Abstract
Homeostatic mechanisms are required to control formation and maintenance of synaptic connections to maintain the general level of neural impulse activity within normal limits. How genes controlling these processes are co-coordinately regulated during homeostatic synaptic plasticity is unknown. MicroRNAs (miRNAs) exert regulatory control over mRNA stability and translation and may contribute to local and activity-dependent posttranscriptional control of synapse-associated mRNAs. However, identifying miRNAs that function through posttranscriptional gene silencing at synapses has remained elusive. Using a bioinformatics screen to identify sequence motifs enriched in the 3′UTR of rapidly destabilized mRNAs, we identified a developmentally and activity-regulated miRNA (miR-485) that controls dendritic spine number and synapse formation in an activity-dependent homeostatic manner. We find that many plasticity-associated genes contain predicted miR-485 binding sites and further identify the presynaptic protein SV2A as a target of miR-485. miR-485 negatively regulated dendritic spine density, postsynaptic density 95 (PSD-95) clustering, and surface expression of GluR2. Furthermore, miR-485 overexpression reduced spontaneous synaptic responses and transmitter release, as measured by miniature excitatory postsynaptic current (EPSC) analysis and FM 1–43 staining. SV2A knockdown mimicked the effects of miR-485, and these effects were reversed by SV2A overexpression. Moreover, 5 d of increased synaptic activity induced homeostatic changes in synaptic specializations that were blocked by a miR-485 inhibitor. Our findings reveal a role for this previously uncharacterized miRNA and the presynaptic protein SV2A in homeostatic plasticity and nervous system development, with possible implications in neurological disorders (e.g., Huntington and Alzheimer's disease), where miR-485 has been found to be dysregulated.
Keywords: activity-dependent development, posttranscriptional gene regulation, presynaptic terminal
Homeostatic synaptic plasticity is the process of regulating synaptic connections to compensate for changes in levels of impulse activity in neural networks over a time course of days, thus maintaining circuit function within an optimal range (1). This homeostasis limits changes in synaptic strength induced by Hebbian synaptic plasticity (2), compensates for nervous system disorders causing hyper- or hypoexcitability, and adjusts excitability of neural circuits during development. How genes involved in this process are coordinately regulated is unknown.
Increasing excitability of hippocampal or cortical neurons in culture using bicuculline (BiC) reduces synaptic strength through both pre- and postsynaptic mechanisms, and decreasing excitability with TTX produces the opposite response (1, 3, 4). Several molecules have been identified that contribute to homeostatic synaptic plasticity over different time scales, including alterations in the α/β Ca2+/CaM-dependent kinase II (CaMKII) ratio (5), astrocytic TNF-α release (6), regulation of cell surface AMPA receptors (7), Arc/Arg3.1 expression (4), and polo-like kinase 2 (8). Structural changes in synaptic connectivity that follow physiological changes in synaptic strength must involve gene regulatory networks controlling synaptic development, maturation, and maintenance. MicroRNAs (miRNAs) rapidly and coordinately regulate stability and translation of sets of mRNAs mediating specific processes (9, 10), suggesting that miRNAs could have an important role in homeostatic synaptic plasticity. In response to increased activity, miRNAs could rapidly destabilize mRNAs and suppress translation of proteins required for synaptic function and development, thereby reducing connectivity homeostatically. There is evidence that translation of mRNAs regulating synaptic strength can be regulated locally at the synapse (11).
Hundreds of miRNAs have been characterized, many of which are enriched in the brain and synapses (12–14), but identifying specific miRNAs contributing to activity-dependent development and plasticity has proven difficult for two reasons (15–18): miRNAs regulating synaptic plasticity are assumed to be relatively rare in comparison to miRNAs regulating more general cellular processes, and plasticity is localized to individual synapses. Thus, potential miRNAs that may regulate mRNA transcript stability in activity-dependent synaptic plasticity may not emerge from global miRNA expression analysis.
To overcome this problem, we used a bioinformatics approach. Microarray analysis showed that the abundance of several mRNA transcripts was rapidly decreased in hippocampal neurons in cell culture after increasing activity. Using motif-based sequence analysis (19), we then determined whether these transcripts shared motifs in the 3′UTRs corresponding to putative binding sites for miRNAs or RNA binding proteins. This approach revealed that many of these rapidly down-regulated transcripts shared sequences in their 3′UTR for several miRNAs that could, in theory, increase degradation of mRNAs that promote synaptic development. One of these motifs was identified as a putative binding site for a rare brain-enriched miRNA (miR-485) for which little is known. Our analysis indicated that several of the genes with putative miR-485 binding sites control synaptic function or development. The following studies were undertaken to determine whether miR-485 regulates homeostatic synaptic plasticity in hippocampal neurons through destabilization of specific gene transcripts regulating synaptic development and function. The results support the hypothesis, and the findings provide a new molecular mechanism regulating homeostatic synaptic plasticity in hippocampal neurons, which is likely to be important in regulating synaptic connectivity during neural development and disease.
Results
Synaptic activity in hippocampal cultures was increased with 50 μM BiC and 500 μM 4-aminopyridine (4-AP) (20); Ca2+ imaging confirmed the increase in repetitive firing in these hippocampal cultures (Fig. S1A). Chronic elevation of synaptic activity with BiC/4-AP for 5 d reduced dendritic spine density by 22 ± 3% (n = 36; P < 0.001) (Fig. S1B), consistent with previously described activity-dependent structural changes (21, 22).
