Abstract
High-resolution protein ground-state structures of proton pumps and channels have revealed internal protein-bound water molecules. Their possible active involvement in protein function has recently come into focus. An illustration of the formation of a protonated protein-bound water cluster that is actively involved in proton transfer was described for the membrane protein bacteriorhodopsin (bR) [Garczarek F, Gerwert K (2006) Nature 439:109–112]. Here we show through a combination of time-resolved FTIR spectroscopy and molecular dynamics simulations that three protein-bound water molecules are rearranged by a protein conformational change that resulted in a transient Grotthuss-type proton-transfer chain extending through a hydrophobic protein region of bR. This transient linear water chain facilitates proton transfer at an intermediate conformation only, thereby directing proton transfer within the protein. The rearrangement of protein-bound water molecules that we describe, from inactive positions in the ground state to an active chain in an intermediate state, appears to be energetically favored relative to transient incorporation of water molecules from the bulk. Our discovery provides insight into proton-transfer mechanisms through hydrophobic core regions of ubiquitous membrane spanning proteins such as G-protein coupled receptors or cytochrome C oxidases.
Keywords: FTIR spectroscopy, functional water molecules, proton-transport chain
The membrane protein bacteriorhodopsin (bR) (1) is an established system for the study of the functional role of protein-bound water molecules and is therefore a favorable model relative to other proton pumps and channels that bind and may also rely on active water molecules (2–4). High-resolution X-ray crystal structures resolving protein-bound water molecules are available for bR in addition to functional studies by time-resolved FTIR spectroscopy. Together they provide a detailed molecular reaction mechanism (1, 5–9).
bR performs a light-driven proton-pumping cycle. Proton release to the extracellular solvent and the involvement of protonated water molecules are well characterized (5, 8–10). However, description of a reprotonation pathway from the cytoplasm to the central proton-binding site, the Schiff base (SB), remains incomplete (for more details, see Fig. 1A). For the proton transfer from Asp96 to SB in the second cycle-half (12), only two water molecules (Wat501 and 502) appear in ground-state crystal structures. The region between these two residues is primarily hydrophobic (1). Two water molecules alone cannot span a Grotthus-type “proton wire” of hydrogen (H)-bonded water molecules (9, 14) between these positions. However, this hydrophobic barrier blocks proton transfer from the protonated SB (PSB) to the cytoplasmic proton uptake site in the release-cycle step, which would counteract the establishment of a proton gradient.
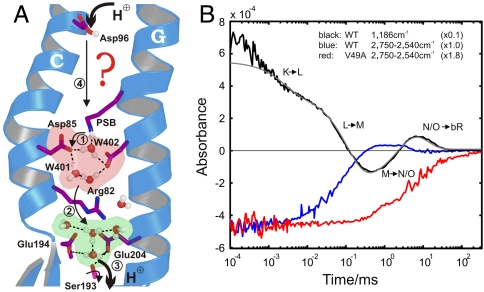
Fig. 1.
Protein-bound water molecules and their IR absorbance changes in bR. (A) Under illumination, bR performs a light-driven proton-pumping cycle including the intermediates J, K, L, M, N, and O (in order of appearance). Following the appearance of M at the end of the first cycle phase, PSB protonates Asp85 (11) (step 1), which cleaves its salt bridge to Arg82. The released Arg82 turns toward Glu194 and Glu204 and pushes the delocalized proton of a protonated water cluster (5) toward these residues (step 2). Transient protonation of Glu194/Glu204 opens the proton diode gate at Ser193, releasing a proton to the extracellular medium (step 3) (9). To reestablish the initial state, SB is reprotonated by Asp96 in N (step 4) (11, 12), which receives a proton from the cytosol in O (12), simultaneous to the isomerization back to all-trans retinal. In the last step Asp85 reprotonates the water cluster at the release site (5, 9). (B) Time-resolved absorbance changes of strong H-bonded water molecules between 2,750 and 2,540 cm-1 for the WT protein are depicted in blue. For comparison, the C14-C15 retinal stretching vibration (13) absorbance change at 1,186 cm-1 is shown in black, reflecting the (unresolved) appearance of the 13-cis isomer, the deprotonation of SB in the L-M transition (step 1), and reprotonation in the M-N transition (step 4). Finally, isomerization back to all-trans retinal takes place in the N-O transition and the absorbance change relaxes to the base line. The N-O transition is not resolved individually due to significant back reactions among the O, N, and M intermediates. The infrared absorbance between 2,750 and 2,540 cm1 in the V49A mutant is depicted in red, which reflects only the change of Wat402. The additional positive signal in M for WT kinetics (blue) reflects the appearance of strong H-bonded water.
