Abstract
The initial step in HIV-1 infection occurs with the binding of cell surface CD4 to trimeric HIV-1 envelope glycoproteins (Env), a heterodimer of a transmembrane glycoprotein (gp41) and a surface glycoprotein (gp120). The design of soluble versions of trimeric Env that display structural and functional properties similar to those observed on intact viruses is highly desirable from the viewpoint of designing immunogens that could be effective as vaccines against HIV/AIDS. Using cryoelectron tomography combined with subvolume averaging, we have analyzed the structure of SOSIP gp140 trimers, which are cleaved, solubilized versions of the ectodomain of trimeric HIV-1 Env. We show that unliganded gp140 trimers adopt a quaternary arrangement similar to that displayed by native unliganded trimers on the surface of intact HIV-1 virions. When complexed with soluble CD4, Fab 17b, which binds to gp120 at its chemokine coreceptor binding site, or both soluble CD4 and 17b Fab, gp140 trimers display an open conformation in which there is an outward rotation and displacement of each gp120 protomer. We demonstrate that the molecular arrangements of gp120 trimers in the closed and open conformations of the soluble trimer are the same as those observed for the closed and open states, respectively, of trimeric gp120 on intact HIV-1 BaL virions, establishing that soluble gp140 trimers can be designed to mimic the quaternary structural transitions displayed by native trimeric Env.
Keywords: viral entry, cryoelectron microscopy, HIV spike, HIV vaccine, AIDS vaccine
The trimeric envelope glycoproteins (Env) that are displayed on human and simian immunodeficiency viruses (HIV and SIV, respectively) are heterodimers of the transmembrane glycoprotein (gp41) and a surface glycoprotein (gp120). The glycoproteins gp120 and gp41 are synthesized initially as a single gp160 polypeptide that is subsequently cleaved to generate the noncovalently associated gp120/gp41 complex. The conformational changes triggered in trimeric Env upon CD4 binding lead ultimately to fusion of the viral and cell membranes and to delivery of the viral core into infected cells (1). Cryoelectron tomographic studies of several HIV-1 and SIV strains have provided insights into the molecular architectures of trimeric Env displayed on native virions in unliganded, antibody-bound, and CD4-liganded states at resolutions of approximately 20 Å (2, 3). These analyses have established that trimeric Env from HIV-1 undergoes a large structural transition from a “closed” unliganded state to an “open” liganded state when complexed to CD4 and 17b (2). This quaternary structural change involves rotation of each gp120 protomer by about 45° around an axis parallel to the central threefold axis, coupled with an out-of-plane rotation of about 15°. In some strains, such as SIV CP-MAC, trimeric Env is already present in this open conformation even in the absence of soluble CD4 (sCD4), providing an explanation for CD4-independent viral entry by this strain (3). Atomic resolution structures are not yet available for trimeric Env in any conformational state, although many sets of coordinates are available from X-ray crystallographic studies for the truncated core of monomeric HIV-1 gp120 (4–7).
The development of soluble versions of trimeric Env that display biochemical and structural properties similar to those observed on infectious viruses is of considerable interest in the context of designing vaccines against HIV/AIDS. The ectodomain of Env is a heterodimer with mass of approximately 140 kDa, composed of the entire gp120 component, and approximately 20 kDa of gp41, which are displayed on the surface of the viral membrane. Soluble versions of trimeric gp140, either cleaved or uncleaved, are being developed as immunogens to elicit a protective humoral immune response against HIV-1 infection. To date, however, several gp120, gp41, or gp140 constructs, whether monomeric or trimeric, have not been able to achieve this goal (8–10). Some of the structural parameters that are considered important for the rational design of a successful HIV-1 Env immunogen are the extent to which its three-dimensional structure mimics that of native trimeric Env, its capability to undergo conformational changes that are known to influence epitope display on the native trimer, and the likelihood that it is capable of displaying conformations that are sufficiently long-lived to elicit antibodies that bind cognate epitopes on infectious viruses.
SOSIP gp140 trimers are soluble, proteolytically cleaved trimers that are stabilized by the presence of an engineered intermolecular disulfide bond between gp120 and gp41 (SOS), combined with a single residue change, I559P, within gp41 (11). Immunogenicity studies in rabbits have shown that SOSIP gp140 trimers derived from the clade A strain KNH1144 are superior at eliciting neutralizing antibodies as compared to gp120 monomers (12), suggesting that further structural investigation of these trimers could be informative for improved immunogen design. Here, we report structural analysis of soluble, cleaved SOSIP gp140 trimers from both KNH1144 and the clade B strain JR-FL using cryoelectron tomography. We compare the structures of unliganded SOSIP trimers with CD4- and 17b-liganded trimers, and compare these structures, in turn, with the corresponding structures derived previously for trimeric Env displayed on intact virions (2).
