Abstract
Precise control of the innate immune response is essential to ensure host defense against infection while avoiding inflammatory disease. Systems-level analyses of Toll-like receptor (TLR)-stimulated macrophages suggested that SHANK-associated RH domain-interacting protein (SHARPIN) might play a role in the TLR pathway. This hypothesis was supported by the observation that macrophages derived from chronic proliferative dermatitis mutation (cpdm) mice, which harbor a spontaneous null mutation in the Sharpin gene, exhibited impaired IL-12 production in response to TLR activation. Systems biology approaches were used to define the SHARPIN-regulated networks. Promoter analysis identified NF-κB and AP-1 as candidate transcription factors downstream of SHARPIN, and network analysis suggested selective attenuation of these pathways. We found that the effects of SHARPIN deficiency on the TLR2-induced transcriptome were strikingly correlated with the effects of the recently described hypomorphic L153P/panr2 point mutation in Ikbkg [NF-κB Essential Modulator (NEMO)], suggesting that SHARPIN and NEMO interact. We confirmed this interaction by co-immunoprecipitation analysis and furthermore found it to be abrogated by panr2. NEMO-dependent signaling was affected by SHARPIN deficiency in a manner similar to the panr2 mutation, including impaired p105 and ERK phosphorylation and p65 nuclear localization. Interestingly, SHARPIN deficiency had no effect on IκBα degradation and on p38 and JNK phosphorylation. Taken together, these results demonstrate that SHARPIN is an essential adaptor downstream of the branch point defined by the panr2 mutation in NEMO.
Keywords: innate immunity, signal transduction, pattern-recognition, ubiquitylation
The innate immune system is critical for host defense but, unchecked, can cause severe inflammatory disease (1–5). Inflammatory sequelae are mitigated at a number of levels. Principal among these is the precise identification of the threat and the appropriate tailoring of the response. Infectious agents are precisely identified by a variety of pattern recognition receptors, including Toll-like receptors (TLRs), which recognize molecular motifs that are specific to the pathogen (6). Although much is known about the mechanisms through which TLRs mediate immune responses, a number of important questions remain unanswered (7). Central to these is a complete knowledge of all of the critical components within the TLR-signaling pathways and how dynamic interactions between them lead to the appropriate coordination of host defense. The precise titration of the response requires multiple levels of regulation that include crosstalk and feedback between various signaling pathways and gene regulatory networks operating on very different spatial and temporal scales. Systems biology provides a framework in which this complexity can be addressed. Systems approaches combine prior knowledge and biological insight with global measurement technologies and computational methods both to reveal regulatory interactions and to place them in context within the innate immune system. We have used these approaches to identify transcription factors that function within regulatory circuits to coordinately amplify and attenuate TLR-mediated responses (8–10). Systems-level analysis can also be used to contextualize and elucidate the function of naturally occurring or induced mutations that impact immune phenotypes. The present work has used this approach to functionally link two mutations, chronic proliferative dermatitis mutation (cpdm) and panr2, in the TLR pathway. cpdm is a spontaneous null mutation in the Sharpin gene (SHANK-associated RH domain-interacting protein) (11), and panr2 is a chemically induced hypomorphic mutation in the Ikbkg gene encoding NEMO (NF-κB Essential Modulator) (12).
By computationally examining transcriptional and epigenomic profiles of macrophages activated with a variety of pathogen-derived agonists, we identified SHARPIN as a potential regulator of TLR responses. SHARPIN was initially described to interact with the Shank family of proteins in the postsynaptic density of excitatory synapses (13) and has subsequently been shown to interact with several other proteins including EYA1 (14) and PTEN (15); however, the functional significance of these interactions remains unknown. A role for SHARPIN in immune regulation was first revealed by the identification of mutations within the Sharpin gene in two lines of mice displaying a Th2-dominated cpdm phenotype (11).
Our systems analysis reported here demonstrates that TLR responses in macrophages are markedly impaired by SHARPIN deficiency and that SHARPIN controls expression of a subset of TLR2-induced and NF-κB– and AP-1–dependent genes that overlaps with those affected by the hypomorphic panr2 mutation in NEMO. It has recently been reported that SHARPIN is a component of the linear ubiquitin chain assembly complex (LUBAC) that modifies NEMO, thereby promoting the activation of NF-κB by multiple receptors (16–18). These data complement our results showing that the panr2 mutation abrogates the interaction between SHARPIN and NEMO, as well as the other LUBAC component RBCK1. Our data demonstrate that SHARPIN controls a branch point in the TLR2/NF-κB/AP-1 pathways that is necessary for the production of proinflammatory cytokines, including the Th1-skewing factor IL-12.
