Abstract
Organophosphonate utilization by Escherichia coli requires the 14 cistrons of the phnCDEFGHIJKLMNOP operon, of which the carbon-phosphorus lyase has been postulated to consist of the seven polypeptides specified by phnG to phnM. A 5,660-bp DNA fragment encompassing phnGHIJKLM is cloned, followed by expression in E. coli and purification of Phn-polypeptides. PhnG, PhnH, PhnI, PhnJ, and PhnK copurify as a protein complex by ion-exchange, size-exclusion, and affinity chromatography. The five polypeptides also comigrate in native-PAGE. Cross-linking of the purified protein complex reveals a close proximity of PhnG, PhnI, PhnJ, and PhnK, as these subunits disappear concomitant with the formation of large cross-linked protein complexes. Two molecular forms are identified, a major form of molecular mass of approximately 260 kDa, a minor form of approximately 640 kDa. The stoichiometry of the protein complex is suggested to be PhnG4H2I2J2K. Deletion of individual phn genes reveals that a strain harboring plasmid-borne phnGHIJ produces a protein complex consisting of PhnG, PhnH, PhnI, and PhnJ, whereas a strain harboring plasmid-borne phnGIJK produces a protein complex consisting of PhnG and PhnI. We conclude that phnGHIJK specify a soluble multisubunit protein complex essential for organophosphonate utilization.
Keywords: oligomerization, Pho regulon, phosphonic acid
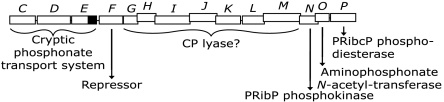
Currently, the Pho regulon of Escherichia coli (i.e., transcription units regulated by the PhoR/PhoB two component system) consists of 31 genes organized in nine transcriptional units (1), although many more genes have been postulated to be regulated by the phosphate supply and may be ascribed to the Pho regulon in the future (2). One of the nine transcriptional units is the 14-cistron phnCDEFGHIJKLMNOP operon, which is involved in the catabolism of organophosphonate (Pn), Fig. 1 (3). Transposon insertion in the phn operon results in two growth phenotypes. Thus, mutants with insertions in phnC, phnD, phnE, phnG, phnH, phnI, phnJ, phnK, phnL, phnM, or phnP are Pn growth deficient, whereas mutants with insertions in phnF, phnN, or phnO are Pn growth proficient (4). Three of the 14 cistrons (phnCDE) encode an ATP-binding cassette transport system with phnD specifying a periplasmic binding protein for Pn (5), phnE specifying a membrane spanning transport protein and phnC specifying an ATP-binding component; i.e., an ABC (3). One cistron (phnF) may specify a repressor of phn operon expression, as evaluated by sequence similarity to Mycobacterium smegmatis phnF, which encodes a repressor of the phosphate transport operon phnCDE (6). Seven cistrons (phnGHIJKLM) have been suggested to specify carbon-phosphorus (CP) lyase, as the formation of methane from methylphosphonate is abolished in strains with mutation in any of these cistrons (4, 7, 8). However, phnK and phnL appear to encode ABCs. Thus, alignments of the deduced amino acid sequence of phnK or phnL with that of hisP, a particularly well-studied ABC (9), reveals a conservation of the important sequence motifs of an ABC including the Walker A and B motifs, the C motif as well as the Q and H loops (3, 11). The reason for three apparent ABCs (PhnC, PhnK, and PhnL) in Pn transport remains to be established. The phnN cistron encodes ribosyl 1,5-bisphosphate phosphokinase (12), the phnO cistron encodes aminophosphonate N-acetyltransferase (13), whereas phnP encodes phosphoribosyl cyclic phosphodiesterase (14, 15). Pn degradation by the CP lyase pathway involves a number of intermediates, which presumably are ribose derivatives. A pathway that involves the compounds 5-phosphoribosyl 1-diphosphate, ribosyl 1,5-bisphosphate, 5-phosphoribosyl 1-phosphonate and 5-phosphoribosyl 1,2-cyclic phosphate has been suggested (15).
Fig. 1.
Organization and gene products of the 14-cistron phnCDEFGHIJKLMNOP operon of E. coli K-12 (3, 10). Transcription occurs from left to right. Each phn cistron is shown as a rectangle above which the cistron is designated in italics. Displacement of a cistron from the previous cistron indicates translational coupling. The function of each gene product is shown below the gene organization. Assignment of phnG to phnM as coding for CP lyase is presumptive (see text for details). The phnE cistron of E. coli K-12 is cryptic due to the presence of an 8-bp duplication, which causes premature termination of translation. The untranslated part of the phnE cistron is shown as a black box. Abbreviations: PRibP, ribosyl 1,5-bisphosphate; PRibcP, 5-phosphoribosyl 1,2-cyclic phosphate.
Pn is abundant as natural components of lipids, proteins, and carbohydrates (16). In addition, large amounts of manmade Pn are released to the environment primarily as detergents and herbicides (17). An important Pn is glyphosate (N-(phosphonomethyl) glycine), the active component of the commercial herbicide Roundup. Microorganisms readily degrade glyphosate either directly by CP lyase to sarcosine and Pi or by a different pathway to aminomethylphosphonic acid (AMPA) (18). AMPA, however, persists in the soil and its further degradation requires CP lyase (19). Although CP lyase is present in many environmentally important microorganisms, E. coli is an ideal organism to study CP lyase structure and mechanism due to the detailed genetic analysis of the E. coli phn operon (3, 4, 18, 20). In addition E. coli is proficient for use of AMPA as a phosphate source (21).
