Abstract
Neural stem and progenitor cells undergo an important transition from proliferation to differentiation in the G1 phase of the cell cycle. The mechanisms coordinating this transition are incompletely understood. Cyclin D proteins promote proliferation in G1 and typically are down-regulated before differentiation. Here we show that motoneuron progenitors in the embryonic spinal cord persistently express Cyclin D1 during the initial phase of differentiation, while down-regulating Cyclin D2. Loss-of-function and gain-of-function experiments indicate that Cyclin D1 (but not D2) promotes neurogenesis in vivo, a role that can be dissociated from its cell cycle function. Moreover, reexpression of Cyclin D1 can restore neurogenic capacity to D2-expressing glial-restricted progenitors. The neurogenic function of Cyclin D1 appears to be mediated, directly or indirectly, by Hes6, a proneurogenic basic helic-loop-helix transcription factor. These data identify a cell cycle-independent function for Cyclin D1 in promoting neuronal differentiation, along with a potential genetic pathway through which this function is exerted.
Keywords: neuronal specification, developmental switch, Notch signaling
The CNS develops from neuroepithelial progenitor cells that generate a large variety of differentiated neuronal and glial progeny (1–3). This process is controlled by complex regulatory mechanisms that coordinate proliferation, fate specification, and differentiation (4–6). A crucial transition, from proliferation to differentiation, occurs during the G1 phase of the cell cycle (7–10). The transition is closely coordinated with cell cycle arrest (11–14), but the mechanisms mediating this coordination are incompletely understood.
The G1 phase is positively regulated by the action of three cyclin D proteins, which link extracellular mitogenic signals to the core cell cycle machinery (15–18). In addition to their positive cell-cycle control function, Cyclin Ds have been implicated in a number of other cellular activities (19), including direct transcriptional regulation (20–22). Nonoverlapping expression of Cyclin D1 and D2 has been reported in the developing mouse forebrain neuroepithelium, suggesting that these isoforms may differentially regulate proliferation and differentiation, respectively (23). This hypothesis has not been tested, however. Here we demonstrate, by loss-of-function (LOF) and gain-of-function (GOF) manipulations in vivo, that Cyclin D1 positively regulates neuronal differentiation in the embryonic spinal cord in a manner independent of its cell cycle regulatory activity.
Results
Cyclin D1, but Not D2, Marks Neuronal Precursors in the Developing Spinal Cord.
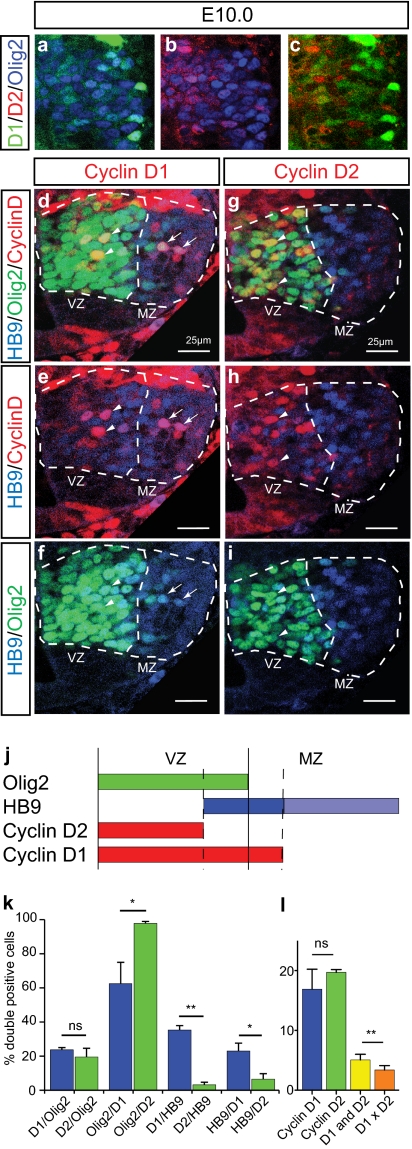
We investigated the functional role of Cyclin Ds in the developing spinal cord, where markers and mechanisms of neurogenesis are well defined (24, 25). Analysis of Cyclin D expression between E9.5 and E13.5 by immunostaining indicated that Cyclin D1 and D2 are expressed in the ventricular zone (VZ) (Fig. S1 A–C), consistent with previous in situ hybridization data (26–28). More detailed analysis was performed in the motoneuron progenitors (pMN) domain, which sequentially generates motoneurons (MNs) at E9.5–E11.5 and oligodendrocytes (29–33) and which expresses the basic helix-loop-helix (bHLH) transcription factor Olig2 (29, 34, 35) (Fig. 1). At E9.5, Cyclin D1 and D2 were expressed in ∼45–50% of Olig2+ cells (Fig. S1 D, H, and P), in a largely overlapping manner (Fig. S1A). By E10.0, the proportion of Olig2+ cells expressing Cyclin D1 or D2 had decreased by approximately half, to ∼20% (Fig. 1K and Fig. S1 E, I, and P) and was mostly nonoverlapping (Fig. 1 A–C and L). However, the magnitude of the small overlap (∼5%) was still significantly greater than chance (3.4%; n = 4; P = 0.0071, paired t test).