A bR conformational change during proton uptake is believed to allow water molecules to invade from bulk cytoplasmic water and build a transient proton-transfer chain (15–17). X-ray structures of various photocycle intermediates based on crystallized, mutated bRs at low temperature, show variability in numbers and positions of water molecules in this domain (15–17) (Fig. S1). To further define how water molecules organize in this space in the WT protein, and to elucidate an active role for protein-bound water molecules in the reprotonation mechanism, we applied time-resolved FTIR spectroscopy to study the WT protein under conditions closer to the physiologically ones.
Results and Discussion
Strongly H-Bonded Waters at the Uptake Site.
IR signatures of water-molecule chains are expected to show up in the spectral regime of strong H bonds, between 3,000 and 1,600 cm-1 (18). In this region we identified a broad spectral signature from 2,750 to 2,540 cm-1, which appears transiently in the M intermediate (Fig. 1B, blue). The origin of this broad absorbance change is complex because absorbance changes of various strongly H-bonded water molecules within the protein overlap in this region (Fig. S2). The early disappearance of this band demonstrates H-bond breakage between Wat402 and the PSB in the pentameric arrangement at the release site (Fig. 1A, red) due to the all-trans to 13-cis retinal isomerization (5, 8). The absorbance of Wat402 is not restored until the last step of the photocycle, during O-BR relaxation, when the pentameric arrangement is recreated by Asp85 deprotonation (5, 9, 11) (Fig. 1A, green). Here we identified an additional absorbance change, which is overlaid in M (Fig. 1B, blue) and indicates the appearance of strongly H-bonded water molecules.
We hypothesized that a water-molecule chain at the uptake site causes the additional IR signature in the M intermediate. To assign this additional feature, we performed FTIR measurements on the V49A mutant (Fig. 1B, red). Val49 is located in the hydrophobic cavity at the proton uptake site just between Asp96 and the SB. The V49A mutation disturbs the proton uptake of the SB and accumulates a greater proportion of the N-like intermediate compared to WT (17). In this mutant the additional infrared absorbance change in the M intermediate is no longer observed when compared to WT. The residual absorbance change in V49A appears to reflect only the H-bond change of Wat402 at the release site, involving cleavage by isomerization followed by restoration of the ground state arrangement. The disappearance of this band in V49A compared to WT indicates that the additional absorbance change (Fig. 1B, blue) reflects the appearance of strong H-bonded water molecules in the M intermediate of WT between Asp96 and the SB.
The Conformational Change at the Uptake Site.