In the present work we analyzed if any of the polypeptides of the proposed CP lyase interact in vivo. Indeed, five of the polypeptides copurified during several purification steps as a soluble protein complex.
Results
Expression of phnGHIJKLM and Identification of Gene Products.
The phnGHIJKLM cistrons were cloned to obtain the plasmid pMN1 as described in Methods. After induction of phn gene expression in strain HO2735 Δphn/pMN1 (phnGHIJKLM), synthesized polypeptides were analyzed by SDS-PAGE. Polypeptides of the eight most prominent bands were identified by tandem mass spectrometry (MS/MS) (Fig. 2A). As shown in Table 1 the seven Phn-polypeptides were unequivocally identified as PhnM (Fig. 2A, lane 1, band 1), PhnI (band 2), PhnJ (band 3 and 4), PhnK (band 5), PhnH (band 6), PhnL (band 7), and PhnG (band 8). We therefore conclude that each of the seven phn cistrons of pMN1 was expressed. Interestingly, PhnJ migrated as two distinct bands, which suggested some heterogeneity and possible modification of this polypeptide. Bands 1, 3, 4, and 7 also contained E. coli elongation factor Tu (band 1), outer membrane protein C and F (band 3), outer membrane protein A and P (band 4), or ribosomal protein L3 (band 7).
Fig. 2.
Characterization of PhnGHIJK protein complex. Position of molecular mass standards are indicated by Roman numerals: i, 14.4; ii, 20.1; iii, 30; iv, 45; v, 66; and vi, 97 kDa. (A) SDS-PAGE of polypeptides specified by pMN1 (phnGHIJKLM). Lane 1, pellet obtained by sonication and centrifugation of cells of strain HO2735/pMN1 followed by dissolving in 6 M urea. The polypeptides of the bands labeled 1 to 8 were identified by MS/MS (Table 1); lane 2, molecular mass standard. (B) SDS-PAGE of selected fractions after elution of PhnGHIJK protein complex by size-exclusion chromatography in Sephacryl S300. Lanes 1 to 7: fraction 18, 20, 22, 24, 26, 28, and 30, respectively. Protein was concentrated by precipitation with trichloroacetic acid; lane 8: molecular mass standard. (C) SDS-PAGE of purified PhnGHIJK protein complex after size-exclusion chromatography and concentration. Lane 1, side fraction; lane 2, peak fraction; lane 3, molecular mass standard. Polypeptides of bands labeled 1 to 7 were identified by MS/MS (with Mowse scores given in parentheses): band 1, PhnI (748); band 2, PhnJ (1281); band 3, PhnJ (1175); band 4, PhnJ (1237); band 5, PhnK (1077); band 6, PhnH (520) and band 7, PhnG (871). (D) SDS-PAGE of purified PhnGHIJK protein complex. Lanes 1 and 2, protein complex purified by ion-exchange and size-exclusion chromatography from strain HO2735 (Δphn)/pMN1 (phnGHIJKLM) and HO2735/pHO571 (phnGHIJK), respectively; lane 3, histidine-tailed protein complex purified by Ni-chelate affinity chromatography from HO2735/pHO575 (phnGHIJK). Protein was concentrated by centrifugation through a Vivaspin column prior to analysis; lane 4, molecular mass standard. (E) Native-PAGE of purified PhnGHIJK protein complex. Polypeptides of the bands labeled 1 to 5 were identified by MS/MS (with Mowse scores given in parenthesis): band 1, PhnI (931), PhnJ (846), PhnK (602), PhnH (330), PhnG (623); band 2, PhnI (1089), PhnJ (1069), PhnK (783), PhnH (338), PhnG (532); band 3, PhnI (977), PhnJ (845), PhnK (698), PhnH (323), PhnG (668); band 4, PhnI (1010), PhnJ (921), PhnK (307), PhnH (286), PhnG (592); band 5, PhnI (1022), PhnJ (927), PhnH (287), PhnG (639). (F) Cross-linking of PhnGHIJK protein complex analyzed by SDS-PAGE followed by silver staining (27). Lanes 1 to 5, protein complex incubated with glutaraldehyde for 0, 1, 3.5, 7, and 24 h, respectively. Phn polypeptides are labeled I, J, K, H, and G; cross-linking products are labeled a, b, c, d, and e.
Table 1.
Identification by MS/MS of phn- polypeptides specified by pMN1
| Band no. | Polypeptide | Mowse score | Sequence coverage, % |
| 1 | PhnM | 379 | 25 |
| 2 | PhnI | 765 | 39 |
| 3 | PhnJ | 522 | 37 |
| 4 | PhnJ | 603 | 34 |
| 5 | PhnK | 566 | 48 |
| 6 | PhnH | 192 | 26 |
| 7 | PhnL | 508 | 52 |
| PhnG | 70 | 9 | |
| 8 | PhnG | 558 | 63 |
Copurification of Phn Polypeptides.