Fig. 1.
Cyclin D1 and D2 expression in the E10.0 pMN domain of the developing mouse spinal cord. (A–C) Triple immunolabeling for Cyclin D1, D2, and Olig2 in the pMN domain at E10.0. C shows a merged view of A and B, with Olig2 omitted for clarity. (D–I) Triple-immunolabeling for HB9, Olig2, and Cyclin D1 (D–F) or D2 (G–I). Dashed lines delineate the Olig2+ pMN domain (VZ) and the MZ containing newly generated HB9+ MNs. Arrowheads indicate Cyclin D/Olig2 double-positive cells, and arrows indicate Cyclin D1/HB9 double-positive cells. (J) Schematic summarizing the relative spatial domains of expression of Olig2, HB9, Cyclin D2, and D1 at E10.0 in the pMN domain. (K) Quantification of the relative proportion of different cell populations, defined by Cyclin D1, D2, Olig2, and HB9 expression, at E10.0. Values are mean ± SEM of between four and six sections from three embryos: *P < 0.05; **P < 0.01 (t test). (L) Quantification of the percentage of Olig2+ cells expressing Cyclin D1, D2, or both at E10.0 in the pMN domain. Values are mean ± SEM of three or four sections from four embryos. D1×D2 indicates predicted overlap assuming expression by independent populations.
The Cyclin D1- and D2-expressing populations also were spatially distinct at E10.0. Virtually all Cyclin D2+ cells expressed Olig2 and were located in the VZ, whereas almost 40% of Cyclin D1+ cells were Olig2− and located in the adjacent marginal zone (MZ) (Fig. 1 D, arrows, G, and K), where differentiating neuronal precursors are located. Indeed, 56% (± 2.8%, n = 6) of Ngn2+ precursors and ∼35% of HB9+ immature MNs (36) were Cyclin D1+, whereas only ∼3% of HB9+ presursors were Cyclin D2+ (Fig. 1 E, arrows, H, and K). These data indicate that Cyclin D1 is initially coexpressed with D2 in undifferentiated pMN precursors at E9.5, but then segregates into differentiating MNs at E10 (Fig. 1J and Fig. S1 D–P).
To generalize this observation, we analyzed the expression of Cyclin Ds throughout the developing spinal cord, using Ngn2 as a marker for neurogenic precursors (32). At E11.5, 86.5% ± 2% of Ngn2+ cells expressed Cyclin D1, whereas only 11% ± 2% expressed Cyclin D2 (n = 7; P < 0.0001, paired t test). Furthermore, Cyclin D1, but not D2, expression could be observed in a subset of differentiating Engrailed-1+ V1 interneurons, as well as in TuJ1+ or NeuN+ proliferating cell nuclear antigen (PCNA)-negative newborn neurons (Fig. S2). Thus, Cyclin D1 expression persists in differentiating neurons throughout the spinal cord.
We next asked whether Cyclin D1 is eventually down-regulated after neurogenesis, which in the pMN domain terminates by E11.5 (31). Indeed, by E11.5, Cyclin D1 expression had virtually disappeared from pMN (Fig. S1 F, G, and P) and became progressively restricted to the dorsal region of the spinal cord (Fig. S1 A–C) following the ventral-to-dorsal gradient of maturation and neurogenesis (37). Cyclin D2 expression persisted in pMN through E13.5 (Fig. S1 H–K and P).
Cyclin D1 Regulates Neurogenesis Independent of the Cell Cycle In Vivo.
To test whether Cyclin D1 plays a causal role in promoting neurogenesis in vivo, we performed LOF and GOF experiments in the embryonic chick spinal cord. In situ hybridization for chick Cyclin D1 (cD1) and cD2 mRNAs revealed a rough spatiotemporal correlation between Cyclin D1 expression and neurogenesis, as in the mouse (Fig. S3), although a more detailed analysis was precluded by a lack of immune reagents.
We first performed LOF experiments using siRNAs to knock down cDs. In ovo electroporation was performed in the E2 neural tube, using a replication-competent avian retrovirus vector (RCAS) (38). The contralateral (nonelectroporated) side of the same embryo and GFP-electroporated embryos served as internal and external controls, respectively. The specificity and efficacy of each siRNA was confirmed by coelectroporating an epitope-tagged cD cDNA expression construct as a surrogate target and performing immunostaining using an antibody to the epitope tag. The effect of these manipulations on proliferation was monitored by in ovo BrdU labeling and measurement of the percentage of BrdU+/Olig2+ cells [labeling index (LI)] (Fig. S4A). To quantify neurogenesis in pMN, we counted the proportion of Olig2+ cells expressing Lim3 (39) or NeuroM (40, 41), two neuronal precursor markers. To quantify generic neurogenesis throughout the spinal cord, we measured the ratio of NeuN+ neurons to PCNA+ proliferating cells.