Results
SHARPIN Deficiency Impairs TLR Responses in Macrophages.
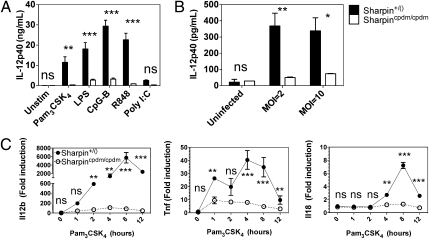
We identified SHARPIN as a potential regulator of macrophage responses over the course of our systems-level transcriptional and epigenomic analyses of combinatorial TLR pathway activation. To evaluate the role of SHARPIN in innate immunity, we analyzed TLR responses in macrophages derived from cpdm mice, which bear a null mutation in the Sharpin gene (11). IL-12p40 production was markedly impaired in response to nearly all TLR ligands evaluated, including Pam3CSK4 (TLR2), LPS (TLR4), CpG-B (TLR9), and R848 (TLR7) (Fig. 1A). The cpdm mutation also strongly attenuated macrophage production of IL-12p40 in response to infection with Listeria monocytogenes, which signals through TLR2, TLR5, and various Nod-like receptor family members (19–21), (Fig. 1B). Because IL-12p40 production in response to Pam3CSK4 was essentially abrogated, we analyzed the effects of SHARPIN deficiency on the response to this ligand in greater detail. Quantitative real-time PCR (qRT-PCR) analysis revealed marked attenuation of Il12b and Tnf mRNA expression as early as 1–2 h poststimulation (Fig. 1C). In addition to Il12b, induction of another Th1-promoting cytokine, Il18, was also abrogated (Fig. 1C). These results demonstrate that SHARPIN plays a major role in proinflammatory cytokine induction in response to TLR activation in macrophages.
Fig. 1.
SHARPIN deficiency impairs TLR responses in macrophages. (A) Bone marrow macrophages (BMM) were derived from homozygous cpdm mice (cpdm/cpdm) and littermate controls [+/()] were stimulated with the indicated TLR ligands for 12 h. Secreted IL-12p40 protein was measured in the supernatant by ELISA. Ligand concentrations were the following: Pam3CSK4 (300 ng/mL), LPS (10 ng/mL), CpG-B (1 μM), R848 (10 μg/mL), and PolyI:C (333 ng/mL complexed with 3 nL/ng Fugene-6). (B) cpdm and control BMM were infected with L. monocytogenes at multiplicity of infection (MOI) of 2 and 10 for 8 h. Secreted IL-12p40 protein was measured by ELISA. (C) cpdm and control BMM were stimulated with Pam3CSK4 (300 ng/mL) for 0–12 h. Il12b, Tnf, and Il18 transcript levels were measured by Taqman qRT-PCR. Error bars indicate mean and SEM from two independent experiments. Significance levels: *P < 0.05, **P < 0.01, ***P < 0.001, and ns (not significant).
Systems Analysis Predicts That SHARPIN Regulates NF-κB and AP-1.
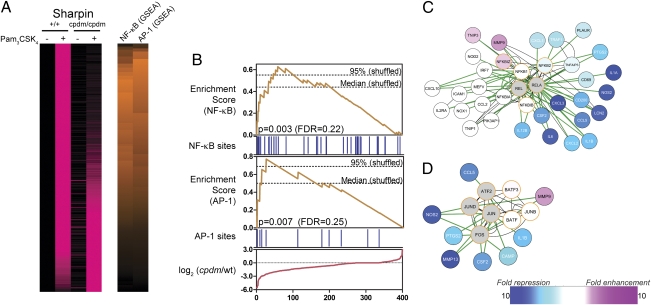
We applied the tools of systems biology to identify the pathways controlled by SHARPIN. Transcriptome analysis of wild-type macrophages identified 400 genes induced threefold or more by a 12-h stimulation with Pam3CSK4 in two independent experiments (Fig. 2A and Dataset S1). SHARPIN deficiency arising from the cpdm mutation resulted in threefold impaired induction of 87 of these genes, including many proinflammatory cytokines (Fig. 2A and Dataset S1). To identify the transcription factors (TFs) that mediate the effect of SHARPIN on macrophage responses, we performed promoter analysis. We used PAINT (22) to scan the proximal promoter sequences of all 400 Pam3CSK4-regulated genes, and we then applied the Gene Set Enrichment Analysis (GSEA) algorithm (23) to determine which TFs were associated with impaired Pam3CSK4 responses. The only TF-binding sites that were over-represented in the promoters of SHARPIN-dependent genes relative to the overall set of 400 Pam3CSK4-induced genes were NF-κB and AP-1 (Fig. 2). This result suggests that SHARPIN may be required for maximal NF-κB and AP-1 activation in response to TLR2 stimulation in macrophages.