Approximately 50% of the Phn polypeptide synthesized by strain HO2735 Δphn/pMN1 (phnGHIJKLM) was found in the soluble fraction. To purify a Phn polypeptide-containing complex from an extract of HO2735/pMN1, advantage was taken of the fact that the presence of a 17-kDa polypeptide, consistent with PhnG, was easily demonstrable due to its position after SDS-PAGE in a region with relatively few bands. After ion-exchange chromatography, fractions containing PhnG were pooled, concentrated by ammonium sulfate precipitation, and submitted to size-exclusion chromatography. Fractions were analyzed by SDS-PAGE, and, as shown in Fig. 2B, five polypeptides coeluted in identical relative amounts over the elution profile. A peak fraction was concentrated, and, following SDS-PAGE, polypeptides of seven bands were identified by MS/MS (Fig. 2C). Each of the seven bands contained Phn polypeptide. PhnJ migrated as three distinct bands (Fig. 2C, lane 2, bands 2, 3 and 4), whereas PhnG (band 7), PhnH (band 6), PhnK (band 5), and PhnI (band 1) each migrated as a single band. No E. coli polypeptides other than Phn polypeptides were detected in bands 1–7.
As the PhnL and PhnM polypeptides did not appear in the PhnGHIJK protein complex, a derivative of pMN1 (phnGHIJKLM) was prepared, pHO571, which was deleted for most of the phnL cistron as well as the entire phnM cistron. A PhnGHIJK protein complex similar to that described above was purified from strain HO2735 Δphn/pHO571 (phnGHIJK) (Fig. 2D, lane 1 and 2). We therefore conclude that assembly of the PhnGHIJK protein complex does not require PhnL, PhnM, or any other phn gene product.
As an additional step in purification and characterization of the PhnGHIJK protein complex a variant of pHO571 (phnGHIJK), pHO575, was constructed. This plasmid contained a phnK variant allele specifying PhnK with a histidine-tail at the carboxy terminus. A PhnGHIJK protein complex could be readily isolated by Ni-chelate affinity chromatography as shown in Fig. 2D (lane 3) in comparison with the PhnGHIJK protein complex isolated from strain HO2735 harboring pMN1 (phnGHIJKLM) or pHO571 (phnGHIJK). As expected, histidine-tailed PhnK migrated at a position slightly above that of native PhnK.
The purity of the PhnGHIJK protein complex was examined by native-PAGE, Fig. 2E. A major molecular form is apparent (band 2) as well as three minor forms located below the major form (band 3, 4, and 5). In addition, several minor forms were located above the major form. Five bands were submitted to MS/MS analysis (Fig. 2E). With the exception of the lower band 5, bands 1 to 4 all contained PhnG, PhnH, PhnI, PhnJ, and PhnK. Band 5 contained the same polypeptides except PhnK, which was absent. The existence of multiple bands containing five Phn polypeptides suggests some heterogeneity among one or more of the Phn polypeptides.
Cross-Linking of Phn Polypeptides.
Purified PhnGHIJK protein complex was incubated with the cross-linker glutaraldehyde and the result was analyzed by SDS-PAGE (Fig. 2F). PhnI, PhnJ, and PhnK quickly disappeared, whereas PhnG only sluggishly disappeared, and PhnH was left untouched by the cross-linker. In contrast, several new bands of higher molecular mass appeared: (a) 125 kDa, (b) 115 kDa, (c) 92 kDa, (d) 78 kDa, and (e) 47 kDa. The individual Phn polypeptides migrated with the following molecular masses (the molecular masses calculated from the deduced amino acid sequence given in parenthesis): PhnI, 43 (38.9) kDa; PhnJ (major form), 38 (31.8) kDa; PhnK, 29 (27.8) kDa; PhnH, 28 (21.0) kDa; and PhnG, 17 (16.5) kDa. After 24 h of incubation all Phn polypeptides, with the exception of PhnG and PhnH were built into very large, inseparable protein complex(es) migrating close to the top of the gel. Overall, the cross-linking analysis suggests that the 47-kDa protein (Fig. 2F, lane 2, band e) very likely is a PhnGK heterodimer, whereas the 92-kDa protein (band c) very likely is a PhnI homodimer.
Molecular Size of PhnGHIJK Protein Complex.
Size exclusion in Superdex 200, performed as described in Methods, revealed two molecular forms of the PhnGHIJK protein complex, both of which eluted as symmetrical peaks. A major form constituting 80% of total protein complex eluted with a molecular mass of approximately 260 kDa (retention time 24.25 min, compare to Methods), whereas a minor form, 20% of total protein complex, eluted with a molecular mass of approximately 640 kDa (retention time 20.50 min). The minor form eluted as a single molecular form of approximately 640 kDa on rechromatography, suggesting a stable complex rather than a dynamic equilibrium of the two molecular forms. Both forms contained PhnG, PhnH, PhnI, PhnJ, and PhnK as analyzed by SDS-PAGE.
Stoichiometry of the PhnGHIJK Protein Complex.
The intensity of the Coomassie Brilliant Blue-stained polypeptide bands was estimated with Quantity One software (BioRad). The relative intensity of the staining of the various bands in a typical lane (Fig. 2B) was: PhnI, 36%; PhnJ (major form), 26%; PhnK, 8%; PhnH, 13%; and PhnG, 17%. With the assumption of the presence of a single copy of PhnK, with the use of the molecular mass of the deduced amino acid sequence of Phn polypeptides and with the use of a molecular mass of the protein complex of 260 kDa the most likely quaternary structure was calculated as PhnG4H2I2J2K, which has an apparent molecular mass of 280 kDa.
Expression of Individually Cloned phnG, phnH, phnI, phnJ, and phnK Genes.