To identify mRNAs that are rapidly reduced by posttranscriptional mechanisms, hippocampal neurons in culture were treated with BiC/4-AP for 5 min in the presence of the transcriptional blocker actinomycin D (25 μM). Microarray profiling identified many rapidly regulated mRNAs under these conditions, and many were synapse- and morphogenetic-associated. A bioinformatics approach was used to examine the 3′UTR of this set of destabilized mRNAs to identify enriched motifs compared with transcripts that were up-regulated or unregulated by increased synaptic activity. If these transcripts are regulated by miRNAs, they would be expected to share miRNA binding sequences that would suppress translation or increase degradation of transcripts. Using motif prediction and analysis tools, Multiple Em for Motif Elicitation (MEME) (19), many of these rapidly destabilized mRNAs were found to share a motif in the 3′UTR corresponding to the binding sequence for miR-485 (Fig. S1C). We then tested whether this miRNA could participate in the homeostatic activity-dependent reduction in synaptic connectivity caused by increased activity.
miR-485 Is Regulated in a Developmental and Activity-Dependent Manner in the Hippocampus.
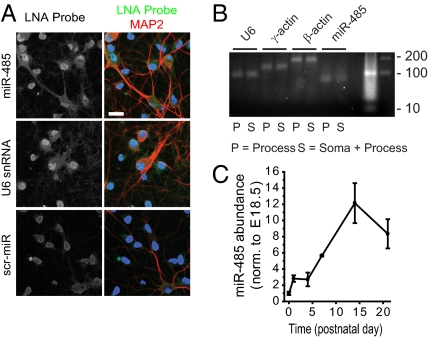
miR-485 was detected in hippocampal neuronal cell bodies and dendrites in culture by FISH (Fig. 1A). These findings were confirmed by localization of miR-485 in neuronal processes by RT-PCR analysis of processes extending through 3-μm pores in transwell cultures (Fig. 1B). Expression levels for miR-485 were low in developing rat hippocampus compared with other brain-enriched miRNAs at embryonic day 18.5 (23, 24) but increased 12.2 ± 2.5-fold (n = 3) by postnatal day 14 (Fig. 1C); in culture, both miR-485 and the precursor miRNA were regulated in an activity-dependent manner, increasing 1.3 ± 0.1-fold (n = 3; P < 0.05) after a 1-h treatment with BiC/4-AP.
Fig. 1.
miR-485 is expressed in hippocampal neurons and developmentally regulated. (A and B) Expression of miR-485 was detected by FISH and RT-PCR. U6 snRNA and scrambled probe are shown as positive and negative controls, respectively. scr-miR, scrambled miR probe. Dendrites (red) were identified by MAP2 immunoreactivity, and nuclei (blue) were stained with Hoechst 33342. (Scale bar = 25 μm.) (B) RT-PCR was performed on neuronal processes growing through 3-μm pores in transwell inserts to isolate from cell soma in the upper compartment. P, process; S, soma + process. Mature miR-485 (based on product size) was present in both fractions. U6 (U6 snRNA) and γ-actin served as reference genes. β-actin was approximately twofold higher in processes, indicating enrichment of neuronal processes. (C) miR-485 transcript is developmentally regulated in the hippocampus. The increase in mature miR-485 (n = 3) at each time point was first normalized to the reference gene U6 snRNA and the abundance expressed relative to embryonic day 18.5. E, embryonic day.
miR-485 Regulates the Presynaptic Protein SV2A.
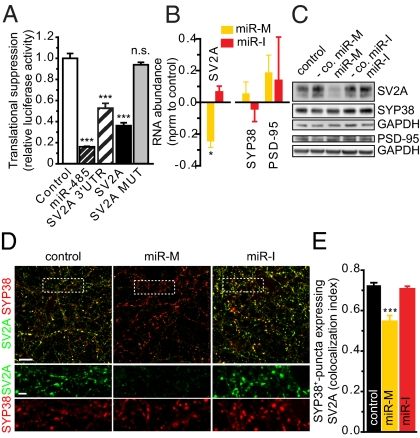
Gene ontology analysis of miR-485–predicted targets (Table S1) showed enrichment in genes functioning in CNS development and disease (e.g., Huntington disease). One of these predicted targets, the synaptic vesicle protein SV2A (Fig. S2), is enriched in presynaptic terminals, where it controls neurotransmitter release (25). Function of predicted miRNA responsive elements (MREs) was tested by luciferase assays on constructs containing either the full-length 3′UTR or the 3′-end of SV2A 3′UTR. Overexpression of a miR-485 mimic (miR-M; a small chemically modified dsRNA designed to mimic the endogenous miRNA) significantly reduced luciferase activity of both constructs (n = 6; P < 0.001) (Fig. 2A), and this was reversed when both MREs were mutated within the 5′-seed region (Fig. S2B). The effects of miR-485 were specific, because cotransfection with a miR-485 inhibitor (miR-I; an antisense RNA oligonucleotide decoy that specifically and competitively binds to and inhibits the function of endogenous miR-485) reduced the suppression of a miR-485 luciferase construct by the miR-M (n = 3; P < 0.01). Furthermore, transcript levels for SV2A 3′UTR constructs were reduced by miR-485 (Fig. S2C), consistent with a degradative mechanism for the miRNA.
