enrichment of AID hotspots in the S region is functionally relevant to CSR has not been directly tested.
AGCT is a palindrome such that the antiparallel DNA strand is also AGCT. Thus, this short sequence has two AID hotspots aligned right across each other on the two DNA strands. Here, we will refer to this sequence motif as an “overlapping AID hotspot.” By this definition, any one of the four WGCW motifs is an overlapping AID hotspot, regardless of whether or not the sequence is a palindrome. There have been speculations that overlapping AID hotspots might facilitate DSB formation because concurrent AID actions on both DNA strands would yield two opposing nicks within 1 bp, resulting in a DSB (15, 23). To directly test this hypothesis, we established a cell line-based CSR assay that allows systematic mutagenesis of S-region sequences. We designed and tested a variety of S-region mutants harboring mutations at AGCT sites. We provide direct evidence that it is not the AGCT sequence per se that is critical but the overlapping AID hotspot motif that is important for CSR.
Results
Cell Line-Based Class-Switch Assay.
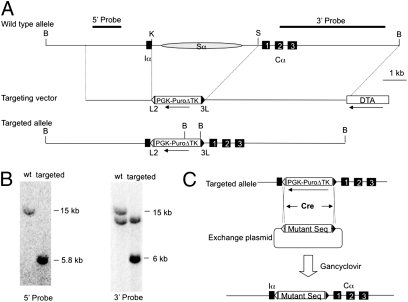
CH12F3 is a mouse B-cell line that is capable of robust cytokine-dependent CSR in vitro (24). To analyze a large number of S-region mutations, we designed a CH12F3-based CSR assay that uses the recombinase-mediated cassette exchange (RMCE) method (25, 26) to efficiently knock-in test sequences into the endogenous Sα locus in CH12F3 cells (Fig. 1). First, we used gene targeting to replace the entire Sα intron, except for elements required for RNA splicing, with a positive-negative selection cassette (PuroΔTK) flanked by a pair of heterologous loxP sites. In the CH12F3 cell line, only one of the two Sα alleles is active for CSR (24). The line (CH12F3-2a) used in this study has a rearranged nonproductive allele and an unrearranged productive allele (27) that is distinguishable by Southern blot analysis (Fig. 1B). Several independent clones were obtained in which the productive Sα allele was correctly targeted. One clone (hereafter called 1F7) was used for all subsequent studies. In the second step, a test sequence was cloned into a plasmid (termed the exchange plasmid) in between the same pair of heterologous loxP sites. When 1F7 cells are cotransfected with an exchange plasmid carrying the test sequence and a Cre-expressing plasmid, Cre-mediated site-specific recombination at the respective loxP sites results in cassette exchange between the chromosome and the exchange plasmid. The use of heterologous loxP sites ensures intermolecular cassette exchange rather than intramolecular deletion. Successful RMCE events were screened by a counter selection against TK using gancyclovir (GANC), resulting in a “marker-free” mutant allele (Fig. 1C).
Fig. 1.
Targeting of RMCE cassette into Sα locus in CH12F3 cells. (A) Genomic organization of the germ-line and targeted Sα locus. The map is drawn to scale. Exons are indicated by filled squares. L2 and 3L are wild-type and mutant loxP sites, respectively. PGK, phosphoglycerate kinase promoter; Puro, puromycin-resistant gene; ΔTK, truncated thymidine kinase gene; DTA, diphtheria toxin A chain. Restriction sites: B, BamHI; K, KpnI; S, SbfI. (B) Southern blot of BamHI-digested genomic DNA from wild-type and targeted cells. The 5′-probe detects only the productive allele and the 3′-probe detects both alleles. (C) RMCE. Exchange plasmid containing floxed mutant sequence and Cre expression plasmid are contransfected into Sα-targeted CH12F3 cells. Successful exchange events are screened by counter selection against TK gene using GANC.