Results
Structure of Unliganded Trimeric gp140.
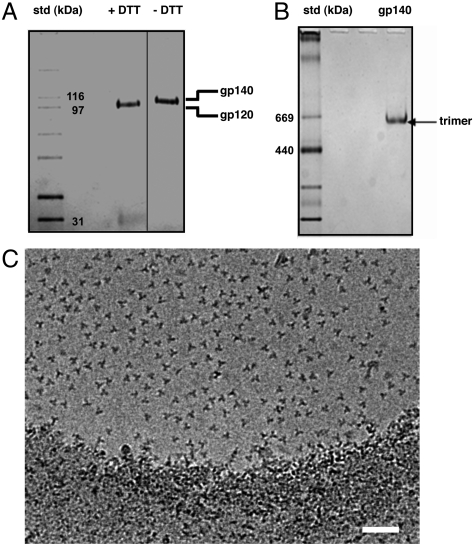
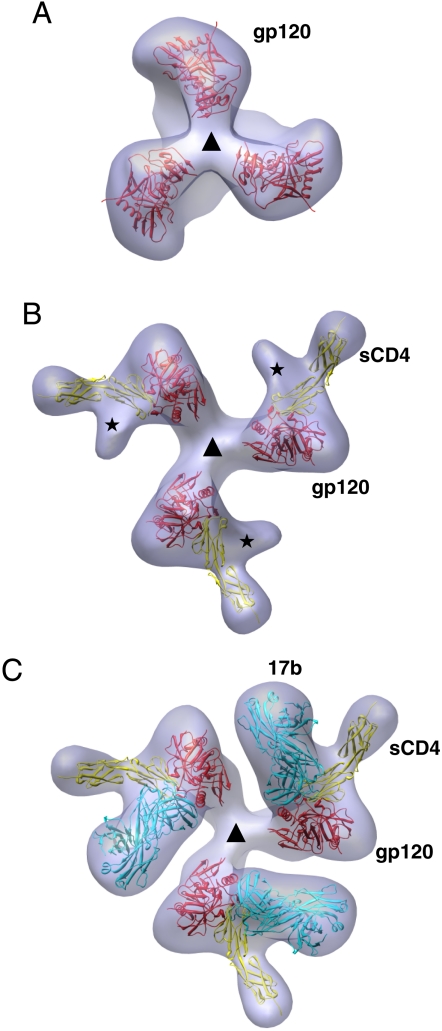
Purified KNH1144 gp140 can be cleaved completely under reducing conditions into its gp120 and gp41 components (Fig. 1A). Native gel electrophoresis analyses show that purified KNH1144 SOSIP trimers are homogeneous in size, with no detectable contamination from monomers, dimers, or high molecular weight aggregates (Fig. 1B). Cryoelectron microscopic images of the purified trimers show that the purified trimers are monodisperse, and they display a characteristic three-bladed propeller appearance in most projection views (Fig. 1C). Using cryoelectron tomography combined with 3D image averaging, we determined the three-dimensional structure of these gp140 trimers at approximately 2-nm resolution. The density map for trimeric KNH1144 gp140, superposed with the previously reported map (2) [EM Data Bank (EMDB) 5018] for native trimeric HIV-1 BaL Env is shown in Fig. 2. The density map includes contributions from gp120, and the gp41 ectodomain near the base, but the region expected to be immediately adjacent to the viral membrane is disordered, most likely because of the absence of the transmembrane domain that anchors the native spike. Similar disordering of this anchoring region has also been observed in the crystal structure of the trimeric spike of Ebola virus (13).
Fig. 1.
Biochemical and cryoelectron microscopic characterization of KNH1144 SOSIP gp140 trimers. (A) SDS-PAGE analysis of purified KNH1144 gp140 under reducing and nonreducing conditions. Lanes: 1, molecular weight standards; 2, gp140, reduced (+DTT); 3, gp140, nonreduced (-DTT). (B) BN-PAGE analysis of purified KNH1144 gp140 trimers. Lanes: 1, molecular weight standards; 2, purified KNH1144 gp140. (C) Projection image of a field of soluble gp140 trimers embedded in vitreous ice. (Scale bar: 50 nm.)