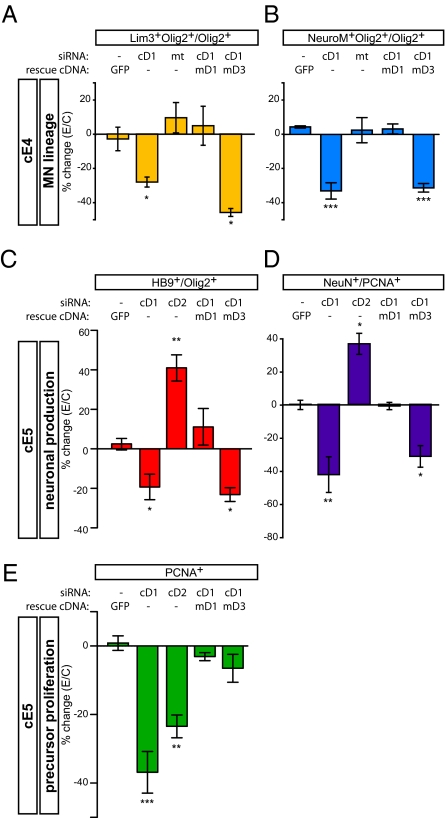
cD1 down-regulation significantly reduced the proportion of Lim3+ or NeuroM+ Olig2+ progenitors at cE4 (Fig. 2 A and B). This phenotype was not observed using a mutant cD1 siRNA (mt; Fig. 2 A and B) and was rescued by coelectroporation of a siRNA-insensitive murine Cyclin D1 (mD1) cDNA. Rescue was not obtained by coelectroporation of mD3 cDNA (Fig. 2 A and B), which on its own strongly promoted proliferation (Fig. S4A). No significant reduction in neurogenic progenitors was observed after cD2 knockdown (Fig. S5 D and E). However, both cD1 and cD2 knockdown significantly reduced the LI in pMN, as well as the total number of Olig2+ cells (Fig. S4 A and B).
Fig. 2.
Cyclin D1 expression is necessary for neurogenesis in the chick spinal cord. Neurogenesis at cE4 (A and B) or cE5 (C and D), and precursor proliferation (E) were analyzed after electroporation of siRNAs for cD1, cD2 or mt mutant control of cD1 siRNA. mD1 and mD3 indicate murine Cyclin D1- and D3-rescuing cDNAs, respectively. Values represent the percent change in the average proportion of marker-positive cells on the electroporated side relative to the controlateral (nonelectroporated) side × 100% (=%change(E/C)). Values are mean ± SEM of between four and eight sections from three to six embryos. One-way ANOVA statistical analysis (Lim3, P = 0.0022; NeuroM, P < 0.0001; #HB9/#Olig2, P = 0.0002; #NeuN/#PCNA, P < 0.0001; #PCNA, P < 0.0001) and Newman–Keuls or Bonferroni posttest comparisons were performed.
In embryos analyzed at cE5, cD1 knockdown significantly reduced the ratio of newly differentiated HB9+ MNs to undifferentiated Olig2+ precursors (Fig. 2C). In contrast, knockdown of cD2 increased this ratio (Fig. 2C), as would be expected for a manipulation that facilitates cell cycle exit (42). Therefore, the effect of cD1 knockdown in inhibiting MN differentiation at cE5 is not likely due to reduced proliferation. Importantly, the effect of cD1 knockdown at cE5 was fully rescued by coexpression of an mD1 cDNA, but not by an mD3 cDNA (Fig. 2C). Thus, knockdown of cD1 decreased both the proportion of MN precursors at cE4 and the proportion of differentiating MNs at cE5. Both phenotypes were rescued by mD1 but not by mD3, and were affected in the opposite direction by knockdown of cD2.
cD1 knockdown also reduced generic neurogenesis in the spinal cord at cE5 (Fig. 2D), similar to the effect on MNs (Fig. 2C). In contrast, cD2 knockdown significantly promoted generic neurogenesis (Fig. 2D). Both cD1 and cD2 knockdown also caused a decrease in VZ size, as monitored by PCNA staining (Fig. 2E), consistent with their effect to reduce to LI. Importantly, although this VZ phenotype of cD1 knockdown was rescued equally well by co-electroporation of mD1 or mD3 (Fig. 2E), the neurogenic phenotype at cE5 was rescued only by mD1 and not by mD3 (Fig. 2 C and D). These data suggest that both Cyclin D1 and D2 contribute to positive regulation of proliferation, but that Cyclin D1 has an additional function to promote neurogenesis.
We next performed GOF experiments by overexpressing different Cyclin Ds. Murine Cyclin D cDNAs were used, to distinguish their expression from that of endogenous cDs. Overexpression of mD1 increased the percentage of Lim3+ and NeuroM+ neuronal progenitors at cE4, although the effect was modest and did not reach significance (20–25%, P = 0.06) (Fig. S5 A and B). However, overexpression of a point-mutant form of mD1, mD1KE, which lacks the ability to interact with CDKs (22, 43) and thus cannot promote proliferation (Fig. S4A), yielded a more robust and statistically significant increase in the percentage of neurogenic progenitors (mD1KE; Fig. S5 A and B). Both WT mD1 and mD1KE significantly enhanced generic neurogenesis in the spinal cord at cE5 (Fig. S5C). The effect of mD1 and mD1KE overexpression was only slightly less than that obtained by overexpression of Ngn2, a potent proneural gene (44, 45) (Fig. S5C).