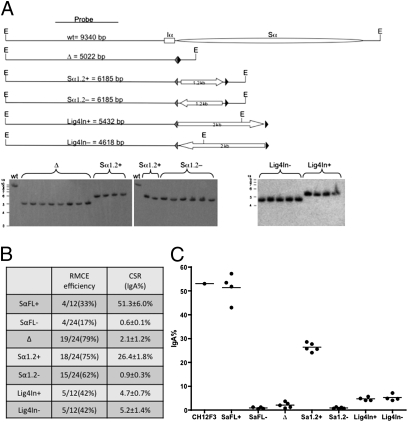
In 1F7 cells, the exchange efficiency can be as high as 80%, depending on the test sequence (Fig. 2B). Almost all GANCR/PuroS clones showed precise sequence exchange without any aberrant rearrangement based on Southern blot analysis (Fig. 2A). Several S- and non–S-region sequences were tested to validate the assay. As expected, deletion of the Sα (RMCE with an empty exchange plasmid) abolishes CSR (< 2% of wild-type level) (Fig. 2 B and C). Targeting with the full-length Sα (∼4 kb) restores CSR to wild-type level. Targeting of a 1.2-kb core Sα fragment from plasmid pSCT(μ,α) (28), which represents the most uniformly repeated region of Sα, restores CSR to 30% to 40% of the wild-type level (Fig. 2 B and C), consistent with a previous report that CSR efficiency is dependent on the length of the S region (29). Neither the core Sα nor the full-length Sα support CSR in the nonphysiological orientation (Fig. 2 B and C). We also targeted an intronic sequence from the DNA ligase IV gene as a non–S-region control, which only confer 4% to 10% of the wild-type CSR level (Fig. 2 B and C), consistent with previous findings that not all sequences can function as an S region (6). All constructs showed minimal clonal variations. These initial tests indicate that this cell line-based CSR assay is robust and suitable for mutagenesis studies of switch-region sequences.
Fig. 2.
Validation of RMCE-based CSR assay in CH12F3 cells. (A) EcoRI-digested genomic DNA from GANC-resistant clones was analyzed by Southern blot with the indicated probe. The expected sizes of the hybridized bands are listed in the diagram. Gray and black triangles indicate wild-type and mutant loxP sites, respectively. Block arrows indicate the orientations of the exchanged fragments. “E” indicates EcoRI site. SaFL indicates full-length Sα. Lig4In indicates an intronic sequence from murine DNA ligase IV gene (AC138397: 142058–144068). Plus and minus indicate physiological and nonphysiological orientations, respectively. (B) RMCE-mediated sequence knock-in at the Sα locus. Exchange efficiency is defined as the percentage of correctly exchanged clones against the total GANC-resistant clones. (C) Class-switch assay. Each dot represents an independent clone generated by RMCE in 1F7 cells. One of at least three independent experiments was shown.
Germ-Line Transcription and Splicing of the Germ-Line Transcript.
It has been reported that either GLT splicing or GLT itself is important for CSR (30, 31). Several earlier studies have indicated that the Iα-Cα GLT uses multiple splice donors (32–34). We confirmed the presence of three alternative splice donor sites (SD1, SD2, and SD3) in CH12F3 cells by RT-PCR (Fig. S1B) and DNA sequencing of the cloned PCR product. In 1F7 cells, the DNA sequences downstream of SD1 were removed as a result of gene targeting; this was necessary for the study of S-region sequence motifs because there are several pentamers in the region between SD1 and SD3. Therefore, in 1F7 cells, there is only one splice donor (SD1) (Fig. S1B, lane 3). We found that the loss of SD2 and SD3 has no adverse effect on CSR as knock-in of the 4-kb deleted intronic sequence without SD2 and SD3 fully restored CSR to wild-type level (Fig. 2C). In addition, adding back SD2 and SD3 to the core Sα neither enhances nor reduces CSR efficiency conferred by the core Sα alone (Fig. S1C).
Mutations of the AGCT Motif: Construction of Synthetic Sα Regions.
