Abstract
Directed cell migration toward spatio-temporally varying chemotactic stimuli requires rapid cytoskeletal reorganization. Numerous studies provide evidence that actin reorganization is controlled by intracellular redistribution of signaling molecules, such as the PI4,5P2/PI3,4,5P3 gradient. However, exploring underlying mechanisms is difficult and requires careful spatio-temporal control of external chemotactic stimuli. We designed a microfluidic setup to generate alternating chemotactic gradient fields for simultaneous multicell exposure, greatly facilitating statistical analysis. For a quantitative description of intracellular response dynamics, we apply alternating time sequences of spatially homogeneous concentration gradients across 300 μm, reorienting on timescales down to a few seconds. Dictyostelium discoideum amoebae respond to gradient switching rates below 0.02 Hz by readapting their migration direction. For faster switching, cellular repolarization ceases and is completely stalled at 0.1 Hz. In this “chemotactically trapped” cell state, external stimuli alternate faster than intracellular feedback is capable to respond by onset of directed migration. To investigate intracellular actin cortex rearrangement during gradient switching, we correlate migratory cell response with actin repolymerization dynamics, quantified by a fluorescence distribution moment of the GFP fusion protein LimEΔcc. We find two fundamentally different cell polarization types and we could reveal the role of PI3-Kinase for cellular repolarization. In the early aggregation phase, PI3-Kinase enhances the capability of D. discoideum cells to readjust their polarity in response to spatially alternating gradient fields, whereas in aggregation competent cells the effect of PI3-Kinase perturbation becomes less relevant.
Keywords: eukaryotic chemotaxis, pseudopod-based motility, gradient sensing, flow chamber
A prime example for combined intracellular feedback mechanisms on fast timescales is chemotaxis, where cells sense, polarize, and migrate toward an external chemical stimulus (1). Chemotactic ability plays a crucial role, for example, in cancer metastasis (2), neuron guidance (3), and inflammatory response (4). Identifying and classifying key proteins in these intracellular signaling events is vital for understanding cellular mechanisms and disease development. Exciting progress has been made by the discovery that actin polymerization dynamics in migrating cells are controlled by PI4,5P2/PI3,4,5P3 gradients, allowing for directed chemotactic migration (5–7). Recent publications introduce parallel pathways like PI3-Kinase-based pseudopod formation and phospholipase A2-induced pseudopod splitting at the front of the cell, which enhance chemotactic efficiency (8). However, profound insight into these signaling networks is complicated by rapid reorganization of protein distributions in living cells (9).
To stimulate eukaryotic cells, various experimental setups generate chemotactic gradients by utilizing micropipettes (10) or diffusion chambers (11, 12). In recent years, microfluidic mixing in labyrinth-like structures and optically manipulated nanocapsules improved definition and manipulation of chemical gradients (13–16), whereas microfluidic T-junctions or photoinduced release of chemotactic stimulants expose individual cells to a rapid, global rise in chemoattractant (17, 18). To evaluate the statistical spread of cellular responses (19), rapidly tunable but spatially homogeneous external stimuli need to be applied to large cell ensembles, while monitoring the upstream and downstream signaling events in single cells by fluorescence (20). The influence of the shape of chemotactic stimulation on intracellular protein redistribution and gradient sensing in general has been established theoretically (21–24), which demonstrate the need to experimentally validate these predictions by systematic, controlled measurements.
Here, we report a microfluidic function generator, producing spatio-temporally tunable but homogeneous gradient fields across 300 μm, to analyze the statistical behavior of many cells simultaneously. Based on hydrodynamic focusing, this microfluidic generator modifies the shape and position of the concentration gradients in a time-dependent manner, which opens unique possibilities for systematic studies of cell polarization response, as a function of switching frequency. Experiments are conducted for a low flow velocity of 100 μm/s at the site of the cell ensemble, permitting a diffusive broadening of the concentration interfaces and leaving cells undisturbed by shear stress. Statistical analysis of single-cell response is performed employing the amoeboid model system Dictyostelium discoideum (25). This fast migrating, chemotactic cell type is ideally suited to test experimental limits of fast gradient switching in our setup. We succeeded in increasing the switching rate up to the point where cell migration is increasingly stalled and finally inhibited. In this chemotactically trapped cell state the external stimulus alternates faster than intracellular feedback is capable to adapt the migration direction. Cell shape analysis is performed to describe the degree of cell polarization, and to elucidate the influence of gradient steepness and PI3-Kinase perturbation on chemotactic response. Furthermore, we used a Lim-GFP label to monitor actin polymerization dynamics on the protein level, analyzing the spatio-temporally changing F-actin distribution.