Fig. 2.
SHARPIN is predicted to regulate TLR2-induced NF-κB and AP-1 activation. (A) cpdm and control BMM were stimulated with Pam3CSK4 (300 ng/mL) for 12 h, and RNA was extracted and analyzed by microarray (Agilent). A total of 400 genes (rows) induced at least threefold by Pam3CSK4 in control BMM in two independent experiments are shown. Genes are sorted according to impairment (top) or enhancement (bottom) of responses in cpdm BMM in two independent experiments. (Left) Pink intensity indicates increasing expression relative to unstimulated wild-type BMM. Values for each gene are scaled relative to the overall maximum value observed for that gene. (Right) Orange intensity indicates increasing GSEA enrichment scores for NF-κB– and AP-1–binding sites. (B) Details of NF-κB and AP-1 GSEA. Genes are ordered according to impairment (left) or enhancement (right) of responses in cpdm BMM. Red line: differences between Pam3CSK4 responses in cpdm and wild-type BMM for ordered genes. Blue bars: presence of NF-κB– or AP-1–binding sites in promoters of ordered genes. Orange lines: GSEA enrichment scores for NF-κB or AP-1, calculated using the effect of cpdm mutation on Pam3CSK4 responses (red line) and binding site information (blue bars). Dashed lines: median and 95% quantile maximum enrichment scores in permuted datasets. (C and D) Cytoscape interaction networks for NF-κB (C) and AP-1 (D). White, blue, and pink nodes (genes) are induced by Pam3CSK4 in a manner not affected, strongly impaired, or enhanced, respectively, by SHARPIN deficiency. Gray nodes: genes not induced by Pam3CSK4. Orange borders: genes predicted to regulate the networks as part of NF-κB (C) or AP-1 (D). Green and black edges (connecting genes and gene products): known protein–DNA and protein–protein interactions, respectively.
To further explore the link between SHARPIN, NF-κB, and AP-1, we performed network analysis using Cytoscape (24) to visualize direct protein–protein and protein–DNA interactions obtained from InnateDB (25) (Fig. 2C). This analysis provides a literature-based context for our TF-binding predictions. Many of the genes with impaired induction in SHARPIN-deficient macrophages are established targets of the NF-κB TFs RELA/p65, NFKB1/p50, and c-REL/REL, including Il12b, Il1a, Il1b, Il6, and Nos2. Notably, several of the genes not affected by SHARPIN deficiency are also known direct targets of NF-κB TFs, including Icam1, Itgal, Mmp9, and Cxcl10. This result suggests that SHARPIN deficiency results in selective inhibition of TLR2-induced NF-κB activation in macrophages. The link between SHARPIN and AP-1 was similarly evaluated (Fig. 2D). Once again, SHARPIN deficiency resulted in the inhibition of a subset of TLR2-induced genes that are known direct targets of AP-1 TFs. Therefore, although the GSEA suggested that loss of SHARPIN significantly impairs activation of both NF-κB and AP-1, it likely does not result in complete ablation of these pathways.
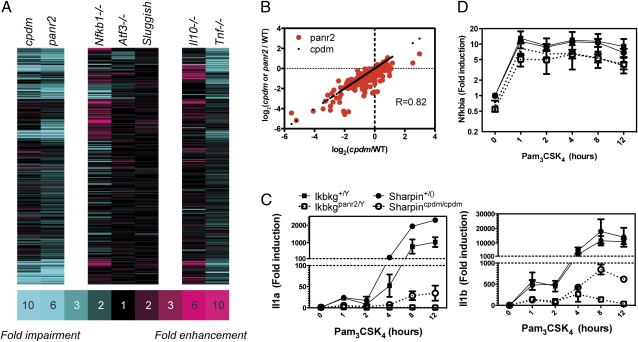
We analyzed the link between SHARPIN, NF-κB, and AP-1 in greater depth by integrating the SHARPIN-dependent gene set defined above with our database of transcriptome responses in mutant macrophages. These included null mutations in the Nfkb1, Tnf, Atf3, and Il10 genes and ENU-induced hypomorphic point mutations in Map3k8 (sluggish) encoding TPL2 (26) and Ikbkg (panr2) encoding NEMO (12). The effects of SHARPIN deficiency on TLR2-activated macrophage transcriptomes did not resemble the effects of Nfkb1, Map3k8, Atf3, and Il10 mutations (Fig. 3A and Dataset S2), indicating that the dominant role of SHARPIN is not specifically associated with these genes. The effects of TNF deficiency were significantly correlated with the effects of SHARPIN deficiency (P < 1 × 10−15), although the correlation coefficient was relatively small (R = 0.52) and the TNF effects were generally twofold less than the effects of SHARPIN (Fig. 3A and Dataset S2). The impaired TNF induction that we observed in cpdm macrophages (Fig. 1C) thus only partially accounts for the overall defect. Contrary to all other mutants examined, the effects of the hypomorphic NEMO mutation panr2 on TLR2 responses were very highly correlated with the effects of SHARPIN deficiency (R = 0.82, P < 1 × 10−15, Fig. 3 A and B), with the mutations tracking each other qualitatively and quantitatively (Fig. 3B). Detailed qRT-PCR temporal analysis in independent experiments confirmed that SHARPIN deficiency and the NEMO panr2 mutation similarly impair Pam3CSK4-induced expression of Il1a and Il1b (Fig. 3C), with the effect of the panr2 mutation being somewhat stronger than that of cpdm. These effects were specific, as Pam3CSK4-induced Nfkbia expression was only marginally affected by either mutation (Fig. 3D). Such remarkable overlap between the effects of these mutations, identified through our systems biology analysis, led us to predict that SHARPIN and NEMO interact functionally in a manner abrogated by the panr2 mutation.