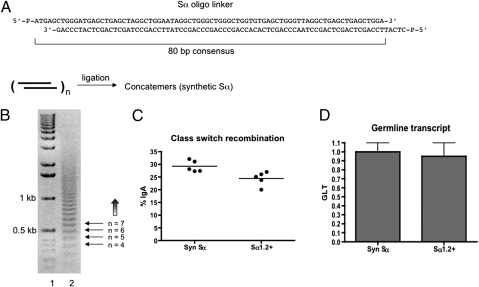
To identify S-region motifs required for CSR, we designed a method to construct artificial Sα regions from synthetic oligonucleotides. A pair of complementary 5′-phosphorylated oligonucleotides were annealed to form a linker (monomer) that can be ligated head to tail in tandem (Fig. 3A). The ligation proceeds in a unidirectional manner as the overhangs of the monomer are not palindromic. After resolving the ligation product on an agarose gel, concatemers differing in unit length (80 bp) can be readily obtained (Fig. 3B). The endogenous Sα has about 19 of the 80-bp repeating unit spanning a region of ∼1.4 kb. We chose to clone the 1.1-kb concatemer consisting of 14 repeats to match the size of the 1.2-kb core Sα fragment used earlier to validate the assay. The size-matched synthetic S region comprised of consensus repeats confers comparable level of GLT (Fig. 3D) but slightly higher CSR than that by the core Sα (Fig. 3C). This finding indicates that the synthetic Sα is fully functional as an S region. All subsequent mutagenesis studies were based on this synthetic Sα.
Fig. 3.
Construction of synthetic switch region. (A) Structure of the 80-bp linker (monomer) reflecting the consensus Sα repeat described in ref. 47. (B) Resolution of ligated concatemers on a 1.2% agarose gel. Lane 1, 1-kb DNA ladder (Invitrogen). Lane 2, ligation products (concatemers). The number of repeats in each band was shown to the right side of the gel. (C) Comparison of class-switch efficiency between the size-matched synthetic and native core Sα region. Each dot represents an independent clone. One of at least three independent experiments was shown. (D) The level of GLTs in stimulated cells. Error bars represent SEM of at least three independent experiments. Each experiment contains at least two independent clones from each construct.
Overlapping AID Hotspot Motifs, Not the Palindrome, Are Important for CSR.
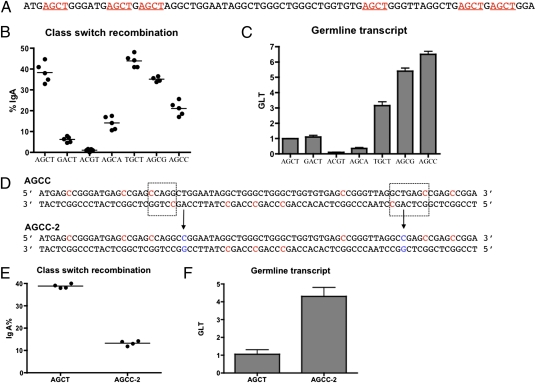
AGCT is an AID hotspot motif that is evolutionarily conserved in all S regions and considered to be functionally important for CSR (6). To directly test the role of the AGCT motif in CSR, mutations were introduced to mutate all six AGCT sites within the 80-bp consensus Sα repeat (Fig. 4A). Without altering S-region nucleotide content, nucleotide positions in the AGCT motif were swapped to create two AGCT mutants: GACT and ACGT. Both mutations inhibit CSR (Fig. 4B). The ACGT mutation completely abrogated CSR (Fig. 4B). However, this mutation also causes an approximately 10-fold reduction in GLT (Fig. 4C), making it difficult to fully ascribe the CSR defect to the loss of AGCT motif. The GACT mutation maintains the wild-type level of GLT (Fig. 4C) but reduces CSR by sixfold (Fig. 4B), suggesting that the AGCT motif is important for efficient CSR.
Fig. 4.
Mutagenesis of AGCT motif in Sα region. (A) The sequence of the consensus Sα repeat. The six AGCT sites are underlined. (B) Class-switch assay of Sα mutations. The name of each mutant reflects the sequence change from the AGCT. Each dot represents an independent clone. One of at least three independent experiments was shown. (C) The level of GLTs in stimulated cells. (D) AGCC-2 mutant. Two point mutations (indicated by arrows) were introduced into the AGCC mutant to disrupt the proximity of AID hotspots (boxed area). Cytosine residues in AID hotspots are labeled red. (E and F) Class-switch (E) and germ-line (F) transcript level in AGCC-2 mutant. Error bars represent SEM of at least three independent experiments.