Overexpression of mD1 also increased the LI by 25.8% (Fig. S4A). Overexpression of mD2, which increased the LI by 21.4% (Fig. S4A), did not increase neurogenesis, but rather showed a trend toward a small decrease (Fig. S5 A–C). Overexpression of mD3, which increased the LI more strongly (by 36.5%), significantly decreased, rather than increased, neurogenesis (mD3; Fig. S5 A and B). These data, along with the neurogenic effect of mD1KE, suggest that the effect of mD1 in enhancing neurogenesis is independent of its effect in promoting proliferation, consistent with the results of our LOF experiments.
Genetic Interactions Between Cyclin Ds and Notch Effectors.
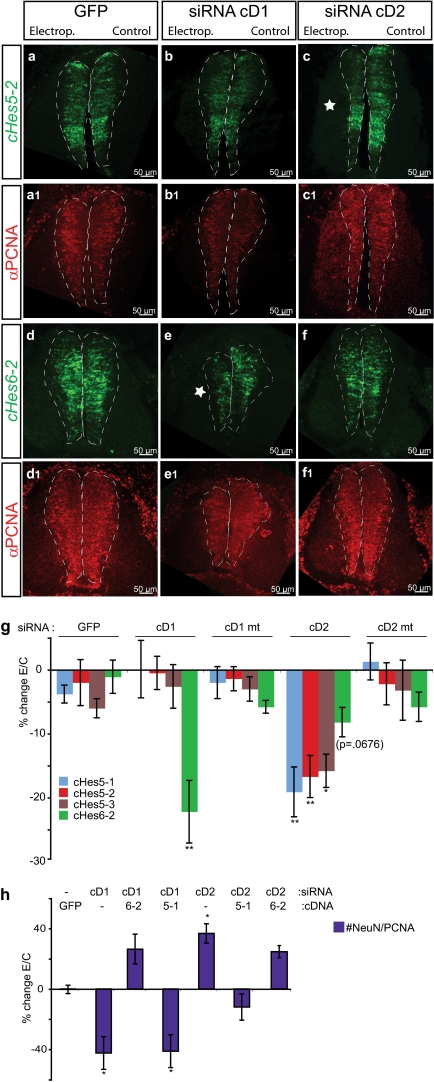
To investigate the genetic pathways mediating the neurogenic function of Cyclin D1, we examined its relationship to Notch signaling, which inhibits neurogenesis and maintains the stem cell or progenitor cell state (46, 47). To do this, we analyzed the impact of cD1 knockdown on Hes genes, which encode bHLH transcription factors (48). We monitored the expression of both chick Hes5 (cHes5), a transcriptional effector of canonical Notch signaling, and cHes6, a noncanonical Notch effector that promotes neurogenesis (49–51). In situ hybridization for Hes transcripts (49) (Fig. 3 A–F and Fig. S6), in combination with PCNA immunostaining (Fig. 3 A1–F1 and Fig. S6), revealed that cD1 knockdown caused a significant decrease in cHes6-2 expression in the VZ relative to PCNA staining, but did not affect Hes5 mRNAs (Fig. 3G, cD1). In contrast, cD2 knockdown significantly decreased Hes5 mRNAs, but not Hes6 mRNAs (Fig. 3G, cD2). Differential cell death cannot explain these phenotypes, because few caspase-3–activated cells were detected in electroporated embryos at cE5 in any condition (Fig. S4C).
Fig. 3.
Genetic interactions between Cyclin Ds and Notch signaling components. (A–F) FISH for chick Hes5-2 (A–C) or Hes6-2 (D–F) combined with immunolabeling of PCNA on E5 spinal cord (A1–F1). Dashed lines delineate the VZ, denoted by PCNA. Asterisks indicate reductions in Hes mRNAs. (G) Quantification of the level of expression of chick Hes5-1, Hes5-2, Hes5-3, and Hes6-2 mRNAs, normalized to the level of PCNA expression, in arbitrary units (%change(E/C)). Values are mean ± SEM of four or five sections from 4–10 embryos. Embryos were electroporated with either GFP or the indicated siRNAs. One-way ANOVA statistical analysis (cHes5-1: P = 0.0067; cHes5-2: P = 0.0010; cHes5-3: P = 0.0185; cHes6-2: P = 0.0011) and Newman–Keuls posttest comparisons were performed. The effect of cD2 knockdown on cHes6-2 level was not significant even on the t test (P = 0.0676). (H) Forced expression of Hes cDNAs is epistatic to cD knockdown phenotypes. Here 6-2 and 5-1 indicate coelectroporation of cDNAs for chick Hes6-2 and Hes5-1, respectively, with the indicated siRNAs. The ratio of NeuN+ to PCNA+ cells was quantified at cE5 (%change(E/C)). Values are mean ± SEM of three sections from three embryos. One-way ANOVA analysis (P < 0.0001) and Bonferroni's posttest comparisons were performed.