Fig. 3.
The effect of SHARPIN deficiency on macrophage TLR2 responses specifically mirrors the NEMOpanr2 mutation. (A) BMM derived from the indicated mutants and respective wild-type (WT) controls were stimulated with Pam3CSK4 (300 ng/mL) for 12 h, and RNA was extracted and analyzed by microarray (Agilent: cpdm, panr2, Nfkb1, Atf3, Il10; Affymetrix: sluggish and Tnf). A total of 251 genes (rows) induced at least twofold in two independent wild-type replicates for each mutant are shown. Genes are colored according to impairment (blue) or enhancement (red) of Pam3CSK4 responses in indicated mutants compared with respective wild type. (B) Correlation between the effects of cpdm mutation (black dots) and panr2 mutation (red dots) on macrophage responses to Pam3CSK4, plotted against the effects of cpdm mutation (R[panr2 vs. cpdm] = 0.82, P < 1 × 10−15). Impairments or enhancements consistently observed in two independent experiments are plotted. (C and D) BMM derived from homozygous cpdm mice, cpdm littermate controls [+/()], hemizygous panr2 mice, and wild-type panr2 littermate controls (+/Y) were stimulated with Pam3CSK4 (300 ng/mL) for 0–12 h. RNA was harvested and reverse-transcribed into cDNA. (C) Il1a and Il1b transcript levels were measured by SYBR Green qRT-PCR. Error bars indicate mean and SEM from two independent experiments. (D) Nfkbia transcript levels were measured by Taqman qRT-PCR.
SHARPIN Interacts with NEMO.
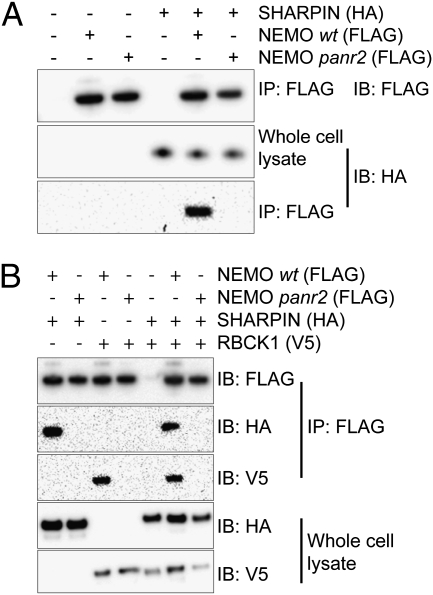
HA-tagged SHARPIN co-immunoprecipated with Flag-tagged wild-type NEMO, confirming our hypothesis that SHARPIN and NEMO interact in cells (Fig. 4A). Expression of NEMO harboring the panr2 mutation, L153P, resulted in abrogation of this interaction (Fig. 4A). Thus, the overwhelming similarity between the effects of SHARPIN deficiency and the NEMO panr2 mutation is likely to result from the specific loss of this interaction.
Fig. 4.
SHARPIN interacts with NEMO in a manner abrogated by the panr2 mutation. Lysates of HEK293T cells expressing tagged SHARPIN, RBCK1, NEMO (wild type), and NemoL153P (panr2) were subjected to immunoprecipitation using anti-FLAG beads and analyzed by Western blot. (A) HA-tagged SHARPIN readily co-immunoprecipitates with FLAG-tagged wild-type (WT) NEMO but not with panr2 NEMO. (B) HA-tagged SHARPIN and V5-tagged RBCK1 readily co-immunoprecipitate with wild-type NEMO but not with panr2 NEMO. Results representative from one of at least two independent experiments are shown.