AGCT is one of four WGCW (W = A or T) sequences that juxtapose two WRC motifs opposite each other across the two DNA strands. It has been suggested that the WGCW motif may facilitate DSB formation if AID catalyzes concurrent cytosine deaminations (15, 23). Such an event would place two opposing nicks within 1 bp after the actions of UNG and APE1 to yield a DSB. If this placement were the case, then changing AGCT to any WGCW motif should have a minimal effect on CSR. On the other hand, if the palindromic nature of AGCT is required, as has been suggested previously (35), disruption of the palindrome would inhibit CSR. To distinguish these possibilities, the AGCT motif was mutated to two nonpalindromic WGCW motifs: TGCT and AGCA. As can be seen, the TGCT mutant switches efficiently. The TGCT mutant has an elevated level of GLT (Fig. 4C) that is likely to be superfluous (see Discussion). Kinetically, CSR in the TGCT mutant is more efficient than that of AGCT (Fig. S2). These data suggest that TGCT is fully compatible, if not better, for CSR than AGCT (Fig. 4B). Substituting AGCT with AGCA reduces CSR by ∼2.5-fold (Fig. 4B), along with a ∼2.5-fold reduction in GLT (Fig. 4C). The reduction of GLT again makes interpretation difficult for this mutant. Nevertheless, the AGCA motif does support a significant level of CSR.
Biochemical studies have shown that, as an exception to the WRC rule, the underlined C in CGC is also an AID hotspot (15, 20). We thereby change AGCT to AGCG, which is rarely present in any switch region. AGCG is a non-WGCW sequence but still has an overlapping AID hotspot structure. Consistent with our hypothesis, the AGCG mutant also confers a high level of CSR (Fig. 4B). Next, AGCT was mutated to AGCC, which has an AID hotspot on the nontemplate strand and a coldspot on the template strand. The AGCC mutation does reduce CSR efficiency, albeit only by twofold (Fig. 4B). We noticed two fortuitous AID hotspots on the template strand that are still in proximity to AID hotspots on the nontemplate strand (Fig. 4D). Therefore, another mutant (AGCC-2) was constructed by two additional point mutations to eliminate these proximal AID hotspots. The AGCC-2 mutant retains robust GLT (Fig. 4F), but further reduces CSR to one-third of that conferred by the AGCT motif (Fig. 4E). These data further support the hypothesis that overlapping AID hotspots are important for CSR efficiency. The remaining CSR activity in the AGCC-2 mutant is more or less expected, given that the maximal difference in AID activities on hot- versus coldspots is only six- to sevenfold in vitro (15, 20). In addition, multiple mechanisms coexist for generating DSBs in switch regions (e.g., MMR) (10, 36).
To find out how alterations of AID hotspot motifs in the S region affect CSR at the DNA level, we analyzed switch junctions from three key constructs (AGCT, GACT, and TGCT), which represent wild-type, abolished, and restored AID hotspot motifs, respectively. We first looked at terminal microhomology and found a decreasing in length in the order of AGCT > TGCT > GACT (Figs. S3 and S4). This finding could be caused by a decrease in the homology in the same order between Sμ and the mutant Sα. We also looked at the positions of potential switch region breaks relative to the AID hotspots in the 80-mer repeat (Fig. S5). The potential break points are scattered and not associated with any particular motif, consistent with previous findings (37). Although the break points appear to be somewhat overrepresented in the region containing overlapping AID hotspots (e.g., in AGCT and TGCT vs. GACT) (Fig. S5), it is not statistically significant (Fig. S5B). Distributions of break points along each switch region do not appear to be markedly different either (Fig. S5C).
CSR Efficiency Correlates with S Region WGCW Density.
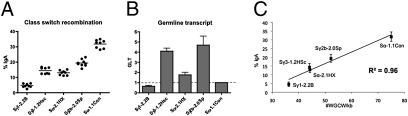
In an attempt to address the question whether S regions of different families can functionally substitute each other, we replaced Sα with restriction fragments of several Sγ regions. Consistent with an earlier study that showed functional substitution of Sγ1 by Sγ3 (38), we found that Sγ regions can also replace Sα for CSR in CH12F3 cells (Fig. 5A). However, the CSR efficiency conferred by different S regions varies dramatically. Although GLT also varied dramatically with each S region (Fig. 5B), the level of GLT and CSR does not correlate (Fig. S6A). Nor does CSR level correlate with the total number of WRC or WGCW sites in the S region (Fig. S6 B and C). CSR level correlates to a certain degree with WRC density (R2 = 0.77) (Fig. S6D), but not nearly as strong as with WGCW density (R2 = 0.96) (Fig. 5C). The extraordinary correlation between CSR and WGCW density further supports our hypothesis about the importance of WGCW motifs in CSR. Obviously CSR efficiency is influenced by a variety of factors, such as GLT and S region length (29). These data nevertheless suggest the WGCW density is a critical determinant of the quality of a switch region.