Fig. 2.
miR-485 decreases SV2A expression in hippocampal neurons. (A) SV2A is a putative target of miR-485. Luciferase assays to validate SV2A as a target of miR-485 were performed on constructs containing either the full-length 3′UTR (SV2A-3′UTR), distal 3′UTR containing two miR-485 binding sites (SV2A), or mutated sites in the 5′-seed of MRE1 and MRE2 (SV2A MUT) as controls (Fig. S2B). Translational suppression by miR-485 was normalized to empty psiCHECK-2 plasmid. Treatment with the miR-M decreased SV2A transcript abundance (B) and protein levels (C) significantly without affecting SYP38 or PSD-95. Treatment with negative controls for either the mimic or inhibitor did not alter transcript or protein levels for SV2A, SYP38, or PSD-95. -co. control. (D) miR-485 overexpression reduced SV2A expression in hippocampal neuron synapses. Cultures were cotransfected with DsRed and miRNAs at 7 days in vitro (DIV) and analyzed by immunocytochemistry at 12 DIV for SV2A and SYP38 expression. Representative examples of untreated (control) and neurons transfected with the miR-M or miR-I showing that SV2A expression is reduced in individual presynaptic terminals. (Lower) Higher magnification is shown. Treatment with negative controls did not alter SV2A or SYP38 expression. (Scale bars: Upper, 10 μm; Lower, 2.5 μm.) (E) Quantitative analysis of randomly chosen dendritic fields showing that the miRNA specifically reduced colocalization of SV2A and SYP38.
We tested if miR-485 controlled SV2A expression in hippocampal neurons by miR-485 overexpression and inhibition. miR-485 overexpression decreased both transcript abundance (n = 3; P < 0.05) (Fig. 2B) and protein expression (Fig. 2C) of SV2A relative to both an untreated control and negative controls for the miR-M and miR-I containing random sequences validated to not affect miR function. Furthermore, the effect on SV2A was specific, because neither transcript nor protein expression of the synaptic markers synaptophysin (SYP38) and postsynaptic density 95 (PSD-95) was altered (Fig. 2 B and C). Neither of these transcripts contains conserved miR-485 binding sites within their 3′UTRs, consistent with a specific effect of the miRNA on genes containing the MRE.
SV2A immunoreactivity at synapses (Fig. 2 D and E and Fig. S3) was specifically reduced by the miR-M. To identify presynaptic terminals, cultures were double-labeled for the presynaptic markers SV2A (green) and SYP38 (red). Colocalization analysis of SV2A+ and SYP38+ puncta showed a reduction of SV2A in cultures overexpressing miR-485 (n = 21; F4,102 = 20.57, P < 0.001 by one-way ANOVA). Colocalization indices for neurons transfected with either control (n = 20, 0.73 ± 0.01) and negative controls for the mimic (n = 25, 0.71 ± 0.01) or inhibitor (n = 20, 0.73 ± 0.01) were not affected. Treatment with the miR-M or miR-I did not affect the number of presynaptic terminals (SYP38+ puncta).
The finding that miR-485 regulated the expression of SV2A, a ubiquitous presynaptic protein that regulates neurotransmitter release (26), suggested that synapses may be altered structurally as a consequence of changes in synaptic function. We therefore tested the function of miR-485 on several morphological properties of hippocampal synapses.
miR-485 Regulates Spine Density and Synapse Morphology.
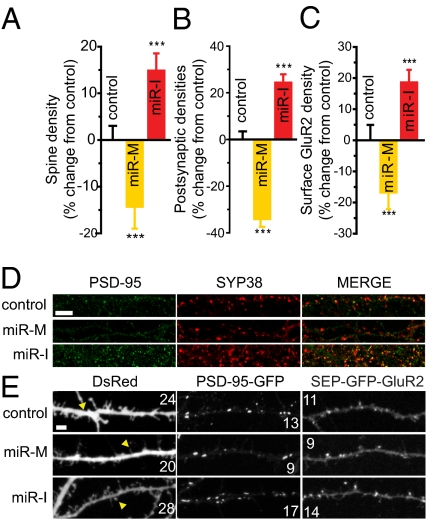
Hippocampal dendritic spine density increases over several weeks in culture and in vivo, and immature spine protrusions gradually become stubby with a distinctive neck and head structure as they mature (27, 28). miR-485 overexpression reduced spine density by 14 ± 5% (n = 19; P < 0.001) and increased the appearance of long and thin immature spines by 35 ± 9% (n = 6; P < 0.01) (Fig. 3 A and E). Conversely, the miR-I increased spine density by 15 ± 4% (n = 19; P < 0.005) and increased the number of short and stubby spines by 46 ± 19% (n = 6; P < 0.05) (Fig. 3E, compare arrowheads in untreated samples vs. miR-M– and miR-I–treated samples). Effects of miR-485 on spine density were highly significant by one-way ANOVA (F4,90 = 9.46, P < 0.001). The magnitude of these changes is consistent with structural changes associated with modulating synaptic activity in culture (22, 29). Furthermore, the miR-I fully reversed the effects of the miR-M, restoring spine density to control levels (Fig. S4). These results show that miR-485 suppresses spine formation and maturation. The effects of miR-485 are consistent with the homeostatic decrease in synaptic connectivity seen after elevating impulse activity and are consistent with the parallel activity-dependent increase in miR-485 levels.