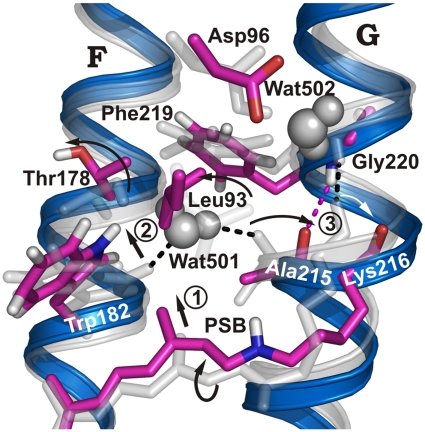
Formation of a deduced water chain depends on protein conformational changes during the photocycle, which would provide sufficient space for the chain. The largest conformational changes (19) occur in late M (20–22) or N (17, 23). A crystal structure showing sufficient space and water molecules between Asp96 and the SB to form a stable Grotthuss connection was presented [Protein Data Bank (PDB) ID code 1P8U] (17), but this structure is of the invasive V49A mutant and may not reflect water distribution within the WT protein in the N intermediate due to steric alterations in V49A in the region of interest (17). In V49A an N′ intermediate is stabilized that shows conformational changes in helices C, F, and G. To obtain the corresponding N intermediate WT structure, we used homology modeling together with molecular dynamics (MD) simulations (Materials and Methods) and mutated Ala49 back to WT valine in silico. When comparing the conformational change in the simulated WT N intermediate to the ground state (9) in Fig. 2, we observed that the retinal was moved up toward Trp182. Additionally, the SB moved away from the release site and toward Asp96 at the uptake site (compare also Fig. S1).
Fig. 2.
Conformational changes in N compared to bR ground state. bR ground-state structure (gray) and simulated N structure (blue/purple). The retinal moves “upward” and the deprotonated SB reorients toward Asp96, causing the twisted conformation of the retinal, especially around the C14-C15 bond, to relax in M (13, 24) (step 1). The movement of the retinal C13-methyl group shifts Trp182 (step 2), which cleaves the H bond to Wat501 and moves helix F by 1 Å. Wat501 is also H-bonded to Ala215 in helix G, where a π-bulge is present in the ground state, resulting in a nonhelical H-bonded Ala215 backbone carbonyl group (1). Retinal relaxation applies a different conformational restraint to the Lys216 backbone. As a result, helical hydrogen bonding changes from Lys216/Gly220 to Ala215/Gly220, and an outward-oriented carbonyl group is found for Lys216 rather than Ala215 (step 3; Fig. S4).
The deprotonated SB must move from the proton release site in an early M state (M1) to the proton uptake site in a late M state (M2) to achieve vectorial proton transfer through the protein. This is called the M1-M2 transition, but early M1 and late M2 absorptions overlap spectrally and are hard to distinguish kinetically (25). The L-M rate constant (IR absorbance changes in Fig. 1B) is dominated by the L-M2 transition, whereas the L-M1 and M1-M2 transitions are kinetically merged and not resolved (25). The low temperature stabilized X-ray structure intermediate of the D96N mutant (PDB ID code 1C8S) (15) shows an upward retinal movement and SB reorientation as expected for late M2, as does the M structure of the E204Q mutant (PDB ID code 1F4Z) (16) (Fig. S1). We assume that our homology model of the WT N intermediate approximates the structure immediately after proton transport to the SB (“early N”), and that this simulated structure is similar to the M2 structure. Asp96 in the homology model presented here is deprotonated as expected for WT N. This is in contrast to the N′ crystal structure of V49A, in which both SB and Asp96 are protonated (17), which does not represent a functionally relevant intermediate.
We hypothesized that the conformational change in the simulated WT N intermediate provides the space required for a water chain between Asp96 and the SB. The movement of the retinal C13-methyl group shifts Trp182 toward the cytoplasmic protein side (Fig. 2, step 1) and pushes the extracellular end of helix F outward by about 1 Å. This is consistent with previous results (19, 21, 22). Trp182 movement in helix F cleaves the H bond to Wat501 (Fig. 2, step 2). The second hydrogen of Wat501 is bonded to the Ala215 backbone carbonyl group in helix G. There is a π-bulge in helix G at this residue so the Ala215 backbone carbonyl group is not H-bonded within the helix as usually expected (1) (Fig. 2). This arrangement is destabilized in M by retinal movement. Furthermore, retinal relaxation applies a different conformational restraint on the Lys216 backbone because of its side chain. As a result, hydrogen bonding within helix G changes from Lys216/Gly220 to Ala215/Gly220, and the carbonyl group of Lys216 moves out of the helix (Fig. 2, step 3 and Fig. S3). Other movements include a side chain rotation of Leu93 away from Ala215 and movement of the Phe219 side chain (Fig. 2). These movements destabilize the previous hydrogen bonding positions for protein-internal water molecules and provide enough space for water molecules to arrange from Asp96 to the SB between helices C, F, and G (Fig. S3). In contrast to other investigations (19–22), the inward movement of Ala215 and outward movement of Lys216 do not move the end of helix G in our simulated structure. As shown previously (22, 26), the outward movement of helix F precedes the inward movement of helix G. Our simulated structure may reflect early F and G helix positioning. Alternatively, our simulation time may be too short to show full helix G movement, which takes place on a microsecond time frame.