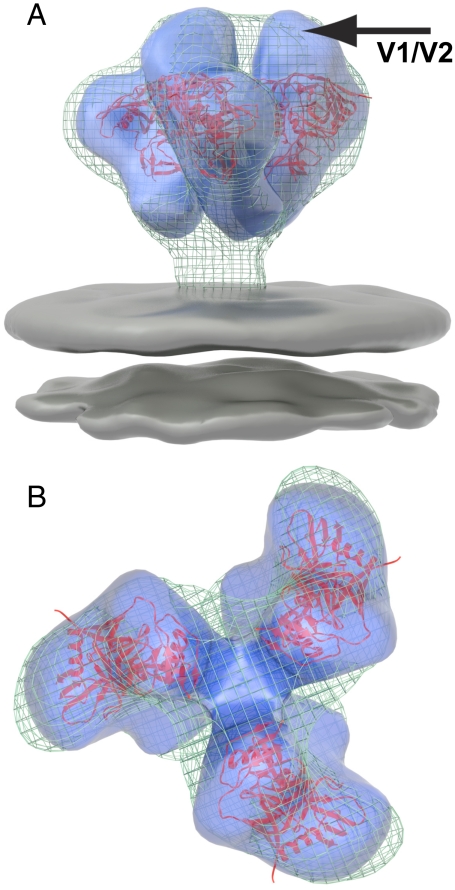
Fig. 2.
Density map and molecular structure of KNH1144 gp140 trimers. (A and B) Transparent isosurface representations of the density map for trimeric KNH1144 gp140 (in blue), shown superposed with the density map (wire mesh) and molecular model for trimeric gp120 (PDB ID 3DNN) derived previously by cryoelectron tomography of native HIV-1 BaL on intact virions (2). The arrow points to the density at the apex of the density map, shown previously to correspond to the location of the V1/V2 loop (3), which is absent in the crystal structure of monomeric gp120. The density representing the lipid bilayer is colored gray in A.
In addition to the close similarity in shape between the maps for trimeric gp140 and native Env, the gp140 density can also be fit with the same coordinates (trimer model 3DNN based on the 1GC1 coordinates for monomeric gp120) that were derived for the molecular structure of trimeric HIV-1 Bal Env on intact virions (2). The coordinate fits place the V1/V2 and V3 loop regions at the top, shown using gp120 coordinates that lack the V3 loop (5) (Fig. 2 A and B) and with gp120 coordinates that include the V3 loop (4) (Fig. S1). Density for the N and C termini of gp120 is visible near the base of the spike, in the same location deduced previously from tomographic studies of native HIV-1 Env (2, 3). The finding that the density map of KNH1144 gp140 trimers can be fit with the coordinates identical to those of the molecular structure of native trimeric Env on HIV-1 BaL establishes that the soluble version mimics the overall quaternary architecture of the native spike at the present resolution of the density maps.
Structures of sCD4- and 17b-bound KNH1144 trimers.
We next addressed structural aspects of the interaction of sCD4 and 17b with gp140 trimers to compare the structures of the complexes with that obtained for sCD4 and 17b bound to native trimeric Env. To this end, we determined structures of gp140 in the presence of sCD4 alone, 17b alone, or in the presence of both sCD4 and 17b. Projection cryoelectron microscopic images recorded from the mixtures show that the presence of the additional density from sCD4 or 17b can be readily visualized even in individual molecular images (Fig. 3 A and B). This result is confirmed in the 3D density maps of the complexes formed by trimeric gp140 with sCD4 (Fig. 3C) or 17b (Fig. 3D). Fitting the density maps with coordinates derived from the X-ray structure of the ternary complex of monomeric gp120, sCD4 and 17b Fab shows that the addition of sCD4 alone is sufficient to convert gp120 trimers in the SOSIP construct from the closed conformation to an open conformation, and that addition of 17b alone is also sufficient to convert the gp120 trimers to the same open conformation. Strikingly, the density maps for both open conformations can be fit with the same coordinates [EMDB entry 5020, Protein Data Bank (PDB) ID 3DNO] derived for the open conformation of trimeric gp120 on intact virions complexed to sCD4 and 17b (2).