Fig. 3.
miR-485 negatively regulates spine density, PSD-95 clustering, and surface GluR2 expression. miR-485 reduces spine density (A), PSD-95 clustering (B), and surface GluR2 expression (C) along dendrites. Spine density, PSD-95, and SEP-GluR2 were 12.7 ± 0.4 spines per 20 μm (n = 18), 9.4 ± 0.3 PSD-95–GFP+ puncta per 20 μm (n = 34), and 6.9 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16) in untreated controls, respectively. These values were not affected by transfection with negative controls for the mimic [12.4 ± 0.3 spines per 20 μm (n = 20), 9.1 ± 0.2 PSD-95–GFP+ puncta per 20 μm (n = 36), and 7.3 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16)] or inhibitor [12.6 ± 0.4 spines per 20 μm (n = 19), 9.3 ± 0.2 PSD-95–GFP+ puncta per 20 μm (n = 35), and 7.3 ± 0.2 SEP-GluR2 puncta per 20 μm (n = 17)]. (D) Endogenous PSD-95 clustering (green) in hippocampal cultures was decreased by transfection with the miR-M and increased by the miR-I. (Scale bar = 10 μm.) (E) Representative examples of neurons transfected with DsRed, PSD-95 (PSD-95–GFP), or surface GluR2 (SEP-GluR2). (Scale bar = 2.5 μm.)
The effects of miR-485 on presynaptic SV2A expression, dendritic spine number, and maturation may have postsynaptic effects because of the loss of mature spines containing postsynaptic density components, such as PSD-95 and AMPA receptors. Consistent with this, miR-485 overexpression reduced PSD-95 clustering and colocalization with SYP38 (Fig. 3D). Blocking endogenous miR-485 increased the number of PSD-95 puncta. Effects of miR-485 on PSD-95 clustering were further examined with a GFP-tagged PSD-95 construct to visualize PSD-95 puncta directly (30). PSD-95–GFP+ puncta density was reduced 34 ± 3% (n = 35; P < 0.001) (Fig. 3 B and E) by miR-485 overexpression. Conversely, inhibition of endogenous miR-485 increased the density of PSD-95–GFP+ puncta by 24 ± 4% (n = 35; P < 0.001). Effects of the miR-M and miR-I were highly significant (F4,170 = 48.73, P < 0.001 by one-way ANOVA). Moreover, effects of the miR-M on PSD-95–GFP+ puncta density were specific because they were reversed by the miR-I (Fig. S4).
AMPA receptor trafficking into the postsynaptic membrane is regulated by synaptic activity to control synaptic strength in a homeostatic manner (7). The role of miR-485 in AMPA receptor trafficking was investigated in hippocampal neurons transfected with pCl–super-ecliptic pH-sensitive (SEP)–GluR2(R) to visualize cell-surface GluR2 receptor clustering (31). miR-485 overexpression significantly reduced SEP-GluR2 density by 17 ± 5% (n = 16; P < 0.01) (Fig. 3 C and E). Conversely, inhibition of endogenous miR-485 significantly increased SEP-GluR2 density by 19 ± 4% (n = 16; P < 0.05). The effects of functional miR-485 on SEP-GluR2 were highly significant (F4,76 = 8.55, P < 0.001 by one-way ANOVA). The morphological changes in synapse number were observable 3 d following miR-485 overexpression. These findings show that miR-485 acts to regulate spine density and maturation, PSD-95 clustering, and surface GluR2 expression negatively in the postsynaptic membrane (Fig. 3).
miR-485 Overexpression Alters Synapse Function.
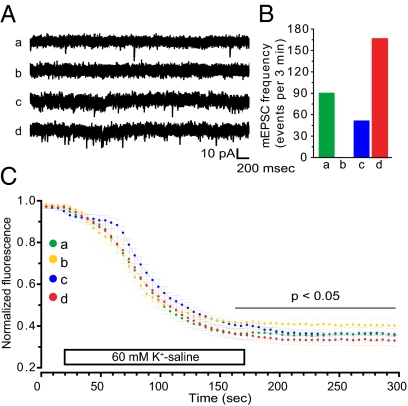
The homeostatic changes in spine density and GluR2 receptor expression regulated by miR-485 were accompanied by corresponding changes in synaptic function. Treatment with the miRNA inhibitor for 6 d increased spontaneous miniature excitatory synaptic current (mEPSC) frequency relative to controls, whereas mEPSC amplitude was not changed [9.1 pA for the negative control compared with 10 pA for the miR-I; χ2(3,313) = 191.984, P < 0.001] (Fig. 4 A and B).
Fig. 4.
miR-485 reduces spontaneous synaptic activity in hippocampal neurons. (A) Representative traces of spontaneous mEPSCs recorded from cultured hippocampal neurons transfected with negative control for the miR-M (a), miR-M (b), negative control for miR-I (c), and miR-I (d) showing a reduction in spontaneous mEPSC frequency after transfection with the mimic (b) and an increase following transfection with the inhibitor (d). (B) Quantitative analysis showing the transfection significantly changed mEPSP frequency [χ2(3,313) = 191.984, P < 0.001]. (C) Synaptic vesicle release (FM 1-43 destaining kinetics) was not affected by miR-485, but the number of recycled vesicles (amount of destaining) was reduced by miR-485 (yellow) and increased by the inhibitor (red) (P < 0.05). Destaining curves for negative controls (n = 8 coverslips with at least 60 boutons each) and for cultures treated with the miR-M (n = 8) and miR-I (n = 11) are shown.