Possible Water Arrangements at the Uptake Site by MD Simulations.
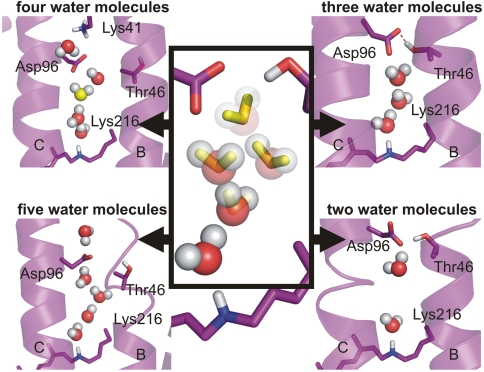
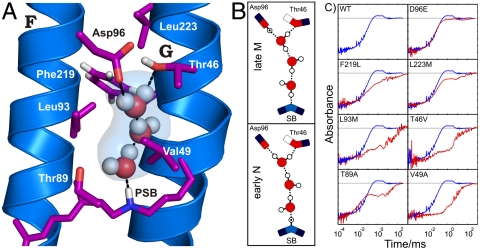
We sought to determine the number of water molecules that are necessary to bridge Asp96 and SB. Intermediate crystal structures demonstrated the movement of water molecules into the hydrophobic barrier in M and N (15–17), but the positions and the number of water molecules differ greatly between these studies (8). In the N′ structure of V49A, alanine occupies less space than valine, such that additional water molecules may be present in the ground state as compared to WT. Using MD simulations we show the WT scenario in the late M/early N intermediate by keeping two to five water molecules between Asp96 and the SB (Fig. 3). In the simulations of the WT N structure, a linear chain of three water molecules provides a stable connection and a continuous proton pathway in late M/early N (Fig. 3). This three-water-molecule arrangement was not observed in the N′ structure of V49A, probably because of steric differences compared to WT.
Fig. 3.
Different number of water molecules in the MD simulated N structure. The start positions are taken from the N′ structure 1P8U (17). For lowering the amount of water molecules included in the simulations, water molecules were deleted from Asp96 down to SB, as indicated in decreasing transparency. Individual simulations of 20-ns length each were run. In the system with five postulated water molecules, one water molecule moved past Thr46 and Asp96, thereby rupturing helix B. In the system with four postulated water molecules, a chain of three to four water molecules between Asp96 and the SB can be observed, but the chain exchanges water molecules with the cytosolic medium. Thr46 does not participate in formation of the chain, which contradicts experimental results from mutation analyses (Fig. 5A). In the system with three postulated water molecules, a stable proton wire connection can be found between Asp96 and SB. This arrangement differs from crystal structure 1P8U. Thr46 is H-bonded to Asp96, thereby modulating its pKa value in agreement with experimental data (16). The remaining two water molecules in the last system cannot form a stable connection between Asp96 and SB. The system with three linear arranged water molecules is most stable.
FTIR Identifies a Third, Highly Mobile Water Molecule.