Results
Microfluidic Device Design.
We control spatio-temporal changes in homogeneous chemotactic gradients to investigate both, single-cell migration and intracellular protein redistribution. Our microfluidic function generator consists of a double T-junction chamber with three separate inlets of variable pressure application (Fig. 1). A central flow without stimulant (CF) and two side flows (SF1 and SF2), containing the chemotactically active agent, are defined by the hydrodynamic focusing effect (26). In the laminar flow regime, diffusive broadening at their interfaces results in homogeneous gradient profiles with reduced gradient steepness downstream the microfluidic chamber (Fig. S1). Fig. 1 A–C show the principle of gradient switching: Initially, cells probe their surroundings by pseudopod extension, a stochastic process inherent to unstimulated cells (t0, Fig. 1A). When a chemotactic stimulus is applied by a controlled displacement of the flow interfaces, all cells are exposed to a homogeneous chemotactic gradient (t1, Fig. 1B). The stimulated cells polarize and initiate directed migration up the chemotactic gradient for sufficiently long lasting stimuli. A change in gradient direction forces the cells to repolarize and to adjust migration in the new gradient direction (t2, Fig. 1C). Chemotactic gradient and cell fluorescence can be visualized simultaneously by the DualView technique, allowing for quantitative measurements during alternating chemotactic exposure in live cell experiments (Fig. 1D). We produce very stable gradients with standard deviations of less than 5% over the course of a 2 h experiment (Fig. S2). Furthermore, we implemented automated image processing to quantitatively analyze the entire cell ensemble on a single cell basis (Fig. 1 E and F).
Fig. 1.
Concept of a microfluidic, bidirectional gradient generator. (A) Schematics of a three-inflow microfluidic chamber, generating alternating chemotactic gradient fields for live cell ensemble imaging. At t0, no chemoattractant (red) is introduced to the cell site (dashed box). Cells form random protrusions, as indicated in green, representing actin polymerization. Chemoattractant concentration is constant at level zero within the cell observation area. The coordinate system is given by the x axis, pointing in flow direction, the y axis perpendicular to the flow, and the z axis perpendicular to the plane of drawing. (B) At t1, SF1 introduces a chemotactic stimulus to the cell ensemble, leading to directed cell migration of the cells in gradient direction to the left. (C) At t2, the chemotactic stimulus is switched to SF2, reversing gradient direction. The cells are forced to switch polarization and to migrate in the opposite direction, now to the right. (D) Fluorescence image overlay of Lim-GFP cells (actin polymerization marker) and of Alexa 568, used as an indicator for the concentration distribution of the chemoattractant gradient field. Scale bar: 20 μm. (E) Zoom out of a single D. discoideum cell, with enhanced actin polymerization at the extending pseudopod, directed toward the chemotactic stimulus. Lim-GFP indicates actin polymerization processes. (F) Concept of automated image processing based on single cell analysis, considering gradient direction and cell fluorescence intensity in the DualView imaging mode.
Spatio-temporally Alternating Chemical Gradients.
Our setup ensures independent control of inflow speeds at all three inflows, which enables us to shape the chemotactic gradients in subtle ways. By alternating inflow speeds of the SFs, the CF is alternatingly shifted sideways and all cells experience transitions between changing gradient directions (Movie S1). We compare measured gradient profiles with numerical finite element calculations (FECs) (SI Text), finding them in remarkable agreement in the stationary state (Fig. S1C), and consistently reproducing the temporal evolution of the concentration gradients within the flow chamber (Movie S2). To enable homogeneous gradients, a low flow velocity of v = 100 μm/s is necessary (Fig. S3), which results in a shear stress of less than 0.01 Pa at the position of the cell ensemble. Therefore, undesirable effects that are known to occur at 1–2 Pa, like shear flow induced migration (27) or gene regulation (28), and reduction of the chemical gradient across the cell diameter (29) can be neglected (Fig. S4). FECs yield a theoretical lower limit of t = 1.5 s as transition times between two opposing gradient directions (Fig. S5). In a typical control experiment, 90% of all cells show directed migration toward a stable gradient, proving the viability of our setup (Fig. S6A). For alternating gradients with an exposure time of tE = 600 s, 11 out of 15 cells exhibit pronounced directed migration, as displayed in our sample plot in Fig. S6B, indicating the statistical spread in cell response.
Chemotactic Trapping of Fast Migrating Cells.