SV2A functions to facilitate synaptic vesicle fusion and neurotransmitter release through interaction with synaptotagmin (32, 33). Therefore, miR-485 should decrease the number of synaptic vesicles released following a stimulus. This was tested by imaging FM 1–43 uptake and release following depolarization. Consistent with this hypothesis, treatment with either the miRNA mimic (30.8 ± 3.0 s, n = 9) or inhibitor (33.2 ± 1.8 s, n = 10) did not alter destaining rates significantly from control (Fig. 4C). However, total dye loss was decreased by treatment with the miRNA mimic and increased by the inhibitor (0.28 ± 0.01 for the miR-I and 0.36 ± 0.02 for the miR-M; P < 0.05) (Fig. 4A). The finding that mEPSC frequency was decreased by the miRNA mimic without effects on mEPSC amplitude or kinetics of FM 1-43 destaining (presynaptic release) supports the morphological findings that miR-485 decreases synapse number.
SV2A Knockdown Mimics the Effects of miR-485 Overexpression.
These results suggest that miR-485 regulation of the presynaptic protein SV2A contributes to the activity-dependent decrease in synaptic connectivity induced by increased neuronal activity. Consistent with this, siRNA-mediated knockdown of SV2A in hippocampal cultures reduced expression of SV2A protein without affecting SYP38 or PSD-95 expression levels (Fig. S5 A and B), and the reduction in SV2A by miR-485 was rescued to levels seen for untreated cultures by SV2A overexpression (pSV2A).
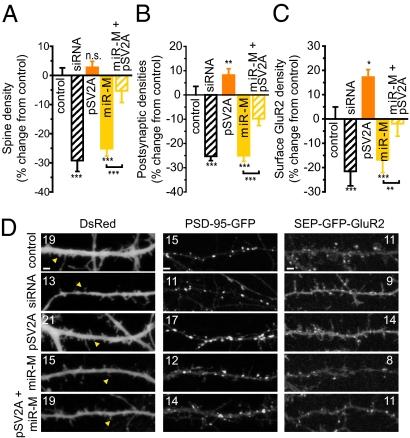
Effects of SV2A knockdown and overexpression on dendritic spines (Fig. 5A), PSD-95 (Fig. 5B), and GluR2 (Fig. 5C) were then tested. Spine density was reduced by 29 ± 4% (n = 20; P < 0.001) (Fig. 5 A and D). A second siRNA targeting a distinct site within the SV2A transcript also reduced spine density by 28 ± 3% (SV2A siRNA-2: n = 19; P < 0.001). SV2A knockdown reduced PSD-95+ GFP puncta by 25 ± 2% (SV2A siRNA-1: n = 30; P < 0.001) (Fig. 5B) and 28 ± 2% (SV2A siRNA-2: n = 34; P < 0.001). SEP-GluR2 density was similarly reduced by 22 ± 6% (SV2A siRNA-1: n = 16; P < 0.001) and 17 ± 3% (SV2A siRNA-2: n = 16; P < 0.001) (Fig. 5C). SV2A overexpression (pSV2A) increased PSD-95+ GFP puncta density by 8 ± 3% (n = 16; P < 0.01) and SEP-GluR2 by 17 ± 3% (n = 17; P < 0.05). Moreover, SV2A overexpression reversed the effects of miR-485 on spine density, PSD-95, and GluR2 (compare the miR-M with miR-M + pSV2A; Fig. 5D). Effects on spines, PSD-95–GFP+ puncta, and SEP-GluR2 were highly significant by one-way ANOVA (F7,178 = 21.96, P < 0.001 for spines; F7,198 = 33.86, P < 0.001 for PSD-95; and F7,120 = 8.98, P < 0.001 for SEP-GluR2). These results demonstrate a new role for SV2A in controlling dendritic spines and postsynaptic specialization.
Fig. 5.
SV2A siRNA-mediated knockdown mimics the effects of miR-485 overexpression on reducing dendritic spine density, PSD-95 clustering, and surface GluR2 expression. SV2A siRNA reduces spine density (A), PSD-95 clustering (B), and surface GluR2 expression (C) along dendrites. Spine density, PSD-95, and SEP-GluR2 were 10.4 ± 0.3 spines per 20 μm (n = 33), 10.3 ± 0.4 PSD-95–GFP+ puncta per 20 μm (n = 20), and 6.9 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16) in untreated controls, respectively. These values were not affected by transfection with negative controls for the siRNA [10.5 ± 0.3 spines per 20 μm (n = 23), 10.3 ± 0.3 PSD-95–GFP+ puncta per 20 μm (n = 27), and 7.3 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16)] or miRNA mimic [10.5 ± 0.3 spines per 20 μm (n = 23), 9.8 ± 0.3 PSD-95–GFP+ puncta per 20 μm (n = 20), and 7.3 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16)]. (D) Representative examples for A–C. (Scale bar = 2.5 μm.)
miR-485 Controls Homeostatic Plasticity of Dendritic Spines and Postsynaptic Specialization.