Given these observations, we asked whether overexpression of Hes cDNAs could rescue the effects of cD knockdown on neurogenesis. Indeed, coexpression of cHes6-2 with the cD1 siRNA rescued the decreased number of NeuN+ cells (Fig. 3H, cD1/6–2), whereas cHes5-1 had no effect (Fig. 3H, cD1/5–1). Conversely, coexpression of cHes5-1, but not of cHes6-2, rescued the effect of cD2 knockdown to increase the proportion of NeuN+ neurons (Fig. 3H, cD2/5–1 vs. cD2/6–2). Thus, forced expression of Hes genes is epistatic to the complementary effects of cD1 and cD2 knockdown on neurogenesis in both directions.
Cyclin D1 Confers Neurogenic Capacity on Transplanted Gliogenic Precursors.
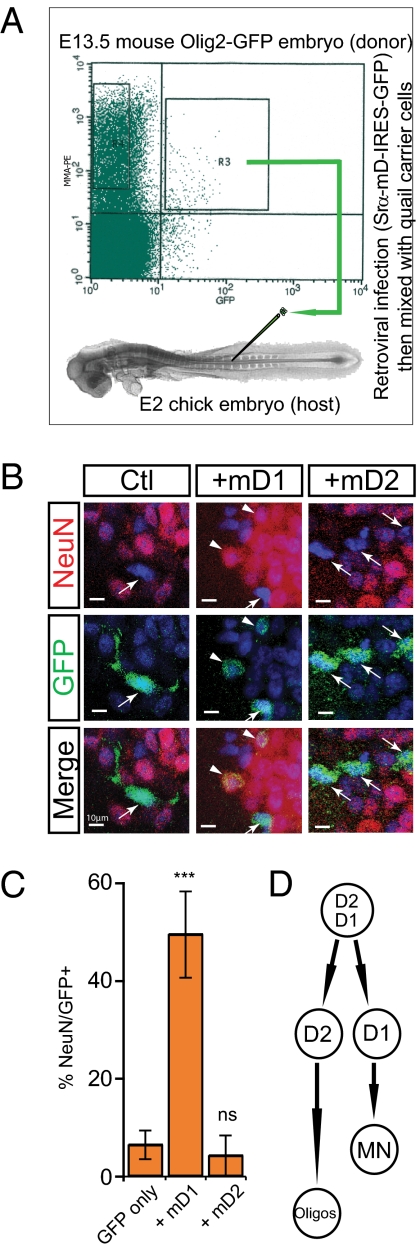
Spinal cord precursors normally undergo a time-dependent restriction in their developmental capacity, in which they lose neurogenic and retain gliogenic potential (52, 53). Using a heterochronic and heterospecific transplantation system (54), we previously demonstrated that Olig2+ progenitors isolated from murine E13.5 spinal cord have lost all neurogenic capacity (54). Because Cyclin D1 is normally down-regulated in pMNs by E13.5 (Fig. S1 G and P), we asked whether forcing its expression in purified E13.5 Olig2+ glial-restricted precursors was sufficient to restore their neurogenic capacity, as assessed by in vivo transplantation.
We transduced FACS-isolated E13.5 murine Olig2+ MMA+ spinal cord progenitors with retroviral vectors encoding either Cyclin D-IRES-GFP or GFP alone, immediately before transplantation into E2 chick neurogenic spinal cord (Fig. 4A). No cell culture incubations were performed during these manipulations. The status of GFP+-transplanted cells was assessed by immunostaining for NeuN at 3 d after transplantation (Fig. 4B). Less than 5% of control GFP-transduced, or mD2-transduced, Olig2+ cells gave rise to neurons (Fig. 4C), consistent with the findings of our previous studies using nontransduced cells (54). In contrast, 50% of mD1-transduced E13.5 Olig2+ cells differentiated to neurons after transplantation (Fig. 4C). These data indicate that forced expression of Cyclin D1, but not of D2, in glial-restricted precursors is sufficient to confer neurogenic capacity on these cells.
Fig. 4.
Forced expression of Cyclin D1 restores neurogenic potential to E13.5 Olig2+ glial-restricted precursors (54). (A) Diagram illustrating the mouse to chick transplantation experimental paradigm. (B) Double-immunolabeling for GFP and NeuN on cE5 spinal cord after transplantation. (C) Quantification of NeuN-expression in GFP+ transplanted cells. Values are mean ± SEM from 11 embryos (GFP-mD1), 3 embryos (GFP-mD2), or 8 embryos (GFP only). (D) Model illustrating the role of Cyclin Ds in pMN development. Fig. S7A provides alternative schemes.
Discussion
Cyclin D1 Promotes Neurogenesis in a Cell Cycle-Independent Manner.