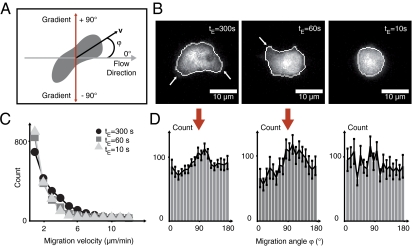
We explore the response times of intracellular signal transduction by reducing gradient switching periods from tE = 300 s to tE = 10 s, investigating 410 gradient switching events out of 42 cells with free GFP label (Movie S3). Cell migration is analyzed by tracking the geometrical center of the cell’s projection onto the focal plane, referred to as “cell center of mass” (SI Text). As defined in Fig. 2A, we investigate the migration angle φ between flow and migration direction. Thus, φ = 0 corresponds to the flow direction, and φ = |90°| to the direction of the chemotactic gradient. For exposure times of tE = 300 s, we observe elongated cells, forming pseudopods toward the chemotactic stimulus (Fig. 2B, Left) and migrating with velocities above 6 μm/ min (Fig. 2C, circles). The distribution of migration angles displays a distinct peak at φ = |90°|, indicating directed migration (Fig. 2D, Left). For reduced exposure times of tE = 60 s, cells respond by less pronounced pseudopod formation (Fig. 2B, Center). We find migration velocities below 6 μm/ min for 95% of the cells (Fig. 2C, squares), but the φ-distribution still exhibits a distinct peak in the direction of the stimulus (Fig. 2D, Center). At tE = 10 s, cells round up (Fig. 2B, Right). Few cells exhibit migration velocities of more than 2 μm/ min (Fig. 2C, triangle), and we find a random distribution of φ (Fig. 2D, Right). We refer to this cell state as chemotactic cell trapping, characterized by stalled cell migration.
Fig. 2.
Decrease in gradient exposure times (from tE = 300 s to tE = 10 s) leads to chemotactically trapped cells (Movie S3). (A) Center of mass motion of migrating cells is analyzed in terms of migration velocity v and migration angle φ, with respect to the flow direction, entailing the direction of the chemotactic gradient at φ = ± 90°. (B) For chemotactic gradient exposure times of tE = 300 s, cells show directed pseudopod formation (arrows, Left) and subsequent migration in gradient direction. For shorter exposure times of tE = 60 s, pseudopod extension is less pronounced (arrow, Center). For more rapid switching (tE = 10 s) cells round up and pseudopod formation is absent (Right). (C) Motion analysis for 410 switching events out of 42 cells with free GFP label reveals a decrease in migration velocities for exposure times of tE = 60 s and tE = 10 s, whereas for an exposure time of tE = 300 s, a large number of cells exhibits migration velocities above 6 μm/ min. (D) Analysis of the cellular migration angle for 410 switching events out of 42 cells with free GFP label: A distinct peak in the direction of the chemotactic gradients at φ = ± 90° is observed for exposure times of tE = 300 s and tE = 60 s (red arrow), whereas a nearly uniform angle distribution is observed for rapid switching with tE = 10 s, indicating a chemotactically trapped cell state, where cells cannot follow the fast gradient switching due to slower internal repolarization times.
Analysis of Intracellular Protein Redistribution.
To correlate the intracellular F-actin redistribution with the reorientation of cell locomotion in spatio-temporally varying chemotactic stimuli, we visualize the fusion protein GFP-LimEΔcc, that has been shown to bind to freshly polymerized actin (30) and allows for a quantitative analysis of protein reorganization during chemotactic cell response. All experiments are performed for exposure times of tE = 600 s, to ensure that cells fully adjust to the respective gradient direction. We find two characteristic types of cellular migration response (Fig. S6 C and D, Movies S4 and S5): The major portion of cells in steep gradients (20–80 pM/μm) show internal actin reorganization by continuous formation of new pseudopods (Fig. 3A). While the new polymerization front forms, the prior leading edge becomes the rear of the cell, reversing cell polarity (Fig. 3A, arrows). In contrast, in shallow gradients (5–15 pM/μm) permanently polarized cells are observed primarily (Fig. 3B). These cells maintain their actin front in response to gradient switching, and no dynamic reorganization of the actin network occurs during the U-turn-like cell motion (Fig. 3B, arrows), confirming previous observations (31, 32). To quantify intracellular reorientation of the actin cytoskeleton architecture, we introduce a fluorescence distribution moment (FDM) as described in SI Text (Fig. 3A, Lower Left and Right). The FDM is the integrated product of the GFP fluorescence intensity FI of each pixel and its displacement from the cell center of mass d. The FDM becomes positive if its vector points parallel to the direction of the gradient and negative if oriented in the opposite direction. A cell, migrating in gradient direction, exhibits pronounced fluorescence intensity at the leading edge, which translates into a large positive FDM value. The amplitude of the FDM vector is displayed over time for a repolymerizing cell during three consecutive gradient switches, well reproducible within tight error margins (Fig. 3A, Center). Temporal analysis of the FDM at the single cell level reveals three distinct regimes of actin polymerization activity: The first phase of cell repolarization is characterized by a delay in response time of tD = 60 s, where initial polarity persists after gradient switching. In the second phase a uniform intensity distribution is found resulting in a reduced FDM. This reorganization time is observed to last for tR = 55 s. In a third phase, the cell adapts to the new stimulus by forming new protrusions in gradient direction about 3 min after gradient switching. To describe the migration type of permanently polarized cells, we refine the migration angle φ as the angle between the major axis of the cell and the flow direction (Fig. 3B, Lower Left and Right). The temporal evolution of φ shows a response delay of tD = 145 s and a turning time of tT = 50 s after a switch in gradient direction (Fig. 3B, Center).