Given that TLR2 responses in panr2 macrophages were slightly more attenuated than those in SHARPIN-deficient cpdm macrophages (Fig. 3), we tested whether interactions between NEMO and the SHARPIN paralog RBCK1 (RBCC protein interacting with PKC 1; also known as HOIL-1L) (11), were similarly abrogated. RBCK1 has recently been shown to interact with and polyubiquitinate NEMO as part of the NF-κB-activating LUBAC (27). V5-tagged RBCK1 readily co-immunoprecipitated with Flag-tagged wild-type NEMO. As in the case of SHARPIN, this interaction was abrogated by the panr2 mutation (Fig. 4B).
SHARPIN Controls a Branch of NEMO-Dependent Signaling.
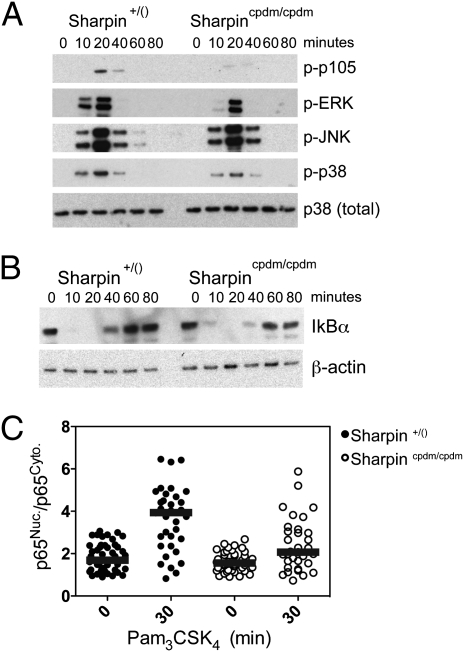
TLR2-induced signaling was affected by SHARPIN deficiency in a manner mirroring, but generally weaker than, the reported effects of panr2 mutation in NEMO. These include impaired phosphorylation of p105 and ERK (Fig. 5A). Phosphorylation of p105 is dependent on I-kappa-B kinase (IKK) complex activation and leads to p105 degradation, TPL2 activation, and ERK phosphorylation (28). Thus, simultaneous impairment in p105 and ERK phosphorylation mutually reinforce each other and suggest a specific abrogation of the p105 kinase activity of the IKK complex in the absence of SHARPIN–NEMO interactions. Neither SHARPIN deficiency nor the NEMO panr2 mutation results in complete ablation of IKK complex activity because IκBα degradation was not impaired by either (Fig. 5B). Thus, SHARPIN–NEMO interactions potentially control a branch point in IKK activity—ablating IKK p105 kinase activity while having no effect on IKK-induced IκBα degradation. In addition to stabilizing TPL2, p105 functions as an IκB itself, sequestering p50 homodimers (29) as well as p65- and c-Rel–containing heterodimers in the cytoplasm (30). This function for p105 in macrophages is supported by our observation that SHARPIN deficiency results in moderately impaired TLR2-induced nuclear localization of p65 (Fig. 5C).
Fig. 5.
SHARPIN controls a specific branch of NEMO-dependent signaling. cpdm and control BMM were stimulated with Pam3CSK4 (300 ng/mL) for the indicated times. Equal amounts of cell lysates were analyzed by immunoblotting for p105, ERK, JNK, and p38 MAPK phosphorylation (A) and IκBα protein levels (B). (C) BMM were stimulated with Pam3CSK4 (300 ng/mL) for 30 min. Cells were fixed, stained with anti-p65, and imaged. Relative nuclear-to-cytoplasmic localization of p65 (p65Nuc./p65Cyto.) for individual BMM was quantified using automated image analysis. Each point represents an individual control macrophage (filled circles) or cpdm BMM (open circles). Representative results from one of two independent experiments are shown; bars indicate median values. Significant differences were observed between stimulated and unstimulated control BMM and between stimulated cpdm and stimulated control BMM (P < 0.001 and P < 0.05, respectively).