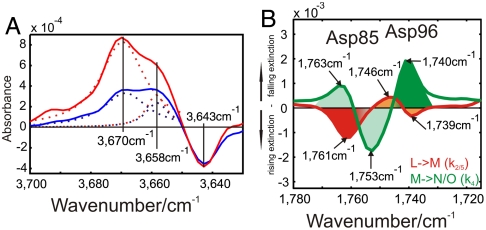
Computationally, three water molecules appear to arrange between Asp 96 and SB during the photocycle. However, in WT ground-state crystal structures, only two water molecules, Wat501 and Wat502, were identified in this region (Fig. S1, yellow). The Wat501 band is indicated at 3,670 cm-1 in M, showing the appearance of a dangling, non-H-bonded water molecule (6) (Fig. 4A). This band has an additional shoulder at 3,658 cm-1, which kinetically behaves differently in M and N (Fig. 4A). Therefore, it must represent two water molecules at the Wat501 position (27) (Fig. S2B). We have named these molecules Wat501a and Wat501b. In agreement, the M1 crystal structure (PDB ID code 1P8H) (17) depicts three water molecules in that region (with only one at Wat501), even before the pronounced helix movements of the late M/early N, when additional water molecules may invade (17).
Fig. 4.
FTIR spectroscopy reveals dangling H bonds and Asp96 early changes. (A) M (red) and N (blue) intermediate spectrum differences resulting from time-resolved measurements. In this spectral condition free OH-stretching vibrations (“dangling bonds”) can be observed. The dangling bond of Wat501 at 3,670 cm-1 exhibits a shoulder with kinetically different behavior at 3,658 cm-1. This identifies two neighboring water molecules, because they are influenced by the same mutations (6). The negative band at 3,643 cm-1 reflects the disappearance of the Wat401 dangling bond in the pentameric arrangement (5) (Fig. 1A). (B) Amplitude spectra of the D115N mutant. This mutant was chosen because Asp115 overlaps with the absorbance change of Asp96 in WT (12). We observe a spectral shift of Asp96 from 1,746 to 1,739 cm-1 and Asp85 protonation at 1,761 cm-1 in the L-M transition (11) (red). A spectral shift of Asp85 from 1,763 to 1,753 cm-1 due to changes in H bonding, and Asp96 deprotonation at 1,740 cm-1 in the M-N/O (green) transition (12) are also observed.
In summary, a highly mobile water molecule (Wat501b) seems unresolved in WT crystal structures. This supports our findings, which show the three water molecules 501a, 501b, and 502 in the ground state of WT bR in the region of interest. These water molecules are rearranged in the late M/early N intermediate by protein conformational changes (Fig. 2) to form the necessary Grotthuss proton wire between Asp96 and SB. The reordering of protein-internal water molecules to form the proton-transport chain appears to be an energetically favored reprotonation mechanism compared to the invasion of water molecules into the hydrophobic region from the protein exterior (15–17).
Experimental Evidence for the Water Chain Between Asp 96 and the SB.
Mutagenesis of each of the amino acids neighboring the proposed transient water chain in our late M/early N simulated structure (Fig. 5A) affects the additional IR signature between 2,750 and 2,540 cm-1 in the M intermediate (Fig. 5C). These data support the presence of the water chain at the proposed position. In contrast to the release-site protonated water cluster, which has a spectral signature between 2,400 and 1,800 cm-1 with decreasing intensity (5), we now observe a signature between 2,750 and 2,540 cm-1 with increasing intensity representing the water chain (Fig. S4A). Quantum mechanics/molecular mechanics (QM/MM) calculations of IR absorbance bands on the release site (10) confirmed the spectral signatures from FTIR measurements (5). We think, that future QM/MM calculations on the linear three-water chain presented here, will confirm the previously undescribed spectral signature as well.
Fig. 5.