Several lines of evidence presented here argue for a proneurogenic function for Cyclin D1 that is unique among the Cyclin D family members examined and is independent of its cell cycle-promoting effect. First, Cyclin D1, but not D2, is expressed by differentiating neuronal precursors and newly generated neurons in the developing spinal cord, consistent with previous observations in the embryonic forebrain (23). Second, knockdown of endogenous cD1, but not of cD2, in the developing chick spinal cord decreased both neuronal specification and differentiation. This phenotype could be rescued by mD1, but not by mD3. Third, overexpression of Cyclin D1 or D1KE enhanced neurogenesis, whereas, conversely, overexpression of mD2 or mD3 inhibited neurogenesis (to varying extents) and enhanced proliferation. Finally, forced expression of Cyclin D1 in glial-restricted progenitor cells restored neurogenic capacity, as assessed by heterospecific in vivo transplantation (54). Although we did not test D1KE in the transplantation assay, the fact that forced expression of Cyclin D2 had no effect suggests that this phenotype is also not due to the proliferation-promoting effect of Cyclin D1.
Direct measurements of apoptosis and DNA synthesis, as well as quantitative considerations, argue that the effects of manipulating Cyclin D1 levels in vivo are not likely due to a primary influence on cell survival or proliferation. Selective death of neurogenic precursors due to Cyclin D1 down-regulation is unlikely, given the small fraction (1%) of cells expressing activated caspase-3 at this stage (Fig. S4C), as reported previously (55). Dilution of neurogenic precursors due to selective enhancement of nonneurogenic (Lim3− Olig2+) precursor proliferation is inconsistent with the overall decrease in the total number of Olig2+ cells and their rate of proliferation (Fig. S4 A and B; SI Text 1). Similar calculations argue that the mD1KE-induced increase in the proportion of neurogenic precursors cannot be explained by a selective inhibition of nonneurogenic precursor proliferation (SI Text 1).
Taken together, these considerations argue for a direct effect of Cyclin D1 manipulation on neurogenesis, independent of any effect on proliferation or cell survival. Nevertheless, the magnitude of these effects was modest. In the case of LOF manipulations, the partial inhibition of neurogenesis may reflect incomplete knockdown of cD1 and/or redundant mechanisms. In the case of GOF manipulations, the modest enhancement of neurogenesis caused by overexpression of mD1KE (as well as Ngn2) may reflect limits on the number of nonneurogenic precursors that can be recruited to produce additional neurons at the stages examined. Consistent with this view, misexpression of Cyclin D1 in a purified population of nonneurogenic Olig2+ precursors isolated from relatively later-stage murine embryos caused a far more robust (ca. 10-fold) increase in neurogenesis.
Although the functional roles of Cyclin D1 and D2 in neural development have been investigated in previous studies, a specific role for Cyclin D1 in promoting neurogenesis has not yet been reported. Forced expression of either Cyclin D1 or D2 in the chick spinal cord was observed to be qualitatively compatible with terminal MN differentiation (27), but quantitative analysis was not performed. In the developing mouse cerebral cortex, coexpression of Cyclin D1 plus CDK4 has been found to stimulate proliferation and to inhibit neuronal production (56). This apparent inconsistency with our GOF results is most likely explained by the fact that we did not include CDK. Indeed, the D1KE mutant, which cannot associate with CDK, was more effective than WT Cyclin D1 in promoting neurogenesis. This suggests that interactions with CDKs may normally antagonize the proneurogenic function of Cyclin D1. If so, then overexpression of Cyclin D1 may titrate out endogenous CDKs, yielding a pool of unbound Cyclin D1 molecules that promote neurogenesis (57). Thus, the transition of Cyclin D1 from a proliferative to a neurogenic role may involve an increase in Cyclin D1 expression, liberation from interactions with CDKs, or both.
Our results further suggest a division of labor between Cyclin D2 and D1 in the developing spinal cord. The former promotes proliferation and maintains the undifferentiated progenitor pool for subsequent glial differentiation, whereas the latter may promote transient proliferation of neurogenic precursors and/or neuronal differentiation (Fig. 4D and Fig. S7AI). However, our data do not formally exclude the possibility that the two Cyclin Ds function in distinct but intermingled neurogenic and nonneurogenic lineages within pMNs (Fig. S7AII). If they indeed function sequentially, then, given the dual roles of Cyclin D1 that we have uncovered here, a transition in the control of proliferation from Cyclin D2 to D1 might be a key step in neurogenic differentiation. Moreover, at later (gliogenic) stages of development, extinction of Cyclin D1 may be part of the developmental program that restricts progenitors from a neuronal fate (54), as suggested by our transplantation experiments.
Cyclin D1 Promotes Neurogenesis Through a Genetic Pathway Involving Hes Genes.