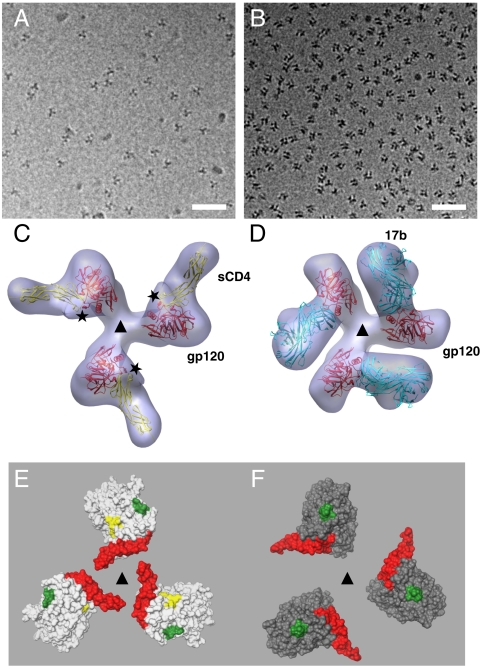
Fig. 3.
Three-dimensional structures of complexes formed by KNH1144 gp140 trimers. (A and B) Projection cryoelectron microscopic images from specimens containing complexes of KNH1144 gp140 trimers with sCD4 (A) or 17b Fab (B). (Scale bar: 50 nm.) (C and D) Top views of density maps and fitted coordinates for KNH1144 gp140 trimers in complex with sCD4 alone (C) or with 17b Fab alone (D). The maps are shown superposed with the molecular coordinates derived previously (PDB ID 3DNO) for native trimeric Env in complex with sCD4 and 17b (2). The “Chinese symbol” type patterns seen in B are characteristic projection views of the gp140-17b complex because most of the trimeric complexes are oriented on their side with the threefold axis perpendicular to the direction of the incident electron beam. Schematic representation of the coordinates of gp120 are in red, Fab fragments are in cyan, and sCD4 are in yellow. The asterisks in C mark the location of the stump of the V1/V2 loop region on gp120. (E and F) Top view, space-filling models for the molecular structure of the closed (E) and open (F) states as seen from the vantage point of the target cell. The truncated V1/V2 loop in the 1GC1 coordinates is shown in red, the stump of the V3 loop is in green, and the residues corresponding to the CD4 binding site are in yellow. The triangles in C–F mark the location of the threefold symmetry axis.
Inspection of the molecular structures of the gp120 trimer in closed and open states (Fig. 3 E and F) highlights the extent and effect of this large conformational change on its organization. The changes result in repositioning the V1/V2 loops toward the outside of the trimer and in significant reorientations of the V3 loop and CD4 binding site on gp120. Because the open conformation observed on native virions was formed in the presence of both sCD4 and 17b, we also tested the effect of simultaneously adding both reagents to soluble gp140 trimers. As shown in Fig. 4, addition of sCD4 and 17b to KNH1144 gp140 trimers results in formation of a ternary complex (Fig. 4 A and B) whose structure is essentially the same as that observed with trimeric Env on intact virions (Fig. 4 C and D), and can be fit with coordinates derived for this native complex. The fact that the same large conformational change in the trimer is observed upon addition of sCD4 alone, 17b alone, or both sCD4 and 17b, strongly suggests that the open conformation is a discrete state of the gp120 trimer that is accessed in each of these complexes, and recapitulates that observed for Env on intact viruses exposed to sCD4 and 17b.
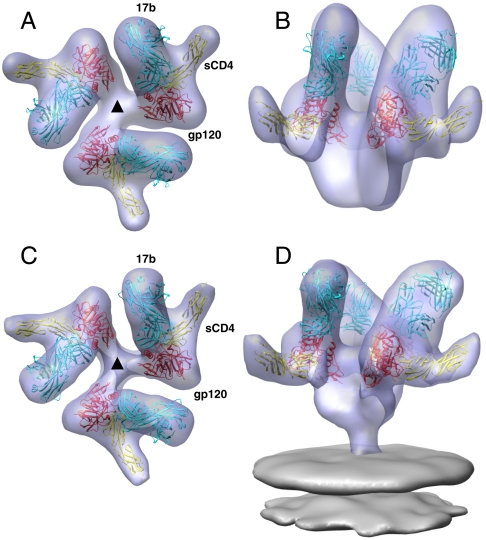
Fig. 4.
Comparison of the ternary complexes formed between soluble gp140 and native Env trimers. (A and B) Top and side views of the structure of ternary complex formed between KNH1144 gp140 trimers, sCD4 and 17b Fab. (C and D) Top and side views of the structure of ternary complex formed between native HIV-1 BaL, sCD4, and 17b. Color scheme for proteins is as in Fig. 3.