Each of the phn cistrons phnG, phnH, phnI, phnJ, and phnK was cloned in an expression vector as alleles, which specify gene products with histidine-tails at the carboxy terminus as described in Methods. Attempts to isolate Phn polypeptide from strain HO2735 (Δphn) harboring any of these five plasmids revealed that only PhnH could be obtained in soluble form, whereas PhnG, PhnI, PhnJ, and PhnK could be demonstrated in the insoluble fraction following cell homogenization and centrifugation. PhnH was cross-linked by glutaraldehyde, Fig. 3. The PhnH polypeptide, molecular mass approximately 28 kDa, reacted with the formation of cross-linked products of molecular mass of approximately 52 and 110 kDa consistent with a PhnH2 homodimer and a PhnH4 homotetramer, respectively (Fig. 3, lanes 1 to 5, band c and b). In addition, a larger cross-linking product appeared (band a). PhnH reacted readily with the cross-linker as the dimeric form was visible a few seconds after initiation of reaction (compare Fig. 3, lanes 1 and 7, time zero sample and untreated sample, respectively). A band corresponding to that labeled c (Fig. 3) was absent in the cross-linking of PhnGHIJK protein complex (Fig. 2F, lane 2 to 5). It is clear from Fig. 3 that PhnH in free form behaved quite differently from PhnH integrated in the PhnGHIJK protein complex during cross-linking (Fig. 2F).
Fig. 3.
Cross-linking of PhnH analyzed by SDS-PAGE followed by silver staining. Lanes 1 to 4, PhnH incubated with glutaraldehyde for 0, 1, 3, and 8 h, respectively; lane 5, same as lane 4, but threefold more loaded; lane 6, molecular mass standard labeled as in Fig. 2; lane 7, untreated PhnH. PhnH monomer is labeled H, whereas cross-linking products are labeled a, b, and c.
Attempts to Demonstrate Enzymatic Activity of the PhnGHIJK Protein Complex.
We employed a number of Pn catabolic pathway intermediates, which accumulate in E. coli phnJ or phnP mutants, as potential substrates for the PhnGHIJK complex. The compounds were isolated in radiolabeled form after cultivating cells in the presence of  and unlabeled methylphosphonate (22). Additionally, we employed the fluorescent compound 3-(5-(dimethylamino)naphthalene-1-sulfonamido)propylphosphonic acid (23). A variety of ribose derivatives, ribosyl 1- or 5-phosphate, ribosyl 1,5-bisphosphate or 5-phosphoribosyl 1-diphosphate, were tested as cosubstrates, and ATP, phosphoenolpyruvate or diphosphate in complex with Mg2+ were tested as potential donors of chemical energy. In addition, the effect of a variety of divalent cations, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, or Zn2+ was tested. The reducing compounds NADH or NADPH were also included. Conversion of the substrates was followed by TLC with the solvent systems previously described (23, 24). Although not all of the possible permuted combinations of substrate, cosubstrate, energizer molecule, and divalent cation were tested with the purified PhnGHIJK protein complex, we were unable to demonstrate any conversion of the substrates. The presence of DTT during protein complex purification had no effect. We conclude, therefore, that the complex under the tested conditions is catalytically incompetent and that catalysis needs additional low-molecular weight cofactor(s) or polypeptide(s).
and unlabeled methylphosphonate (22). Additionally, we employed the fluorescent compound 3-(5-(dimethylamino)naphthalene-1-sulfonamido)propylphosphonic acid (23). A variety of ribose derivatives, ribosyl 1- or 5-phosphate, ribosyl 1,5-bisphosphate or 5-phosphoribosyl 1-diphosphate, were tested as cosubstrates, and ATP, phosphoenolpyruvate or diphosphate in complex with Mg2+ were tested as potential donors of chemical energy. In addition, the effect of a variety of divalent cations, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, or Zn2+ was tested. The reducing compounds NADH or NADPH were also included. Conversion of the substrates was followed by TLC with the solvent systems previously described (23, 24). Although not all of the possible permuted combinations of substrate, cosubstrate, energizer molecule, and divalent cation were tested with the purified PhnGHIJK protein complex, we were unable to demonstrate any conversion of the substrates. The presence of DTT during protein complex purification had no effect. We conclude, therefore, that the complex under the tested conditions is catalytically incompetent and that catalysis needs additional low-molecular weight cofactor(s) or polypeptide(s).
Phn Protein Complexes Specified by phn Deletions.
Each of the phnGHIJK cistrons of pHO571 (phnGHIJK) was deleted individually as described in Methods. The ΔphnG, ΔphnH, ΔphnI, and ΔphnJ alleles were in-frame deletions. Expression of the various phn cistrons of these five plasmid constructions was first analyzed. The data of Fig. 4A show that in general strain HO2735 (Δphn) harboring a plasmid with a phn deletion synthesized the products of the four other cistrons [i.e., strain HO2735 (Δphn)/pFM32 (phnGΔHIJK) synthesized PhnG, PhnI, PhnJ, and PhnK, but not PhnH, whereas HO2735/pFM33 (phnGHΔIJK) synthesized PhnG, PhnH, PhnJ, and PhnK, but not PhnI; HO2735/pFM34 (phnGHIΔJK) synthesized PhnG, PhnH, PhnI, and PhnK, but not PhnJ; and HO2735/pHO572 (phnGHIJΔK) synthesized PhnG, PhnH, PhnI, and PhnJ, but not PhnK]. In contrast to this expected gene expression pattern of strains harboring deletions of phnH, phnI, phnJ, or phnK, the phn gene expression pattern of HO2735/pFM31 (phnΔGHIJK) was different. Very little, if any at all of phn-specified polypeptide could be detected by SDS-PAGE. Three additional clones, which appeared in the transformation together with pFM31, were also analyzed. The pattern of synthesis of Phn polypeptides of these three additional clones was identical to that of pFM31. Isolation of plasmid DNA of strain HO2735 harboring pFM31 (phnΔGHIJK), pFM32 (phnGΔHIJK), pFM33 (phnGHΔIJK), pFM34 (phnGHIΔJK), or pHO572 (phnGHIJΔK) revealed identical plasmid DNA content in the five strains, and, thus, the pattern of synthesis of Phn polypeptides in the strain harboring pFM31 could not be attributed to a low copy number.