Discussion
Systems biology approaches have the capacity to unravel the biological complexity that underlies the exquisite precision of innate immune responses. This precision is achieved by a large number of signaling networks that influence each other by subtle feed-forward and feedback mechanisms. Systems approaches usually begin with large-scale measurements of transcriptomes or proteomes, and the data are then computationally analyzed to provide testable hypotheses that are evaluated by more traditional approaches. Measurement technologies are now robust and sensitive; however, biological inference technologies are still being developed. We have developed a number of computational tools that have enabled us to identify transcriptional control mechanisms governing innate immune responses. For example, we used genomic tools and computational methods to first predict and then to confirm that the transcription factor ATF3 functions as a negative regulator of a subset of NF-κB–dependent genes that are induced by TLR4 (8). A follow-up systems analysis further refined our understanding of this process by demonstrating the subtle interplay between ATF3 and C/EBPδ in fine-tuning the response: NF-κB acting as an initiator, ATF3 acting as an attenuator, and C/EBPδ acting as an amplifier (9). Further studies showed that the interactions within this regulatory circuit occur at the epigenetic level. The in vivo relevance of this network was confirmed in a mouse sepsis model (9, 31).
Perhaps the most powerful tool in unraveling the immune response has been genetic analysis of the mouse. This analysis has been enabled by targeted gene deletion studies, chemical- or radiation-induced mutations as well as mutations that arose spontaneously. Whereas gene-targeting experiments are often initiated on the basis of a priori assumptions about predicted gene function within established pathways, phenotypic screens of mutagenized mice can reveal unique and unpredicted components of such pathways. However, it is difficult and labor intensive to establish mechanisms linking mutations to their respective phenotypes. Systems biology may be of assistance in this process. Massively parallel short-read sequencing will permit rapid identification of the genes affected by mutations causing phenotypes of interest. It will also enable accelerated breeding strategies to develop mice bearing mutations in desired genes. As illustrated in the present paper, systems approaches may then be used to analyze immune cells from mice harboring mutations that cause interesting immune phenotypes. A reference library of networks, such as those that we defined for NF-κB, ATF3, and C/EBPδ, which are generated in a highly standardized manner, could be used as a comparator to identify signaling pathways that are functionally associated with mutated genes of interest. For example, a gene associated with an innate immune phenotype could be compared with a compendium of TLR-induced signaling and gene regulatory networks; if the network generated overlaps with the network triggered by a known gene, one could infer that the new mutation functions in the same or in an associated pathway. The fact that these networks are generated using thousands of data points (e.g., entire transcriptomes) makes it far less likely that such an overlap occurs by chance.
Such a comparative transcriptomic approach was applied in the present study to link the cpdm mutation in SHARPIN to pathways known to regulate TLR responses. The SHARPIN-dependent genes specifically overlapped with genes regulated by the panr2 hypomorphic mutation in NEMO (12), and the extraordinarily strong association between the effects of these mutants suggested that SHARPIN might interact with NEMO. This prediction was confirmed; SHARPIN and NEMO interact, and interestingly, we demonstrated that the panr2 mutation in NEMO impairs this interaction (Fig. 4). Although qualitatively and quantitatively associated, the effects of the panr2 mutation on macrophage responses appeared stronger than the effects of SHARPIN deficiency; there is a residual level of proinflammatory cytokine induction in SHARPIN-deficient macrophages that is not observed in panr2 macrophages (Fig. 3). This suggested that the panr2 mutation was also able to impair a SHARPIN-independent pathway. The SHARPIN paralog, RBCK1/HOIL-1L (13), which recently was shown to interact with NEMO as part of the LUBAC complex (27), was an attractive candidate to mediate the SHARPIN-independent pathway. This hypothesis was reinforced by our observation that the panr2 mutation ablates the RBCK1–NEMO interaction as well (Fig. 4B). This contention is strengthened by a recent observation that SHARPIN and RBCK1 are present in distinct LUBAC complexes that are both capable of polyubiquitinating NEMO (16–18).
Given the interaction between SHARPIN and NEMO, we examined whether the known signaling pathways that are impaired by the panr2 mutation (12) were similarly affected by SHARPIN deficiency. To facilitate the interpretation of the results, we have constructed a model that is presented in Fig. S1. Like panr2, TLR-induced phosphorylation of p105 was ablated in SHARPIN-deficient macrophages with the consequential interruption of NF-κB p65 translocation to the nucleus (Fig. 5). Similarly, ERK phosphorylation was significantly decreased, although to a lesser extent than was observed for panr2 (Fig. 5). This suggests that p105 phosphorylation is mediated exclusively through SHARPIN and that SHARPIN-dependent ERK phosphorylation may occur via the p105-dependent TPL2 pathway (Fig. S1A). It is possible that the ERK phosphorylation that occurs in the absence of SHARPIN, but is nevertheless impaired by the panr2 mutation, involves the SHARPIN homolog RBCK1 (Fig. S1A). These signaling defects were observed in the absence of any effects on IκBα degradation or p38 and JNK phosphorylation.