Fig. 5.
CSR efficiency correlates with switch region WGCW density. (A) CSR assay of Sα replacement constructs. Sγ1–2.2B, 2,172 bp BamHI-BamHI fragment of mouse Sγ1 (D78344: 5824–8000). Sγ3–1.2HSc, 1,181 bp of HindIII-SacI fragment of mouse Sγ3 (48). Sα-2.1HX, 2,076 bp HindIII-XbaI fragment of mouse Sα (AC160982: 124768–122693). Sγ2b-2.0Sp, 2,051 bp SpeI-SpeI fragment of mouse Sγ2b (D78344: 29538–31593). Sα-1.1Con, 1,120 bp of Sα made by ligation of 14 repeats of consensus 80 mer (Fig. 3). (B) The level of GLTs in stimulated cells. Error bars represent SEM of at least three independent experiments. Each experiment contains at least two independent clones from each construct. (C) CSR correlates with S region WGCW density. Error bars represent SD of three independent experiments.
Discussion
CSR is a region-specific DNA recombination directed by long and repetitive S-region sequences. CSR breakpoints are promiscuously distributed without obvious association with any specific DNA sequence. Understanding what distinguishes S regions from other DNA sequences in the genome is important for our knowledge about the mechanisms of many CSR-associated oncogenic chromosome translocations. In this study, we provided direct evidence that a short sequence motif (AGCT) conserved in all S regions is functionally important for CSR. Our results suggest that AGCT exerts its function via its overlapping AID hotspot structure, instead of its palindrome or the sequence itself. The most likely role of AGCT is to allow juxtaposition of two AID-initiated nicks opposite to each other to create a DSB. In addition, this study highlights the CH12F3 cell line as a valuable experimental system for genetic dissection of not only transacting protein factors (39, 40), but also cis-acting DNA sequences in CSR.
Cell Line-Based CSR Assay.
To identify critical elements in the complex S regions, an efficient CSR assay is required for analysis of a large number of mutant S-region sequences. In this study, a cell line-based CSR assay was established that allows S-region sequence variations to be examined at an endogenous locus, which was previously only possible in mouse models. Establishing this cell line-based assay has many advantages: it is fast and technically simple, and allows mutant sequences to be tested in a physiological setting; clonal switch junctions are easy to obtain (39); the high efficiency of RMCE allows systematic mutagenesis of the DNA sequences; and the robust CSR in CH12F3 cells makes it possible to detect small changes in CSR efficiencies.
AID Hotspots and Palindromic Sequences in S Regions.
The AID site preference in vitro nicely explains SHM hotspots found in vivo, but whether it is relevant to CSR has not been determined. Here we demonstrated that AID hotspot motifs that are highly enriched in S regions in the form of AGCT play a central role in CSR. Complete disruption of AID hotspot motifs markedly reduces CSR efficiency. This finding is in accordance with a recent report showing reduced CSR in several AID active-site mutants with altered sequence preference (41).
These data also provide a plausible explanation for the mechanism by which AGCT facilitates CSR. AGCT is important because of the overlapping AID hotspot structure but not the palindrome or its sequence per se. In fact, CSR can occur efficiently without a single AGCT in the S region (e.g., TGCT). Although AGCT is highly enriched in all S regions (22), some Sγ regions contain abundant AGCA. Our study shows that non-AGCT sequences conforming to the overlapping AID hotspot motif (e.g., AGCA, TGCT or AGCG) can functionally substitute for AGCT in CSR.