Our genetic epistasis data suggest involvement of Hes6, a bHLH protein that facilitates neurogenesis (50, 51, 58) and antagonizes the Notch pathway (49), in the promotion of neurogenesis by Cyclin D1. Because of the lack of antibodies to cHes6, it is technically difficult to distinguish whether cD1 knockdown reduces Hes6 gene expression or reduces the proportion of Hes6+ cells within the VZ population. However, the rescue of the effect of cD1 knockdown by cHes6 to inhibit neurogenesis is suggestive of an effect on gene expression (Fig. S7BI). Nevertheless, we cannot exclude the possibility that this rescue reflects a cellular or molecular “bypass” effect of Hes6 to promote neuronal precursor formation, rather than a role for Hes6 as a true effector of Cyclin D1’s proneurogenic activity (Fig. S7BII; SI Text 2). Although indirect evidence suggests that Hes6 and Cyclin D1 are likely to be coexpressed, at least transiently (51) (Fig. S8), a “molecular bypass” of cD1 knockdown by Hes6 could still occur within the same precursor population.
Despite these uncertainties, the fact that cD2 knockdown decreases Hes5-2 expression while enhancing neurogenesis, and that coexpression of Hes5 cDNAs (but not of Hes6) rescues this phenotype, suggests that Cyclin D1 and Cyclin D2 may exert their opposing influences on neurogenesis via regulation (either direct or indirect) of Hes6 and Hes5, respectively. Such a model would fit well with the fact that these two Hes factors antagonize each other's activity (49) and would link Cyclin D1 to regulation of the Notch pathway.
The detailed molecular mechanisms through which Cyclin D1 promotes neurogenesis remain to be unraveled. Given the increasing biochemical and molecular evidence indicating that Cyclin D1 can regulate transcription (19–22, 47), it is attractive to think that a transcriptional function underlies its neurogenic influence. The identification of an experimentally tractable developmental system in which Cyclin D1 plays a clear role in neuronal cell fate determination independent of its cell cycle function should provide a valuable setting in which to investigate the biological relevance of the transcriptional regulatory activities that have been uncovered for this multifunctional regulatory protein (20). This in turn should lead to deeper insight into the general mechanisms that control the balance between self-renewal and lineage commitment in stem cells and progenitor cells.
Materials and Methods
Generation of RCAS siRNA.
siRNA constructs directed against cD1 and cD2 were generated as described previously (38). Specificity and efficiency were assessed by analyzing the down-regulation of a co-electroporated HA-tagged chick cD and the impact on cell cycle kinetics (Fig. S4 A and B).
Chick Electroporation.
Chicken embryos were electroporated at cE2 with RCAS(B) replication-competent avian retroviruses using established methods (30). For LI calculation, 50 μL of a 5-mg/mL BrdU-PBS solution was added directly to the embryo at cE4, 30 min before fixation.
In Situ Hybridization.
Nonradioactive FISH on frozen chicken embryos was performed as described previously (30), and immunolabeling for PCNA was performed sequentially. Double-FISH for cD1 and cD2 (15, 27) was performed as described previously (59).
Immunohistochemistry.
Immunohistochemistry analyses were performed on frozen mouse and chick embryonic tissue. The antibodies used are listed in SI Materials and Methods.
Microscopy and Quantification of Protein Levels.
All pictures were obtained using the Leica TCS SP confocal system. Further analysis of intensity levels was done using Adobe Photoshop software.
In Vivo Transplantation of Single-Cell Suspensions.
Olig2-GFP+ MMA+ precursors from E13.5 mouse spinal cord were FACS-isolated as described previously (54). Before being mixed with quail career cells and transplanted to a cE2 spinal cord, FACS-isolated cells were incubated for 2 h at 32 °C with ecotopic retrovirus Srα coding for mouse Cyclin D and/or nuclear GFP cDNA.
Supplementary Material
Acknowledgments
We thank D. Perez and R. Diamond for assistance with the FACS experiments; S. Pease, B. Kennedy, J. Alex, L.C. Sandoval, and the staff of the Transgenic Animal Facility at California Institute of Technology for assistance with mouse breeding and care; M. Martinez for genotyping mouse lines; R. Ho for technical assistance; G. Mosconi for laboratory management; and G. Mancuso for administrative assistance. We thank Dr. F. Pituello (Université Toulouse III) for sharing the chick cD1 and cD2 cDNA; Dr. D. Henrique (Faculdade de Medicina de Lisboa) for sharing the cHes5-1, cHes5-2, cHes5-3, and cHes6-2 in situ hybridization probes and cHes5-2 and cHes6-2 cDNA; and Dr. D. Rowitch (University of California, San Francisco) for sharing his NeuroM antibody. AMV, Engrail-1, and QCPN hybridomas were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa. This work was supported by funding from Fondation Cino del Duca, Amyotrophic Lateral Sclerosis Association, the Howard Hughes Medical Institute, the Pritzker Neurogenesis Consortium, the California Institute for Regenerative Medicine postdoctoral training fellowship, and the National Institutes of Health (Grant RO1-NS23476). D.J.A. is an investigator for the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106230108/-/DCSupplemental.