Comparison Between KNH1144 and JR-FL gp140 Trimers.
Our experiments show that soluble KNH1144 SOSIP gp140 trimers display a quaternary structure comparable to native, virion-associated Env, and that the gp140 trimer can sample at least two discrete conformations. Hence, the additional disulfide bond engineered between gp120 and gp41 to help stabilize the soluble trimer does not constrain functionally relevant conformational changes as suggested by previous biochemical experiments (14). To test whether this conformational flexibility is also observed with soluble gp140 trimers from a different strain, we analyzed the structure of SOSIP JR-FL gp140 trimers bound to sCD4 and 17b. The 3D structures of the unliganded JR-FL gp140 trimer show a closed conformation similar to that observed for trimeric KNH1144 gp140 (Fig. 5A). In the presence of sCD4, an open conformation is observed (Fig. 5B) whose structure is the same as that found for the corresponding sCD4 complex with KNH1144 gp140 trimers. A more prominent density for the V1/V2 loop region is observed in the JR-FL trimers as compared to KNH1144 trimers, suggesting these loops may be more ordered in the sCD4-bound state of JR-FL gp140. However, in contrast to KNH1144, addition of 17b alone did not result in a significant change in structure of the trimer. However, simultaneous addition of both sCD4 and 17b to JR-FL gp140 trimers resulted in formation of a ternary complex (Fig. 5C) with the same open conformation as observed for the ternary KNH1144 gp140/sCD4/17b complex. These results show that the JR-FL gp140 trimers do not show significant binding to 17b unless sCD4 is also present, whereas KNH1144 gp140 trimers are able to bind 17b even in the absence of sCD4. Nevertheless, the trimeric gp120 components of both KNH1144 and JR-FL trimers in the ternary sCD4/17b-bound complex have the same open conformation as that observed for native trimeric Env complexed to sCD4 and 17b (Fig. 4D). This difference in the ability of the trimers from KNH1144 and JR-FL to bind 17b in the absence of sCD4 could reflect structural differences in gp120 from the two strains, and may be related in turn to functional properties of these viruses.
Fig. 5.
Structure and structural transitions in JR-FL gp140 trimers. (A) Top view of density map for unliganded JR-FL gp140 trimers with fitted coordinates for trimeric gp120 (PDB ID 3DNN). (B and C) Top view of density map from JR-FL gp140 trimers complexed to sCD4 alone (B) or with sCD4 and 17b Fab (C) fitted with the same gp120 and sCD4 coordinates derived for the ternary complex formed between sCD4, 17b, and native trimeric HIV-1 BaL Env (PDB IDs 3DNO and 1GC1). The density for the V1/V2 loop region in B is marked by an asterisk; this region appears to be more ordered in sCD4-complexed JR-FL trimers as compared to sCD4-complexed KNH1144 trimers. Color scheme for coordinates is as in Fig. 3.
Discussion
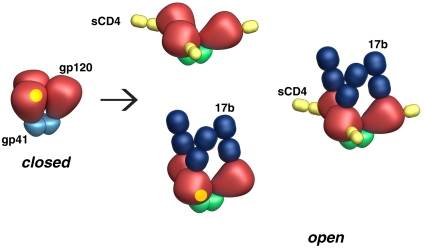
A schematic depiction of the unliganded, CD4-liganded, and antibody-bound conformational states that are sampled by SOSIP gp140 trimers is presented in Fig. 6. A central conclusion from our analyses is that sCD4/17b binding to either KNH1144 or JR-FL SOSIP trimers results in an open conformation which has the same quaternary structure as that previously established for the sCD4/17b-bound state of trimeric Env on intact virions (2). One way to interpret the surprising finding that 17b binding to KNH1144 SOSIP trimers can stabilize the same open conformation as that observed with sCD4 binding is that the binding of either sCD4 or 17b is sufficient to trigger the opening of the trimer. An alternative possibility is that unliganded gp140 trimers exist in a dynamic equilibrium that allows sampling of at least two discrete conformations where trimeric gp120 is in either the closed or open state, and that sCD4 and 17b can independently shift the equilibrium in favor of the open conformation (15). Irrespective of which of these models is the better way to account for the results, the unequivocal conclusion from our studies is that both the clade A KNH1144 and the clade B JR-FL gp140 SOSIP trimers are capable of displaying at least two discrete conformations: a closed state similar to unliganded trimeric Env and an open state that resembles the open CD4/17b-bound state on native Env.