Structure of the Grotthuss-type proton wire of three water molecules. (A) Simulated water chain in the N structure. The linear chain of three water molecules provides a proton pathway from Asp96 to SB. Our simulated structure probably resembles an early N intermediate. The molecules are most likely the rearranged water molecules 501a, 501b, and 502 in the ground state. (B) Model for the proton shift along the transport chain between Asp96 and SB. In the late M intermediate, the upper part, the proton is still located on Asp96, and in the early N intermediate, the lower part, is shifted along the H bonds to SB. This transfer is similar to the proposed proton transfer within carbon nanotubes (28). (C) Experimental evidence for the linear water chain from mutagenesis of residues surrounding the predicted water chain (see A). All mutations affect the overlaid time-resolved IR absorbance between 2,750 and 2,540 cm-1 (red; see also Fig. 1B). For comparison, the WT absorbance changes are also shown (blue). This clearly confirms the localization of the transient water chain between Asp96 and SB. The D96E mutation exerts low impact on the absorbance change, due to the conservative nature of this mutation.
For a linear water chain in a hydrophobic environment, both strong H bonds and dangling H-bond absorbance bands should appear in the FTIR spectra (27–29). The observed dangling vibrational band of Wat501a and 501b in M and N (Fig. 4A) fit well into the calculated spectral range of dangling waters in a protein (30). Mutagenesis of neighboring residues along the chain influenced these dangling bonds (6). This constitutes additional supporting evidence for the path of the linear water chain between Asp96 and SB.
Coincidently, with the formation of a water chain as early as M, we observe an H-bond change at Asp96 from 1,746 to 1,739 cm-1 in the L-M transition (Fig. 4B). We deduced that this reflected the participation of Asp96 in chain formation at M, but the connection extending to SB through H bonding was not established before the late M/early N intermediate. As soon as SB bonds to the nearby water molecule, a proton is transferred from Asp96 to reestablish PSB. Late M and early N differ particularly in this proton shift along the water chain H bonds as expected for Grotthus proton transfer (Fig. 5B), and the excess proton is located either on Asp96 by late M or on SB by early N.
In conclusion, we used time-resolved FTIR spectroscopy and MD simulations to resolve a transient, linear, proton-transport water chain in bR. Water molecules are stored at inactive storage locations in the first half of the pump cycle during proton release, thus preventing proton backflow. The cofactor-twist relaxation due to deprotonation of the SB in the M intermediate repacks amino acids between helices C, F, and G. This causes the inactive water molecules to rearrange and form a functional chain between Asp96 (proton donor) and the central proton-binding site in the proton uptake phase of the cycle. Together with the “proton diode” at the release site (9), this switch-like mechanism at the uptake site ensures efficient, directional, cross-membrane proton transport. The key event is the precise relocation of protein-bound internal water molecules, which appears to be energetically favorable to the invasion and extrusion of water molecules from the protein exterior because the necessary conformational and entropy changes are much smaller. We present a concept of protein-bound water molecules being actively involved in protein function as a model that may be applicable to proton-transfer mechanisms known to occur through other membrane proteins, including G-protein coupled receptors, cytochrome C oxidase (3), photosystem II (2), and hydrogenases (31).
Materials and Methods
bR Mutation, Expression, and Purification.
The mutagenesis, expression, isolation, and purification of bR in Halobacterium salinarum are described elsewhere (32–34).
Sample Preparation and FTIR Measurements.
A suspension of 600 μg of purified purple membrane sheets in 1 M KCl and 100 mM Tris at pH 7 was centrifuged at 200,000 × g for 2 h. The resulting pellet was squeezed between CaF2 windows and transferred into an air- or vacuum-tight cuvette. Measurements were performed with a modified Bruker IFS66V (35) or modified Bruker Vertex 80v spectrometer, recording data with 4 cm-1 spectral resolution and a time resolution of down to 30 ns. The sample was kept at 20 °C during measurements. The photocycle was triggered by an excimer laser (Lambda Physics 305i) driven dye laser (Coumarin153).
MD Simulations.