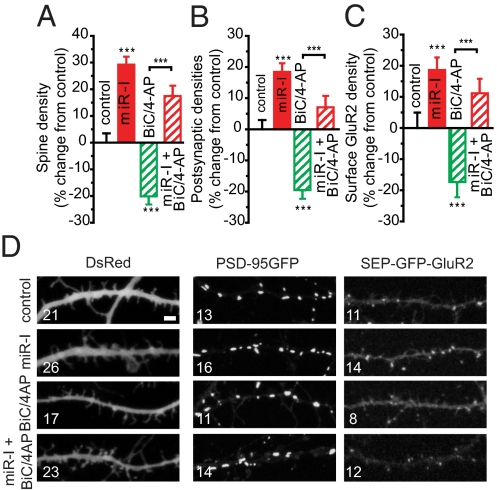
The effects of miR-485 on synapse-associated transcripts, including SV2A (Table S1); spine density; and postsynaptic specialization are consistent with a possible role of this miRNA in homeostatic synaptic plasticity acting to reduce transcripts rapidly and/or suppress translation to decrease synaptic connectivity under conditions of persistent hyperactivity. Consistent with this, chronic BiC/4-AP treatment for 5 d reduced SV2A transcript levels by 20 ± 6% (n = 4; P < 0.05). This hypothesis was tested by increasing synaptic activity with BiC/4-AP for 5 d and blocking endogenous miR-485 to determine whether the homeostatic changes in spines, PSD-95 clustering, and surface GluR2 expression observed following chronic activity are impaired when miR-485 function is blocked. The results support this conclusion. Spine density (−20 ± 3%, n = 32; P < 0.001) (Fig. 6A), PSD-95–GFP+ puncta (−20 ± 3%, n = 35; P < 0.001) (Fig. 6B), and SEP-GluR2 puncta (−17 ± 5%, n = 16; P < 0.001) (Fig. 6C) were significantly reduced after increasing synaptic activity with BiC/4-AP for 5 d. Consistent with the hypothesis, inhibiting endogenous miR-485 blocked the homeostatic reduction in spine density (Fig. 6A), PSD-95–GFP+ puncta density (Fig. 6B), and SEP-GluR2 expression (Fig. 6C) induced by BiC/4-AP. Effects of BiC/4-AP and miR-485 inhibition on dendritic spine (n = 32), PSD-95 clustering (n = 35), and SEP-GluR2 (n = 16) were highly significant by one-way ANOVA (F4,166 = 31.19, P < 0.001 for spines; F4,157 = 24.66, P < 0.001 for PSD-95–GFP+; and F4,75 = 9.99, P < 0.001 for SEP-GluR2) and Kruskal–Wallis post hoc tests. These findings indicate a role for miR-485 in homeostatic plasticity in hippocampal neurons acting through morphological changes in dendritic spines and postsynaptic specialization to decrease synaptic connectivity and function.
Fig. 6.
BiC/4-AP for 5 days in vitro (DIV) induces a homeostatic reduction in synaptic connectivity that is reversed by blocking endogenous miR-485. Hippocampal neurons transfected with a miR-I and constructs to visualize changes in synaptic properties were treated with 50 μm BiC plus 500 μm 4-AP for 5 d. BiC/4-AP reduces spine density (A), PSD-95 clustering (B), and surface GluR2 expression (C) along dendrites that are reversed by the miR-I. Spine density, PSD-95, and SEP-GluR2 were 10.8 ± 0.4 spines per 20 μm (n = 35), 9.3 ± 0.3 PSD-95–GFP+ puncta per 20 μm (n = 29), and 6.9 ± 0.3 SEP-GluR2 puncta per 20 μm (n = 16) in untreated controls. These values were not affected by transfection with negative controls for the miRNA inhibitor [11.2 ± 0.4 spines per 20 μm (n = 30), 9.1 ± 0.2 PSD-95–GFP+ puncta per 20 μm (n = 32), and 7.3 ± 0.2 SEP-GluR2 puncta per 20 μm (n = 17)]. (D) Representative examples for A–C. (Scale bar = 2.5 μm.)
Discussion
The results identify an activity-regulated miRNA, miR-485, that functions in a homeostatic manner to control synaptic development and plasticity. Individual miRNAs can target up to several hundred genes (10, 34) to destabilize sets of mRNA transcripts that mediate complex cellular processes, such as development and plasticity, in a coordinated manner. This provides a rapid posttranscriptional mechanism regulating cellular function that can be localized to specific subcellular domains, thus making this mechanism well suited to function at synaptic terminals undergoing activity-dependent homeostasis (9). miRNA-485 is expressed in the hippocampus and cerebral cortex (23, 24), and the results of this study show that this miRNA potently suppresses dendritic spine development, structure, and function. We found that miR-485 did not alter release properties at individual presynaptic terminals but, instead, altered the number of functional synapses (Fig. 4). A gene ontology analysis shows that many predicted targets of miR-485 are implicated in neural development and morphogenesis (Table S1), including the target, SV2A, which we confirmed as a target regulated by both translational suppression (Fig. 2) and mRNA degradation (Fig. S2). Furthermore, SV2A expression affects spine maturation, PSD-95 clustering, and surface GluR2 expression. Chronic synaptic activity induced by BiC/4-AP treatment reduced the density of spines and PSD-95 and GluR2 puncta, and this activity-dependent regulation was reversed by the miR-I.