Fig. 6.
Schematic representations of the closed and open gp140 conformational states based on the density maps obtained by cryoelectron tomography. Color scheme is with gp120 in red, the closed and open states of gp41 in light-blue and green, respectively, 17b Fab in dark-blue, and with sCD4 in yellow. The yellow patch denotes the location of the CD4 binding site on gp120.
We note that other groups working toward structure determination of detergent-solubilized HIV-1 spikes (16) and uncleaved gp140 trimers (17) using cryoelectron microscopy have reported density maps for the ectodomain of trimeric Env that differ from each other, and also differ from the maps that we have obtained for cleaved gp140 trimers (this work) and native SIV (3) and HIV-1 (2) envelope glycoprotein trimers.
Although the studies we present here describe the conformational transitions in soluble gp140 immunogens, it is interesting to compare these findings with conformational changes of trimeric Env as displayed on intact virions. Compared to pseudotyped viruses expressing JR-FL Env, pseudotyped viruses expressing KNH1144 have been shown to be significantly more sensitive to CD4-IgG2 (11). This result is consistent with the structural findings that an open conformation of the trimer is not observed for JR-FL trimers with 17b unless sCD4 is also present. It is thus plausible that the activation barrier for trimeric Env to transition to the open state may be related to the neutralization sensitivity of the virus with trimeric Env on neutralization-sensitive strains being more prone to make the transition to an open state than their resistant counterparts. However, there can be considerable clone- or isolate-dependent variation in sensitivity and, because there are very few reagents that can react with viruses across different clades because of antigenic diversity, much more systematic structural and virological analyses need to be carried out with many different strains and reagents to generalize these results. Interestingly, structures of trimeric Env that have been determined so far by cryoelectron tomography include the laboratory-adapted, neutralization-sensitive HIV-1 strains HIV-1 BaL and HIV-1 R3A, both of which display trimeric Env in the closed conformation (2, 3). Thus, at least in these two instances, one could conclude that neutralization sensitivity does not require that trimeric Env is locked in the open conformation, and it is probably adequate that the barrier to transition between the two states is sufficiently low.
The answer to what in fact does correlate with the open conformation comes from recent cryoelectron tomographic studies with SIV, which show that trimeric, membrane-associated Env from the CD4-independent strain SIV CP-MAC is expressed in a constitutively open conformation (3) with the same molecular structure for gp120 trimers as that seen in the open conformations reported here for SOSIP gp140 trimers. In sharp contrast to the neutralization-resistant strain SIVmac239, which displays trimeric Env in the closed conformation, the neutralization-sensitive SIV CP-MAC strain is capable of infecting cells even in the absence of cell surface CD4, and displays efficient binding to a 17b-like antibody (7D3) that recognizes the coreceptor binding site. These findings support the idea (18) that the joint requirement of CD4 and chemokine receptor CCR5 represents an evolutionary adaptation of CCR5-dependent HIV and SIV to use CD4-dependent spike opening to cloak conserved antigenic sites on gp120 that represent targets for neutralizing antibodies until virus-cell contact, where antibody access is sterically restricted. The fact that both closed and open conformations of trimeric Env can be displayed on intact viruses establishes clear functional connections between CD4 binding and quaternary structural changes in both soluble gp140 and native, virion-bound trimers.
Env-based immunogens are important HIV-1 vaccine candidates, but neither monomeric gp120 nor trimeric gp140 proteins of any design have yet proved capable of eliciting broadly neutralizing antibodies at useful titers in humans or animals (8–10). Most studies that have compared the two forms using the same Env genotype have shown that gp140 trimers are superior to gp120 monomers at inducing neutralizing antibodies (19–21). However, the improvement in immunogenicity is not yet good enough to conclude that any forms of gp140 trimers, in their current iteration, are good enough to use in practical HIV-1 vaccines. The challenge is to determine what structural and other factors limit the immunogenicity of gp140 trimers and design ways to overcome any problems that are identified (22). Structural studies are of critical importance in this regard. Whereas our work establishes the broad structural similarities between SOSIP gp140 trimers and native Env trimers at approximately 2-nm resolution, there could be numerous subtle differences in conformation that we have not been able to decipher but that may become apparent at a higher resolution. Nevertheless, the differential ability of KNH1144 and JR-FL trimers to bind antibodies such as 17b could be useful for immunogen design, and variant gp140 trimers preferentially stabilized in distinct intermediate states could potentially be engineered and then mixed to create a composite vaccine, with the goal of increasing the breadth of the neutralizing antibody response. Any variations in the extent to which different regions on gp120 and gp41 are accessible in the open state may also increase the likelihood of inducing broadly neutralizing antibodies, such as b12 (23), PG9/16 (24), VRC01 (25), or HJ16 (26).