Fig. 3.
Alternating gradient fields produce characteristic cell polarization types in steep versus shallow gradients. (A) In steep gradients (20–80 pM/μm), cells preferably repolarize in response to gradient switching. The displayed cell shows initial polarization toward the preceding stimulus (Upper Left). At t = 62 s after gradient switching, a uniform intensity distribution is observed (Upper Center) before the cell repolarizes toward the new gradient (Upper Right). This transient polarization is analyzed by an FDM, the integrated product of the GFP fluorescence intensity FI of each pixel and its displacement from the cell center of mass d. The FDM becomes positive if its vector points parallel to the direction of the gradient and negative if orientated in the opposite direction. The FDM is averaged over three consecutive gradient switches in a single cell (graph, Center). (B) In shallow gradients (5–15 pM/μm), cells preferably show a permanent polarization front and follow switches in gradient direction by performing a U-turn. The displayed cell is polarized toward the preceding gradient (Upper Left), before performing a U-turn (Upper Center) and redirecting toward the new gradient (Upper Right). This cell behavior is analyzed by the angle φ between the major axis of an ellipse, fit to the cell shape, and the flow direction (Lower Left and Right). The temporal evolution of a single U-turn event shows a response delay of tD = 145 s, and a turning time of tT = 50 s (graph, Center). (C) Cell eccentricity is introduced as a measure for cell polarization. Highly polarized cells exhibit eccentricities ecc > 0.9. (D) Mean ratio of cells with ecc > 0.9, for advancing cell starvation, averaged over 150 cells in steep gradients and for 154 cells in shallow gradients. Error bars display the variance.
Influence of Gradient Steepness on Cell Polarization During Starvation.
In D. discoideum a starvation time of 6 h is perceived as the onset of chemotactic aggregation (33). We distinguish between preaggregating cells, starved for 5–6 h, and aggregation competent cells, starved for 6–7 h. Monitoring cells before they enter the aggregation phase defines precise starting conditions in our experiments. This approach enables us to distinguish molecular mechanisms, which initiate a chemotactic response, from cellular behavior, which is influenced by an already existent polarization. We quantify changes in the ratio of transiently polarized cells (Fig. 3A) to highly elongated cells with stable polarization fronts (Fig. 3B), in steep and shallow alternating gradients for 150 and 154 cells, respectively (Fig. S7 A and B). Therefore, we determine cell eccentricity (ecc) as the ratio between the major axis of an ellipse, fitted to the cell perimeter, and the foci distance (Fig. 3C). Cells with a stable polarization front yield ecc > 0.9 (Fig. 3D).
In steep gradients (20–80 pM/μm), 6% of preaggregating cells show ecc > 0.9. In shallow gradients (5–15 pM/μm), 13% of the cells show ecc > 0.9 after 5 h of starvation and 23% at 6 h of starvation. In aggregation competent cells, the percentage of cells with ecc > 0.9 rises to 13% (after 6 h of starvation) and to 26 % (at 7 h of starvation) in steep gradients. In shallow gradients, we find 34% (after 6 h) and 32% (at 7 h). Overall, more highly elongated cells are observed in the shallow gradient regime. With ongoing starvation, an increase in the ratio of highly polarized cells is found for both gradient regimes.