Activity-dependent control of miR-485 and its targets could function as a mechanism to rapidly down-regulate other transcripts enriched at the pre- and postsynaptic terminals that mediate a decrease in synaptic connectivity to adapt to chronically elevated excitation. The experiments blocking these homeostatic changes with the miR-I support this conclusion. Mature miR-485 levels were elevated by increased excitatory activity in hippocampal neurons, and other studies show that the miRNA processing enzyme Dicer is regulated in a Ca2+-dependent manner (35), suggesting that pre-miRNAs targeted to synapses may undergo processing into mature miRNAs in an activity-dependent manner.
SV2A regulation by miR-485 and synaptic activity in the context of homeostasis are important for several reasons. SV2A is highly expressed in all presynaptic terminals of the hippocampus, where it facilitates neurotransmitter release through the readily releasable pool (25, 26, 32). Mice lacking SV2A develop seizures by postnatal day 7 and die (26), demonstrating the importance of SV2A in postnatal brain development and synaptic function. Moreover, SV2A is down-regulated following seizures (36, 37), suggesting that miR-485 may participate in a homeostatic response to hyperexcitation during seizure through posttranscriptional regulation of SV2A to reduce neurotransmitter release. More importantly, we found that SV2A knockdown mimicked the morphological effects of the miR-M and SV2A overexpression reversed the effects of the miR-M, supporting a downstream role of this protein in homeostatic synaptic plasticity.
These results differ in some respects from previous studies. Studies in microisland cultures (32) and hippocampal slices (26) did not find differences in minifrequency and synapse number in SV2A KO mice. Other targets of miR-485 (Table S1) may contribute to the decrease in minifrequency observed in response to increasing miR-485 levels (Fig. 4).
We have altered activity for longer than most other studies investigating homeostatic mechanisms to study the morphological consequences of increased synaptic activity on development of synaptic connections. The results show changes in postsynaptic specialization attributed to miR-485 acting through SV2A at presynaptic terminals. Pre- and postsynaptic sites undergo homeostatic changes through both the size of the readily releasable pool of synaptic vesicles and scaling of synaptic strength (38–40). Furthermore, our findings are in agreement with transcriptional requirements for homeostatic plasticity occurring presynaptically as well as postsynaptic morphological changes in synaptic density (3, 39).
Homeostatic synaptic plasticity is also important in adjusting neural circuit connectivity and excitation in neurological disease and in response to brain trauma. A survey of public databases from miRNA expression studies reveals that miR-485 is regulated in a number of neurological diseases and after brain trauma; miR-485 is down-regulated in Huntington disease (41), Alzheimer's disease (42), and traumatic brain injury (43) and is up-regulated in stroke (44). These disease studies, together with our findings on the effects of miR-485 on spines and synaptic development, suggest that miR-485 may contribute to pathophysiology and adaptive responses to several neurodegenerative diseases disrupting normal levels of neural network activity. Down-regulating miR-485 function increased spine density and accompanying postsynaptic changes, suggesting the feasibility of using a miR-I therapeutically to promote regeneration and synaptogenesis.
In conclusion, these results identify miR-485 as an important regulator of synaptic development in an activity-dependent manner, and they extend the presently identified mechanisms controlling homeostatic synaptic plasticity to include a new posttranscriptional mechanism involving miRNAs. The ability of miRNAs to regulate large sets of mRNAs controlling complex cellular processes in a coordinated manner rapidly and locally within different regions of a neuron makes the activity-dependent effects of miR-485 well suited to participate in homeostatic regulation of synaptic circuits under many different conditions in pathology and in the normal development and function of neural circuits.
Materials and Methods
Animals and Cell Culture.
Timed-pregnant albino Sprague–Dawley rats were used throughout this study. Dissociated hippocampal cultures were prepared as previously described (45). HEK293 cells were maintained in DMEM plus 10% (vol/vol) FBS at 5% (vol/vol) CO2. All procedures conformed to National Institutes of Health animal welfare guidelines and approved animal study protocols. Details are provided in SI Materials and Methods.
Drug Treatments and Reagents.
Cultures were treated by replacing medium with fresh medium containing drugs at the following final concentrations: 50 μM BiC methiodide, 1 μM TTX Na+ citrate (Sigma), 500 μM 4-AP, 50 μM d-(−)-2-amino-5-phosphonopentanoic acid (d-APV), 40 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), 50 μM picrotoxin (Tocris Biosciences), and 25 μM actinomycin D (Invitrogen). Short-term treatments of cultures were in sterile-filtered saline containing 145 mM NaCL, 4.5 mM KCl, 0.8 mM MgSO4, 1.8 mM CaCl2, 10 mM Hepes, and 10 mM glucose, adjusted to 320 mOsm with sucrose at pH 7.3.
Transfections.
Neurons were cotransfected at 7 days in vitro (DIV) with 2 μg of DsRed-C1 monomer (Clontech); PSD-95–GFP (30) or pCl-SEP-GluR2(R) (31); and 25 pmol of the miR-M, miR-I, miR-M negative control, miR-I negative control, or distilled H2O (Ambion) with Lipofectamine 2000 (Life Technologies). siRNA-mediated knockdown of SV2A was performed using 25 pmol of Silencer Select siRNA targeting SV2A (s138846 and s138848) (Invitrogen) and siRNA negative controls (Invitrogen). Transfected cultures were analyzed by electrophysiological recordings, immunocytochemistry, live imaging, luciferase assays, RT-PCR, and Western blotting as described in SI Materials and Methods.