Fig. 4.
Characterization of Phn protein complex specified by phn deletions. Molecular mass standard is labeled as in Fig. 2. (A) Synthesis of polypeptides specified by plasmids containing various phn deletions. After sonication of plasmid-harboring cells of strain HO2735 and centrifugation, pellet was dissolved in 6 M urea and analyzed by SDS-PAGE. Cells contained: lane 1, pFM31 (phnΔGHIJK); lane 2, pFM32 (phnGΔHIJK); lane 3, pFM33 (phnGHΔIJK); lane 4, pFM34 (phnGHIΔJK); lane 5, pHO572 (phnGHIJΔK); lane 6, pHO571 (phnGHIJK); lane 7, pUHE23-2; i.e., empty vector. The position of each Phn polypeptide is indicated in lane 6. (B) PhnGHIJ and PhnGI protein complexes of phnK and phnH deletions analyzed by SDS-PAGE. Lane 1, PhnGHIJK protein complex purified from strain HO2735 (Δphn)/pHO571 (phnGHIJK); lane 2, PhnGI protein complex purified from HO2735/pFM32 (phnGΔHIJK); lane 3, PhnGHIJ protein complex purified from HO2735/pHO572 (phnGHIJ); lane 4, molecular mass standard. (C) Cross-linking of PhnGI protein complex analyzed by SDS-PAGE and silver staining. Lane 1, molecular mass standard; lanes 2 to 7, protein complex incubated with glutaraldehyde for 0, 2, 5, 30, 60, and 180 min, respectively. Phn polypeptides are indicated by I and G, cross-linking product is indicated by a. (D) Native-PAGE of PhnGHIJ and PhnGI protein complexes. Lane 1, PhnGHIJK protein complex identical to that shown in Fig. 2E; lane 2, PhnGHIJ protein complex; lane 3, PhnGI protein complex. Polypeptides of bands labeled 1–5 were identified by MS/MS (with Mowse scores given in parenthesis): band 1, PhnI (935), PhnJ (971), PhnH (289), PhnG (610); band 2, PhnI (940), PhnJ (947), PhnH (273), PhnG (574); band 3, PhnI (928), PhnJ (807), PhnH (275), PhnG (649); band 4, elongation factor G (1681); band 5, PhnI (1105), PhnG (639).
Attempts to purify Phn protein complex from the phnH, phnI, phnJ, and phnK deletion strains described above were conducted by the same procedure as that described for the PhnGHIJK protein complex; i.e., ion-exchange chromatography followed by size-exclusion chromatography. In the absence of phnK a PhnGHIJ protein complex was found. Size-exclusion chromatography revealed a molecular mass of approximately 210 kDa (retention time 20.52 min) consistent with the loss of one or two copies of PhnK polypeptide. In contrast, the absence of phnH resulted in formation of a PhnGI protein complex of molecular mass of approximately 120 kDa (retention time 27.41 min). This value is consistent with a PhnG2I2 heterotetramer. SDS-PAGE analysis of PhnGHIJ and PhnGI protein complexes in comparison with PhnGHIJK protein complex is shown in Fig. 4B. Cross-linking of PhnGI protein complex resulted in the rapid formation of a PhnI-dimer of molecular mass 84 kDa (Fig. 4C, lanes 5–7, band a), whereas the PhnG polypeptide only sluggishly if at all reacted as described above for the PhnG polypeptide of the PhnGHIJK protein complex. PhnGHIJ and PhnGI protein complexes were submitted to native-PAGE (Fig. 4D). The PhnGHIJ protein complex revealed two bands at approximately the same position as the two lower bands of the PhnGHIJK protein complex as well as a number of bands located above these. The three bands, which were analyzed by MS/MS, each contained PhnG, PhnH, PhnI, and PhnJ. Two bands of the lane containing PhnGI protein complex were also analyzed by MS/MS. Indeed, one of these bands consisted of PhnG and PhnI polypeptides (Fig. 4D, lane 3, band 5), whereas the other consisted of elongation factor G (band 4).
Phn protein complex could not be detected in extracts of strain HO2735 containing pFM31 (phnΔGHIJK), pFM33 (phnGHΔIJK) or pFM34 (phnGHIΔJK). With extracts of strain HO2735/pFM33 and HO2735/pFM34 the PhnG polypeptide could be identified by SDS-PAGE following ion-exchange chromatography. However, after size-exclusion chromatography of the pooled fractions there was no coelution of PhnG with other Phn polypeptides. With strain HO2735/pFM31 the Phn polypeptide content was so low that we were unable to detect coeluted Phn protein.