Starvation Time-Dependent Influence of PI3-Kinase on Cell Eccentricity.
D. discoideum cells rapidly orient toward the chemotactic source by establishing a PIP2/PIP3 gradient, inducing the formation of new pseudopods. To gain molecular insight into this intracellular control mechanism, we incorporated a pharmacological perturbation of PIP2 phosphorylation. The drug LY 294002 disturbs multiple isoforms of PI3-Kinases (34), having a strong impact on PIP2/PIP3 conversion. We perform measurements on 150 untreated cells and 185 drug treated cells in steep gradients (20–80 pM/μm) for starving times from 5–7 h, constituting the transition period from preaggregating cells to aggregation competent cells (Fig. S7 A and C). In preaggregating cells, perturbation of PI3-Kinase results in 18% of highly elongated cells with ecc > 0.9 after 5 h of starvation and 17% at 6 h of starvation (Fig. 4A). In contrast, we only observe 6% for unperturbed cells. In aggregation competent cells (6–7 h), the influence of PI3-Kinase perturbation on cell polarization is strongly reduced. We observe 13% (after 6 h) and 26% (at 7 h) strongly polarized cells with PI3-Kinase fully functional, and 20% (after 6 h) and 30% (at 7 h) with PI3-Kinase perturbation. The perturbation of PIP2 phosphorylation by PI3-Kinase results in more stably polarized cells at the start of our experiments, before cells enter the aggregation phase.
Fig. 4.
Impact of PI3-Kinase perturbation on cell polarization and repolymerization dynamics for advancing starvation time. (A) Changes in ratio of permanently polarized cells (ecc > 0.9), depending on starvation times from 5–7 h. We average over 185 cells treated with the drug LY 294002 perturbing PI3-Kinase (gray), and over 150 untreated cells (black). Error bars display the variance. (B) Temporal evolution of the FDM for 150 untreated cells (black) and for 185 cells treated with LY 294002 (gray) during alternating gradient exposure for tE = 10 min. The number of gradient switches analyzed in each plot is given by n. In cells starved for 5–5.5 h, only unperturbed cells show a slight repolymerization behavior. After 5.5–6 h of starvation, both cell types show initial repolymerization, but drug treated cells loose stable polarization after 10 min. After 6 h of starvation, polarization of both cell types is pronounced and comparable. After 6.5 h of starvation, drug treated cells repolarize faster, whereas untreated cells show delayed response, reaching comparable polarization states after 10 min of chemotactic gradient exposure. (C) Mean FDM values after tE = 10 min of chemotactic exposure in alternating gradient fields, as indicated by the arrows in B.
Starvation Time-Dependent Influence of PI3-Kinase on Chemotactically Induced Cell Repolarization.
In repolarizing cells (ecc < 0.9), rearranging their actin cortex in response to switches in gradient direction, the FDM is investigated for advancing starvation times (5–7 h) (Fig. 4B). In preaggregating cells, we find that PI3-Kinase perturbation impedes cell repolarization, whereas significant actin reorganization is found in unperturbed cells (Fig. 4B, Left). After 6 h of starvation, the influence of PI3-Kinase perturbation ceases and both cell types show pronounced repolarization behavior (Fig. 4B, Center Right). Untreated cells show a slowing down in readjustment to the new gradient direction at 7 h of starvation, eventually reaching comparable FDM values after 10 min (Fig. 4B, Right). For clarity, Fig. 4C shows FDM values after 10 min of chemotactic stimulus (as indicated by the black and gray arrows in Fig. 4B). More than 40 switches in chemotactic gradient exposure are averaged for each cell type and starvation period.
Discussion
We developed a microfluidic device for controlled application of alternating chemotactic gradient fields to large cell ensembles. This setup exhibits key innovative features: (i) control over rapidly alternating but homogeneous gradients, and (ii) exposure of entire cell populations to homogeneous gradients, extending across 300 μm. We study subtle differences of cell shapes and the fluorescence distribution of actin polymerization activity FDM in D. discoideum cells by evaluating a large number of cells. All cells are subjected to the same homogeneous gradient, as compared to reported experiments (16, 17) addressing cells one by one. The steady, reproducible flow in our microfluidic setup prevents distortions of the gradient profile by remnants of previous chemotactic stimuli or by a global rise in background chemoattractant concentration (9, 16). Our reproducible and stable gradients can switch directions with a theoretical lower limit of t = 1.5 s. Insight into the control of cell locomotion by cytoskeleton reorganization, which is mediated by intracellular signaling, is gained by combined observation (i) of polarized actin gelation via microfluorescence and (ii) of cell shape changes during migration in chemotactic gradients.