Data and Statistical Analysis.
All values are reported as the mean ± SE of the mean of at least three biological replicates per experiment. In all figures, statistical significance is presented as *P < 0.05, **P < 0.01, and/or ***P < 0.001. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Drs. David Bredt and Robert J. Wenthold for PSD-95–GFP and Robert Malinow for pCI–SEP-GluR2(R) used in this study. This work was supported by the National Institute of Child Health and Human Development and the National Eye Institute Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017576108/-/DCSupplemental.
References
- 1.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 2.Hebb DO. The Organization of Behavior: A Neuropsychological Theory. New York: Wiley; 1949. [Google Scholar]
- 3.Thiagarajan TC, Lindskog M, Tsien RW. Adaptation to synaptic inactivity in hippocampal neurons. Neuron. 2005;47:725–737. doi: 10.1016/j.neuron.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiagarajan TC, Piedras-Renteria ES, Tsien RW. alpha- and betaCaMKII. Inverse regulation by neuronal activity and opposing effects on synaptic strength. Neuron. 2002;36:1103–1114. doi: 10.1016/s0896-6273(02)01049-8. [DOI] [PubMed] [Google Scholar]
- 6.Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-alpha. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- 7.Hou Q, Zhang D, Jarzylo L, Huganir RL, Man HY. Homeostatic regulation of AMPA receptor expression at single hippocampal synapses. Proc Natl Acad Sci USA. 2008;105:775–780. doi: 10.1073/pnas.0706447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 10.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton MA, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kye MJ, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 18.Wayman GA, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 20.Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- 21.Piccoli G, et al. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J Proteome Res. 2007;6:3203–3215. doi: 10.1021/pr0701308. [DOI] [PubMed] [Google Scholar]
- 22.Verpelli C, et al. Synaptic activity controls dendritic spine morphology by modulating eEF2-dependent BDNF synthesis. J Neurosci. 2010;30:5830–5842. doi: 10.1523/JNEUROSCI.0119-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pena JT, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajjalieh SM, Frantz GD, Weimann JM, McConnell SK, Scheller RH. Differential expression of synaptic vesicle protein 2 (SV2) isoforms. J Neurosci. 1994;14:5223–5235. doi: 10.1523/JNEUROSCI.14-09-05223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crowder KM, et al. Abnormal neurotransmission in mice lacking synaptic vesicle protein 2A (SV2A) Proc Natl Acad Sci USA. 1999;96:15268–15273. doi: 10.1073/pnas.96.26.15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: Implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papa M, Bundman MC, Greenberger V, Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal M, Kreher U, Greenberger V, Braun K. Is fragile X mental retardation protein involved in activity-induced plasticity of dendritic spines? Brain Res. 2003;972:9–15. doi: 10.1016/s0006-8993(03)02410-7. [DOI] [PubMed] [Google Scholar]
- 30.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 31.Kopec CD, Li B, Wei W, Boehm J, Malinow R. Glutamate receptor exocytosis and spine enlargement during chemically induced long-term potentiation. J Neurosci. 2006;26:2000–2009. doi: 10.1523/JNEUROSCI.3918-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Custer KL, Austin NS, Sullivan JM, Bajjalieh SM. Synaptic vesicle protein 2 enhances release probability at quiescent synapses. J Neurosci. 2006;26:1303–1313. doi: 10.1523/JNEUROSCI.2699-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao J, Nowack A, Kensel-Hammes P, Gardner RG, Bajjalieh SM. Cotrafficking of SV2 and synaptotagmin at the synapse. J Neurosci. 2010;30:5569–5578. doi: 10.1523/JNEUROSCI.4781-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 36.Gorter JA, et al. Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J Neurosci. 2006;26:11083–11110. doi: 10.1523/JNEUROSCI.2766-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Vliet EA, Aronica E, Redeker S, Boer K, Gorter JA. Decreased expression of synaptic vesicle protein 2A, the binding site for levetiracetam, during epileptogenesis and chronic epilepsy. Epilepsia. 2009;50:422–433. doi: 10.1111/j.1528-1167.2008.01727.x. [DOI] [PubMed] [Google Scholar]
- 38.Murthy VN, Schikorski T, Stevens CF, Zhu Y. Inactivity produces increases in neurotransmitter release and synapse size. Neuron. 2001;32:673–682. doi: 10.1016/s0896-6273(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 39.Han EB, Stevens CF. Development regulates a switch between post- and presynaptic strengthening in response to activity deprivation. Proc Natl Acad Sci USA. 2009;106:10817–10822. doi: 10.1073/pnas.0903603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wierenga CJ, Walsh MF, Turrigiano GG. Temporal regulation of the expression locus of homeostatic plasticity. J Neurophysiol. 2006;96:2127–2133. doi: 10.1152/jn.00107.2006. [DOI] [PubMed] [Google Scholar]
- 41.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cogswell JP, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 43.Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 45.Cohen JE, Fields RD. Activity-dependent neuron-glial signaling by ATP and leukemia-inhibitory factor promotes hippocampal glial cell development. Neuron Glia Biol. 2008;4:43–55. doi: 10.1017/S1740925X09000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.