Discussion
We have isolated a soluble protein complex consisting of five polypeptides PhnG, PhnH, PhnI, PhnJ, and PhnK. Bacterial strains containing chromosomal knockout alleles of any of the cistrons phnG, phnH, phnI, phnJ, or phnK are Pn growth-deficient (4). In addition, bacterial strains lacking any of these five cistrons are unable to produce methane from methylphosphonate (7, 8). We therefore conclude that the PhnGHIJK protein complex is essential for CP bond cleavage and Pn utilization. Although we have not yet been able to demonstrate a biochemical function of the PhnGHIJK protein complex, it is tempting to postulate that the complex constitutes CP lyase. The chemical structure of the substrate for CP lyase has not been determined, but it has been suggested to be 5-phosphoribosyl 1-phosphonate and the product has been suggested to be 5-phosphoribosyl 1,2-cyclic phosphate (15). The putative substrate, 5-phosphoribosyl 1-phosphonate, is presently unavailable in sufficient amount and purity for enzymatic analysis. At any rate, it is plausible that PhnGHIJK comprises an enzyme complex. A suggested pathway for methylphosphonic acid catabolism is shown in Fig. 5. It is based on a previously suggested pathway with the inclusion of an additional, still undiscovered reaction in the conversion of ribosyl 1,5-bisphosphate to 5-phosphoribosyl 1-phosphonate (15). We suggest that the PhnGHIJK protein complex constitutes CP lyase, and that the complex catalyzes reaction i. The possibility also exists, however, that the protein complex catalyzes the reactions of the entire cycle apart from that catalyzed by phnP-specified phosphoribosyl cyclic phosphodiesterase (Fig. 5, reaction ii). Formation of methane from methylphosphonate has been previously demonstrated in vitro and was found to be stimulated by ATP and NADH, although no intermediates were identified (25).
Fig. 5.
Proposed reactions for mehylphosphonic acid catabolism by the CP lyase pathway. Compounds: a, 5-phosphoribosyl 1-methylphosphonate; b, 5-phosphoribosyl 1,2-cyclic phosphate; c, ribosyl 1,5-bisphosphate; d, a proposed, unknown intermediate. The existence of intermediates b and c, as well as a dephosphorylated derivative of intermediate a have been confirmed (15). Reactions: i, CP lyase, presumably identical the PhnGHIJK protein complex; ii, phnP specified phosphoribosyl cyclic phosphodiesterase. Reactions iii and iv have not yet been defined. Both of these may be exchange reactions involving an unidentified cosubstrate, x. Omitted from the scheme is the reaction catalyzed by phnN-specified ribosyl 1,5-bisphosphate phosphokinase, which may serve to initiate the reaction cycle by supplying ribosyl 1,5-bisphosphate (intermediate c).
A detailed study of the architecture of the PhnGHIJK protein complex has not yet been conducted, but certain conclusions may be drawn. The three-dimensional structure of PhnH has been determined, the quaternary structure being a homodimer (8). PhnH is the only protein among PhnG, PhnH, PhnI, PhnJ, and PhnK, which is soluble when synthesized individually (i.e., in the absence of the other four polypeptides) in large amount in E. coli. It is therefore tempting to postulate that PhnH constitutes a scaffold on which the PhnGHIJK protein complex is assembled. Interestingly, the PhnH subunit of the PhnGHIJK protein complex was left untouched in cross-linking, whereas free PhnH readily reacted with the cross-linker reagent. This behavior suggests that the reactive amino acid residues of PhnH are inaccessible, when PhnH participates in the complex, whereas in a free PhnH dimer the reactive amino acid residues freely react. Thus, although protein complex-bound PhnH did not react in the cross-linking experiment, PhnH is clearly part of the protein complex, which is further substantiated by the demonstration of the presence of PhnH in the protein complexes analyzed by MS/MS following native-PAGE.
When the phnH cistron was deleted a reduced PhnGI protein complex was formed, which demonstrates an interaction between PhnG and PhnI. The cross-linking experiment of PhnGHIJK protein complex indicated that a PhnGK heterodimer might be formed, suggesting interaction of these two polypeptides. By native-PAGE of the PhnGHIJK protein complex we discovered a small subfraction, which consisted of a PhnGHIJ protein complex; i.e., lacking PhnK. It is possible therefore that PhnK may be loosely associated to the other part of the protein complex, permitting the formation of a stable protein complex without PhnK. This is furthermore substantiated by isolation of a PhnGHIJ protein complex in a strain deleted for phnK. The phnK gene has been previously suggested to encode an ABC. ABCs bind to transmembrane transporters and, thus, provide energy for the transport process. How this apparent function of PhnK as a member of a transport system relates to that of the PhnGHIJK protein complex remains to be established. However, the possibility exists that PhnK serves to physically attach the PhnGHIJK complex to the Pn transport system, which may cause tight regulation or coupling of transport and catabolic reactions. Perhaps such a physical association of PhnGHIJK protein complex and Pn transporter is necessary to make the protein complex catalytically competent, which may explain the lack of demonstrable enzymatic activity of the soluble protein complex. If the PhnGHIJK protein complex, indeed, is CP lyase, and phnL encodes an ABC only phnM remains to have assigned a biochemical function to its product.