For gradient switching frequencies of 0.02 Hz, we observe directed cellular motion toward the new direction of chemotactic stimulus, whereas at 0.1 Hz cells are trapped chemotactically. The external chemotactic gradient field alternates faster than intracellular signaling is capable to respond by cell reorientation. This observation agrees well with previously reported F-actin dynamics in D. discoideum cells, induced by global chemotactic stimulation (5, 17). They show that cAMP induces an actin polymerization peak at approximately 10 s (0.1 Hz) and persistent actin polymerization after roughly 1 min (0.02 Hz), in good agreement with our values for the onset of frustrated cell migration, and therefore, chemotactic cell trapping in alternating gradient fields. When correlating actin repolymerization dynamics with cell migration after a switch in gradient direction, we observe an adaptation time of approximately 3 min for D. discoideum cells to completely rearrange their actin polymerization activity before initiating migration in the reversed direction. This adaption time is in good agreement with an average periodicity of approximately 3 min, observed for repetitive elongated and cringed motion states in migrating cells without external stimuli (35, 36).
Analysis of cell shape and FDM reveal fascinating results, implying that the impact of PI3-Kinase strongly depends on starvation time. PI3-Kinase is known to stimulate the formation of new pseudopods in steep chemotactic gradients (37). Yet, in shallow gradients, perturbation of PI3-Kinase influences D. discoideum chemotaxis weakly (32). Hence, parallel molecular mechanisms are proposed for D. discoideum chemotaxis signaling (8). Now our results imply that the balance between the PI3-Kinase pathway, which allows for fast reorientation of the cell, and the phospholipase A2 (Pla2) pathway, which mediates persistent cell migration, strongly depends on gradient steepness as well as on starvation time. Because of the intrinsic behavior of cells and those parallel pathways, an absolute threshold, deciding which pathway is preferred cannot be expected. Instead both response behaviors are observed in parallel, but with a changing ratio between repolymerizing cells and cells that perform U-turns. In steep gradients we primarily find PI3-Kinase based formation of new pseudopods after 5 h of starvation. This effect decreases with ongoing starvation time, as well as with a reduction of gradient steepness. The shift away from a PI3-kinase-based intracellular signaling to a persistent cell polarization, as proposed by the Pla2 pathway, is also observed when PI3-Kinase is perturbed through the addition of drugs. Here, we explicitly find a strong dependence of PI3-Kinase perturbation on starvation time, as shown in Fig. 4. We identify a higher ratio of elongated cells in early starvation but a decreased impact for starvation times of more than 6 h. One possible explanation for this starvation time dependence could be that a rapid change in migration direction, as mediated by PI3-Kinase, is important for D. discoideum cells during early starvation, whereas it is more important for aggregating cells to persistently migrate toward the initial chemotactic source.
Consecutive experiments will help to advance the understanding of the dynamic and spatial redistribution of other chemotactic key regulators like Rac GTPase and PI3 phosphatase PTEN. In principle, this microfluidic gradient generator allows for the local stimulation of single cells or whole developing organisms (2, 38) with specific molecular gradients, which is necessary in a broad range of research areas, notably in cell and developmental biology. Ensembles of cells can be subjected simultaneously to chemotactic gradients and biochemical agents, by large scale bidirectional chemical gradients. The spatial, temporal, and chemical flexibility makes it applicable not only to gain insights into complex intracellular feedback schemes, but it may also lay the ground for advanced assays in drug delivery, inflammatory response, embryogenesis, and angiogenesis.
Materials and Methods
Microfluidic Function Generator.
We use μ-slide 3-in-1 microfluidic chambers (Ibidi) with three 0.4 × 1.0 mm2 inflows, that converge under an angle of α = 32° to the main channel of dimension 0.4 × 3.0 × 23.7 mm3. Both SFs are connected to reservoirs, built from two 50 mL syringes (Braun Melsungen AG), separately connected to a customized suction control pressure pump (Nanion). Two micrometer valves (Upchurch Scientific) reduce the flow velocities at the SFs. The CF is connected to an infusion syringe pump (TSE Systems), which generates a stable flow of 1 mL/h. Measurements were performed with an Axiovert 135 TV microscope (Zeiss), with LD Plan-Neofluar objectives 20x/0.50 N.A. and 40x/0.75 N.A. (Zeiss) in combination with a DV2 DualView system (Photometrics).