The importance of the WGCW motif has several implications. It implies that UNG and APE1-mediated BER is the major pathway of processing AID-generated uracils into DSBs in S regions. This finding is consistent with the fact that UNG deficiency poses a larger impact on CSR than that of an MSH2-deficiency (10–12). The data also imply that deamination events at WGCW sites may be coordinated across the two DNA strands. Otherwise, the chance of landing two opposing nicks close enough to generate a DSB would be rare if AID-catalyzed deamination is scattered across the S region. Our data further imply a plausible multimeric AID complex that carries out concurrent deaminations at WGCW sites. Although the stoichiometry of AID has not been solved, several lines of evidence are consistent with AID being a dimer or multimer (14, 42).
Short sequences like AGCT are common in the genome, including transcribed regions. Therefore, it is unlikely that these short sequences alone can dictate where AID acts. There has been some evidence that Ig 3′ enhancers in coupling with the transcription machinery play a role in recruiting AID to the S regions (43). Once AID is recruited, the high density of WGCW motif in the S region would be the next important factor to facilitate DSB formation. Of course, two opposing nicks at the WGCW site is not the only mechanism for generating DSBs in S regions. It has been suggested that the mismatch repair pathway could convert distally located nicks on opposite DNA strands into a DSB via exonuclease 1 (23, 36, 44, 45).
GLT and CSR.
Splicing of GLT or the transcript itself might also play a role in CSR (30, 31). Our data showed that reducing splice donor sites from three to one at Iα has no detrimental effect on CSR. Although this is by no means a test of the importance of splicing, it nevertheless indicates that having multiple splicing donors does not enhance CSR.
We constructed numerous S-region mutants that reduced both CSR and GLT. In such cases, the results are difficult to interpret because the CSR defect could be attributable to either the loss of certain sequence motifs or the reduction in GLT. Although it is clear that CSR cannot occur without GLT, it is not clear to what extent CSR correlates with the level of GLT. It is known that at least for the γ1 locus, the endogenous level of GLT is in large excess of what is needed for CSR. For example, a point mutation in the Iγ1 promoter that reduces Iγ1-Cγ1 GLT by fivefold had no effect on CSR to IgG1 (46). The lack of correlation between GLT and CSR frequently observed in this study certainly agrees with that point. Therefore, although some level of GLT is required for CSR, more is not necessarily better. Without knowing the threshold level of Iα-Cα GLT needed for CSR in CH12F3 cells, we were obligated to limit our analysis on S-region mutations that do not reduce GLT. Mutations that markedly reduce GLT (5 ∼10-fold), including several G-cluster mutations (GGG to GGC or AGG) and AGCT mutations (AGCT to ACGT, GGCT, AACT), are therefore excluded.
One interesting observation is that for TGCT, AGCG, and AGCC mutants, the levels of GLT are very high even without cytokine stimulation (Fig. S7). The molecular basis for this basal level of GLT is not clear.
Materials and Methods
Cell Culture and CSR Assay.
CH12F3 cells were cultured in RPMI1640 medium supplemented with 10% FBS and 50 μM of β-mercaptoethanol. For CSR assay, healthy CH12F3 cells were seeded at 5 × 104 cells/mL in the presence of 1 μg/mL anti-CD40 antibody (eBioscience 16–0402-86), 5 ng/mL of IL-4 (R&D Systems), and 0.5 ng/mL TGF-β1 (R&D Systems), and grown for 72 h. Cells were stained with a FITC-conjugated anti-mouse IgA antibody and analyzed by flow cytometry. CSR efficiency was defined by the percentage of IgA cells.
Targeting of the Sα Locus.
A 15-kb BamHI fragment containing the entire IgHα locus was isolated from BAC clone RP23-351J19 (GenBank accession no. AC160982) to build homology blocks for gene targeting. The entire intron between Iα and Cα (Kpn I to Sbf I), except elements required for RNA splicing, was removed and replaced with a RMCE cassette (a positive-negative selection marker, PuroΔTK, flanked by a wild-type and a mutant loxP site) (Fig. 1A). The resulting targeting vector was linearized and transfected into CH12F3 cells by electroporation. Puromycin (1 μg/mL) was added 48 h later and resistant clones were obtained after 7 to 8 d. Gene-targeting events were first screened by PCR, followed by Southern blot analysis. Detailed procedure of gene targeting in CH12F3 cells has been previously described (39).
Construction of Synthetic Sα Region.