The N model was created based on the N′ crystal structure of Schobert et al. (PDB ID code 1P8U) (17). Ala49 was mutated back to valine, thus recreating the WT sequence. Missing residues in the E–F loop were inserted based on the WT ground-state structure (PDB ID code 1QHJ) (36). These residues are also missing from other crystal structures, including 1C3W (1), 1M0M (37), and 1F4Z (16). The late M structure (PDB ID code 1C8S) (15) lacks more residues in this region. In silico mutation, followed by side chain minimization, was carried out with the MOBY program package (38). The applied protonations of amino acid chains matched the N intermediate. A bR trimer consisting of three ground-state molecules used as the starting structure previously (9, 39) was recreated from the crystallographic information using PyMOL (Schrödinger, LLC.). The N′ intermediate may accumulate to a maximum of 40% in protein samples (17), so we replaced one of the ground-state structures with our N model by superposition of the transmembrane helix Cα atoms. The resulting trimer was then placed into a palmitoyloleoylphosphatidylcholine membrane/saline environment and trajectories 20 ns in length were recorded, and water densities in the last 5 ns were analyzed, as described previously (39, 40).
Supplementary Material
Acknowledgments.
We thank Gabi Smuda for the preparation of bR mutants, and the John von Neumann Institute for Computing Jülich (Project hbo26) and Regionales Rechenzentrum Köln for providing computing time. This work is funded by the Deutsche Forschungsgemeinschaft (SFB642). S.W. is funded by a Chinese Academy of Sciences Fellowship for Young International Scientists. K.G. acknowledges a fellowship from the Mercator Research Center Ruhr. Molecular figures were prepared by PyMOL (Schrödinger, LLC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104735108/-/DCSupplemental.
References
- 1.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 angstrom resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 3.Belevich I, Verkhovsky MI, Wikstrom M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- 4.Sharma M, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garczarek F, Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 6.Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460:177–191. doi: 10.1016/s0005-2728(00)00138-9. [DOI] [PubMed] [Google Scholar]
- 7.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz-Fonfria VA, Furutani Y, Kandori H. Active internal waters in the bacteriorhodopsin photocycle. A comparative study of the L and M intermediates at room and cryogenic temperatures by infrared spectroscopy. Biochemistry. 2008;47:4071–4081. doi: 10.1021/bi7024063. [DOI] [PubMed] [Google Scholar]
- 9.Wolf S, Freier E, Potschies M, Hofmann E, Gerwert K. Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: A proton diode. Angew Chem Int Ed Engl. 2010;49:6889–6893. doi: 10.1002/anie.201001243. [DOI] [PubMed] [Google Scholar]
- 10.Mathias G, Marx D. Structures and spectral signatures of protonated water networks in bacteriorhodopsin. Proc Natl Acad Sci USA. 2007;104:6980–6985. doi: 10.1073/pnas.0609229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerwert K, Souvignier G, Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1990;87:9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerwert K, Hess B, Soppa J, Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci USA. 1989;86:4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerwert K, Siebert F. Evidence for light-induced 13-cis, 14-s-cis isomerization in bacteriorhodopsin obtained by FTIR difference spectroscopy using isotopically labelled retinals. EMBO J. 1986;5:805–811. doi: 10.1002/j.1460-2075.1986.tb04285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agmon N. The Grotthuss mechanism. Chem Phys Lett. 1995;244:456–462. [Google Scholar]
- 15.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 16.Luecke H, et al. Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin. J Mol Biol. 2000;300:1237–1255. doi: 10.1006/jmbi.2000.3884. [DOI] [PubMed] [Google Scholar]
- 17.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base. J Mol Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 18.Emsley J. Very strong hydrogen-bonding. Chem Soc Rev. 1980;9:91–124. [Google Scholar]