The progress we report here on structural analysis of soluble gp140 trimers using cryoelectron tomography represents an important technical advance in the use of tomographic methods (27) to determine the structures of protein complexes. In conventional cryoelectron microscopy, structure determination is based on assembling the 3D structure by averaging the structural information present in a large number of individual 2D projection images, whose relative orientations are determined iteratively starting from an initial 3D model at lower resolution. In contrast, structure determination using cryoelectron tomography relies on averaging of 3D molecular images, with the model building steps combined with assignment of relative orientations (28). Earlier cryoelectron tomographic studies of the structure of native Env displayed on intact virions (2, 3) were helped by the fact that the bilayer membrane provides a reference point to define the approximate initial orientation of the spike in the tomograms, thereby restricting the initial search of relative orientations of individual spikes to two angular parameters. The structural findings we report here represent a validated generalization of this 3D classification and averaging approach (28) to soluble protein complexes where there is no prior information on the orientation, and demonstrates its utility for the structural analysis of small protein complexes, such as HIV-1 gp140 trimers.
Materials and Methods
Antibodies and CD4 Reagents.
Monoclonal antibody 17b was a gift from Dennis Burton (The Scripps Research Institute). Fab fragments were prepared by digestion with papain. Two-domain soluble CD4 (sCD4, amino acids 1–183) was provided by the National Institutes of Health AIDS Research and Reference Reagent Program.
SOSIP gp140 Constructs.
KNH1144 SOSIP gp140 (11) was modified to contain A662E, S668N, and S676T substitutions relative to the wild-type KNH1144 membrane proximal external region sequence (Genbank accession no. AY736812). JR-FL SOSIP gp140 constructs have been described previously (29). Codon-optimized SOSIP gp140 genes were subcloned into the PPI4 vector (11).
SOSIP gp140 Preparation.
JR-FL SOSIP gp140 trimers were produced according to published methods (30). For production of KNH1144 SOSIP gp140 trimers, 293 T cells were seeded into triple flasks (Corning Life Sciences) in DMEM with 10% fetal bovine serum and supplements 24 h prior to transfection. Cells were cotransfected with SOSIP-PPI4 and furin-pcDNA3.1, using polyethylenimine (Polysciences, Inc.). Following transfection, cells were cultured for 48 h in DMEM. Supernatants were then harvested and clarified. The clarified supernatant was concentrated approximately fivefold by tangential flow filtration over a 100 kDa molecular weight cutoff (MWCO) membrane (GE Healthcare). Concentrated supernatant was captured on a Galanthus nivalis agglutinin lectin affinity column (Vector Laboratory). Impurities, such as monomers, dimers, and α2-macroglobulin, were removed by two sequential DEAE Sepharose Fast Flow columns. In the second column, 0.05% Tween 20 was added to convert higher order aggregates to homogeneous trimers. The trimers were then concentrated using 100 kDa MWCO spin filter (Sartorius) and purity was further refined by passage over a Superdex™ 200 size exclusion column (GE Healthcare). The concentration of purified trimer was determined using the bicinchoninic acid assay (Bio-Rad Laboratories, Inc.).
Analysis of Purified SOSIP gp140 Trimers.
Trimers were analyzed by Blue Native polyacrylamide gel electrophoresis (BN-PAGE) using NativePAGE 4–16% Bis-Tris gels and by SDS-PAGE using NuPAGE 4–12% Bis-Tris gels (Invitrogen). NativeMark™ were used as molecular weight standards in BN-PAGE, and Mark12™ were used in SDS-PAGE. Gels were stained with Colloidal Blue Stain Kit (Invitrogen). Protein purity was determined by densitometric analysis of the stained gels using ImageQuant software (GE Healthcare).
Cryoelectron Microscopy.