The 5′-phosphorylated 80-mer oligonucleotides were purified on a denaturing polyacrylamide gel. A pair of purified complementary oligos were annealed at a final concentration of 5 μM. A 50-μL ligation mixture containing 40 μL (200 pmoles) of annealed linker (monomer) was incubated overnight at room temperature and ligation products were separated on a 1.2% agarose gel. The 1.1-kb fragment corresponding to 14 repeats of the monomeric unit was recovered from the gel, blunt-ended by klenow fill-in, and cloned into exchange vector (pLH28) at SnaBI site. The orientation and sequence accuracy of the insert was determine by DNA sequencing (Mclab).
Recombinase-Mediated Cassette Exchange.
Five microgram of exchange vector and 1 μg of Cre-expression vector were cotransfected into 1F7 cells by electroporation. Transfected cells were resuspended in 1 mL of prewarmed medium. Ten microliters (1% of the total) of cells were resuspended in 10 mL of prewarmed medium and seeded to a 96-well plate at 100 μL per well. The 96-well plate was returned to incubator for 72 h before GANC was added to a final concentration of 2 μg/mL About 30 to ∼50 GANC colonies can be obtained after 6 to ∼8 d. Typically, 24 colonies were screened by PCR followed by Southern blot confirmation. Five to six colonies were kept for CSR assays.
Quantitation of Germ-Line Transcripts.
Cells were cultured to a density around 1 × 106 cells/mL before cytokines (CIT: anti-CD40, IL4, and TGF-β) were added. Four hours after the addition of cytokines, total cellular RNA was harvested using the TRIzol reagent (Invitrogen). Two microgram of RNA was reverse-transcribed into cDNA using the high capacity RT kit (ABI) according to the manufacturer's instructions. One-tenth of the RT reaction was used in a Taqman-based real-time PCR (Taqman gene expression master mix from ABI). β-Actin was used as an internal control. Each sample, including the β-actin control, was done in triplicates. Cycling and data collection were done on an ABI StepOnePlus apparatus. See Table S1 for a list of oligonucliotides used.
Switch Junction Analysis.
Switch junctions were amplified from genomic DNA of stimulated cells by nested PCR (20 cycles each round). PCR products were cloned in Topo-TA vector pCR2.1 (Invitrogen) and sequenced at the Michigan State University genomic core facility. See Table S1 for a list of oligonucliotides used.
Supplementary Material
Acknowledgments
We thank Dr. Geoffrey Wahl for the recombinase-mediated cassette exchange selection cassette and Dr. Wesley Dunnick for mouse switch-region sequences. This work is supported by National Institute of Health Grant R01 AI081817 (to K.Y.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018726108/-/DCSupplemental.
References
- 1.Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair (Amst) 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, Alt FW. Class-switch recombination: Interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 3.Gritzmacher CA. Molecular aspects of heavy-chain class switching. Crit Rev Immunol. 1989;9(3):173–200. [PubMed] [Google Scholar]
- 4.Kataoka T, Miyata T, Honjo T. Repetitive sequences in class-switch recombination regions of immunoglobulin heavy chain genes. Cell. 1981;23:357–368. doi: 10.1016/0092-8674(81)90131-8. [DOI] [PubMed] [Google Scholar]
- 5.Mills FC, Brooker JS, Camerini-Otero RD. Sequences of human immunoglobulin switch regions: Implications for recombination and transcription. Nucleic Acids Res. 1990;18:7305–7316. doi: 10.1093/nar/18.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrin AA, et al. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 8.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 9.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 10.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Rada C, et al. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 12.Imai K, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 13.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 15.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 16.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang FT, et al. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang FT, Yu K, Hsieh CL, Lieber MR. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci USA. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 20.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424(6944):103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro GS, Aviszus K, Murphy J, Wysocki LJ. Evolution of Ig DNA sequence to target specific base positions within codons for somatic hypermutation. J Immunol. 2002;168:2302–2306. doi: 10.4049/jimmunol.168.5.2302. [DOI] [PubMed] [Google Scholar]
- 22.Hackney JA, et al. DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv Immunol. 2009;101:163–189. doi: 10.1016/S0065-2776(08)01005-5. [DOI] [PubMed] [Google Scholar]