PhnJ consistently migrated as two or three polypeptide bands by SDS-PAGE. Repeatedly, the fastest migrating band was the most abundant, although some alteration of this distribution was seen. This electrophoretic pattern suggests that more than one posttranslational modification of PhnJ occurs. The structure of modified PhnJ remains to be established. This apparent heterogeneity of PhnJ may contribute to the formation of multiple PhnGHIJK bands observed in native-PAGE, although other possibilities exist such as side chain alteration. Heterogeneities of this type would not be revealed by size-exclusion chromatography.
There is a remarkable evolutionary conservation of PhnJ amino acid sequences compared to that of other Phn amino acid sequences. Thus, alignment of PhnJ of E. coli and Rhizobium meliloti reveals 68% identity (97% similarity), whereas for example for PhnH the values are 37% identity (73% similarity) (26). Although PhnGHIJK polypeptides of the two organisms obviously share homologous functions, the value for similarity of PhnJ amino acid sequences indicates a special function of this polypeptide, for example as part of a catalytic mechanism.
Lack of phnG resulted in a completely different pattern of synthesis of Phn polypeptides, as phn specified polypeptides could be hardly detected, compared to the pattern of synthesis of Phn polypeptides generated by deletion of phnH, phnI, phnJ, or phnK. Consequently, the formation of a phn specified protein complex in the absence of PhnG remains unresolved.
The determined molecular mass of the PhnGHIJK protein complex (approximately 260 kDa) is sufficient to contain two or more copies of one or more of the five PhnGHIJK polypeptides as the expected molecular mass of a complex containing a single copy of each of the five polypeptides is 136 kDa. Assuming similar efficiency in staining of the five polypeptide bands by Coomassie Brilliant Blue a best estimate of the quaternary structure was PhnG4H2I2J2K. Interestingly, there is translational coupling between the open reading frames of phnG and phnH, of phnH and phnI, of phnI and phnJ, and of phnJ and phnK, whereas this is not the case with phnK and phnL (compare Fig. 1). This translational coupling may serve to maintain a correct molar relationship of the five polypeptides, and appears consistent with the presence of two copies of each of PhnH, PhnI, and PhnJ subunits, although there was a decrease in the number of polypeptides relative to the distance of the cistron from the promotor; i.e., four G subunits, two each of the H, I and J subunits and one K subunit.
In summary, our results shed light on a longstanding question concerning CP lyase: Why are so many gene products required to cleave a CP bond? A partial answer is that they constitute the components of a holoenzyme, the core of which is formed by PhnGHIJK. It remains to be seen if each component of this holoenzyme has an active site performing a distinct reaction related to and including CP bond cleavage, or whether there is a mix of scaffold, regulatory, and catalytic roles. Opportunities may now be available to perform structural studies and to reconstitute the CP lyase reaction in vitro with substrates isolated from cells or synthesized chemically (15).
Methods
E. coli Strains, Growth Conditions, and Preparation of Cell Extracts.
Strain HO2735 (Δ(lac)C74 Δ(phnCDEFGHIJKLMNOP)33–30/F lacIq zzf∷Tn10) (8) was used as host for the various phn-harboring plasmids. Other strains and microbial procedures were used (SI Text). The expression of phn genes was induced in cells of strain HO2735 carrying the various plasmids as follows: At an OD600 of approximately 0.5, 200 mL of a culture growing exponentially at 37 °C with aeration by shaking was cooled in ice for 30 .min, at which time isopropyl β-d-1-thiogalactoside was added to 0.5 mM and incubation was continued with shaking at 26–28 °C for 3–4 h. Cells were harvested by centrifugation, resuspended in 10 mL of 50 mM Tris/HCl pH 8.0, 1 mM EDTA and broken with a Labsonic Ultrasonic Homogenizer (model M, Sartorius Stedim Biotech) at 70% amplitude for 2 × 1 min. Cell debris was removed by centrifugation in a Sorvall SS-34 rotor at 10,000 rpm for 10 min to obtain a cleared lysate.
DNA Technology.
A DNA fragment containing the phnG-M genes was amplified by PCR with the oligodeoxyribonucleotides GupMN and Mdw (SI Text) as primers, chromosomal DNA of strain HO1429 as the template, the four deoxyribonucleoside triphosphates, and the Long PCR Enzyme Mix (Fermentas). The resulting PCR product was restricted by EcoRI and HindIII, and the liberated 5,647-bp DNA fragment was ligated to similarly restricted DNA of pUHE23-2 to generate pMN1. Sequencing of the insert of pMN1 revealed five nucleotide alterations (SI Text). Other procedures for manipulation of DNA were used (SI Text). Plasmids containing deletions of phnG, phnH, phnI, phnJ, phnK, and phnLM as well as plasmids containing phn alleles specifying histidine-tailed Phn polypeptides were also constructed (SI Text).
Protein Methods.
Standard procedures were used for PAGE (SI Text). Protein bands were visualized by staining with Coomassie Brilliant Blue R-250 unless otherwise stated. Other protein chemistry procedures as well as procedures for purification of PhnGHIJK, PhnGHIJ, and PhnGI protein complexes, as well as procedures for cross-linking and protein identification are described in SI Text.
Supplementary Material
Acknowledgments.
We are grateful to Ditlev E. Brodersen (Aarhus University) for assistance with molecular mass determination. Financial support was provided by the Danish Council for Independent Research, Natural Sciences, by the Natural Sciences and Engineering Research Council of Canada (NSERC), and by the Danish National Research Foundation.
Note.