The expanded model suggests a bifurcation in the MyD88 pathway that occurs at NEMO. On the one hand, there are the signals that are ablated by the L153P/panr2 mutation (Fig. S1A). We have shown that SHARPIN controls part of this branch. This branch is essential for maximal induction of many proinflammatory cytokines, including IL-12, IL-1α, IL-1β, IL-18, and TNF. On the other hand, there are the signals that are unaffected by the L153P/panr2 mutation. This branch controls the induction of a different set of genes (Fig. S1B). Interestingly, NF-κB and AP-1 are effector TFs for both branches of the pathway. This suggests a previously unappreciated specificity in NF-κB and AP-1 activities. It is possible that different members of these TF families mediate the differential responses. This specificity may also arise at the level of the IKK complex itself, given that IKK-dependent phosphorylation of p105 (32) is SHARPIN-dependent and IKK-dependent phosphorylation of IκBα (32) is not.
The etiology of hyper-eosinophilic skin inflammation in SHARPIN-deficient mice remains unclear. Reciprocal engraftment and hematopoietic reconstitution experiments indicate that disease initiation is dependent upon SHARPIN deficiency within the skin (33). However, disease progression is associated with an imbalanced Th2-dominated T-cell response (34, 35). Thus, both skin-intrinsic and -extrinsic mechanisms contribute to disease. Exogenous IL-12 attenuates the severity of disease in SHARPIN-deficient mice, suggesting that IL-12 insufficiency contributes to disease pathogenesis (34). Notably, we have shown that SHARPIN is essential for the induction of two myeloid-derived cytokines important for Th1-polarized immune responses, IL-12 and IL-18, in response to TLR2 activation. The panr2 mouse does not display the skin phenotypes associated with SHARPIN-deficient mice. Although differences in genetic backgrounds may contribute to these phenotypic distinctions, it is also possible that the panr2 mutation is permissive for those pathways critical for dermal/epidermal homeostasis. Additionally, impaired TNF-induced activation of NF-κB within keratinocytes is responsible for skin inflammation associated with epidermal lesions in NF-κB pathway components (36) and with SHARPIN deficiency (18). On the basis of these findings, it is likely that TNF receptor signaling within the epidermis is less affected by panr2 than by SHARPIN deficiency. Future efforts will be directed toward a comprehensive analysis of receptor-specific and cell type-specific requirements for SHARPIN in NF-κB– and AP-1–dependent gene regulation.
Materials and Methods
Mice.
Sharpincpdm, C57BL/KaLawRij controls for Sharpincpdm, Il10−/−, Nfkb1−/−, and Tnf−/− mice were obtained from the Jackson Laboratory. Atf3−/−, Ikbkgpanr2, and Map3k8sluggish mice have been described (8, 12, 26). For microarrays, macrophages derived from female homozygous Sharpincpdm mice were compared with age- and sex-matched wild-type controls. For all other experiments involving Sharpincpdm, macrophages derived from homozygous mutants were compared with littermate controls. For all experiments involving Ikbkgpanr2, macrophages derived from male hemizygous mutants were compared with wild-type littermate control males. For all other strains, macrophages derived from mutant strains were compared with macrophages derived from age- and sex-matched C57BL/6 controls (Jackson Laboratory). All work was approved by the Institute for Systems Biology Institutional Animal Care and Use Committee (IACUC), which implements the National Institutes of Health Guide for Care and Use of Laboratory Animals as its standard.
Bone Marrow-Derived Macrophage Cultures.
Bone marrow was collected from femurs and cultured for 7–10 d in complete RPMI containing 10% heat-inactivated FCS (HyClone Laboratories), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 50 ng/mL recombinant human Macrophage Colony-Stimulating Factor (rhM-CSF) (Peprotech).
Ligands.
TLR ligands were obtained as follows: Pam3CSK4 (EMC microcollections GmbH), Salmonella minnesota R595 (Re) ultra-pure LPS (List Biological), CpG-ODN1826 (CpG-B; Invivogen), R848 (GL Synthesis), PolyI:C (Amersham Biosciences), and Fugene-6 (Roche).
Macrophage Infections.
Bone marrow-derived macrophages were plated in six-well cell-culture plates at 1 × 106 cells/well. The following day, wild-type Listeria monocytogenes 10403s (a generous gift from Dan Portnoy, University of California, Berkeley, CA) was added at multiplicities of infection (MOIs) of 2 and 10. Macrophages were then incubated for 1 h at 37 °C before changing to cell culture media containing 15 μg/mL gentamicin to kill all extracellular bacteria. Cells were then incubated for an additional 7 h before supernatants were harvested.
ELISA.