- 23.Min IM, Rothlein LR, Schrader CE, Stavnezer J, Selsing E. Shifts in targeting of class switch recombination sites in mice that lack mu switch region tandem repeats or Msh2. J Exp Med. 2005;201:1885–1890. doi: 10.1084/jem.20042491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura M, et al. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- 25.Wong ET, et al. Reproducible doxycycline-inducible transgene expression at specific loci generated by Cre-recombinase mediated cassette exchange. Nucleic Acids Res. 2005;33(17):e147. doi: 10.1093/nar/gni145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toledo F, Liu CW, Lee CJ, Wahl GM. RMCE-ASAP: A gene targeting method for ES and somatic cells to accelerate phenotype analyses. Nucleic Acids Res. 2006;34(13):e92. doi: 10.1093/nar/gkl518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono SJ, et al. Identification of a stimulus-dependent DNase I hypersensitive site between the Ialpha and Calpha exons during immunoglobulin heavy chain class switch recombination. FEBS Lett. 2000;467:268–272. doi: 10.1016/s0014-5793(00)01151-0. [DOI] [PubMed] [Google Scholar]
- 28.Lee CG, et al. Quantitative regulation of class switch recombination by switch region transcription. J Exp Med. 2001;194:365–374. doi: 10.1084/jem.194.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarrin AA, Tian M, Wang J, Borjeson T, Alt FW. Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci USA. 2005;102:2466–2470. doi: 10.1073/pnas.0409847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. doi: 10.1126/science.8438159. [DOI] [PubMed] [Google Scholar]
- 31.Hein K, et al. Processing of switch transcripts is required for targeting of antibody class switch recombination. J Exp Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaff C, Grumont RJ, Gerondakis S. Transcriptional regulation of the germline immunoglobulin C alpha and C epsilon genes: implications for commitment to an isotype switch. Int Immunol. 1992;4:1145–1151. doi: 10.1093/intimm/4.10.1145. [DOI] [PubMed] [Google Scholar]
- 33.Lebman DA, Nomura DY, Coffman RL, Lee FD. Molecular characterization of germ-line immunoglobulin A transcripts produced during transforming growth factor type beta-induced isotype switching. Proc Natl Acad Sci USA. 1990;87:3962–3966. doi: 10.1073/pnas.87.10.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radcliffe G, Lin YC, Julius M, Marcu KB, Stavnezer J. Structure of germ line immunoglobulin alpha heavy-chain RNA and its location on polysomes. Mol Cell Biol. 1990;10:382–386. doi: 10.1128/mcb.10.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tashiro J, Kinoshita K, Honjo T. Palindromic but not G-rich sequences are targets of class switch recombination. Int Immunol. 2001;13:495–505. doi: 10.1093/intimm/13.4.495. [DOI] [PubMed] [Google Scholar]
- 36.Min IM, et al. The Smu tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity. 2003;19:515–524. doi: 10.1016/s1074-7613(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 37.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarrin AA, Goff PH, Senger K, Alt FW. Sgamma3 switch sequences function in place of endogenous Sgamma1 to mediate antibody class switching. J Exp Med. 2008;205:1567–1572. doi: 10.1084/jem.20080451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J Exp Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han L, Masani S, Yu K. Cutting edge: CTNNBL1 is dispensable for Ig class switch recombination. J Immunol. 2010;185:1379–1381. doi: 10.4049/jimmunol.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207(1):141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai K, et al. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol. 2005;115:277–285. doi: 10.1016/j.clim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Wuerffel R, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luby TM, Schrader CE, Stavnezer J, Selsing E. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J Exp Med. 2001;193(2):159–168. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 46.Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire H chain constant region locus: effect of a mutation in a STAT6 binding site in the gamma 1 promoter. J Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- 47.Arakawa H, et al. The complete murine immunoglobulin class switch region of the alpha heavy chain gene-hierarchic repetitive structure and recombination breakpoints. J Biol Chem. 1993;268:4651–4655. [PubMed] [Google Scholar]
- 48.Szurek P, Petrini J, Dunnick W. Complete nucleotide sequence of the murine gamma 3 switch region and analysis of switch recombination sites in two gamma 3-expressing hybridomas. J Immunol. 1985;135:620–626. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.