A solution of 1 μM Alexa Fluor 568 hydrazide (Invitrogen) was used to characterize the concentration profile of cAMP (Sigma-Aldrich) because of their comparable molecular weight.
Cell Culture.
All cells are derived from the D. discoideum AX2 strain. Cells with free GFP expressed in AX2 background (39) are used for cell tracking, whereas cells with LimEΔcc-GFP expressed in a LimEΔcc-null AX2 background (40) are used for FDM analysis (SI Text). LimEΔcc mutant cells are grown in AX2 nutrition medium, containing 10 μg/mL Geneticin 418 disulfate salt (G418) (Sigma-Aldrich) and 10 μg/mL Blasticidine S hydrochloride (Sigma-Aldrich), whereas free GFP mutants are treated with 20 μg/mL G418.
Live Cell Experiment.
Cells are concentrated to c = 5 × 106 cells/mL in shaking culture (150 rpm). Five hours prior to the experiment, cells are washed with 17 mM K-Na PBS pH 6.0 (Sigma-Aldrich). Cells are introduced into the microfluidic chamber at c = 2.5 × 105 cells/mL. cAMP at c = 1 × 10-6 M is added as chemoattractant for measurements displayed in Fig. 2 and Movies S1, S3, S4, S5, and at c = 2 × 10-4 M for measurements displayed in Fig. 3 and Fig. 4. PI3-Kinase was inhibited using 50 μM (0.17% DMSO) LY 294002 (L9908, Sigma-Aldrich). Measurements are performed with cells starved for 5–7 h. Cell center of mass is extracted using the CellEvaluator algorithm (SI Text). Velocity and angle distributions in Fig. 2 are calculated over 10 frames, with a frame rate of (2.5 s)-1. A frame rate of (2 s)-1 is used for FDM and eccentricity analysis in Fig. 3 and Fig. 4. Cell eccentricity is obtained using the regionprops function in Matlab 2010a (Mathworks).
Supplementary Material
Acknowledgments.
We acknowledge support from the Statistical and Biological Physics Chair , E. Frey, at the Arnold Sommerfeld Center of Theoretical Physics at Ludwig-Maximilians-Universität in Munich and we thank G. Gerisch (Max Planck Institute of Biochemistry) for providing us with D. discoideum cells. This work was funded by the German Excellence Cluster Nanosystems Initiative Munich, Deutsche Forschungsgemeinschaft Grant HE 5958/2-1 (to B.M. and D.H.), a Microsoft Research European PhD scholarship (S.Y.), and Volkswagen Foundation Grant I/85100.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014853108/-/DCSupplemental.
References
- 1.Weiner OD. Regulation of cell polarity during eukaryotic chemotaxis: The chemotactic compass. Curr Opin Cell Biol. 2002;14:196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 3.Gundersen RW, Barrett JN. Neuronal chemotaxis—chick dorsal-root axons turn toward high-concentrations of nerve growth-factor. Science. 1979;206:1079–1080. doi: 10.1126/science.493992. [DOI] [PubMed] [Google Scholar]
- 4.Martin P, Leibovich SJ. Inflammatory cells during wound, repair: The good, the bad and the ugly. Trends Cell Biol. 2005;15:599–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, et al. Two phases of actin polymerization display different dependencies on PI(3,4,5)P-3 accumulation and have unique roles during chemotaxis. Mol Biol Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Insall RH, Weiner OD. PIP3, PIP2, and cell movement—similar messages, different meanings? Dev Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sackmann E, Keber F, Heinrich D. Physics of cellular movements. Annu Rev Condens Matter Phys. 2010;1:257–276. [Google Scholar]
- 8.Boosgraf L, van Haastert PJM. Navigation of chemotactic cells by parallel signaling to pseudopod persistence and orientation. PloS One. 2009;4(8):e6842. doi: 10.1371/journal.pone.0006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samadani A, Mettetal J, van Oudenaarden A. Cellular asymmetry and individuality in directional sensing. Proc Natl Acad Sci USA. 2006;103:11549–11554. doi: 10.1073/pnas.0601909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futrelle RP. Dictyostelium chemotactic response to spatial and temporal gradients—theories of the limits of chemotactic sensitivity and of pseudo-chemotaxis. J Cell Biochem. 1982;18:197–212. doi: 10.1002/jcb.1982.240180207. [DOI] [PubMed] [Google Scholar]
- 11.Zicha D, Dunn GA, Brown AF. A new direct-viewing chemotaxis chamber. J Cell Sci. 1991;99:769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 12.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of gradients having complex shapes using microfluidic networks. Anal Chem. 2001;73:1240–1246. [Google Scholar]