IL-12p40 protein levels were measured in supernatants using Duoset DY499 according to the manufacturer's instructions (R&D). Tests for significant differences were performed by Bonferonni posttests of repeated measures two-way ANOVA (Graphpad PRISM).
qRT-PCR, Microarrays and Analysis, Expression Constructs and Cloning, Immunoprecipitation and Immunoblotting, and Immunofluorescence.
Detailed methods are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Contract HHSN272200700038C (to A.A.), Grants 5R01AI032972 and 5R01AI025032 (to A.A.), and by Academy of Finland application no. 213462, Finnish Programme for Centres of Excellence in Research 2006–2011 (A.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Microarray data from this study have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE29947).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107577108/-/DCSupplemental.
References
- 1.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi KS, Flavell RA. Shielding the double-edged sword: Negative regulation of the innate immune system. J Leukoc Biol. 2004;75:428–433. doi: 10.1189/jlb.0703321. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 7.Zak DE, Aderem A. Systems biology of innate immunity. Immunol Rev. 2009;227:264–282. doi: 10.1111/j.1600-065X.2008.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. and correction (2008) 451:1022. [DOI] [PubMed] [Google Scholar]
- 9.Litvak V, et al. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramsey SA, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLOS Comput Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seymour RE, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 12.Siggs OM, et al. A mutation of Ikbkg causes immune deficiency without impairing degradation of IkappaB alpha. Proc Natl Acad Sci USA. 2010;107:3046–3051. doi: 10.1073/pnas.0915098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim S, et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci. 2001;17:385–397. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 14.Landgraf K, et al. Sipl1 and Rbck1 are novel Eya1-binding proteins with a role in craniofacial development. Mol Cell Biol. 2010;30:5764–5775. doi: 10.1128/MCB.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L, Ingram A, Rybak AP, Tang D. Shank-interacting protein-like 1 promotes tumorigenesis via PTEN inhibition in human tumor cells. J Clin Invest. 2010;120:2094–2108. doi: 10.1172/JCI40778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga F, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 18.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 19.Zenewicz LA, Shen H. Innate and adaptive immune responses to Listeria monocytogenes: A short overview. Microbes Infect. 2007;9:1208–1215. doi: 10.10110/2/076/j.micinf.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren SE, et al. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. J Immunol. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leber JH, et al. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: A promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn DJ, et al. InnateDB: Facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao N, et al. The Tpl2 mutation Sluggish impairs type I IFN production and increases susceptibility to group B streptococcal disease. J Immunol. 2009;183:7975–7983. doi: 10.4049/jimmunol.0902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 28.Beinke S, Robinson MJ, Hugunin M, Ley SC. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IkappaB kinase-induced proteolysis of NF-kappaB1 p105. Mol Cell Biol. 2004;24:9658–9667. doi: 10.1128/MCB.24.21.9658-9667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sriskantharajah S, et al. Proteolysis of NF-kappaB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat Immunol. 2009;10:38–47. doi: 10.1038/ni.1685. [DOI] [PubMed] [Google Scholar]
- 31.Gilchrist M, et al. A key role for ATF3 in regulating mast cell survival and mediator release. Blood. 2010;115:4734–4741. doi: 10.1182/blood-2009-03-213512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C. Shared pathways of IkappaB kinase-induced SCF(betaTrCP)-mediated ubiquitination and degradation for the NF-kappaB precursor p105 and IkappaBalpha. Mol Cell Biol. 2001;21:1024–1035. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gijbels MJ, HogenEsch H, Bruijnzeel PL, Elliott GR, Zurcher C. Maintenance of donor phenotype after full-thickness skin transplantation from mice with chronic proliferative dermatitis (cpdm/cpdm) to C57BL/Ka and nude mice and vice versa. J Invest Dermatol. 1995;105:769–773. doi: 10.1111/1523-1747.ep12325599. [DOI] [PubMed] [Google Scholar]
- 34.HogenEsch H, et al. Increased expression of type 2 cytokines in chronic proliferative dermatitis (cpdm) mutant mice and resolution of inflammation following treatment with IL-12. Eur J Immunol. 2001;31:734–742. doi: 10.1002/1521-4141(200103)31:3<734::aid-immu734>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 35.HogenEsch H, Dunham A, Seymour R, Renninger M, Sundberg JP. Expression of chitinase-like proteins in the skin of chronic proliferative dermatitis (cpdm/cpdm) mice. Exp Dermatol. 2006;15:808–814. doi: 10.1111/j.1600-0625.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 36.Wullaert A, Bonnet MC, Pasparakis M. NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res. 2011;21:146–158. doi: 10.1038/cr.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.