The nucleotide sequence straddling the fusion point of the ΔphnG allele of pFM31 was CCGCGAACGGTCGCGG. Transcription of this sequence might result in the formation of an mRNA stem-loop structure, which contains five guanylate-cytidylate pairs. Such a stem-loop structure may severely affect translation of the ΔphnG allele as well as all the downstream cistrons, phnH, phnI, phnJ, and phnK, thus explaining the poor yield of Phn polypeptide of strain HO2735/pFM31 (28).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104922108/-/DCSupplemental.
References
- 1.Hsieh YJ, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. 2010;13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol. 1996;178:4344–4366. doi: 10.1128/jb.178.15.4344-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CM, Ye QZ, Zhu ZM, Wanner BL, Walsh CT. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990;265:4461–4471. [PubMed] [Google Scholar]
- 4.Metcalf WW, Wanner BL. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using Tn phoA' elements. J Bacteriol. 1993;175:3430–3442. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizk SS, Cuneo MJ, Hellinga HW. Identification of cognate ligands for the Escherichia coli phn D protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci. 2006;15:1745–1751. doi: 10.1110/ps.062135206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gebhard S, Cook GM. Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: Identification of PhnF, a repressor of the phnDCE operon. J Bacteriol. 2008;190:1335–1343. doi: 10.1128/JB.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yakovleva GM, Kim SK, Wanner BL. Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli. Appl Microbiol Biotechnol. 1998;49:573–578. doi: 10.1007/s002530051215. [DOI] [PubMed] [Google Scholar]
- 8.Adams MA, et al. Crystal structure of PhnH: An essential component of carbon-phosphorus lyase in Escherichia coli. J Bacteriol. 2008;190:1072–1083. doi: 10.1128/JB.01274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PM, George AM. Subunit interactions in ABC transporters: Towards a functional architecture. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 10.Makino K, Kim SK, Shinagawa H, Amemura M, Nakata A. Molecular analysis of the cryptic and functional phn operons for phosphonate use in Escherichia coli K-12. J Bacteriol. 1991;173:2665–2672. doi: 10.1128/jb.173.8.2665-2672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hove-Jensen B, Rosenkrantz TJ, Haldimann A, Wanner BL. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): Dual role in phosphonate degradation and NAD biosynthesis pathways. J Bacteriol. 2003;185:2793–2801. doi: 10.1128/JB.185.9.2793-2801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Errey JC, Blanchard JS. Functional annotation and kinetic characterization of PhnO from Salmonella enterica. Biochemistry. 2006;45:3033–3039. doi: 10.1021/bi052297p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podzelinska K, et al. Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation. J Biol Chem. 2009;284:17216–17226. doi: 10.1074/jbc.M808392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hove-Jensen B, McSorley FR, Zechel DL. Physiological role of phnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway. J Am Chem Soc. 2011;133:3617–3624. doi: 10.1021/ja1102713. [DOI] [PubMed] [Google Scholar]
- 16.Wanner BL. Molecular genetics of carbon-phosphorus bond cleavage in bacteria. Biodegradation. 1994;5:175–184. doi: 10.1007/BF00696458. [DOI] [PubMed] [Google Scholar]
- 17.Ternan NG, Grath JW, McMullan G, Quinn JP. Organophosphonates: Occurence, synthesis, and biodegradation by microorganisms. World J Microbiol Biotechnol. 1998;14:635–647. [Google Scholar]
- 18.Kononova SV, Nesmeyanova MA. Phosphonates and their degradation by microorganisms. Biochemistry. 2002;67:184–195. doi: 10.1023/a:1014409929875. [DOI] [PubMed] [Google Scholar]
- 19.Borggaard OK, Gimsing AL. Fate of glyphosate in soil and the possibility of leaching to ground and surface waters: A review. Pest Manag Sci. 2008;64:441–456. doi: 10.1002/ps.1512. [DOI] [PubMed] [Google Scholar]
- 20.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 21.Avila LZ, Loo SH, Frost JW. Chemical and mutagenic analysis of aminomethylphosphonate biodegradation. J Am Chem Soc. 1987;109:6758–6764. [Google Scholar]
- 22.Hove-Jensen B, Rosenkrantz TJ, Zechel DL, Willemoës M. Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli. J Bacteriol. 2010;192:370–374. doi: 10.1128/JB.01131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He SM, Luo Y, Hove-Jensen B, Zechel DL. A fluorescent substrate for carbon—phosphorus lyase: Towards the pathway for organophosphonate metabolism in bacteria. Bioorg Med Chem Lett. 2009;19:5954–5957. doi: 10.1016/j.bmcl.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 24.Houlberg U, Hove-Jensen B, Jochimsen B, Nygaard P. Identification of the enzymatic reactions encoded by the purG and purI genes of Escherichia coli. J Bacteriol. 1983;154:1485–1488. doi: 10.1128/jb.154.3.1485-1488.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kononova SV, Trutko SM, Laurinavichus KS. Detection of C—P lyase activity in a cell-free extract of Escherichia coli. Appl Biochem Micro. 2007;43:394–398. [PubMed] [Google Scholar]
- 26.Parker GF, Higgins TP, Hawkes T, Robson RL. Rhizobium (Sinorhizobium) meliloti phn genes: Characterization and identification of their protein products. J Bacteriol. 1999;181:389–395. doi: 10.1128/jb.181.2.389-395.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA, and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 28.Hall MN, Gabay J, Debarbouille M, Schwartz M. A role for secondary structure in the control of translation initiation. Nature (London) 1982;295:616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.