- 14.Irimia D, et al. Microfluidic system for measuring neutrophil migratory responses to fast switches of chemical gradients. Lab Chip. 2006;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamholz AE, Yager P. Theoretical analysis of molecular diffusion in pressure-driven laminar flow in microfluidic channels. Biophys J. 2001;80:155–160. doi: 10.1016/S0006-3495(01)76003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krees H, et al. Cell stimulation with optically manipulated microsources. Nat Methods. 2009;6:905–909. doi: 10.1038/nmeth.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beta C, Wyatt D, Rappel W, Bodenschatz E. Flow photolysis for spatio-emporal stimulation of single cells. Anal Chem. 2007;79:3940–3944. doi: 10.1021/ac070033y. [DOI] [PubMed] [Google Scholar]
- 18.Kuczenski B, Ruder WC, Messner WC, LeDuc PR. Probing cellular dynamics with a chemical signal generator. PloS One. 2009;4(3):e4847. doi: 10.1371/journal.pone.0004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzzy D, van Oudenaarden A. Quantitative time-lapse fluorescence microscopy in single cells. Annu Rev Cell Dev Biol. 2009;25:301–327. doi: 10.1146/annurev.cellbio.042308.113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Y, Huang C-H, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci USA. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 23.van Haastert PJM, Postma M. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 2007;93:1787–1796. doi: 10.1529/biophysj.107.104356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Endres RG, Wingreen NS. Accuracy of direct gradient sensing by single cells. Proc Natl Acad Sci USA. 2008;105:15749–15754. doi: 10.1073/pnas.0804688105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janetopoulos C, Firtel RA. Directional sensing during chemotaxis. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee GB, Chang C-C, Huang S-B, Yang R-J. The hydrodynamic focusing effect inside rectangular microchannels. J Micromech Microeng. 2006;16:1024–1032. [Google Scholar]
- 27.Decave E, Garrivier D, Bréchet Y, Fourcade B, Bruckert F. Shear flow-induced detachment kinetics of Dictyostelium discoideum cells from solid substrate. Biophys J. 2002;82:2383–2395. doi: 10.1016/S0006-3495(02)75583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papadaki M, Eskin SG. Effects of fluid shear stress on gene regulation of vascular cells. Biotechnol Prog. 1997;13:209–221. doi: 10.1021/bp970029f. [DOI] [PubMed] [Google Scholar]
- 29.Beta C, Fröhlich T, Bödeker HU, Bodenschatz E. Chemotaxis in microfluidic devices—a study of flow effects. Lab Chip. 2008;8:1087–1099. doi: 10.1039/b801331d. [DOI] [PubMed] [Google Scholar]
- 30.Prassler J, et al. DdLim is a cytoskeleton-associated protein involved in the protrusion of lamellipodia in Dyctiostelium. Mol Biol Cell. 1998;9:545–559. doi: 10.1091/mbc.9.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–463. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- 32.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nat Cell Bio. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 33.Fisher PR, Merkl R, Gerisch G. Quantitative analysis of cell motility and chemotaxis in Dictyostelium discoideum by using an image processing system and a novel chemotaxis chamber providing stationary chemical gradients. J Cell Biol. 1989;108:973–984. doi: 10.1083/jcb.108.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Haastert PJM, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyotelium discoideum chemotaxis. J Cell Biol. 2007;177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schindl M, et al. Cell-substrate interactions and locomotion of Dictyostelium wild-type and mutants defective in three cytoskeletal proteins: A study using quantitative reflection interference contrast microscopy. Biophys J. 1995;68:1177–1190. doi: 10.1016/S0006-3495(95)80294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etzrodt M, et al. Time-resolved responses to chemoattractant, characteristic of the front and tall of Dictyostelium cells. FEBS Lett. 2006;580:6707–6713. doi: 10.1016/j.febslet.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 37.Loovers HM, et al. Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: A quantitative in vivo Aanalysis by inhibition of PI3-Kinase. Mol Biol Cell. 2006;17:1503–1513. doi: 10.1091/mbc.E05-09-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregor T, Wieschaus EF, McGregor AP, Bialek W. Tank DW stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabriel D, et al. The contractile vacuole network of Dictyostelium as a distinct organelle: Its dynamics visualized by a GFP marker protein. J Cell Sci. 1999;112:3995–4005. doi: 10.1242/jcs.112.22.3995. [DOI] [PubMed] [Google Scholar]
- 40.Bretschneider T, et al. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.