Specimens of purified gp140 trimers were applied to holey carbon films, thinned by blotting, and vitrified by plunging into liquid ethane cooled to approximately -180° C in a liquid nitrogen environment. To prepare complexes, Fab fragments of monoclonal antibody 17b (3.3 mg/mL) was mixed with KNH1144 (420 μg/mL) or JR-FL (238 μg/mL) gp140 trimers on ice for 2 h at molar ratios of ca. 12 Fab fragments per gp140 trimer. When sCD4 was included, it was at a molar ratio of approximately 10 relative to gp140 trimers unless indicated otherwise. Plunge-frozen grids were transferred into the column of the electron microscope designed to maintain the specimen at approximately -180° C. Projection microscopic images were obtained on an FEI Titan Krios electron microscope, using a 4,000 × 4,000 CCD camera (Gatan), at an accelerating voltage of 80 kV, pixel size of 1.45 Å at the specimen plane, at doses of approximately 20 electrons/Å2, and nominal defocus values ranging from ca. 0.7 to 3.8 μm. Three-dimensional structures of unliganded gp140 trimers and associated complexes were obtained using cryoelectron tomography combined with classification and 3D subvolume averaging implementing procedures described in Bartesaghi et al. (28) and White et al. (3). Illustration of image classes obtained for unliganded and liganded gp140 trimers are presented in Figs. S2 and S3. All maps were obtained using data collected on an FEI Polara G2 microscope, equipped with a Gatan GIF 2002 imaging filter and 2,000 × 2,000 CCD camera, at a pixel size of 4.1 Å in the specimen plane. Typically, about 4,000 subvolumes of each complex were averaged. Map resolution was estimated by determining the resolution at which the Fourier shell correlation between two randomly chosen half-datasets had a value of 0.5. Using this criterion, the resolutions of the maps were determined to be approximately 2 nm, comparable to that obtained for native spikes (3). The coordinate fits shown for unliganded maps are the same as those reported for native trimeric Env (EMDB entry 5018, PDB ID 3DNN), and the fits for all of the binary and ternary complexes with sCD4 and/or 17b are from those deposited (EMDB entry 5020, PDB ID 3DNO) for the native gp120-17b-sCD4 ternary complex.
Supplementary Material
Acknowledgments.
We thank Steven Fellini and colleagues for assistance with use of the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH) (http://biowulf.nih.gov); Dr. Dennis Burton (The Scripps Research Institute) for providing 17b antibody; and A.M. Cupo, A.A. Ouattara, and L. Pagel for excellent technical assistance. This work was supported by the Center for Cancer Research at the National Cancer Institute, NIH and by NIH Grants AI36082 and 45463; by the International AIDS Vaccine Initiative; and by NIH HIV Vaccine Research and Design Grant P01 AI-082362.
Footnotes
Conflict of interest statement: Progenics has a proprietary interest in SOSIP gp140. K.K., A.J.M., and W.C.O. have equity interests in Progenics.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101414108/-/DCSupplemental.
References
- 1.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: Current approaches and future directions. Curr Opin Immunol. 22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 10.Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 11.Beddows S, et al. Construction and characterization of soluble, cleaved, and stabilized trimeric Env proteins based on HIV type 1 Env subtype A. AIDS Res Hum Retroviruses. 2006;22:569–579. doi: 10.1089/aid.2006.22.569. [DOI] [PubMed] [Google Scholar]
- 12.Kang YK, et al. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine. 2009;27:5120–5132. doi: 10.1016/j.vaccine.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore JP, Trkola A, Sattentau QJ, Cao Y, Ho DD. Studies on the interactions of monoclonal antibodies with HIV-1 isolates from clades A–F. Neuvieme Colloque Des Cent Gardes. 1994:151–155. [Google Scholar]
- 16.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moscoso CG, et al. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proc Natl Acad Sci USA. 2011;108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman TL, et al. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beddows S, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360:329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75:1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bower JF, Yang X, Sodroski J, Ross TM. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78:4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klasse P, Sanders R, Cerutti A, Moore J. How can HIV-type-1 Env immunogenicity be improved to facilitate antibody-based vaccine development? AIDS Res Hum Retroviruses. 2011 doi: 10.1089/aid.2011.0053. 10.1089/aid.2011.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 24.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartesaghi A, Subramaniam S. Membrane protein structure determination using cryo-electron tomography and 3D image averaging. Curr Opin Struct Biol. 2009;19:402–407. doi: 10.1016/j.sbi.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartesaghi A, et al. Classification and 3D averaging with missing wedge correction in biological electron tomography. J Struct Biol. 2008;162:436–450. doi: 10.1016/j.jsb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beddows S, et al. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2005;79:8812–8827. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.