- 19.Subramaniam S, Henderson R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 20.Dencher NA, Dresselhaus D, Zaccai G, Buldt G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc Natl Acad Sci USA. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dencher NA, Sass HJ, Buldt G. Water and bacteriorhodopsin: Structure, dynamics, and function. Biochim Biophys Acta. 2000;1460:192–203. doi: 10.1016/s0005-2728(00)00139-0. [DOI] [PubMed] [Google Scholar]
- 22.Radzwill N, Gerwert K, Steinhoff HJ. Time-resolved detection of transient movement of helices F and G in doubly spin-labeled bacteriorhodopsin. Biophys J. 2001;80:2856–2866. doi: 10.1016/S0006-3495(01)76252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vonck J. A three-dimensional difference map of the N intermediate in the bacteriorhodopsin photocycle: Part of the F helix tilts in the M to N transition. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 24.Schulten K, Tavan P. A mechanism for the light-driven proton pump of Halobacterium halobium. Nature. 1978;272:85–86. doi: 10.1038/272085a0. [DOI] [PubMed] [Google Scholar]
- 25.Hessling B, Herbst J, Rammelsberg R, Gerwert K. Fourier transform infrared double-flash experiments resolve bacteriorhodopsin’s M1 to M2 transition. Biophys J. 1997;73:2071–2080. doi: 10.1016/S0006-3495(97)78237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rink T, Pfeiffer M, Oesterhelt D, Gerwert K, Steinhoff H-J. Unraveling photoexcited conformational changes of bacteriorhodopsin by time resolved epr spectroscopy. Biophys J. 2000;78:1519–1530. doi: 10.1016/S0006-3495(00)76704-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandt C, Gerwert K, Schlitter J. Water dynamics simulation as a tool for probing proton transfer pathways in a heptahelical membrane protein. Proteins. 2005;58:528–537. doi: 10.1002/prot.20343. [DOI] [PubMed] [Google Scholar]
- 28.Cao Z, et al. Mechanism of fast proton transport along one-dimensional water chains confined in carbon nanotubes. J Am Chem Soc. 2010;132:11395–11397. doi: 10.1021/ja1046704. [DOI] [PubMed] [Google Scholar]
- 29.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 30.Baer M, et al. Spectral signatures of the pentagonal water cluster in bacteriorhodopsin. Chemphyschem. 2008;9:2703–2707. doi: 10.1002/cphc.200800473. [DOI] [PubMed] [Google Scholar]
- 31.Stripp ST, et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc Natl Acad Sci USA. 2009;106:17331–17336. doi: 10.1073/pnas.0905343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrando E, Schweiger U, Oesterhelt D. Homologous bacterio-opsin-encoding gene expression via site-specific vector integration. Gene. 1993;125:41–47. doi: 10.1016/0378-1119(93)90743-m. [DOI] [PubMed] [Google Scholar]
- 33.Needleman R, et al. Properties of Asp212----Asn bacteriorhodopsin suggest that Asp212 and Asp85 both participate in a counterion and proton acceptor complex near the Schiff base. J Biol Chem. 1991;266:11478–11484. [PubMed] [Google Scholar]
- 34.Oesterhelt D, Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 35.Rammelsberg R, Hessling B, Chorongiewski H, Gerwert K. Molecular reaction mechanisms of proteins monitored by nanosecond step-scan FT-IR difference spectroscopy. Appl Spectrosc. 1997;51:558–562. [Google Scholar]
- 36.Belrhali H, et al. Protein, lipid and water organization in bacteriorhodopsin crystals: A molecular view of the purple membrane at 1.9 A resolution. Structure. 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 37.Lanyi JK, Schobert B. Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: The switch in the bacteriorhodopsin photocycle. J Mol Biol. 2002;321:727–737. doi: 10.1016/s0022-2836(02)00682-4. [DOI] [PubMed] [Google Scholar]
- 38.Höweler U. Altenberge, Germany: CHEOPS; 2007. MAXIMOBY 8.1 and MOBY 3.0. [Google Scholar]
- 39.Wolf S, Freier E, Gerwert K. How does a membrane protein achieve a vectorial proton transfer via water molecules? Chemphyschem. 2008;9:2772–2778. doi: 10.1002/cphc.200800703. [DOI] [PubMed] [Google Scholar]
- 40.Kandt C, Schlitter J, Gerwert K. Dynamics of water molecules in the bacteriorhodopsin trimer in explicit lipid/water environment. Biophys J. 2004;86:705–717. doi: 10.1016/S0006-3495(04)74149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.