References
- 1.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 2.Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 3.O'Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- 4.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 5.Huttner WB, Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr Opin Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Ulloa F, Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- 7.Dehay C, Savatier P, Cortay V, Kennedy H. Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. J Neurosci. 2001;21:201–214. doi: 10.1523/JNEUROSCI.21-01-00201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukaszewicz A, Savatier P, Cortay V, Kennedy H, Dehay C. Contrasting effects of basic fibroblast growth factor and neurotrophin 3 on cell cycle kinetics of mouse cortical stem cells. J Neurosci. 2002;22:6610–6622. doi: 10.1523/JNEUROSCI.22-15-06610.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calegari F, Huttner WB. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci. 2003;116:4947–4955. doi: 10.1242/jcs.00825. [DOI] [PubMed] [Google Scholar]
- 10.Pilaz LJ, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci USA. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budirahardja Y, Gönczy P. Coupling the cell cycle to development. Development. 2009;136:2861–2872. doi: 10.1242/dev.021931. [DOI] [PubMed] [Google Scholar]
- 12.Cremisi F, Philpott A, Ohnuma S. Cell cycle and cell fate interactions in neural development. Curr Opin Neurobiol. 2003;13:26–33. doi: 10.1016/s0959-4388(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 13.Humbert PO, Brumby AM, Quinn LM, Richardson HE. New tricks for old dogs: Unexpected roles for cell cycle regulators revealed using animal models. Curr Opin Cell Biol. 2004;16:614–622. doi: 10.1016/j.ceb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Ohnuma S, Philpott A, Harris WA. Cell cycle and cell fate in the nervous system. Curr Opin Neurobiol. 2001;11:66–73. doi: 10.1016/s0959-4388(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 15.Lobjois V, Benazeraf B, Bertrand N, Medevielle F, Pituello F. Specific regulation of cyclins D1 and D2 by FGF and Shh signaling coordinates cell cycle progression, patterning, and differentiation during early steps of spinal cord development. Dev Biol. 2004;273:195–209. doi: 10.1016/j.ydbio.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Kerkhoff E, Rapp UR. Cell cycle targets of Ras/Raf signaling. Oncogene. 1998;17:1457–1462. doi: 10.1038/sj.onc.1202185. [DOI] [PubMed] [Google Scholar]
- 17.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 18.Massagué J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 19.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview. Cyclin D1: Normal and abnormal functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 20.Bienvenu F, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu M, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280:29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 22.Ratineau C, Petry MW, Mutoh H, Leiter AB. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J Biol Chem. 2002;277:8847–8853. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 23.Glickstein SB, Alexander S, Ross ME. Differences in cyclin D2 and D1 protein expression distinguish forebrain progenitor subsets. Cereb Cortex. 2007;17:632–642. doi: 10.1093/cercor/bhk008. [DOI] [PubMed] [Google Scholar]
- 24.Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- 25.Tanabe Y, Jessell TM. Diversity and pattern in the developing spinal cord. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Nornes HO, Neuman T. Expression of Rb, E2F1, cdc2, and D, and B cyclins in developing spinal cord. Neurosci Lett. 1995;190:49–52. doi: 10.1016/0304-3940(95)11497-k. [DOI] [PubMed] [Google Scholar]
- 27.Lobjois V, Bel-Vialar S, Trousse F, Pituello F. Forcing neural progenitor cells to cycle is insufficient to alter cell-fate decision and timing of neuronal differentiation in the spinal cord. Neural Dev. 2008;3:4. doi: 10.1186/1749-8104-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- 29.Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 31.Richardson WD, et al. Oligodendrocyte lineage and the motor neuron connection. Glia. 2000;29:136–142. doi: 10.1002/(sici)1098-1136(20000115)29:2<136::aid-glia6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 32.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 33.Briscoe J, Ericson J. Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 34.Lu QR, et al. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 36.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 37.Altman J, Bayer SA. The development of the rat spinal cord. Adv Anat Embryol Cell Biol. 1984;85:1–164. doi: 10.1007/978-3-642-69537-7. [DOI] [PubMed] [Google Scholar]
- 38.Deneen B, et al. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchida T, et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 40.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 41.Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- 42.Gui H, Li S, Matise MP. A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Dev Biol. 2007;301:14–26. doi: 10.1016/j.ydbio.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 45.Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- 46.Louvi A, Artavanis-Tsakonas S. Notch signaling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 47.Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 48.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 49.Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Dev Biol. 2005;281:318–333. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 50.Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–2943. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- 51.Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127:4203–4216. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- 52.Rao MS, Mayer-Proschel M. Glial-restricted precursors are derived from multipotent neuroepithelial stem cells. Dev Biol. 1997;188:48–63. doi: 10.1006/dbio.1997.8597. [DOI] [PubMed] [Google Scholar]
- 53.Rao MS, Noble M, Mayer-Pröschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci USA. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukouyama YS, et al. Olig2+ neuroepithelial motoneuron progenitors are not multipotent stem cells in vivo. Proc Natl Acad Sci USA. 2006;103:1551–1556. doi: 10.1073/pnas.0510658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuida K, et al. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 56.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 57.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 58.Jhas S, et al. Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J Neurosci. 2006;26:11061–11071. doi: 10.1523/JNEUROSCI.1358-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White PM, Anderson DJ. In vivo transplantation of mammalian neural crest cells into chick hosts reveals a new autonomic sublineage restriction. Development. 1999;126:4351–4363. doi: 10.1242/dev.126.19.4351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.